Significance
Prior work has identified many sex differences in the brain, including during brain aging and in neurodegenerative diseases. Notably, many of these studies are performed by comparing age-matched females and males. Evolutionary theorists have predicted that females might have more youthful brains (neoteny) as compared with males, but until now findings in support of this theory have been limited to postmortem transcriptional analysis, some of which is contradictory. To test this hypothesis in vivo, we analyzed sex differences in a unique brain PET dataset in over 200 normal human adults across the adult life span. We find that in terms of brain metabolism, the adult female brain is on average a few years younger than the male brain.
Keywords: brain aging, brain metabolism, sex differences, neoteny, machine learning
Abstract
Sex differences influence brain morphology and physiology during both development and aging. Here we apply a machine learning algorithm to a multiparametric brain PET imaging dataset acquired in a cohort of 20- to 82-year-old, cognitively normal adults (n = 205) to define their metabolic brain age. We find that throughout the adult life span the female brain has a persistently lower metabolic brain age—relative to their chronological age—compared with the male brain. The persistence of relatively younger metabolic brain age in females throughout adulthood suggests that development might in part influence sex differences in brain aging. Our results also demonstrate that trajectories of natural brain aging vary significantly among individuals and provide a method to measure this.
Human brain aging is characterized by varying trajectories. Some individuals succumb to rapid cognitive decline, whereas other individuals retain their cognitive abilities as they age beyond the typical human life span. Accordingly, it is important to understand the factors that influence brain aging, particularly in the context of an aging population. In humans, normal aging is associated with a decline in brain metabolism (1–4). Our recent multitracer PET brain imaging data demonstrate that as the brain ages, its resting metabolism gradually shifts away from a mixture of nonoxidative and oxidative use of glucose to predominantly oxidative metabolism (4, 5). This occurs even in a cognitively normal, amyloid-negative cohort, suggesting that neurodegeneration alone is unlikely to explain this metabolic shift. However, the reasons for this metabolic brain aging phenomenon are currently unknown.
One potential view of the normal metabolic changes in the aging brain is that it reflects a gradual cessation of ongoing developmental and maturational processes. In comparison with other primates, the human brain has a high degree of neoteny, i.e., a prolonged persistence/maintenance of developmental characteristics, based on ex vivo transcriptomic, metabolic, and microstructural data, as well as in vivo metabolic brain imaging data (5–9). Some regions of the brain show a heightened degree of transcriptional and metabolic neoteny (5), and these same regions show the most loss of nonoxidative glucose use, i.e., aerobic glycolysis, with typical aging (4).
Thus, factors that influence brain development, such as sexual differentiation, might also be critical in determining the trajectory of brain aging. Sex differences influence structural aspects of brain development in children and during early adulthood (10) and also delay developmental changes in cerebral blood flow during early adulthood (11), raising the possibility that the adult female brain retains more youthful, i.e., neotenous, features compared with the adult male brain. Transcriptomic data seem to support this view: aging-related gene expression changes are regionally and genetically sexually dimorphic. Specifically, in females there is less aging-related change in genes related to energy production and protein synthesis (12), though increased aging-related gene expression related to the immune system and reactive changes to environmental stressors (13). In a more recent study, peak changes in a brain transcriptome were found to occur later in females than in males in both humans and mice (14).
Here we apply our multiparametric metabolic brain imaging data to examine the influence of sex on brain aging in vivo. These data derive from a cohort of 205, cognitively normal, individuals, including 40 that were brain amyloid positive, across the adult life span (4). We developed a machine learning algorithm on these data to derive “metabolic brain age.” By comparing this metabolic brain age to an individual’s actual age, the machine learning algorithm can determine whether an individual’s brain appears metabolically younger or older than it should. Hence, this provides a single measure of accelerated or decelerated metabolic brain aging based on multiparametric and multiregional metabolic data, which thereby allows us to determine whether females differ from males with respect to their aging brain.
Results
Participants and Data.
