Significance
Voltage-gated sodium channels play a central role in cellular excitability and are key targets for drug development. Recent breakthroughs in high-resolution cryo-electron microscopy protein structure determination, Rosetta computational protein structure modeling, and multimicrosecond molecular dynamics simulations are empowering advances in structural biology to study the atomistic details of channel−drug interactions. We used Rosetta structural computational modeling and molecular dynamics simulations to study the interactions of antiarrhythmic and local anesthetic drugs with cardiac sodium channel. Our results provide crucial atomic-scale mechanistic insights into the channel–drug interactions, necessary for the rational design of novel modulators of the human cardiac sodium channel to be used for the treatment of cardiac arrhythmias.
Keywords: sodium channel, local anesthetics, antiarrhythmic drugs, docking, molecular dynamics
Abstract
The human voltage-gated sodium channel, hNaV1.5, is responsible for the rapid upstroke of the cardiac action potential and is target for antiarrhythmic therapy. Despite the clinical relevance of hNaV1.5-targeting drugs, structure-based molecular mechanisms of promising or problematic drugs have not been investigated at atomic scale to inform drug design. Here, we used Rosetta structural modeling and docking as well as molecular dynamics simulations to study the interactions of antiarrhythmic and local anesthetic drugs with hNaV1.5. These calculations revealed several key drug binding sites formed within the pore lumen that can simultaneously accommodate up to two drug molecules. Molecular dynamics simulations identified a hydrophilic access pathway through the intracellular gate and a hydrophobic access pathway through a fenestration between DIII and DIV. Our results advance the understanding of molecular mechanisms of antiarrhythmic and local anesthetic drug interactions with hNaV1.5 and will be useful for rational design of novel therapeutics.
Voltage-gated sodium channels (NaV) are transmembrane proteins that give rise to action potential generation and propagation in excitable cells. There are nine human NaV (hNaV) channel subtypes expressed in neuronal, cardiac, and muscle cells (1). The cardiac NaV channel (NaV1.5) plays a central role in congenital and acquired cardiac arrhythmias and has been an important target for antiarrhythmic drug development (2–4). Long-standing failures in drug treatment of heart rhythm disturbances and many other syndromes stem from a persistent failure to predict the effective or harmful action of drugs. For example, the Cardiac Arrhythmia Suppression Trial (5) and Survival with Oral D-Sotalol (6) clinical trials showed that common antiarrhythmic drugs, such as encainide and flecainide, increased mortality and risk of sudden cardiac death in patients. Thirty years later, there is still no effective preclinical methodology to differentiate useful or potentially harmful drugs at the molecular level. Mechanistic understanding of drug interactions with NaV channels at the atomic scale is needed to develop and screen novel drugs to reveal the mechanisms of drug failure or efficacy for treatment of cardiovascular and other disorders and to minimize side effects.
NaV channels respond dynamically to changes in cell membrane voltage and adopt distinct conformational states: open (conducting), closed (nonconducting), and inactivated (nonconducting). NaV channels contain four homologous domains (DI to DIV), with each domain consisting of a voltage-sensing domain (VSD) containing transmembrane segments S1 to S4 and a pore domain (PD) containing transmembrane segments S5 and S6 connected by a loop region with P1 and P2 helices forming selectivity filter (SF). Each VSD senses changes in membrane potential that lead to movement of its S4 segment which can, in turn, trigger channel activation (pore opening) or channel deactivation (pore closing) at the intracellular gate. The intracellular linker between DIII and DIV contains a hydrophobic isoleucine−phenylalanine−methionine (IFM) motif, which contributes to a fast inactivation gating mechanism, resulting in rapid termination of Na+ conduction subsequent to the channel opening (7–9). This inactivation process plays critical roles in NaV channel function and drug binding (10, 11).
Forty years ago, Hille (12) proposed two distinct access pathways for local anesthetics to the central binding site: the hydrophobic pathway through the membrane and the hydrophilic pathway through the intracellular gate. Many antiarrhythmic and local anesthetic drugs are weak bases that exist in equilibrium between both neutral and charged forms at physiological pH. Neutral drugs may access the pore lumen binding site through both hydrophobic and hydrophilic pathways (13), but charged drugs are much more likely to access the pore binding site through the hydrophilic pathway, due to a large energetic penalty for traversing a lipid membrane (14). Extensive electrophysiological and site-directed mutagenesis experiments have identified a key receptor site for antiarrhythmic and local anesthetic drugs within the eukaryotic NaV channel pore lumen (15–18). Mutations of two conserved aromatic residues in the DIV segment S6 (DIVS6) of NaV channels, F1760 and Y1767 (hNaV1.5 numbering), significantly reduce antiarrhythmic and local anesthetic drug binding (15, 16). Other key residues for drug binding within the pore lumen have been identified in DIS6 and DIIIS6 segments (17, 18). In addition, mutations within the NaV channel SF region can affect drug binding, either through enhancement of slow inactivation or formation of alternative access pathway (19–21).
Structural studies have advanced our mechanistic understanding of NaV channel−drug interaction mechanisms. The first crystal structure of the bacterial NaV channel NaVAb revealed open fenestrations within the PD (22), which supported the hypothesis that drugs can access the binding site within the pore lumen through the hydrophobic pathway. Crystal structures of NaVMs and voltage-gated calcium (CaV) Ab channels have been determined with drugs bound near the fenestration regions or in the pore lumen, suggesting the possibility of similar drug binding receptor sites in eukaryotic NaV channels (23, 24). The first high-resolution structures of eukaryotic NaV channels have recently been resolved using cryo-electron microscopy (cryoEM). The American cockroach NaVPaS channel structures have been solved in a closed state (25, 26), and electric eel NaV1.4 (eeNaV1.4) channel structure has been solved in a partially open and presumably inactivated state (27). These structures have unlocked new opportunities to study drug interactions with eukaryotic NaV channels at the atomic scale.
