Significance
Time-series transcriptomes of a biological process are useful for identifying the regulators of the process. To analyze such data, we developed a method that avoids time-point alignment and normalization between two conditions. We applied it to analyze time-series transcriptomes of developing maize leaves under light–dark cycles and under total darkness and obtained a time-ordered gene coexpression network (TO-GCN) that likely includes all genes involved in the development of Kranz anatomy, a structure crucial for the high efficiency of C4 photosynthesis. Using this TO-GCN, we predicted and experimentally validated a regulatory cascade of Kranz anatomy development. Moreover, we applied the method to compare transcriptomes from maize and rice leaf segments and identified regulators of C4 enzyme genes, demonstrating its broad utility.
Keywords: comparative transcriptomics, gene coexpression, Kranz anatomy, C4 enzymes
Abstract
Time-series transcriptomes of a biological process obtained under different conditions are useful for identifying the regulators of the process and their regulatory networks. However, such data are 3D (gene expression, time, and condition), and there is currently no method that can deal with their full complexity. Here, we developed a method that avoids time-point alignment and normalization between conditions. We applied it to analyze time-series transcriptomes of developing maize leaves under light–dark cycles and under total darkness and obtained eight time-ordered gene coexpression networks (TO-GCNs), which can be used to predict upstream regulators of any genes in the GCNs. One of the eight TO-GCNs is light-independent and likely includes all genes involved in the development of Kranz anatomy, which is a structure crucial for the high efficiency of photosynthesis in C4 plants. Using this TO-GCN, we predicted and experimentally validated a regulatory cascade upstream of SHORTROOT1, a key Kranz anatomy regulator. Moreover, we applied the method to compare transcriptomes from maize and rice leaf segments and identified regulators of maize C4 enzyme genes and RUBISCO SMALL SUBUNIT2. Our study provides not only a powerful method but also novel insights into the regulatory networks underlying Kranz anatomy development and C4 photosynthesis.
Transcriptomes obtained from the same tissue under different conditions can reveal differentially expressed genes that underlie condition-specific responses. Moreover, transcriptomes from time-series experiments can provide data that inform the dynamic regulation of developmental processes over time (1). Such 3D (gene expression, condition, and time) data are very useful for studying gene regulatory networks as well as the dynamics of biological processes. Note that one can replace “conditions” with “species” or “strains,” and “time series” with “tissues” or “sources.” Although methods have been developed for analyzing 3D data (2, 3), they do not deal with the full complexity of time-series data. For example, one approach is to replace the time-series expression levels of a gene in an experiment with representative values, such as the mean or maximum (3). This approach loses the temporal information. Another approach is to fuse multiple time-series datasets into one time series (2). This method simplifies the analysis but produces clusters of gene expression patterns that do not exist in the original data. As advances in sequencing technology have created an exponentially increasing influx of transcriptome data, there is a strong demand for a method capable of extracting valuable information from 3D data.
In this study, we developed a comparative, time-ordered gene coexpression network (TO-GCN) method to analyze 3D data. To illustrate its formulation and application, we used two sets of time-series transcriptomes of developing maize leaves from 0 h (T00, dry seed) to 72 h (T72) post imbibition under the natural light–dark (LD) cycle (4) and under total darkness (TD; obtained in this study). Because the maize gene expression dynamics under TD differs greatly from that under LD, it is difficult to directly compare gene expression profiles between the two conditions. Our approach overcame this challenge, and its application to the above two time-series datasets led to eight TO-GCNs, one of which is light-independent. Since Kranz anatomy develops under both LD and TD (SI Appendix, Fig. S1), the light-independent TO-GCN likely includes all genes involved in Kranz anatomy development, which is crucial for the high efficiency of C4 photosynthesis. Using this TO-GCN, we inferred and experimentally validated an upstream regulatory cascade of a key Kranz anatomy regulator, maize SHORTROOT1 (ZmSHR1, Zm00001d021973) (5). Finally, to show the broad utility of our method, we used it to compare the transcriptomes from different segments of developing maize and rice leaves (6) and identified regulators of genes encoding key maize C4 enzymes and RUBISCO SMALL SUBUNIT2 (ZmRBCS2, Zm00001d004894). Thus, our study provides not only a powerful method but also novel insights into the regulatory networks underlying the development of Kranz anatomy and C4 photosynthesis.
Results
Early Maize Leaf Development Under LD or TD.
To illustrate the need for a new method of comparing time-series transcriptome data, we chose to study the developmental progression of maize seedlings under the natural light–dark cycle (13 h light/11 h darkness; daylight from ∼6 AM to ∼7 PM) and under total darkness (SI Appendix, Fig. S1A). Under LD, seedlings undergo photomorphogenesis and are characterized by short radicle, coleoptile, leaves, and mesocotyl, followed by greening of coleoptile (66 to 72 h post imbibition). In contrast, under TD, seedlings undergo skotomorphogenesis and are characterized by etiolated and elongated coleoptile, leaves, mesocotyl, and radicle. As is well-known, maize seedlings growing under LD and TD both develop vascular tissues with Kranz signatures (SI Appendix, Fig. S1B), and therefore the comparison of LD and TD data helps to eliminate the massive numbers of light-regulated genes that are not involved in the development of Kranz anatomy.
Maize Leaf Transcriptomes.
Liu et al. (4) obtained 13 time-course transcriptomes of developing embryonic leaves every 6 h from dry seeds (0 h) to 72 h post imbibition under LD (SI Appendix, Fig. S1A). In this study, we obtained a parallel set of 12 time-course transcriptomes of developing embryonic leaves from T06 to T72 under TD (7) (SI Appendix, Table S1). By sharing the transcriptome at T00 (dry seeds), we possess two sets of 13 transcriptomes under LD and TD. The processing and mapping of sequence reads and the normalization of RPKMs (reads per kilobase of transcript per million mapped reads) are described in Methods. We defined a gene as expressed if its RPKM >1 in at least 2 of the 25 transcriptomes. In total, 25,489 genes, including 1,718 TF (transcription factor) genes, were considered expressed and used for subsequent analysis.
Methodological Considerations for Analyzing 3D Transcriptome Data.