Regional PET imaging data from a cohort of 205, 20- to 82-y-old, cognitively normal adults participating in six different studies at our institution were obtained and processed, as described previously (4). These PET data include measurements of regional total glucose use (CMRGlc), oxygen consumption (CMRO2), and cerebral blood flow (CBF). In addition, we calculated regional aerobic glycolysis (AG)—the fraction of glucose use presumably not accounted for by oxidative metabolism—from these data, defined as the absolute molar difference between age-normalized CMRGlc and CMRO2. These data were further quantile normalized for each metabolic parameter individually (here CBF is included as a “metabolic” parameter to simplify our nomenclature), equalizing the mean and statistical distribution of brain metabolism across 79 brain regions for each of 184 PET imaging sessions in 165 cognitively normal, young or amyloid-negative individuals, including 19 repeat sessions. Thus, what differentiates the normalized brain metabolism data in one individual from another is primarily the rank order of brain regions for each metabolic parameter.
Calculation of Metabolic Brain Age.
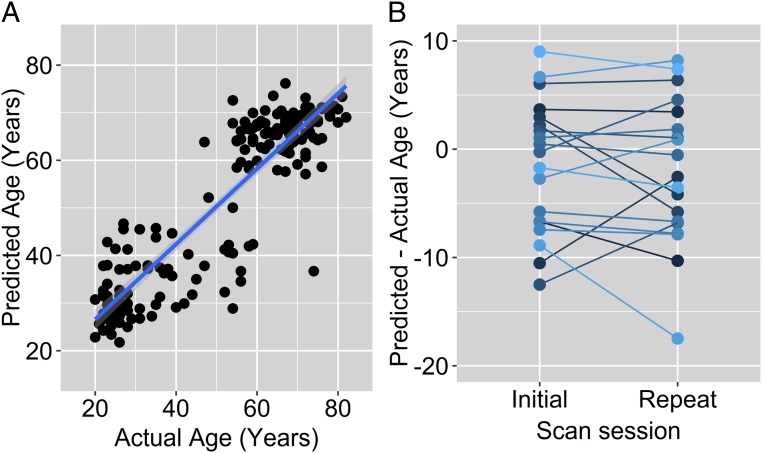
A supervised machine learning algorithm, random forest regression with bias correction (15), was applied to the quantile normalized brain metabolism data and trained and tested against the actual chronological age of the participants. Ten-fold cross-validation demonstrates that the predicted age based on this algorithm—defined as metabolic brain age—closely matches the actual chronological age of the participants (Pearson’s r = 0.88–0.90 over 10 runs) (Fig. 1A); the difference between metabolic brain age and actual chronological age varied across participants with a typical SD of 8.6 y, median absolute deviation of ∼5.4 y, and ranging from −18 to +16 y. A bootstrap analysis with randomly assorted actual ages confirmed that this result was not due to spurious feature detection by the random forest algorithm (for n = 100 null permutations, Pearson’s r ranged from −0.26 to 0.25).
Fig. 1.
Machine learning predicts participant age from normalized metabolic brain PET data. (A) Random forest regression with bias correction was trained on 184 quantile normalized metabolic brain PET data to predict participant age. Ten-fold validation was performed, and the dots represent the collated test cases. The resulting predicted age—described as metabolic brain age—correlates highly with actual age (Pearson’s r = 0.89, bootstrap Z score >3.8, P < 0.0001). (B) Nineteen participants underwent repeat PET imaging 1–2 y after their initial PET. The difference between their metabolic brain age and actual age, while variable among individuals, remained largely consistent within individuals between repeat tests (Pearson’s r = 0.80, P < 0.0002).
To assess the influence of each metabolic parameter (AG, CMRGlc, CMRO2, or CBF) on the accuracy of the machine learning algorithm, we recalculated metabolic brain age after removing each of the metabolic parameters from the dataset. This resulted in only minor changes in accuracy between metabolic brain age and actual age (Pearson’s r = 0.87–0.90), with removing AG causing the largest loss of accuracy (SI Appendix, Fig. S1A). Further, the BorutaR package, which identifies features that are important to maximize random forest accuracy, found that at least 59 of 316 features involving all four metabolic parameters are important for determining metabolic brain age (SI Appendix, Fig. S2); thus we use all of the metabolic data when subsequently calculating metabolic brain age.
To further validate metabolic brain age, we assessed the stability of this metric over time within individuals. The 184 PET imaging sessions included repeat sessions 1–2 y later in 19 participants (Fig. 1B). The mean difference in metabolic brain age between the two sessions was 1.1 y (actual mean time difference 1.6 y). There was a high correlation between initial and subsequent metabolic brain age after correcting for actual age (n = 19 pairs of PET scans, Pearson’s r = 0.80, P < 0.0002), suggesting that metabolic brain age remains consistent within individuals over a short period of time.
Sex Differences in Metabolic Brain Age.