The Rosetta computational modeling software (28, 29) has been used to study conformational changes in NaV, voltage-gated potassium (KV), CaV, and TRPV1 channels (30–33) and peptide toxin interactions with NaV, KV, and TRPV1 channels (34–36). RosettaLigand flexible docking (37) has been used to study small-molecule interactions with NaV, TRPV1, and calcium-activated K+ channels (38–41). Molecular dynamics (MD) simulations have previously revealed drug binding and access pathways to bacterial NaV channels (13, 42–44). Molecular docking of antiarrhythmic, local anesthetic, and anticonvulsant drugs with homology models of a eukaryotic NaV1.4 channel based on bacterial NaVMs channel in an open state has recently revealed electroneutral and cationic drug interactions with the phenylalanine in the DIVS6 segment (F1760 in hNaV1.5) and SF region (45). Differences in binding of neutral and charged local anesthetics have been recently studied using the bacterial NaVMs channel in an open state and the eukaryotic NaVPaS channel in a closed state (46). Structural, experimental, and modeling studies have all provided a better understanding of drug interactions with bacterial NaV channels and models of eukaryotic NaV channels in open or closed states. However, atomistic details remain elusive for antiarrhythmic and local anesthetic drug access pathways, specific binding sites, and stoichiometry of binding to eukaryotic NaV channels in an inactivated state, which forms a high-affinity drug binding site (47).
In this study, we used Rosetta to build a model of the hNaV1.5 channel in a partially open and presumably inactivated state based on the cryoEM structure of the eeNaV1.4 channel and conducted a docking study to investigate the interactions of antiarrhythmic and local anesthetic drugs—lidocaine, QX-314, etidocaine, flecainide, and ranolazine—with hNaV1.5. The results revealed that both antiarrhythmic and local anesthetic drugs share a receptor site formed by the S6 segments from DIII and DIV. Multimicrosecond unbiased MD simulations of neutral lidocaine interacting with hNaV1.5 using the Anton 2 supercomputer revealed a hydrophilic access pathway through the intracellular gate, and a hydrophobic access pathway through a fenestration between DIII and DIV. Distinct binding sites were identified in the pore region for both neutral and charged lidocaine. We also observed that the channel can accommodate up to two lidocaine molecules binding at the same time. Our results reveal the high-resolution structural determinants of drug block of hNaV1.5 in an inactivated state. They also serve as initial steps toward linking of structural determinants of channel−drug interactions to the modification of hNaV1.5 function.
Results and Discussion
A Structural Model of the hNaV1.5 Channel Based on eeNaV1.4 Channel Structure.
To study the state-dependent molecular mechanisms of high-affinity binding of antiarrhythmic and local anesthetic drugs to hNaV channels at the atomic scale, high-resolution structures of eukaryotic NaV channels in open and inactivated states are needed. The cryoEM structure of the eeNaV1.4 channel in a partially open and presumably inactivated state (Protein Data Bank ID code 5XSY) (27) provides an atomic-level accuracy structural template for modeling of hNaV channels. The sequence identity between hNaV1.5 and eeNaV1.4 is ∼84% in the pore-forming transmembrane region (SI Appendix, Fig. S1), which is within an atomic-level accuracy homology modeling range (48), allowing us to generate an accurate model of hNaV1.5 in a partially open and presumably inactivated state.
The hNaV1.4 structure was published in September 2018 (9), when this study was already completed. The sequence identity between hNaV1.5 and hNaV1.4 is ∼87%, which is similar to the sequence identity between hNaV1.5 and eeNaV1.4 (∼84%) and suggests that eeNaV1.4 and hNaV1.4 structures are within the same range of accuracy for modeling of hNaV channels. The overall root-mean-square deviation (RMSD) between hNaV1.4 and eeNaV1.4 structures is less than 1 Å (9) and RMSD over the PD segments S5 and S6 and P1 and P2 helices is less than 0.7 Å, which suggests very similar conformations of the PD structure—the main focus of this study.
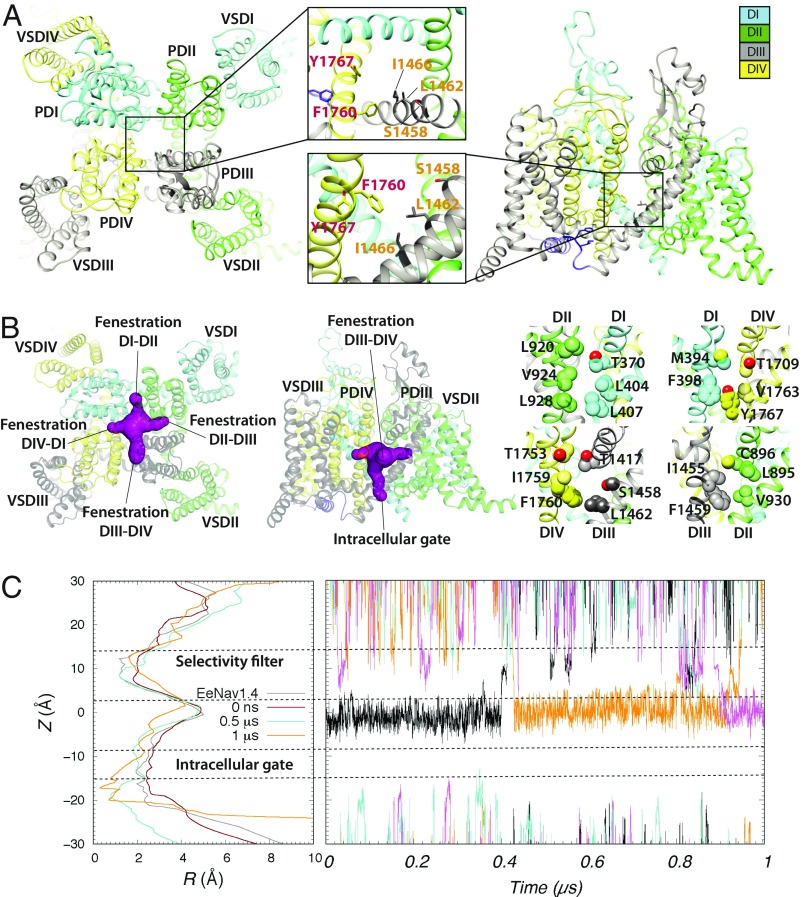
The eeNaV1.4 structure (27) has the following distinct features: (i) a partially open state of the PD intracellular gate, which is different from a closed state of the intracellular gate in NaVPaS structure (25) [notably, the PD intracellular gate can be stabilized in a partially open state by local anesthetic binding to an open and inactivated state (49)]; (ii) activated states of DIII and DIV VSDs with S4 in DIII positioned farther “up” compared with its position in NaVPaS structure in a closed state (25) [remarkably, the DIII and DIV VSDs can be stabilized in activated states by local anesthetic binding to the PD in an open and inactivated state (49, 50)]; and (iii) an inactivation gate (IFM motif in DIII–DIV linker) binding to the DIII and DIV S4–S5 linkers and DIVS6, which agree with an inactivated state of the channel (51–54). DIII–DIV linker conformation is different in eeNaV1.4 structure compared with NaVPaS structure in a closed state (25). Based on these structural observations and related experimental data, we suggest that the eeNaV1.4 structure is consistent with a partially open and inactivated state, which has high affinity for antiarrhythmic and local anesthetic drugs (15, 16). We used the Rosetta structural modeling software with the eeNaV1.4 channel structure as a template to build a homology model of hNaV1.5 channel in a partially open, inactivated state as described in SI Appendix, SI Methods (Fig. 1). Key hNaV1.5 residues forming the putative antiarrhythmic and local anesthetic drug binding site in DIIIS6 and DIVS6 segments (15–18) are F1760 and Y1767 in the DIVS6 segment and L1462 and I1466 in the DIIIS6 segment (Fig. 1A). The hNaV1.5 residues forming the drug access pathway at the fenestration between the DIIIS6 and DIVS6 segments are T1753 and I1759 in the DIVS6 segment and L1413 in the P1 helix of DIII (Fig. 1B and SI Appendix, Figs. S2–S4) (16, 55).