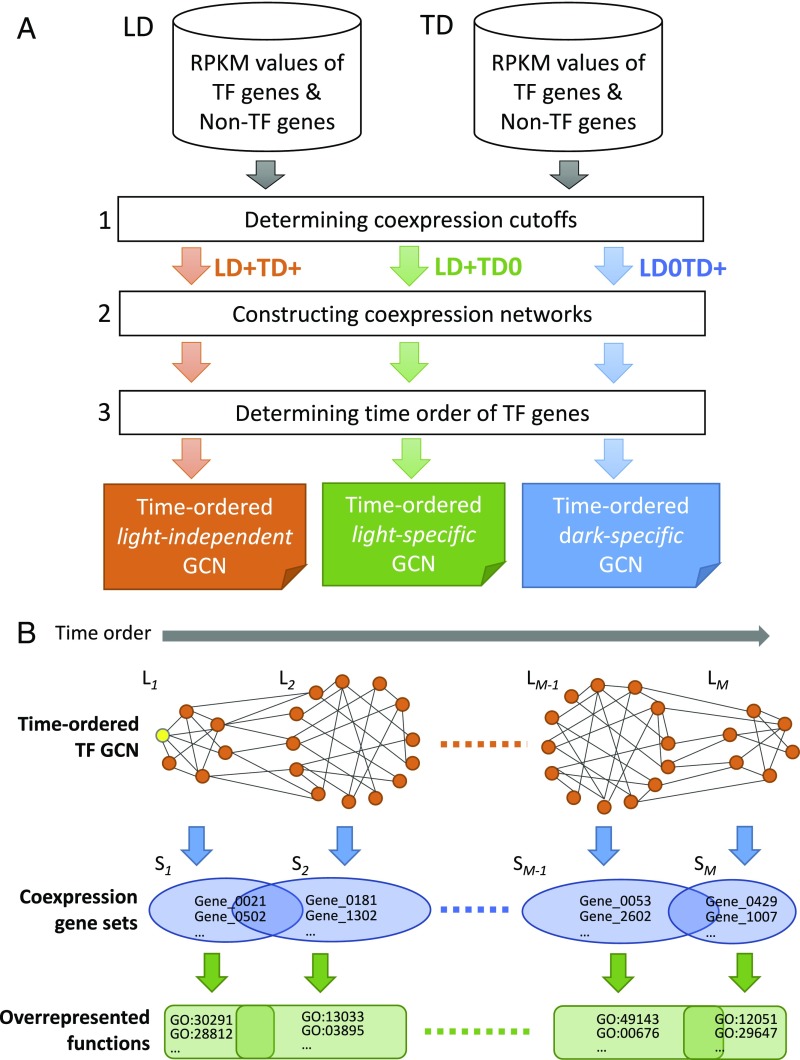
Embryonic leaves initially develop faster under TD than LD (SI Appendix, Fig. S1A), so genes tend to be up- or down-regulated earlier under TD than under LD (see examples in SI Appendix, Fig. S2). For example, ZmSCR1 (SCARECROW; Zm00001d052380), a key TF in maize Kranz leaf anatomy development (8), was up-regulated at T54 under LD but at T48 under TD. The expression profile of a gene under TD may show a time shift, even though it is similar to that under LD (SI Appendix, Fig. S2). Thus, it is important to consider the time-shift effect when comparing the two sets of transcriptomes obtained from two different developmental programs. However, this is not simple, because the degree of time shift varies from gene to gene (SI Appendix, Fig. S2). To overcome this, our method first considers the coexpression of genes over each set of time-series transcriptomes separately and then compares the coexpression patterns under LD with those under TD (Fig. 1A). Specifically, we consider whether two coexpressed genes under LD are also coexpressed under TD, and vice versa. Then we use the gene coexpression relationships to construct GCNs. Moreover, as our data are time-course data, our method can also determine the time order of the nodes in a GCN (Fig. 1B). A time-ordered GCN can reveal the dynamics of gene functions and the temporal transition of biological processes (Fig. 1B). Below, we explain how we compute gene coexpression relationships and then construct TO-GCNs.
Fig. 1.
Flowchart of the comparative transcriptomics method. (A) Three steps in the construction of the TF time-ordered gene coexpression networks. The light-independent (LD+TD+), dark-specific (LD0TD+), and light-specific (LD+TD0) GCNs are shown as three examples. +/−, positively/negatively coexpressed. (B) The levels (L1 to LM) in a TO-GCN representing the up-regulation time order of TF genes, the coexpressed gene sets (S1 to SM) (including non-TF genes) corresponding to different levels, and the overrepresented functions. The yellow node in L1 represents the initial node. Genes in a set may be coexpressed with TFs in multiple levels, so they may belong to multiple sets.
Computing Gene Coexpression Relationships and Networks.
For simplicity, we first focus on TF genes. We define the coexpression relationships between two TF genes, or a TF and a non-TF gene, as follows. First, we calculate the Pearson correlation coefficients (PCCs) of the raw RPKM values for all pairs of the 1,718 expressed TF genes under LD and TD separately and find that the probability for the PCC between any two TF genes to exceed 0.84 is P < 0.05 (SI Appendix, Fig. S3). For coexpression between TF and non-TF genes, the threshold is similar (i.e., P < 0.05 for PCC ≥0.83). Therefore, we define two genes as positively coexpressed (denoted LD+ or TD+ depending on the dataset) if PCC ≥0.84. Also, we define two genes as not coexpressed (denoted LD0 or TD0) if −0.5≤ PCC <0.5, and negatively coexpressed (denoted LD− or TD−) if PCC <−0.75. Jointly considering the coexpression states under LD and TD, we say that two genes belong to the set of LD+TD+ relationships if they are positively coexpressed under both LD and TD. Similarly, we say that two genes belong to LD+TD0 (or LD0TD+) if they are positively coexpressed only under LD (or TD) but not under TD (or LD). SI Appendix, Table S2 shows the total numbers of genes and TF–gene pairs (gene here can be TF or non-TF) in each of the eight sets of gene coexpression relationships; the set of noncoexpressed genes (LD0TD0) is not of interest to us. Below, we examine only LD+TD+, which likely includes all key genes involved in Kranz anatomy development. The other sets will be discussed in a follow-up study.