Self-reported participant sex was gathered consistently in each of the six studies that comprise the whole dataset; we acknowledge that genetically determined sex was not available in this retrospectively collected data, but the frequency of discrepancies between genetic versus reported sex is likely to be too low to significantly influence our results.
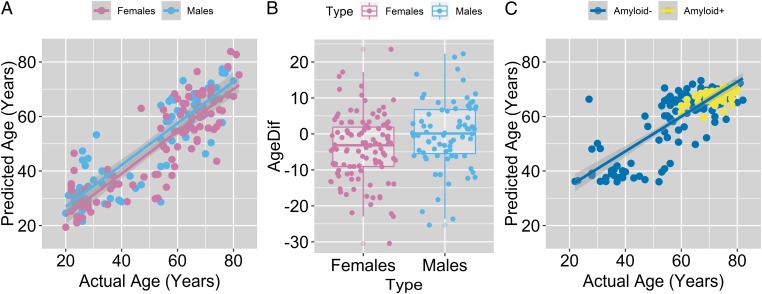
Using both males and females to train the random forest algorithm in calculating metabolic brain age would necessarily obscure sex differences since the machine learning algorithm would be inclined to account for these differences in maximizing its accuracy. On the other hand, if female brains are truly more neotenous than male brains, then they should look “younger” when the algorithm is trained on males only. The random forest regression with bias correction algorithm was thus trained first on the male cohort only, and then “tested” on the female cohort. For both the training (males) and test data (females), metabolic brain age again correlated strongly with actual chronological age (Pearson’s r > 0.88) (Fig. 2A). However, the mean metabolic brain age (minus actual age) was on average 3.8 y less for females compared with males (n = 108 females and 76 males, 95% CI 1.0–6.6 y, P < 0.010 t test, Cohen’s d > 0.40) (Fig. 2B, see also SI Appendix, Fig. S3A). To confirm that the female–male difference in metabolic brain age was not specific to training the data on males, we also trained the algorithm with female data only and found the predicted metabolic brain age for males to be 2.4 y older compared with females (P < 0.038 t test, one-sided; in a combined bootstrap analysis, the likelihood that female metabolic brain age would be younger than male metabolic brain age by 3.8 y in either of the two tests was P < 0.014). Despite these sex differences in metabolic brain aging, random forest was, surprisingly, relatively poor at distinguishing males from females using the brain metabolism data (n = 184, gender predicted accurately in 66% of instances).
Fig. 2.
Metabolic brain age is significantly lower in females. (A) To identify sex differences in metabolic brain age without allowing sex-related age imbalances to bias the machine learning algorithm, random forest regression was first performed on males and then tested on females. Each dot represents a different individual PET session and lines represent best fits. Note that as a group, females across the life span have a lower predicted versus actual age compared with males. (B) Metabolic brain age for both groups correlated with their actual age, but the difference between the predicted and actual age was lower for females compared with males (mean difference females versus males −3.8 y, n = 108 females and 76 males, 95% CI 1.0–6.6 y, P < 0.01 t test); the boxplot hinges represent the mean and the 1st and 3rd quartiles of the data. (C) Metabolic brain age (“predicted age”) was assessed in 40 amyloid brain PET imaging-positive individuals after training the random forest regression algorithm on the 184 PET sessions in young and/or amyloid-negative individuals. This revealed no significant difference in metabolic brain age between the two groups. Subtle differences between metabolic brain age (predicted age) between the figures in A and C are likely due to different cohorts being used to train the random forest algorithm.
It is possible that this sex difference in metabolic brain age could be driven by a handful of brain regions and/or metabolic parameters. The procedure of calculating a difference between female and male metabolic brain age was thus repeated on a random selection of 60 of the 79 brain regions in 1,000 permutations; in all cases females demonstrate a mean metabolic brain age (minus actual age) that was younger than males (ranging 1.4–7.9 y younger across all 1,000 permutations, mean 4.7 y younger, see SI Appendix, Fig. S3B), demonstrating that this finding cannot be ascribed to a small subgroup (<20) of brain regions. To determine how female versus male differences in metabolic brain age depended on specific metabolic parameters, we recalculated the sex-based metabolic brain age difference after removing each metabolic parameter (AG, CMRGlc, CMRO2, or CBF) separately. Removing either CMRGlc or AG decreased the sex-based age difference slightly (2.7 and 3.3 y, respectively), but removing CBF increased the difference further (5.3 y), suggesting that female–male metabolic brain age differences are more dependent on brain glucose use than CBF or CMRO2 (SI Appendix, Fig. S1B).