Fig. 1.
Rosetta model of the hNaV1.5 channel. (A) (Left) Extracellular and (Right) transmembrane views of the hNaV1.5 model shown in ribbon representation. (Insets) Zoom-in views of putative drug binding residues within hNaV1.5 pore lumen. Each domain is colored individually and labeled. In Insets, DIII residues are labeled orange, whereas DIV residues are labeled red. (B) (Left) Extracellular and (Center and Right) transmembrane views of all four hNaV1.5 fenestrations using molecular surface representation (shown in purple in Left and Center). In Right, fenestration-facing residue sidechains are labeled and shown in space-filling representations using corresponding domain colors, with O atoms shown in red. (C) (Left) The hNaV1.5 pore lumen radius (R) profile changes during molecular dynamic simulation at time 0 (colored red), at 0.5 μs (colored cyan), and at 1 μs (colored orange). A pore lumen R profile for a cryoEM eeNaV1.4 structure is also shown in gray for comparison. (Right) Sodium ion trajectories within the PD during a 1-μs molecular dynamic simulation of hNaV1.5.
To determine whether the hNaV1.5 channel model represents a conductive or nonconductive open state, we performed MD simulations of the hNaV1.5 model as described in SI Appendix, SI Methods. The Rosetta hNaV1.5 model and the eeNaV1.4 structure both have a ∼2.5-Å pore radius within the intracellular gate region (Fig. 1C, Left) (27). During the MD simulation of the hNaV1.5 model, the intracellular gate radius decreased from ∼2.5 Å at the start of the simulation to ∼2.0 Å after 0.5 μs and then to ∼1.0 Å to 2.0 Å after 1 μs (Fig. 1C). While we observed several Na+ ions passing up and down between the SF region and the pore lumen, we did not detect any Na+ ions passing through the intracellular gate of the pore during the 1-μs simulation (Fig. 1C, Right). Based on these results, we assume our hNaV1.5 model to be in a nonconductive inactivated state.
Modeling of Antiarrhythmic and Local Anesthetic Drugs Interaction with hNaV1.5 Channel Using RosettaLigand.
To study high-affinity binding of antiarrhythmic and local anesthetic drugs to the hNaV1.5 pore in the nonconductive inactivated state at atomic scale, we used RosettaLigand (56, 57) as described in SI Appendix, SI Methods.
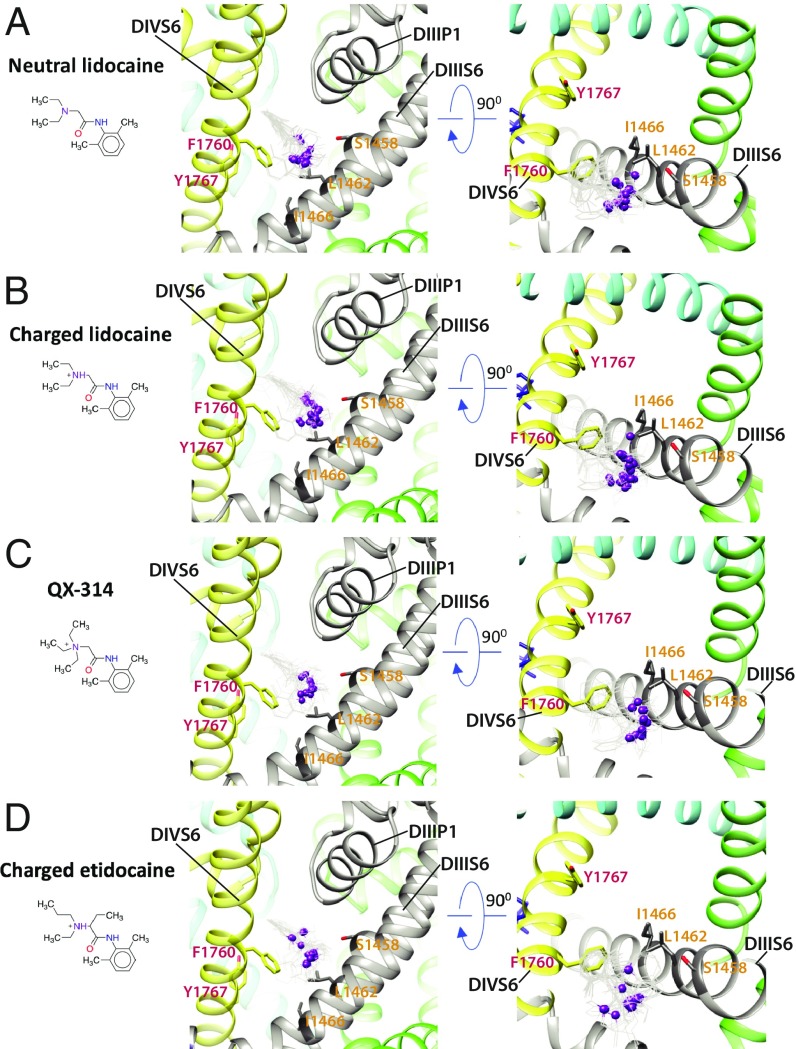
Lidocaine is a local anesthetic and class Ib antiarrhythmic drug used for the treatment of ventricular arrhythmias (58). Experimental data suggest that phenylalanine and tyrosine residues in the DIVS6 segment of mammalian NaV channels (F1760 and Y1767 in hNaV1.5) play a key role in antiarrhythmic and local anesthetic drug binding (15). The most frequently sampled lowest binding energy RosettaLigand models of neutral or charged lidocaine interacting with hNaV1.5 indicate that the region above F1760 in the DIVS6 segment forms a “hot spot” for lidocaine binding (Fig. 2 A and B and SI Appendix, Figs. S5 and S6). This hot spot extends from the fenestration between the DIIIS6 and DIVS6 segments into the pockets under the SF region in DIII and DIV. The tertiary amine group of neutral and charged lidocaine is positioned above F1760 (Fig. 2 A and B). The phenyl ring of neutral and charged lidocaine is observed in multiple different orientations near F1760 (Fig. 2 A and B and SI Appendix, Figs. S5 and S6). We observed only one neutral and one charged lidocaine pose among