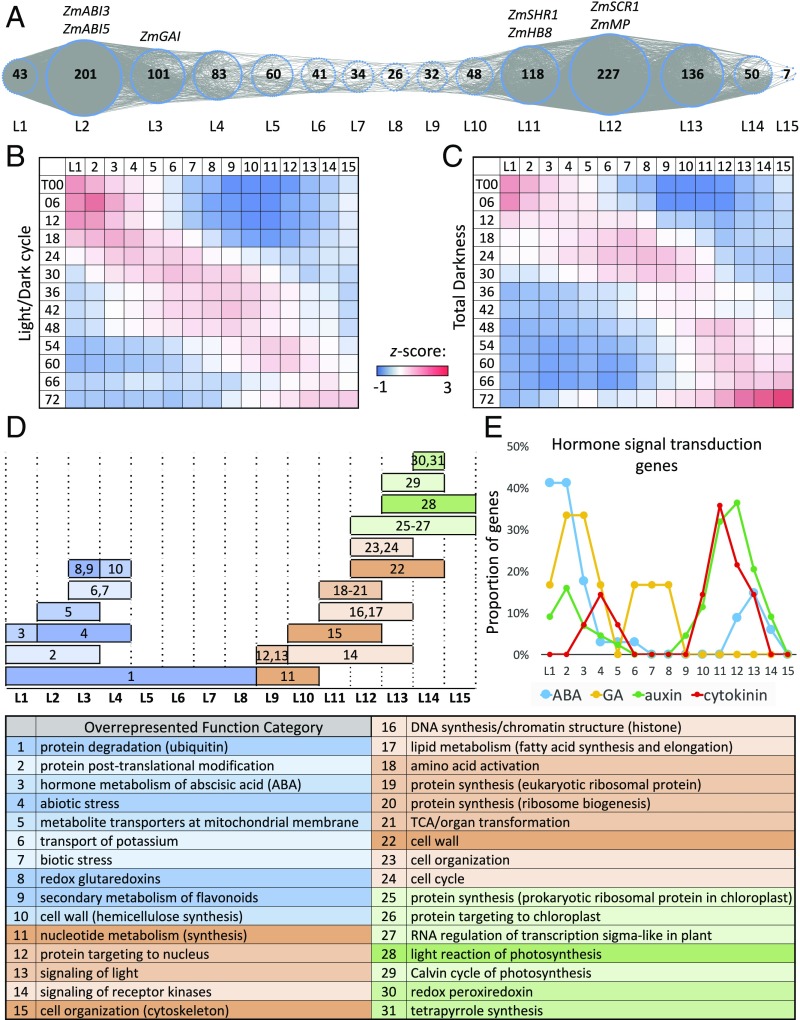
In the LD+TD+ set, the coexpression relationships are independent of light effect, allowing us to narrow down the candidate regulators of Kranz anatomy. Among the 1,275 TF genes in LD+TD+, 1,207 form a large, major TF GCN with the nodes of TF genes connected by coexpression relationships. This GCN is called the light-independent TF GCN, and is visualized in SI Appendix, Fig. S4. The remaining 68 TF genes form 28 GCNs, each with <10 TF genes. Since the connected TF genes in a GCN have similar time points of up- or down-regulation in the time course, we can infer the expression time order during leaf development for all TF genes in the major GCN. For this purpose, we select ZmARF2-1 (AUXIN RESPONSE FACTOR2-1; Zm00001d041056) as the initial node, because it is in the first coexpression module with peak expression at T00 under LD (4), that is, its expression level was very high (RPKM 96) at time 0 and monotonically decreased until T72. In addition, ZmARF2-1 is an auxin response factor, and auxin is an important plant hormone in cell division and seedling development. These observations match our hypothesis that ZmARF2-1 plays a role in the germination process. We then apply the breadth-first search (BFS) algorithm (9) to assign the time-ordered levels for all TF genes in the major GCN (Methods). We refer to this major GCN as the light-independent TF TO-GCN, which consists of 15 time-ordered levels (denoted L1 to L15 in Fig. 2A).
Fig. 2.
Light-independent TF TO-GCN and normalized gene expression profiles of TF genes at different levels. (A) The TO-GCN structure with TF genes as nodes (blue dotted circles). The number in a circle indicates the TF gene number at that level. (B and C) The heatmaps of average normalized RPKMs (z scores) at each time point of TF genes at each level of the TO-GCN under LD and TD, respectively. For each TF gene, the RPKMs over time points are normalized to z scores first. For each level, the z scores of all TFs are averaged in the heatmaps. (D) Overrepresented MapMan functions for coexpressed genes at each level. The numbers in the plot (Top) correspond to the index number of overrepresented functions listed in the table (Bottom). The blue, orange, and green colors represent the stage of germination, leaf development, and photosynthesis, respectively. (E) The proportions of genes in four hormone signal transduction pathways at each TO-GCN level (Dataset S2).
The Light-Independent TF TO-GCN.
As mentioned above, the TF genes in the light-independent TF TO-GCN are assigned to 15 levels (Fig. 2A). These assigned levels match the expression time order of the TF genes over the 13 time points under LD and TD, as revealed by the red squares (high expression levels) along the diagonal in each of the two heatmaps of mean normalized RPKMs (z scores) (Fig. 2 B and C). Moreover, the high-expression time periods overlap between consecutive levels, indicating that TF genes at a level might be regulators of TF genes at the next level. In addition, most of the TF genes at the same level are up- or down-regulated earlier under TD than under LD (Fig. 2 B and C). For example, the TF genes at L1 are down-regulated at T12 under LD but at T06 under TD (Fig. 2 B and C). This observation is consistent with the faster developmental pace under TD (SI Appendix, Fig. S1A).
In the light-independent TF TO-GCN, many TFs related to seed germination or breaking dormancy appear at the earlier levels (Fig. 2A and Dataset S1). For example, ZmABI3 (ABSCISIC ACID INSENSITIVE3; Zm00001d042396) and ZmABI5 (Zm00001d012296), which play key roles during embryogenesis (10, 11), belong to L2. ZmGAI (GIBBERELLIC ACID INSENSITIVE PROTEIN; Zm00001d013465), which promotes germination (12), belongs to L3. These observations indicate that our method can effectively reveal key regulators of seed germination. On the other hand, all known positive regulators of vascular tissue development are found at later levels, including the two ZmMP genes (MONOPTEROS; Zm00001d001945 and Zm00001d026540) and ZmHB8 (HOMEOBOX GENE 8; Zm00001d008869) (13), which belong to L12 and L11, respectively. Furthermore, the bundle sheath (BS) development-related positive regulator ZmSHR genes (5) (Zm00001d021973, Zm00001d029607, and Zm00001d006721) and ZmSCR1 (8) belong to L11 and L12, respectively. According to the TF gene expression profiles of L11 in the heatmaps (Fig. 2 B and C), the development of vascular tissues and peripheral cells likely begins at T30 and T24 under LD and TD, respectively. Therefore, the key regulators of Kranz anatomy formation, which is closely related to the development of vascular tissues and BS cells, should belong to L8, L9, or L10.
Light-Independent Functions.
By identifying the overrepresented functional categories among the coexpressed genes at each level of the light-independent TO-GCN (Fig. 2A and Dataset S2), we find a clear developmental-stage transition between L8 and L9 (Fig. 2D). Throughout the first eight levels, protein degradation mediated by ubiquitination is overrepresented. At the first four levels, nine other functions are also overrepresented, including, for example, protein posttranslation modification, hormone metabolism of abscisic acid (ABA), biotic and abiotic stresses, metabolite transporters on the mitochondrial membrane, and transport of potassium (Fig. 2D), some of which are related to seed germination (14). By mapping the first eight levels to time points in the heatmaps (Fig. 2 B and C), the germination stage is found to end at T48 and T30 under LD and TD, respectively. Compared with L1 to L8, genes at L9 to L15 mostly function in processes relevant to cell proliferation, indicating a shift to leaf and root development after germination. The overrepresented functional categories include nucleotide synthesis at L9 and L10, cytoskeleton of cell organization from L10 to L12, DNA synthesis from L11 to L13, and cell cycle from L12 to L13 (Fig. 2D). In addition to cell proliferation, functions related to chloroplast development and photosynthesis emerge at the last four levels. This observation indicates that even under total darkness, maize embryonic leaf cells are programmed to develop into cells capable of conducting photosynthesis upon illumination.
Plant hormones are essential for triggering developmental reprogramming. We find genes related to ABA signal transduction tend to emerge at the first three levels but diminish later, consistent with the maintenance of seed dormancy by ABA until the start of germination (Fig. 2E). In contrast to ABA, most genes related to gibberellin (GA) signal transduction emerge at L2 and L3, consistent with GA’s antagonistic functions to ABA for the induction of the germination process. Most genes related to auxin and cytokinin (CK) signal transduction emerge at L9 to L15 and L10 to L13, respectively (Fig. 2E). This is also in line with previous studies that these two hormones together play the major role in regulating leaf cell proliferation and differentiation (15).