Brain Amyloid and Metabolic Brain Age.
One possible confounding source of interindividual variability in metabolic brain age might be brain amyloid deposition, which has been reported to trend slightly more frequently in aging women compared with men above the age of 70 y (16). After using the initial 184 PET dataset in cognitively normal, young or amyloid-negative individuals to train the bias-corrected random forest regression algorithm as per above, we applied the trained algorithm to new PET imaging sessions obtained in a separate cohort of cognitively normal participants who were amyloid imaging positive (n = 40, ages 60–80, 10 underwent two PET sessions). Compared with age-matched amyloid-negative participants, asymptomatic amyloid-positive participants did not have a significantly different metabolic brain age (mean metabolic brain age: amyloid negative = 66.5 y, amyloid positive = 67.1 y; P = 0.27, two-tailed t test), suggesting that brain amyloid status does not significantly account for interindividual variability in metabolic brain age (Fig. 2C).
Discussion
Our results demonstrate that from an in vivo metabolic view, throughout the adult life span the typical female brain is more youthful, i.e., metabolically neotenous, than the male brain. There is no indication in the data that this sex difference in metabolic brain age varies between young and older adults, though it should be noted that our study contains relatively fewer data points in middle-aged individuals (35–50 y). Nonetheless, our findings are consistent with other data showing in females less loss of cerebral blood flow following puberty (11), more brain glycolysis during young adulthood (17), less loss of protein synthesis-related gene expression during aging (12), and a delay in the peak transition point of brain gene expression (14). Our results suggest that female brain neoteny is present in young adults and persists throughout the adult life span, suggesting that sex differences during brain development set the stage for subsequent trajectories of brain aging.
The implication of increased metabolic neoteny in the female brain with regards to neurodegenerative diseases warrants further investigation. An intriguing hypothesis is that higher neoteny in the female brain might provide some degree of resilience to aging-related changes. This is supported by data from a previously reported cohort that found evidence for less memory decline and hippocampal atrophy in aging females compared with males (16). Similarly, in the Baltimore Longitudinal Study of Aging, noncognitively impaired women outperformed males on most cognitive tests and on a few had less steeper decline than males (18). Heightened glycolysis in the adult female brain might explain some of this resilience, since brain aerobic glycolysis is involved in learning and neurite growth (19, 20). However, it is likely that sex influences on brain aging are multifactorial and complex since evolution and natural selection effectively perform combinatorial optimization of multiple biological processes simultaneously (21–23). Further, sex differences in the development and progression of pathological processes, such as amyloid or tau deposition, and in the clinical recognition of cognitive impairment will likely further complicate how sex influences the development of dementia (24).
The reasons for sex differences in brain metabolism are not yet well elucidated. Hormones might be a specific mediating factor; estrogens enhance synaptic plasticity in rodent models and thereby may help to maintain more youthful brain metabolism (25, 26). A prior study found significantly lower CMRGlc in several brain regions in postmenopausal women compared with premenopausal women (27). We did not have sufficient data in this retrospective cohort to accurately assess menopausal status in our study participants, although the sex differences in metabolic brain age appear to persist beyond the age of 60, suggesting that this finding is unlikely to be entirely due to menopausal status. On the other hand, the effects of hormones on brain metabolism might occur at a younger age, “setting” in females a younger baseline metabolic brain age during or soon after puberty at the start of adulthood. Indeed, cerebral blood flow patterns are less affected by puberty in females compared with males (11), which might allow females to begin adulthood with a more “youthful” pattern of brain metabolism as seen here.
Additional sex differences may also be relevant. Intrinsic cellular, metabolic, and immune system sexual dimorphism have been implicated in various other organ systems (28, 29) as well as tumor metabolism (30); similar factors might explain sex differences in brain metabolism (31). Prolongation of metabolic youth in the female brain also parallels increased female longevity in humans, though mechanisms underlying the latter are similarly not well understood (32).
Note that this study cannot separate the effects of sex—a biologically determined characteristic—from gender, which includes societal influences. Societal effects on the environment and lifestyle of females, including their decision to participate in brain imaging studies, might have influenced our results due to sex-specific cohort effects. As the effect size of sex on metabolic brain age was relatively modest, our results require further testing in an independent cohort to test for generalizability and reproducibility. Also, investigations into how sex determines brain metabolism and aging are needed, particularly to identify sex-related mechanisms that might increase or decrease the risk of pathologic or accelerated trajectories of brain aging.