the lowest-energy models near Y1767 (SI Appendix, Figs. S5 and S6), potentially reflecting a lower-affinity binding site near this residue and in agreement with a weaker impact of Y1767 mutations on drug binding compared with F1760 mutations (15, 16). Experimental data suggest that leucine and isoleucine residues in the DIIIS6 segment of mammalian NaV channels (L1462 and I1466 in hNaV1.5) also form receptor sites for antiarrhythmic and local anesthetic drug binding (18, 59). The L1462 residue is positioned near F1760 in our model (Fig. 2 A and B). However, I1466 is not in direct contact with lidocaine in any of the top neutral and charged lidocaine models, suggesting an allosteric effect of mutations at this position on drug binding. We validated the robustness of the RosettaLigand modeling of lidocaine binding to hNaV1.5 by exploring two well-studied lidocaine variants: QX-314 and etidocaine (Fig. 2 C and D and SI Appendix, SI Results and Discussion and Figs. S7 and S8). Our models confirmed that the hot spot for QX-314 and etidocaine binding is similar to the hot spot observed in our lidocaine–hNaV1.5 models.
Fig. 2.
Rosetta models of hNaV1.5 channel interaction with antiarrhythmic and local anesthetic drugs. Close-up (Left) transmembrane and (Right) extracellular views of hNaV1.5 interactions with (A) neutral lidocaine, (B) charged lidocaine, (C) QX-314, and (D) charged etidocaine. Drug molecules are shown in the wireframe representations, with basic N atoms depicted as purple balls. The hNaV1.5 DI is colored in blue, DII is colored in green, DIII is colored in gray, and DIV is colored in yellow. Sidechains of key residues forming the receptor site in DIIIS6 and DIVS6 segments are shown in stick representation and labeled in orange and red, respectively.
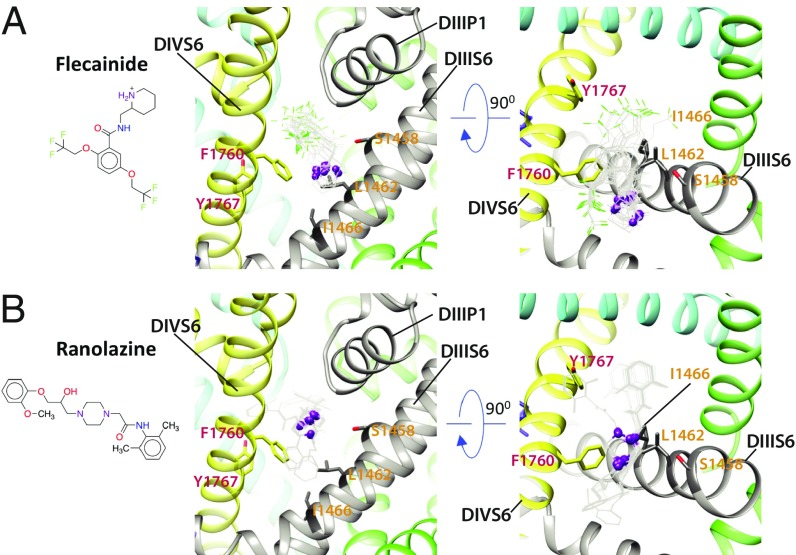
Flecainide is a class 1c antiarrhythmic drug used to prevent and treat tachyarrhythmias, which also may have unpredictable proarrhythmic effects (60, 61). Experimental data suggest that flecainide preferentially binds to NaV channels in an open state and that phenylalanine and tyrosine residues in the DIVS6 segment (F1760 and Y1767 in hNaV1.5) play an important role in its binding (15, 60, 62). The most frequently sampled and lowest binding energy RosettaLigand models of flecainide binding to hNaV1.5 are consistent with binding of the other drugs to the region above F1760 in the DIVS6 segment (Fig. 3A and SI Appendix, Fig. S9). However, the larger and branched structure of flecainide compared with lidocaine, etidocaine, and QX-314 results in a greater surface area of interaction that spans from the fenestration region between the DIII and DIV to the ion conduction pathway under the SF region (Fig. 3A).
Fig. 3.
Rosetta models of hNaV1.5 channel interaction with antiarrhythmic and local anesthetic drugs. Close-up (Left) transmembrane and (Right) extracellular view of hNaV1.5 interactions with (A) flecainide and (B) ranolazine. Drug molecules are shown in the wireframe representations, with flecainide F atoms colored in green and basic N atoms of both drugs depicted as purple balls. The hNaV1.5 DI is colored in blue, DII is colored in green, DIII is colored in gray, and DIV is colored in yellow. Sidechains of key residues forming the receptor site in DIIIS6 and DIVS6 are shown in stick representation and labeled in orange and red, respectively.