Upstream Regulators of a Key Kranz Anatomy Regulator.
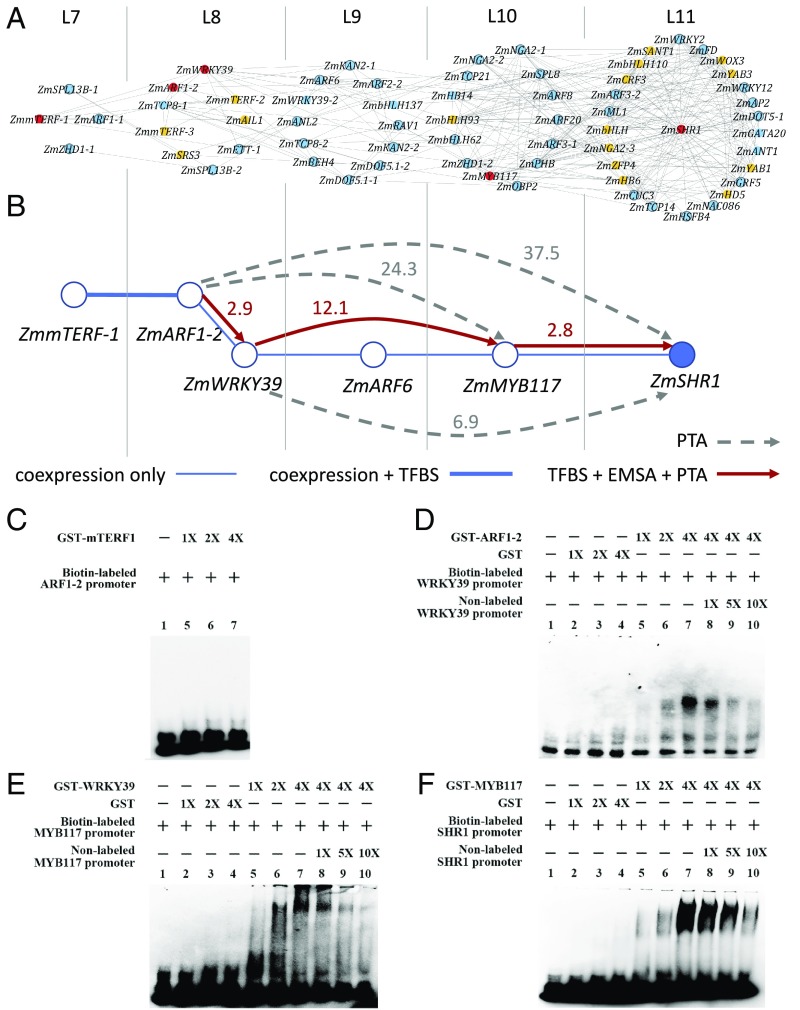
ZmSHR1 has been shown to play an important role in the BS cell development of maize Kranz anatomy (8, 16, 17), but its upstream regulators remain unknown. Thus, we chose ZmSHR1 as an example to show how our approach can be used to identify upstream regulators of a specific gene. Specifically, we used the light-independent TF TO-GCN to design a three-step workflow (Fig. 3) to identify an upstream regulatory pathway that modulates ZmSHR1 expression.
Fig. 3.
Inference and experimental validation of candidate upstream regulators of the ZmSHR1 gene in maize leaf development. (A) The first- to fourth-order candidate upstream regulators of ZmSHR1 inferred from the light-independent TF TO-GCN (SI Appendix, Table S3). ZmSHR1 is placed at the center of level 11 of this subnetwork. TFs without known TFBS are shown in orange. Known TFBSs of TFs (blue color) are used to check the presence of their mapped sites in the promoter sequences of their candidate downstream genes. The TFs in the predicted regulatory pathway with TFBS support are shown in red and are selected for experimental validation. (B) The upstream regulatory network of ZmSHR1 targeted for experimental validation. The number beside a directed solid line or dashed line with an arrow represents the fold increase in the expression level of the potential direct or indirect target gene in the PTA experiment. Red lines: the presence of the TFBS in the promoter of the target gene (indicated by arrows) and validated by both PTA and EMSA. Dashed gray lines: with PTA support but no TFBS found in the promoter. (C–F) EMSA experiments testing interactions between predicted TFBSs in the promoters of putative targets and GST-ZmmTERF1 (C), GST-ZmARF1-2 (D), GST-ZmWRKY39 (E), or GST-ZmMYB117 (F). In each case: lane 1: biotin-labeled probe alone; lanes 2 to 4: with increasing amounts of purified GST (except for C); lanes 5 to 7: with increasing amounts of GST-TF; and lanes 8 to 10: with increasing amounts of the nonlabeled probe (except for C).
First, we used the TO-GCN to predict candidate direct regulators of ZmSHR1, which should be coexpressed with ZmSHR1 at the same level as or at one level earlier than ZmSHR1 (i.e., L11 and L10 in Fig. 3A). (A TF can be an upstream regulator of another TF at the same level because only one transcriptome was taken every 6 h.) We call these TFs the first-order candidate regulators. Similarly, we infer the second-, third-, and fourth-order candidate regulators at L9, L8, and L7, respectively (Fig. 3A). These candidate regulators are listed in SI Appendix, Table S3.
Second, for each candidate regulator with unknown TFBS (transcription factor binding site), we predict its TFBS. Using the method of Yu et al. (18) and the TFBS databases for Arabidopsis TFs (Methods), we are able to predict the TFBSs of ZmmTERF-1 (Zm00001d031533, L7), ZmARF1-2 (Zm00001d003601, L8), ZmWRKY39 (Zm00001d013307, L8), and ZmMYB117 (Zm00001d032194, L10) (indicated by red color in Fig. 3A). The predicted TFBSs are shown in SI Appendix, Fig. S5. For those TFs indicated in orange color in Fig. 3A, we are unable to predict their TFBSs because either the DNA-binding domains are too divergent between maize and Arabidopsis or no Arabidopsis data are available.
Third, for each TF with known or predicted TFBS, we checked the presence of the TFBS in the promoter region of each candidate target gene. This procedure further simplifies the network in Fig. 3A to that in Fig. 3B.
We next used the electrophoretic mobility-shift assay (EMSA) and protoplast transient assay (PTA) to validate our predictions of the candidate regulators and their hypothesized target genes. Our EMSA experiments provide evidence for the binding of ZmARF1-2 to the promoter of ZmWRKY39, the binding of ZmWRKY39 to the promoter of ZmMYB117, and the binding of ZmMYB117 to the promoter of ZmSHR1 (Fig. 3 B and D–F), though not for the binding of ZmmTERF-1 to the promoter of ZmARF1-2 (Fig. 3C). Our PTA experiments show that overexpression of ZmARF1-2 up-regulates the expression of ZmWRKY39, ZmMYB117, and ZmSHR1; overexpression of ZmWRKY39 up-regulates the expression of ZmMYB117 and ZmSHR1; and overexpression of ZmMYB117 up-regulates the expression of ZmSHR1 (Fig. 3B). Thus, our EMSA and PTA experiments both support the upstream regulatory cascade indicated by the red arrows in Fig. 3B; that is, ZmARF1-2 is a direct upstream regulator of ZmWRKY39, which in turn is a direct upstream regulator of ZmMYB117, which then acts as the direct upstream regulator of ZmSHR1.