The absence of significant metabolic brain age differences due to brain amyloid deposition is intriguing. It is possible that amyloid deposition does not appreciably affect brain metabolism until a certain threshold of neurodegeneration is met. Alternatively, the cohort of cognitively normal brain amyloid-positive participants in this study might be enriched with individuals who are to some degree “metabolically resilient” to the effects of amyloid deposition, thereby canceling out potential amyloid effects on metabolic brain age. Also, the relationship between biomarkers of AD pathology and brain metabolism may be complex. For example, in a subset of amyloid-positive individuals with tau PET scans available, we found an association between brain AG and higher tau deposition but not in amyloid-negative individuals (33). Further prospective and longitudinal studies will help to investigate these possibilities.
Our results demonstrate the value of applying machine learning to assess factors that influence brain aging (34). Also, whereas prior work on sex differences in the adult brain often compared age-matched females to males, our current results argue that some of these differences might simply arise from females having more neotenous-appearing brains. Measures such as metabolic brain age might be useful in predicting the risk of cognitive decline and in identifying other factors that could potentially improve or worsen the trajectory of human brain aging. Further investigations are now needed to validate metabolic brain age in other cohorts, determine its predictive potential, and answer why sex might affect it.
Methods
Participants and PET Data.
The data for this study have been previously analyzed and reported (4). For full details, please refer to this prior publication. Briefly, 205 cognitively normal individuals (59% women, self-reported sex/gender) aged 20–82 y were recruited from six different studies performed at the Washington University School of Medicine. Participants were screened for neurological, psychiatric, and systemic medical illnesses, and excluded if they had contraindications to MRI. All participants reported normal cognitive status, and nearly all above the age of 60 underwent amyloid imaging (n = 157) of which 40 were brain amyloid imaging positive. Of the nonamyloid-positive participants, 19 underwent repeat metabolic brain PET imaging within 1–2 y (mean 1.6 y). All assessments and imaging procedures were approved by the Human Research Protection Office and Radioactive Drug Research Committee at our institution. Written consent was provided from each participant.
PET Imaging.
All individuals underwent metabolic brain PET and structural 3T MRI for registration and regional assessment, as described previously (4). Briefly, 18F-FDG and 15O-O2, -CO, and -H2O PET scans were performed on all of the participants while they were in the awake, eyes-closed state. PET images were coregistered to individual MRI T1 weighed sequences. The registered PET data were used to derive regional maps of CMRGlc, CMRO2, CBF, and AG after partial volume correction using the regional spread function approach based on FreeSurfer-based subcortical gray and white matter segmentation and Desikan-Killiany Atlas-based cortical regions of interest (35–37). The final data were normalized to whole brain age-normative data and summarized according to these regions of interest for each individual session. A subset of participants also underwent 11C-labeled Pittsburgh compound B brain PET imaging, which was defined as being “positive” when mean cortical (from predefined regions of interest) standard uptake value ratios compared with the cerebellum was >1.42, a value commensurate with a mean cortical binding potential of 0.18 and consistent with prior studies from our group (33, 38, 39).
Machine Learning Algorithm.
The metabolic PET data were quantile normalized across individuals independently for each metabolic parameter according to brain regions. Random forest regression was performed using the randomForest package in R (v3.4.1). Since random forest regression biases values on either end of the range toward the mean, bias correction was performed using smooth spline fits (span = 1) on the random forest regression results (15); altering the span value from 0.5 to 1.5 did not affect the direction of our results. Investigating feature importance for machine learning algorithms remains an active area of development; we applied the Boruta package in R to the normalized dataset to identify which of the 316 (79 regions per the Desikan-Killiany Atlas × 4 metabolic parameters) were most important in maximizing random forest accuracy (35). Similarly, we performed random forest regression with bias correction using multiple subsets of the data to determine how specific aspects of the data (regional or metabolic) influence our results, as described above.
Statistics.
All statistics were performed using R (v3.4.1). The differences between metabolic brain age and actual age is approximately normally distributed across and within groups, but bootstrap analyses (n = 1,000 random tests in each analysis) were used to confirm significant findings from t tests to account for potential nonparametric effects from the machine learning algorithm.
Data and Software Availability.