Ranolazine is an antianginal drug that inhibits late NaV current. Experimental data suggest that ranolazine binds to NaV channels in an open state and that phenylalanine in the DIVS6 segment (F1760 in hNaV1.5) plays a key role in its binding (63, 64). The most frequently sampled and lowest binding energy RosettaLigand models of ranolazine show that the same region above F1760 in the DIVS6 segment forms the hot spot for ranolazine binding (Fig. 3B and SI Appendix, Fig. S10). Ranolazine has a flexible linear rather than branched structure and interacts via the same modality as flecainide over a larger surface area that spans from the fenestration region between the DIII and DIV to the ion conduction pathway under the SF region (Fig. 3B).
Overall, although drug molecules can adopt different orientations in the channel (SI Appendix, Figs. S5–S10), localization of the drugs between the DIIIS6 and DIVS6 segments in our models is in agreement with the Chanda laboratory structural hypothesis that local anesthetics may stabilize primarily VSDIII and partially VSDIV in activated “up” states (49). The position of the drugs under the SF region in DIII and DIV is notable with respect to several mutations in this region that have been shown to significantly affect the slow inactivation of NaV channels (65–68). We hypothesize that, upon binding above F1760 in DIVS6 and under the SF region in DIII and DIV, the antiarrhythmic and local anesthetic drugs may induce conformational changes that may enhance slow inactivation of NaV channels, in agreement with experimental data (69, 70). We also propose that, since the antiarrhythmic drugs ranolazine and flecainide have more extensive interactions with the channel compared with lidocaine and its derivatives in our models (Figs. 2 and 3), their effect on channel gating might be more prominent as well. In fact, our recent multiscale kinetic modeling and experimental study examined lidocaine and flecainide interactions with NaV1.5 and their consequence on proarrhythmia proclivities (4). We found, for example, that cardiac-safe lidocaine has faster channel unbinding kinetics, resulting in more facile recovery of channels from drug blockade, and lower incidence of reentrant arrhythmias at a cardiac tissue and a whole-heart level compared with flecainide.
Neutral and Charged Lidocaine Partitioning into the Membrane.
The molecular docking calculations, described in Modeling of Antiarrhythmic and Local Anesthetic Drugs Interaction with hNaV1.5 Channel Using RosettaLigand, provided us with atomistic structural models of convergent binding poses of several antiarrhythmic and local anesthetic drugs in the hNaV1.5 pore. However, static molecular models cannot reveal how a drug accesses the binding site and whether such drug−channel interactions are long-lived or transient. Such information can be provided by atomistic MD simulations of a channel embedded in a hydrated lipid membrane with one or multiple drug molecules present. To perform such simulations, we developed atomic-resolution structural models for charged and neutral forms of lidocaine, compatible with biomolecular all-atom Chemistry at Harvard Macromolecular Mechanics (CHARMM) force fields as described in SI Appendix, SI Methods. Lidocaine was chosen over larger NaV1.5 blockers such as flecainide and ranolazine because our previous MD simulation study of drug–bacterial NaV channel interactions suggested that we can more efficiently predict entry and egress pathways for a smaller drug (13). Indeed, experimental data indicate that lidocaine has faster NaV1.5 association and dissociation kinetics than the larger flecainide (4). Moreover, in aqueous solution, lidocaine exists as a mixture with substantial fractions of both charged (∼78% at pH = 7.4) and neutral forms (∼22% at pH = 7.4) which have different membrane permeabilities and can differently affect NaV channel function (4, 45, 46). Therefore, in this study, we have explored charged and neutral lidocaine–lipid membrane and NaV1.5 interactions via all-atom MD simulations.
The derived parameters were validated by performing MD simulations of charged and neutral lidocaine partitioning across a 1-palmitoyl-2-oleoyl-phosphatidylcholine lipid membrane and computing the water−membrane distribution coefficient log D = 1.25 ± 0.32, which agrees reasonably well with the experimental values 1.4 (71) and 1.76 (72) (see SI Appendix, SI Results and Discussion for more information). We also found that a charged drug has more favorable interfacial binding (−3 kcal/mol vs. −1 kcal/mol for neutral) but much slower transition rate through the membrane (38.9 s−1 vs. 21.1 ms−1 for neutral) due to a higher barrier (see SI Appendix, SI Results and Discussion, Figs. S11–S13, and Tables S1–S3 for more information). Since charged lidocaine is the dominant form at a physiological pH (71), we primarily expect its accumulation at water−membrane interfaces, in agreement with recent solid NMR experiments (73). However, deeper into the hydrophobic membrane core, neutral lidocaine is expected to be more prevalent. This indicates that we need to study interactions of both forms with hNaV1.5 to assess hydrophobic and hydrophilic channel pore access pathways.
MD Simulations Reveal Neutral Lidocaine Access Pathways to the Binding Site via the Intracellular Gate and Fenestration Between DIII and DIV.