Identifying Regulators of C4 Enzyme Genes.
To demonstrate the broad utility of our method, we analyzed the 3D data of Wang et al. (6), which are two series of transcriptomes from 15 and 11 segments of the third developing leaf in maize (C4) and rice (C3), respectively (Dataset S3). Note that here we are considering spatial points instead of time points. As all key C4 enzyme genes are preferentially expressed in bundle sheath or in mesophyll (M) cells, we aimed to identify the upstream regulators of key C4 enzyme genes, each of which has a strong cell-type preference of expression in maize leaf (19, 20). Following the method in Fig. 1A, we first determine the cutoffs of positive and no coexpression for any TF–gene pair in maize (denoted Zm+ and Zm0) and in rice (denoted Os+ and Os0) as PCC ≥0.93 (P < 0.05) and 0.5≥ PCC >−0.5, respectively. Second, we construct a maize-specific GCN with Zm+OS0 coexpression relationships. Third, we build a spatially ordered GCN based on the GCN constructed in the second step and identify the TF genes coexpressed with those key C4 enzyme genes (Table 1). Finally, the candidate TF–targets are validated by EMSA.
Table 1.
Predicted regulators of C4 enzyme genes
| Target C4 enzyme gene* | TF gene name | TF gene ID | | Ri |† | Available‡/purchased | Purified§/EMSA |
| BS: ZmNADP-ME | ZmGATA12 | Zm00001d037605 | 0.76 | ✓/✓ | ✓/✓ |
| BS: ZmNADP-ME | ZmbHLH43 | Zm00001d033267 | 0.98 | ✓/✓ | ✓/✓ |
| BS: ZmNADP-ME | ZmERF | Zm00001d052229 | 0.83 | ✓/✓ | |
| BS: ZmNADP-ME | ZmNAC | Zm00001d050893 | 0.76 | ✓/ | |
| BS: ZmPCK | ZmMYB48 | Zm00001d041576 | 1.00 | ✓/✓ | ✓/✓ |
| BS: ZmPCK | ZmMYB88 | Zm00001d048623 | 1.00 | ✓/✓ | ✓/✓ |
| BS: ZmPCK | ZmMYB56 | Zm00001d030678 | 1.00 | ✓/✓ | ✓/✓ |
| BS: ZmPCK | ZmbHLH118 | Zm00001d038357 | 0.72 | ✓/✓ | ✓/✓ |
| BS: ZmRBCS2 | ZmMYBr17 | Zm00001d044409 | 0.91 | ✓/✓ | ✓/✓ |
| M: ZmCA | ZmABI33 | Zm00001d011639 | 0.97 | ||
| M: ZmCA | ZmRAV | Zm00001d043782 | 0.91 | ||
| M: ZmPEPC | ZmRAV | Zm00001d043782 | 0.91 |
BS: preferred expression in bundle sheath cells. M: preferred expression in mesophyll cells.
Degree of cell-type preference of gene i is defined as | Ri | = |mi − bi|/max(mi, bi), where mi and bi represent the RPKM value of gene i in the M and BS RNA samples, respectively (20).
Available in the Maize TFome Collection; the ZmNAC clone was not ordered because this candidate TF was predicted after we sent out the purchase order of TF clones.
Expression of the ZmERF clone failed.
For each TF–candidate target gene pair, we determined whether the TFBS can be found in the promoter region of the candidate target gene. Using the method of Yu et al. (18) and the Arabidopsis TFBS databases, we predict 12 TF–target gene pairs (Table 1 and SI Appendix, Table S4). We find nine TFs that may regulate BS-preferred C4 enzyme genes, including four TFs (ZmGATA12, ZmbHLH43, ZmERF, and ZmNAC) for ZmNADP-ME (NADP-MALIC ENZYME, Zm00001d000316), four TFs (ZmMYB48, ZmMYB88, ZmMYB56, and ZmbHLH118) for ZmPCK (PHOSPHOENOLPYRUVATE CARBOXYKINASE, Zm00001d028471), and one TF (ZmMYBr17) for ZmRBCS2 (Table 1). The four TFs we identified for ZmNADP-ME are different from the two TFs, ZmbHLH128 (Zm00001d054038) and ZmbHLH129 (Zm00001d014995), identified by Borba et al. (21), which showed no strong preferential expression in BS or M cells (20). For M-preferred C4 enzyme genes, we find two TFs (ZmABI33 and ZmRAV) that may up-regulate ZmCA (CARBONIC ANHYDRASE, Zm00001d044099) and that ZmRAV may also up-regulate ZmPEPC (PHOSPHOENOLPYRUVATE CARBOXYLASE, Zm00001d046170) (Table 1).
For EMSA validation, seven recombinant TF proteins (ZmGATA12, ZmbHLH43, ZmMYB88, ZmMYB56, ZmMYB48, ZmbHLH118, and ZmMYBr17) are successfully expressed, purified, and used to assess the direct TF–target gene interactions for the three candidate target genes (ZmNADP-ME, ZmPCK, and ZmRBCS2) (Table 1). Our EMSA experiments validate the binding of ZmGATA12 and ZmbHLH43 to ZmNADP-ME (SI Appendix, Fig. S6 A and B), the binding of ZmMYB88, ZmMYB56, ZmMYB48, and ZmbHLH118 to ZmPCK (SI Appendix, Fig. S6 C–F), and the binding of ZmMYBr17 to ZmRBCS2 (SI Appendix, Fig. S6G). The binding of ZmMYB88, ZmMYB56, and ZmMYB48 to the same TFBS in ZmPCK supports the view that TFs with similar DNA-binding domains bind similar DNA sequence motifs (22).
Discussion
A major contribution of this study is a method for comparing 3D transcriptomes obtained under different conditions (or tissues/organs or species) and time points (or spatial points). Our method has the following advantages. First, it can readily identify coexpressed gene pairs in each condition and then find out which coexpression relationships have been conserved among conditions. Second, there is no need to normalize the RPKM values among conditions. Normalization can be difficult if the developmental dynamics are very different between conditions. Third, there is no need to align the time or spatial points between two conditions. Thus, the number of sample points studied can differ between conditions. The application of our method to the two series of transcriptomes from 15 and 11 segments of developing maize and rice leaves provided such an example. Fourth, our approach can reduce batch effects, because coexpression is defined within each set of transcriptomes before the construction of GCNs. For this reason, this approach can be applied to do meta-analysis of heterogeneous transcriptome datasets from different laboratories. Fifth, and most importantly, our method provides TO-GCNs, which can reveal temporal dynamics of gene expression underlying developmental transitions as influenced by environmental conditions. Moreover, TO-GCNs provide a convenient way to infer candidate upstream regulators of any gene of interest, if the gene is in at least one of the TO-GCNs.