CSV files are provided as Dataset S1, which contains regional, age-normalized, partial volume corrected values for CMRGlc, CMRO2, CBF, and AG and additional results required for processing. An R script that reproduces the results presented here is also provided.
Supplementary Material
Acknowledgments
We are continually grateful for our participants’ time and effort in this and other studies. The data presented here were the result of several independently funded efforts including grants from the Barnes-Jewish Hospital Foundation, Charles F. and Joanne Knight, the James S. McDonnell Foundation, the McDonnell Center for Systems Neuroscience (22-3922-26239N), and the National Institutes of Health (NS06833, NS057901, P50 AG05681, P01 AG003991, R01 AG053503, R01 AG057536, and UF1AG032438).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815917116/-/DCSupplemental.
References
- 1.Kety SS. Human cerebral blood flow and oxygen consumption as related to aging. J Chronic Dis. 1956;3:478–486. doi: 10.1016/0021-9681(56)90146-1. [DOI] [PubMed] [Google Scholar]
- 2.Kuhl DE, Metter EJ, Riege WH, Phelps ME. Effects of human aging on patterns of local cerebral glucose utilization determined by the [18F]fluorodeoxyglucose method. J Cereb Blood Flow Metab. 1982;2:163–171. doi: 10.1038/jcbfm.1982.15. [DOI] [PubMed] [Google Scholar]
- 3.Martin AJ, Friston KJ, Colebatch JG, Frackowiak RS. Decreases in regional cerebral blood flow with normal aging. J Cereb Blood Flow Metab. 1991;11:684–689. doi: 10.1038/jcbfm.1991.121. [DOI] [PubMed] [Google Scholar]
- 4.Goyal MS, et al. Loss of brain aerobic glycolysis in normal human aging. Cell Metab. 2017;26:353–360.e3. doi: 10.1016/j.cmet.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goyal MS, Hawrylycz M, Miller JA, Snyder AZ, Raichle ME. Aerobic glycolysis in the human brain is associated with development and neotenous gene expression. Cell Metab. 2014;19:49–57. doi: 10.1016/j.cmet.2013.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Somel M, et al. Transcriptional neoteny in the human brain. Proc Natl Acad Sci USA. 2009;106:5743–5748. doi: 10.1073/pnas.0900544106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Somel M, Rohlfs R, Liu X. Transcriptomic insights into human brain evolution: Acceleration, neutrality, heterochrony. Curr Opin Genet Dev. 2014;29:110–119. doi: 10.1016/j.gde.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Petanjek Z, et al. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc Natl Acad Sci USA. 2011;108:13281–13286. doi: 10.1073/pnas.1105108108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu X, et al. Rapid metabolic evolution in human prefrontal cortex. Proc Natl Acad Sci USA. 2011;108:6181–6186. doi: 10.1073/pnas.1019164108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gennatas ED, et al. Age-related effects and sex differences in gray matter density, volume, mass, and cortical thickness from childhood to young adulthood. J Neurosci. 2017;37:5065–5073. doi: 10.1523/JNEUROSCI.3550-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Satterthwaite TD, et al. Impact of puberty on the evolution of cerebral perfusion during adolescence. Proc Natl Acad Sci USA. 2014;111:8643–8648. doi: 10.1073/pnas.1400178111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berchtold NC, et al. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci USA. 2008;105:15605–15610. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan Y, Chen YP, Boyd-Kirkup J, Khaitovich P, Somel M. Accelerated aging-related transcriptome changes in the female prefrontal cortex. Aging Cell. 2012;11:894–901. doi: 10.1111/j.1474-9726.2012.00859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skene NG, Roy M, Grant SG. A genomic lifespan program that reorganises the young adult brain is targeted in schizophrenia. eLife. 2017;6:e17915. doi: 10.7554/eLife.17915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang GY, Lu Y. Bias-corrected random forests in regression. J Appl Stat. 2012;39:151–160. [Google Scholar]