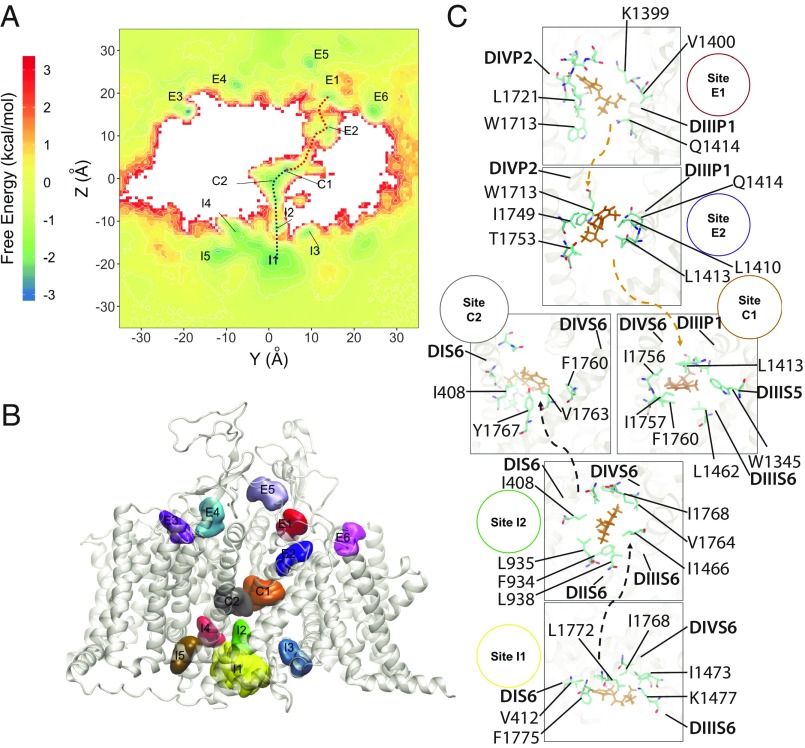
To explore the lidocaine access pathways to its binding site within the hNaV1.5 channels, we ran multimicrosecond MD simulations on the Anton 2 supercomputer (74), with neutral or charged lidocaine, as described in SI Appendix, SI Methods. The MD simulations of neutral lidocaine revealed that it can access its binding site within the NaV channel pore lumen either through an opening formed by the intracellular gate (hydrophilic pathway) or through a path formed between the lipids, the P1 helix in DIII, the P2 helix in DIV, and the fenestration region between DIII and DIV (hydrophobic pathway) (Fig. 4 and Movies S1 and S2). The hydrophilic pathway is formed by the following residues at the intracellular gate (see sites I1 and I2 in Fig. 4 A and C): L404, I408, and V412 (DIS6); L931, F934, L935, and L938 (DIIS6); L1462, I1466, and I1470 (DIIIS6); and V1764, Y1767, I1768, and I1771 (DIVS6). Notably, all of the residues lining the intracellular gate in hNaV channels are hydrophobic and highly conserved. The hydrophobic pathway between DIII and DIV is formed by the following residues (see sites E1, E2, C1, and C2 in Fig. 4 A and C): L1338, L1342, and W1345 (in DIIIS5); L1410, L1413, and Q1414 (in P1 helix of DIII); L1462 and F1465 (in DIIIS6); W1713, L1717, and L1721 (in P2 helix of DIV); and I1749, T1753, I1756, and I1757 (in DIVS6). Remarkably, lidocaine molecules that accessed the pore binding sites (C1 and C2 sites) are not those partitioned from lipid membrane. Lidocaine accessed the fenestration between DIII and DIV from the extracellular side by going through the cleft formed between P1 DIII and P2 DIV (E1 and E2 sites). Furthermore, F1760 (in DIVS6) and L1462 (in DIIIS6) are the first residues that lidocaine encounters as it enters the pore lumen through the fenestration region; both of these residues are forming the hot spot for all of the drugs simulated using RosettaLigand (Figs. 2 and 3). Moreover, neutral lidocaine was found to access the receptor site via the fenestration between DIII and DIV, but not through the fenestrations between the other domains. We hypothesize that specific amino acid differences between the residues forming the fenestration between DIII and DIV versus residues forming fenestrations between all other domains are preventing lidocaine from accessing the receptor site through the other fenestrations (SI Appendix, Figs. S3 and S4). Sequence alignment of hNaV1.5 domains in the P1 helix from DIII and P2 helix from DIV regions revealed that only the sidechain of L1413 in the P1 helix from DIII is significantly different from the larger sidechain of Phenylalanine at corresponding position in other domains (SI Appendix, Figs. S3 and S4). However, other residues facing the lipid environment at the interface between the P1 helix from DIII and P2 helix from DIV are similar in size and hydrophobicity between all four domains in the hNaV1.5 channel (SI Appendix, Fig. S3). Remarkably, L1413 is positioned near T1753 in DIVS6 at the vertical channel−lipid interface region (SI Appendix, Fig. S4). We suggest that residues at corresponding positions in all NaV channel domains may form barriers for drug access at the entry to the fenestrations. This is the barrier that neutral lidocaine molecule is passing in our simulations when it travels from site E2 to site C1 at the fenestration between DIII and DIV (Fig. 4). The corresponding amino acid pairs that form barriers at the entry to other fenestrations are bulkier and more hydrophobic: F366 (P1 helix in DI) and L920 (DIIS6), F892 (P1 helix in DII) and V1451 (DIIIS6), and F1705 (P1 helix in DIV) and M394 (DIS6) (SI Appendix, Figs. S3 and S4).
Fig. 4.
MD simulation of the hNaV1.5 channel interaction with neutral lidocaine reveals two drug access pathways. (A) Free energy surface of neutral lidocaine binding projected on the y−z plane (with z corresponding to a transmembrane axis). Binding sites for neutral lidocaine, identified from free energy minima, are labeled as intracellular I1-5, channel pore C1-2, and extracellular E1-6. (B) Transmembrane view of the channel, with neutral lidocaine binding sites represented as colored surfaces. Colors and sizes are for clarity, not actual binding properties. (C) Close-up view of binding sites forming the hydrophobic (orange arrows) and hydrophilic (gray arrows) binding pathways. Lidocaine molecules (orange) and interacting residues on the channel (cyan for C, red for O, and blue for N) are shown using stick representation.