We tested the level-order stability when a TF gene other than ZmARF1-2 is used as the initial node to construct the TO-GCN. We randomly chose 10 different TF genes in level 1 of the original TO-GCN and tested them one by one. We calculated the differences in level number for each tested TF gene against the original one. The results showed that on average ∼12.5% of TFs in the original TO-GCN were assigned to a different level (SI Appendix, Table S5). However, the average and SD of the overall level change for each new TO-GCN with a different seed is very small (SI Appendix, Table S5), indicating that the new ordered TO-GCNs are very similar to the original one.
Besides providing a method, our study contributes to a better understanding of the light-independent process in early maize leaf development. More than 1,200 TF genes are assigned to the light-independent TO-GCN, providing a global picture of light-independent gene regulatory relations. In general, it is much more difficult to predict an upstream regulator than a downstream target gene. In this study, we showed that our method can successfully identify an upstream regulatory cascade of key Kranz anatomy regulators from ZmARF1-2 (L8) to ZmSHR1 (L11), which is consistent with the fact that auxin plays an important role in Kranz anatomy development (17). In addition, we compared the expression profiles of ZmARF1-2, ZmWRKY39, ZmMYB117, and ZmSHR1 in maize foliar and husk developments. These comparisons may reveal how these TF genes have evolved in the C4 and C3 plants under study, because in maize the foliar and husk leaves exhibit C4 and C3 photosynthesis, respectively. Wang et al. (3) obtained the transcriptomes in six developmental stages of foliar and husk (P1, P2, P3/4, P5, I, and E; SI Appendix, Fig. S7). We found that ZmARF1-2, ZmWRKY39, and ZmMYB117 have similar expression profiles and are expressed much higher in foliar than in husk leaves at the early embryonic stages (P1, P2, and P3/4 in SI Appendix, Fig. S7), which are the stages during which Kranz anatomy develops. This observation indicates that the regulation of these three TF genes have changed during the evolution of C4 leaves. The similarities in the expression profiles of ZmARF1-2, ZmWRKY39, and ZmMYB117 to that of ZmSHR1 in foliar and their dissimilarities in husk (SI Appendix, Fig. S7) suggest that the three TFs may regulate ZmSHR1 in foliar (C4) but not in husk (C3). This observation is consistent with our proposal that ZmARF1-2, ZmWRKY39, and ZmMYB117 are upstream regulators of ZmSHR1. Some direct and indirect target genes of ZmSHR1 had been reported in the study of Arabidopsis root vasculature development (23). We found that ZmMGP/NUC (MAGPIE/NUTCRACKER; Zm00001d009030) and ZmSCR1, two direct targets of ZmSHR1, are at L11 and L12, respectively, and that ZmSCL3 (SCR-LIKE 3; Zm00001d011881), two paralogs of ZmSNE (SNEEZY; Zm00001d028159 and Zm00001d048185) and two paralogs of ZmRLK (RECEPTOR-LIKE KINASE; Zm00001d005298 and Zm00001d046626), and five indirect targets of ZmSHR1 are at L11, L12, or L13, suggesting that the regulatory pathway of ZmSHR1 in dicot roots is largely conserved in monocot leaves. In addition to the time-series data, we applied our approach to compare another 3D gene expression dataset of leaf developmental series between maize and rice, leading to the identification of a “spatially ordered” GCN as well as novel regulator–target relations, further supporting the value of our approach.
In summary, we have demonstrated the utility of a robust method for analyzing 3D datasets. It can be applied to contrast gene coexpression profiles in a wide range of contexts. Considering the rapid influx of gene expression data with increasing complexity in experimental design, our approach provides a means for mining these expression data to obtain biological insights. Through application of our approach to the temporal expression data under LD and TD in maize leaves and to the leaf developmental transcriptomes from maize and rice, TO-GCNs were inferred, providing a wealth of regulatory interaction predictions. Combined with experimental validation, we further revealed the regulatory cascade that is key to the leaf vein development in C4 photosynthesis. These findings not only highlight the quality of the regulatory interaction predictions by our method but also provide much needed information on the regulatory basis of C3–C4 evolution transition, paving the way for genetic engineering of C3 crops with the capacity of C4 photosynthesis in the future (24, 25).
Methods
RNA Sequencing and Read Processing.
Seeds of Zea mays cv. White Crystal, a glutinous maize cultivar, were purchased from a local supplier. For germination, seeds were imbibed in distilled water at 6:00 PM, shaken for 10 min at 200 rpm, and then germinated on wet filter paper on Petri dishes in the dark room at 30 °C and with 60% humidity. Plumules were collected every 6 h under dim green light in the dark room, the coleoptiles were removed within an hour, and the embryonic leaves were frozen in liquid nitrogen and stored at −80 °C.
Total RNA was extracted by using TRIzol reagent (Invitrogen). To remove traces of DNA contamination, 1 µL TURBO DNase (Ambion) per 10 μg RNA was added and the reaction was incubated for 30 min at 37 °C, followed by phenol:chloroform extraction. The RNA samples were quantified and their qualities were examined by the BioAnalyzer RNA 6000 Nano Kit (Agilent). RNA-sequencing libraries were constructed using TruSeq RNA Library Prep Kit v2 (Illumina). The adaptor-ligated reactions were selected for two size ranges (∼300 and ∼400 bp). The purified libraries were then amplified by 12 cycles of PCR and cleaned up using AMPure XP beads (Beckman Agencourt). The libraries were assayed using the Qubit HS DNA Kit and BioAnalyzer HS DNA Kit (Agilent), and the molar concentrations were normalized using KAPA Library Quantification Kit Illumina Platforms (Kapa Biosystems). Paired-end 2 × 101-nt sequencing was conducted on the Illumina HiSeq 2000 at the NGS High Throughput Genomics Core Facility at Academia Sinica. Raw reads were deposited in the Short Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession no. SRP140487).
The read-processing procedure was the same as in Liu et al. (4). The processed reads were mapped to the maize genome (B73 RefGen_v4) using TopHat (26) (v2.0.10) and its embedded aligner Bowtie2 (27) (v2.1.0). The expression level (RPKM) of each gene was estimated using Cufflinks (28) (v2.1.1). To compare the RPKMs of the selected genes across time points in a set of transcriptomes, we applied the upper-quartile normalization procedure (29).
Construction of GCNs.