- 16.Jack CR, Jr, et al. Age, sex, and APOE ε4 effects on memory, brain structure, and β-amyloid across the adult life span. JAMA Neurol. 2015;72:511–519. doi: 10.1001/jamaneurol.2014.4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aanerud J, Borghammer P, Rodell A, Jonsdottir KY, Gjedde A. Sex differences of human cortical blood flow and energy metabolism. J Cereb Blood Flow Metab. 2016;37:2433–2440. doi: 10.1177/0271678X16668536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarrey AC, An Y, Kitner-Triolo MH, Ferrucci L, Resnick SM. Sex differences in cognitive trajectories in clinically normal older adults. Psychol Aging. 2016;31:166–175. doi: 10.1037/pag0000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon BJ, et al. Brain aerobic glycolysis and motor adaptation learning. Proc Natl Acad Sci USA. 2016;113:E3782–E3791. doi: 10.1073/pnas.1604977113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segarra-Mondejar M, et al. Synaptic activity-induced glycolysis facilitates membrane lipid provision and neurite outgrowth. EMBO J. 2018;37:e97368. doi: 10.15252/embj.201797368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kauffman SA. Spin Glasses and Biology. World Scientific; Singapore: 1992. pp. 61–100. [Google Scholar]
- 22.Forrest S. Genetic algorithms: Principles of natural selection applied to computation. Science. 1993;261:872–878. doi: 10.1126/science.8346439. [DOI] [PubMed] [Google Scholar]
- 23.Muhlenbein H, Gorgesschleuter M, Kramer O. Evolution algorithms in combinatorial optimization. Parallel Comput. 1988;7:65–85. [Google Scholar]
- 24.Ferretti MT, et al. Women’s Brain Project and the Alzheimer Precision Medicine Initiative Sex differences in Alzheimer disease–The gateway to precision medicine. Nat Rev Neurol. 2018;14:457–469. doi: 10.1038/s41582-018-0032-9. [DOI] [PubMed] [Google Scholar]
- 25.Gould E, Woolley CS, Frankfurt M, McEwen BS. Gonadal steroids regulate dendritic spine density in hippocampal pyramidal cells in adulthood. J Neurosci. 1990;10:1286–1291. doi: 10.1523/JNEUROSCI.10-04-01286.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarthy MM. Estradiol and the developing brain. Physiol Rev. 2008;88:91–124. doi: 10.1152/physrev.00010.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mosconi L, et al. Perimenopause and emergence of an Alzheimer’s bioenergetic phenotype in brain and periphery. PLoS One. 2017;12:e0185926. doi: 10.1371/journal.pone.0185926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson LR, et al. Type 2 diabetes, obesity, and sex difference affect the fate of glucose in the human heart. Am J Physiol Heart Circ Physiol. 2015;308:H1510–H1516. doi: 10.1152/ajpheart.00722.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadkhodayan A, et al. Sex affects myocardial blood flow and fatty acid substrate metabolism in humans with nonischemic heart failure. J Nucl Cardiol. 2017;24:1226–1235. doi: 10.1007/s12350-016-0467-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen GK, Mellnick VM, Yim AK, Salter A, Ippolito JE. Synergy of sex differences in visceral fat measured with CT and tumor metabolism helps predict overall survival in patients with renal cell carcinoma. Radiology. 2018;287:884–892. doi: 10.1148/radiol.2018171504. [DOI] [PubMed] [Google Scholar]
- 31.McCarthy MM, Pickett LA, VanRyzin JW, Kight KE. Surprising origins of sex differences in the brain. Horm Behav. 2015;76:3–10. doi: 10.1016/j.yhbeh.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Austad SN, Fischer KE. Sex differences in lifespan. Cell Metab. 2016;23:1022–1033. doi: 10.1016/j.cmet.2016.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vlassenko AG, et al. Aerobic glycolysis and tau deposition in preclinical Alzheimer’s disease. Neurobiol Aging. 2018;67:95–98. doi: 10.1016/j.neurobiolaging.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cole JH, Franke K. Predicting age using neuroimaging: Innovative brain ageing biomarkers. Trends Neurosci. 2017;40:681–690. doi: 10.1016/j.tins.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 35.Desikan RS, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 36.Fischl B. FreeSurfer. Neuroimage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su Y, et al. Dominantly Inherited Alzheimer Network Partial volume correction in quantitative amyloid imaging. Neuroimage. 2015;107:55–64. doi: 10.1016/j.neuroimage.2014.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vlassenko AG, et al. Imaging and cerebrospinal fluid biomarkers in early preclinical Alzheimer disease. Ann Neurol. 2016;80:379–387. doi: 10.1002/ana.24719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mintun MA, et al. [11C]PIB in a nondemented population: Potential antecedent marker of Alzheimer disease. Neurology. 2006;67:446–452. doi: 10.1212/01.wnl.0000228230.26044.a4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.