We found this observation of the hydrophobic pathway very intriguing. Although early work on local anesthetics and quaternary derivatives provided compelling evidence for a hydrophobic pathway as a result of drug partitioning into lipid membrane (12, 75), variants of different channel isoforms appeared to have a specific residue-dependent external access pathway. Membrane-impermeant QX-314 was shown to block the cardiac isoform NaV1.5 in rats (rNaV1.5) when applied from either side of the membrane. The blocking effect of extracellular QX-314 was reduced by substitution of DIVS6 T1755 in cardiac rNaV1.5 (equivalent to T1753 in hNaV1.5) to valine in brain rNaV1.2 (55). T1753 is the unique polar residue along the drug access pathway at the entry to the DIII−DIV fenestration (see discussion above and SI Appendix, Figs. S3 and S4). Similarly, mutation of the equivalent residue C1572 in muscle rNaV1.4 to threonine in cardiac rNaV1.5 also allowed QX-222 to block the channel from the extracellular side (76). In addition, mutations of I1575 in DIVS6 of muscle rNaV1.4 or equivalent residue I1760 in brain rNaV1.2 (I1756 in hNaV1.5) to alanine (relatively small amino acid) created an external access pathway for QX-222 (77). Remarkably, these residues (T1753 and I1756 in DIVS6 in hNaV1.5) are part of the E2 and C1 binding sites forming the hydrophobic pathway in our simulations (Fig. 4). We hypothesize that equivalent positions in other NaV channels could form a hydrophobic pathway for drug access from the extracellular environment for both neutral and charged drugs. While neutral drugs may pass along the hydrophobic pathway to access the binding site within the pore lumen, charged drugs may pass along this pathway only if polar or small sidechain amino acids are present in this critical region to lower the energy barrier for drug access. Results from previously published experimental data provide structural explanations for the ultrafast blocking kinetics of extracellularly applied neutral drugs on NaV channels (12). This hydrophobic drug access pathway in our simulations also revealed another interesting observation. Neutral lidocaine is climbing down the vertical lipid–channel interface formed by the P1 helix in DIII, P2 helix in DIV, and DIII−DIV fenestration (Movie S2). Since neutral lidocaine is amphipathic, this could be considered to be an energetically favorable pathway. It is tempting to speculate that other ion channels and transmembrane proteins can adopt a similar amphipathic drug access pathway at the interface between lipid and protein environments. However, our results may not exclude the classic hydrophobic drug access pathway from the lipid membrane to the fenestration laterally, which can potentially be observed with significantly longer MD simulations.
MD Simulations Reveal Two Neutral Lidocaines Simultaneously Binding Within the hNaV1.5 Channel Pore Lumen.
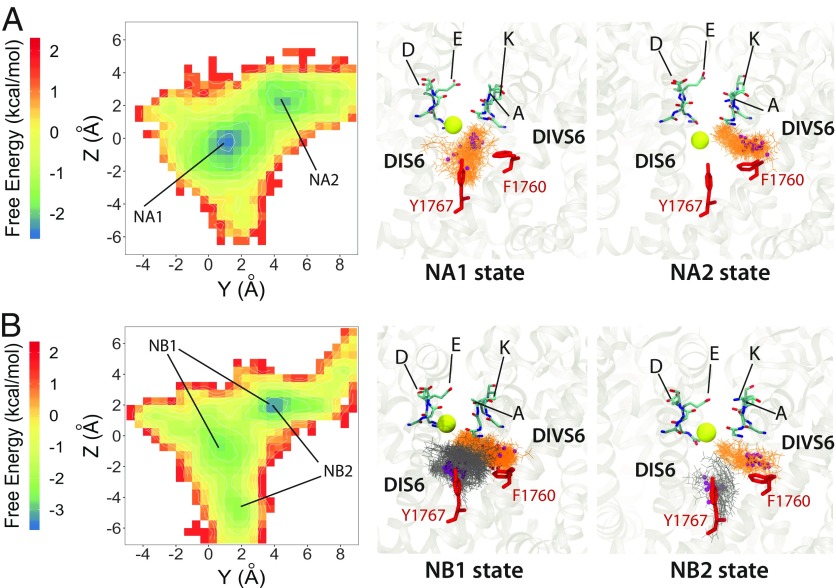
Our unbiased simulations of neutral lidocaine revealed up to two lidocaine molecules binding within the channel pore lumen (Fig. 5). When there is one molecule in the pore, neutral lidocaine is localized at two distinct binding sites, NA1 and NA2. NA1 is the binding site at the center of the pore, involving Y1767 and other residues from the S6 segment of all four domains. There are limited contacts of neutral lidocaine with F1760 in the NA1. The NA2 binding site is positioned on top of F1760, near the DIII−DIV fenestration and under the P1 helix in DIII, which is similar to the most frequent and lowest interface energy pose for neutral lidocaine observed by RosettaLigand (Fig. 2A). Both the amine group and the phenyl ring of lidocaine form interactions with F1760, L1462, and I1466. Two lidocaine molecules in the pore can occupy both the NA1 and NA2 binding sites, which are sampled by a sole lidocaine molecule in the pore (Fig. 5). The first neutral lidocaine in our model is positioned in a binding site formed by a region above F1760 and under the P1 helix in DIII, and fenestration region between DIII and DIV, i.e., a site equivalent to NA2 for one lidocaine binding in the pore (Fig. 5A). The second neutral lidocaine is positioned between F1760 and Y1767 in the central pore, resembling a single lidocaine NA1 binding site. We classify them, in general, as DIVS6 F1760 binding site and central pore binding site. While the lidocaine binding at F1760 is unchanged during simulations, lidocaine binding at the central pore can shift up and down, thus creating two states of binding, NB1 and NB2 (Fig. 5). These observations from our simulations are in agreement with experimental data showing that F1760 and Y1767 in hNaV1.5 play key roles in lidocaine binding (15). It is also noticeable that the DIII SF region residue K1419 is part of the aspartate, glutamate, lysine, and alanine (“DEKA”) motif and plays an important role in NaV channel selectivity (78, 79). Mutations of K1419 to serine or glutamate enhance slow inactivation of NaV channels (68). It is possible that, while binding at the central pore can provide a simple steric blocking mechanism, lidocaine binding at F1760 and the P1 helix in DIII may directly interfere with the normally conductive state of the SF region and induce a conformational change that may promote transition to the slow inactivated state. Remarkably, cooperative binding of multiple lidocaine molecules to NaV channels has been previously suggested based on dose–response of inhibition with a Hill coefficient value greater than 1 (80). Furthermore, N-linked lidocaine dimers have been previously shown to bind to NaV channels with 10- to 100-fold higher affinity than lidocaine monomers (81). These experimental observations agree with our MD simulation results and suggest that lidocaine may have at least two hot spots for binding within the NaV channel pore lumen formed between the P1 helix from DIII, F1760, and Y1767. Precise neutral lidocaine poses in our models are different from lidocaine poses in recent models based on NaVMs and NaVPaS structures (46). These disparities most likely arise from differences in key residues forming the S6 receptor site and specific channel states in our hNaV1.5 model based on eeNaV1.4 structure versus NaVMs and NaVPaS structures.
Fig. 5.