Our comparative transcriptomics method was designed to analyze time-course transcriptomes that may have different numbers of time points under two or more conditions. The method consists of three steps: determining coexpression cutoffs, constructing GCNs, and determining the time order of TF gene expression (Fig. 1). In this study, the inputs were two time series of transcriptomes from maize embryonic leaves under LD and TD with the maize TF gene list in Dataset S4 [updated from Lin et al. (30)]. First, the Pearson correlation coefficient values of all TF–TF gene pairs were calculated under LD and TD separately and used to determine the cutoffs of positive coexpression (denoted as LD+ or TD+), negative coexpression (denoted as LD− or TD−), and no coexpression (denoted as LD0 or TD0) (for examples of cutoff points, see Results). Second, using these three types of relationships, we determined all types of GCNs (SI Appendix, Table S2). In this study, we focus on the major GCN with LD+TD+ coexpression relationships with 1,207 nodes; the other LD+TD+ GCNs have <10 nodes. This GCN is light-independent, because all of the coexpression relationships in this GCN hold regardless of the presence or absence of light. Third, the time order of TF genes in each GCN was assigned by the breadth-first search algorithm (9) initiated from a selected node which should be the first up-regulated TF in the GCN. BFS is an algorithm for searching a network graph. It starts with an initial seed and searches all its neighbors (nodes with connecting edges) to form a set of nodes (level 1). Then, the process proceeds from all nodes in level 1 and searches their neighbors (excluding level 1 nodes) to form another set of nodes (level 2) and so on, until all nodes in the network are assigned. Computer programs for the method are available at https://github.com/petitmingchang/TO-GCN (31).
As mentioned earlier, for the light-independent GCN, ZmARF2-1 was selected as the initial node. According to the BFS algorithm, ZmARF2-1 and all nodes coexpressed with it were assigned to level 1 (denoted as L1). Then all nodes coexpressed with any nodes at L1 were assigned to L2, all nodes coexpressed with any nodes in L2 were assigned to L3, and so on, until all nodes in the GCN were assigned.
Coexpressed Gene Sets and Overrepresented Functions in Each TO-GCN Level.
For the TF genes at each level of a TO-GCN, a corresponding set of coexpressed genes can be identified with the same coexpression relationship for adding the genes to the TO-GCN. Since a gene may be coexpressed with TFs in multiple levels, two neighboring gene sets will have some overlapping genes (Fig. 1B).
For each set of genes corresponding to a level in a TO-GCN, the functional enrichment analysis was conducted with the background set of all expressed genes in this study. Fisher’s exact test with false discovery rate (FDR) <0.05 (32) was applied with functional annotations from MapMan (https://mapman.gabipd.org) (Fig. 1B).
Predicting Binding Sites of Maize TF Genes and Their Target Genes.
To predict binding sites of maize TF genes, we collected known TFBSs (position weight matrices; PWMs) of Arabidopsis TF genes from TF databases and literature, including CIS-BP (22), JASPAR (33), Plant Cistrome (34), Franco-Zorrilla et al. (35), and Sullivan et al. (36). If a TF had multiple PWMs from different sources, we took the PWM with the highest information content. As similar DNA-binding domains (DBDs) of TFs have similar DNA sequence preferences, for a maize TF we used its DBD to identify homologous Arabidopsis DBDs with known TF–TFBS pairs in Arabidopsis. Using method 2 of Yu et al. (18), we found coexpressed genes of a maize TF with PCC >0.8 under LD and TD and subjected the genes to gene set enrichment analysis. For those promoters of genes which were enriched in a gene set (FDR < 0.05), we identified overrepresented motifs (PWMs) and tested the conservation among four reference species (Brachypodium distachyon, Oryza sativa, Sorghum bicolor, and Setaria italica) with P value < 10−5. Then the passed PWMs were considered putative TFBMs (transcription factor binding motifs, PWMs). Among the putative TFBMs identified for a TF, the TFBM most similar to the known TFBM of the Arabidopsis TF (DBD sequence similarity with the maize TF DBD sequence >70%) was regarded as the major TFBM for that TF. We then applied the major TFBMs of maize TFs to predict TF target genes by examining the conservation of the TFBSs (location mapped to by TFBMs) in the promoter regions as described in Yu et al. (18). The promoter region of a gene is defined as the 1-kb sequence upstream of the transcription start site of a gene. For example, in the identification of regulators of C4 enzyme genes, we found that the DBD of ZmbHLH43 has 79% identity with the DBD of Arabidopsis PIF3 (AT1G09530), which has a known DNA-binding motif (ID M2863 1.02) in CIS-BP. The promoter of ZmNADP-ME included the DNA sequence which passed our conservation test in three NADP-ME promoters of the four reference species.
Vector Construction.
For EMSA, the full-length cDNA of a predicted TF was cloned into a pet42a vector with the designed primer pair (SI Appendix, Tables S6 and S7). The plasmid was used for Escherichia coli Rosetta (DE3) transformation. For protoplast transfection, genes for GFP and full-length cDNAs of the predicted TFs in maize under the control of the maize ubiquitin 1 promoter were cloned into a pBI221 vector with the designed primer pairs (SI Appendix, Table S6). The plasmid DNA purified using the Maxi Plasmid Kit (Qiagen) was used for transfection.
EMSA Validation.
The procedure was as described in Yu et al. (18) with minor modifications. The biotin-labeled probes (SI Appendix, Tables S8 and S9) were incubated with 1- (∼50 ng), 2-, or 4-fold of GST or recombinant TF protein expressed in and purified from E. coli Rosetta (DE3) for 20 min at 22 °C. Competition experiments were performed with 1- (10 ng), 5-, or 10-fold of unlabeled probes as competitors. The EMSA mixture was separated by a 3.75% polyacrylamide native gel and transferred to a Hybond N+ membrane (GE) by semidry transfer cell (Bio-Rad). The biotin-labeled probe and the TF–probe complexes were detected by streptavidin–HRP conjugates (Life Technologies) with substrates from ECL Plus (GE). The chemiluminescent signals were visualized by the BioSpectrum Imaging System (UVP).
Protoplast Transient Assay.
Mesophyll protoplasts were isolated from leaves of young etiolated maize seedlings as in Chang et al. (20) with minor modifications. Cell concentration was adjusted to 5 × 105 per mL, and 200 μL protoplasts was mixed with plasmid DNA (5 μg pBI221-GFP with 10 μg pBI221 or 10 μg pBI221-TF). Equal volumes of PEG solution (0.6 M mannitol, 0.1 M CaCl2, and 40% PEG 4000) were added, and the tubes were gently inverted to mix the mixture. After incubation for 20 min at room temperature, the protoplasts were collected by centrifugation (150 × g for 2 min) and resuspended in 500 μL incubation solution (0.6 M mannitol, 4 mM KCl, and 4 mM Mes, pH 5.7). The transfected protoplasts were transferred to Falcon culture plates and incubated at 26 °C in the dark for 6 h (37). Protoplasts were harvested by centrifugation at 150 × g for 2 min and used for total RNA extraction.
RNA Extraction and qRT-PCR.
Total RNA was isolated from protoplasts following the procedure of Chang et al. (20), and its quality was examined by NanoDrop (Thermo Scientific) and formaldehyde gel electrophoresis. The first-strand cDNAs were synthesized from 0.5 μg of RNA using the SuperScript III First-Strand Synthesis SuperMix Kit (Invitrogen). qRT-PCR was conducted using 0.01 μg of the cDNA on a LightCycler 480 Instrument System (Roche) with KAPA SYBR FAST qPCR Master Mix and with an initial denaturing step at 95 °C for 5 min, followed by 55 cycles of 95 °C for 10 s, 60 °C for 20 s, and 72 °C for 5 s. The PCR primers used for qRT-PCR are listed in SI Appendix, Table S10. First, the difference in the cycle threshold (Ct) values between the actin gene and a target gene was calculated as deltaCtcontrol (pBI221-GFP with pBI221) or deltaCtTF (pBI221-GFP with pBI221-TF) treatments. Then, the difference between these values was calculated as 2deltaCT(TF − control).
Data Availability.
The Illumina reads were deposited in the Sequence Read Archive under https://www.ncbi.nlm.nih.gov/sra (accession no. SRP140487).
Code Availability.
The source code, documentation, and test datasets are available at https://github.com/petitmingchang/TO-GCN.
Supplementary Material
Acknowledgments
This work was supported by Academia Sinica, Taiwan Grants AS-106-TP-L14-1, -2, and -3.
Footnotes
The authors declare no conflict of interest.
Data deposition: The Illumina reads reported in this paper have been deposited in the Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession no. SRP140487). Computer programs for the methods have been deposited in GitHub, https://github.com/petitmingchang/TO-GCN.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817621116/-/DCSupplemental.
References
- 1.Bar-Joseph Z, Gitter A, Simon I. Studying and modelling dynamic biological processes using time-series gene expression data. Nat Rev Genet. 2012;13:552–564. doi: 10.1038/nrg3244. [DOI] [PubMed] [Google Scholar]
- 2.Jung I, et al. TimesVector: A vectorized clustering approach to the analysis of time series transcriptome data from multiple phenotypes. Bioinformatics. 2017;33:3827–3835. doi: 10.1093/bioinformatics/btw780. [DOI] [PubMed] [Google Scholar]
- 3.Wang P, Kelly S, Fouracre JP, Langdale JA. Genome-wide transcript analysis of early maize leaf development reveals gene cohorts associated with the differentiation of C4 Kranz anatomy. Plant J. 2013;75:656–670. doi: 10.1111/tpj.12229. [DOI] [PubMed] [Google Scholar]
- 4.Liu WY, et al. Anatomical and transcriptional dynamics of maize embryonic leaves during seed germination. Proc Natl Acad Sci USA. 2013;110:3979–3984. doi: 10.1073/pnas.1301009110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slewinski TL, et al. Short-root1 plays a role in the development of vascular tissue and Kranz anatomy in maize leaves. Mol Plant. 2014;7:1388–1392. doi: 10.1093/mp/ssu036. [DOI] [PubMed] [Google Scholar]
- 6.Wang L, et al. Comparative analyses of C4 and C3 photosynthesis in developing leaves of maize and rice. Nat Biotechnol. 2014;32:1158–1165. doi: 10.1038/nbt.3019. [DOI] [PubMed] [Google Scholar]
- 7.Chang YM, et al. 2019 Time-course transcriptomes from maize embryonic leaves grown under total darkness. Sequencing Reads Archive. Available at https://www.ncbi.nlm.nih.gov/sra/SRP140487. Deposited April 2, 2018.
- 8.Slewinski TL, Anderson AA, Zhang C, Turgeon R. Scarecrow plays a role in establishing Kranz anatomy in maize leaves. Plant Cell Physiol. 2012;53:2030–2037. doi: 10.1093/pcp/pcs147. [DOI] [PubMed] [Google Scholar]
- 9.Knuth DE. The Art of Computer Programming. Addison-Wesley; Reading, MA: 1997. [Google Scholar]
- 10.Tanaka M, Kikuchi A, Kamada H. The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 2008;146:149–161. doi: 10.1104/pp.107.111674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002;32:317–328. doi: 10.1046/j.1365-313x.2002.01430.x. [DOI] [PubMed] [Google Scholar]
- 12.Lee S, et al. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 2002;16:646–658. doi: 10.1101/gad.969002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donner TJ, Sherr I, Scarpella E. Regulation of preprocambial cell state acquisition by auxin signaling in Arabidopsis leaves. Development. 2009;136:3235–3246. doi: 10.1242/dev.037028. [DOI] [PubMed] [Google Scholar]
- 14.Bewley JD. Seed germination and dormancy. Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su YH, Liu YB, Zhang XS. Auxin-cytokinin interaction regulates meristem development. Mol Plant. 2011;4:616–625. doi: 10.1093/mp/ssr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fouracre JP, Ando S, Langdale JA. Cracking the Kranz enigma with systems biology. J Exp Bot. 2014;65:3327–3339. doi: 10.1093/jxb/eru015. [DOI] [PubMed] [Google Scholar]
- 17.Slewinski TL. Using evolution as a guide to engineer Kranz-type C4 photosynthesis. Front Plant Sci. 2013;4:212. doi: 10.3389/fpls.2013.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu CP, et al. Transcriptome dynamics of developing maize leaves and genomewide prediction of cis elements and their cognate transcription factors. Proc Natl Acad Sci USA. 2015;112:E2477–E2486. doi: 10.1073/pnas.1500605112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li P, et al. The developmental dynamics of the maize leaf transcriptome. Nat Genet. 2010;42:1060–1067. doi: 10.1038/ng.703. [DOI] [PubMed] [Google Scholar]
- 20.Chang YM, et al. Characterizing regulatory and functional differentiation between maize mesophyll and bundle sheath cells by transcriptomic analysis. Plant Physiol. 2012;160:165–177. doi: 10.1104/pp.112.203810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borba AR, et al. Synergistic binding of bHLH transcription factors to the promoter of the maize NADP-ME gene used in C4 photosynthesis is based on an ancient code found in the ancestral C3 state. Mol Biol Evol. 2018;35:1690–1705. doi: 10.1093/molbev/msy060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weirauch MT, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levesque MP, et al. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 2006;4:e143. doi: 10.1371/journal.pbio.0040143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Caemmerer S, Quick WP, Furbank RT. The development of C4 rice: Current progress and future challenges. Science. 2012;336:1671–1672. doi: 10.1126/science.1220177. [DOI] [PubMed] [Google Scholar]
- 25.Hibberd JM, Sheehy JE, Langdale JA. Using C4 photosynthesis to increase the yield of rice-rationale and feasibility. Curr Opin Plant Biol. 2008;11:228–231. doi: 10.1016/j.pbi.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 26.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C, et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin JJ, Yu CP, Chang YM, Chen SC, Li WH. Maize and millet transcription factors annotated using comparative genomic and transcriptomic data. BMC Genomics. 2014;15:818. doi: 10.1186/1471-2164-15-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang YM, et al. 2019 Pipeline of time-ordered gene coexpression network (TO-GCN) construction. Github. Available at https://github.com/petitmingchang/TO-GCN. Deposited August 15, 2018.
- 32.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 33.Khan A, et al. JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46:D260–D266. doi: 10.1093/nar/gkx1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Malley RC, et al. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell. 2016;165:1280–1292. doi: 10.1016/j.cell.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franco-Zorrilla JM, et al. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci USA. 2014;111:2367–2372. doi: 10.1073/pnas.1316278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullivan AM, et al. Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Rep. 2014;8:2015–2030. doi: 10.1016/j.celrep.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 37.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Illumina reads were deposited in the Sequence Read Archive under https://www.ncbi.nlm.nih.gov/sra (accession no. SRP140487).