MD simulation of the hNaV1.5 channel interaction with neutral lidocaine reveal two binding poses: (A) states NA1 and NA2 for one lidocaine bound in the pore lumen and (B) NB1 and NB2 for two lidocaine molecules binding in the pore lumen at the same time. (Left) Free energy surfaces projected on the y−z plane, with binding sites identified from free energy minima and labeled. (Center and Right) Close-up transmembrane views of molecular models of charged lidocaine binding. In the close-up views, lidocaine molecules (orange and dark gray) and interacting residues on the channel (red) as well as SF DEKA motif (cyan for C, blue for N, and red for O) are shown using stick representation. Lidocaine basic N atoms are shown as small purple spheres, and an SF bound Na+ atom is shown as a yellow sphere.
MD Simulations Reveal Two Unique Hot Spots for Binding of Charged Lidocaine in the hNaV1.5 Channel Pore Lumen.
Unbiased MD simulations of high concentrations of charged lidocaine molecules placed in aqueous solution have shown that the drug did not pass either through the opening formed by the hydrophobic intracellular gate or through the fenestration between DIII and DIV during 1 μs simulation. Combined with results from our calculation of charged lidocaine membrane partitioning described in Neutral and Charged Lidocaine Partitioning into the Membrane, we suspect that those events may not be effectively sampled in a few microseconds of simulation time. To further understand interactions of charged lidocaine with the hNaV1.5 channel, we explored potentially unique binding poses by starting simulations with one or two charged lidocaines in the pore lumen, as described in SI Appendix, SI Methods.
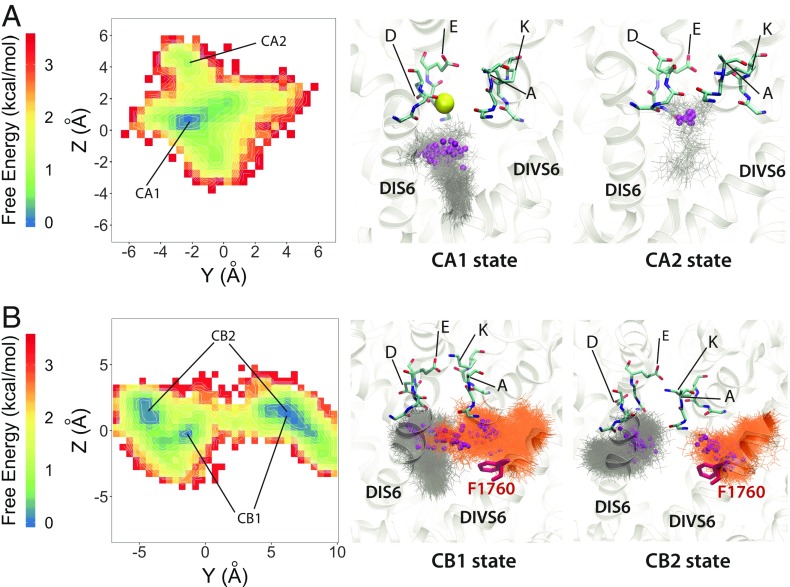
Simulation of one charged lidocaine revealed two highly convergent binding states lining along the vertical pore axis with the protonated amine (i.e., cationic ammonium) group of lidocaine in close proximity to the DI and DII SF region and the phenyl group of lidocaine pointing down into the lumen (see CA1 and CA2 states in Fig. 6A). The CA1 state represents binding of charged lidocaine at the central pore with the protonated amine group attracted to the electron negative region below the SF. Interestingly, most of the time during the simulation, lidocaine binding in CA1 appeared to have a sodium ion binding in the SF, right above the protonated amine group, whereas, in the absence of sodium binding in the CA2 state, charged lidocaine binds directly to the SF with the sodium binding site being taken by the protonated amine. We found that this result agrees with a variety of functional, structural, and computational data suggesting that the SF region may form a part of local anesthetic drug binding (19, 23, 45, 46). However, compared with the single neutral lidocaine binding result, we did not see the involvement of F1760 in binding of one charged lidocaine. We assume this is a result of limited sampling from 1 μs of unbiased simulation, although a similar result was observed in the simulation of charged lidocaine with open NaVMs and closed NaVPaS channel using an enhanced sampling technique of replica exchange with solute tempering (46).
Fig. 6.
MD simulation of hNaV1.5 channel interaction with charged lidocaine reveal two binding poses: (A) states CA1 and CA2 for one lidocaine bound in the pore lumen and (B) CB1 and CB2 for two lidocaine molecules binding in the pore lumen at the same time. (Left) Free energy surface projected on the y−z plane, with binding sites identified from free energy minima and labeled. (Middle and Right) Close-up transmembrane views of molecular models of charged lidocaine binding. SF DEKA motif residues are shown in stick representation and colored in cyan for C, blue for N, and red for O. Sodium ions are shown as spheres and colored in yellow. Lidocaine molecules are shown in stick representation and colored in gray or orange. The nitrogen atoms of the tertiary ammonium groups on charged lidocaine molecules are shown as small spheres and colored in purple. The F1760 sidechain is shown in stick representation and colored in red.
Simulation of two charged lidocaines revealed two localized binding sites, a DIVS6 F1760 binding site and a central pore binding site, similar to the case of neutral lidocaine. While lidocaine binding at the F1760 site is relatively stable, binding to the central pore can be shifted, creating two highly convergent states, CB1 and CB2 (Fig. 6). The first highly converged state (CB1) has one charged lidocaine lining along the vertical pore axis, with the protonated amine group in close proximity to the DI and DII SF region and the phenyl group pointing down into the lumen (see CB1 state in Fig. 6B), the same orientation as for one lidocaine molecule (CA1 state in Fig. 6A). Another charged lidocaine at the DIVS6 F1760 site has the protonated amine group forming cation−π interactions with F1760 and the phenyl group pointing into the fenestration region between DIII and DIV (see CB1 state in Fig. 6B). Notably, the cation−π interaction is dominant during the simulation. We rarely observed π−π stacking interactions between the phenyl ring of charged lidocaine and F1760. This agrees with experimental data suggesting that interactions between charged lidocaine and F1760 are cation−π interactions, not π−π interactions (82). The second highly converged binding state (CB2) has the central pore localized charged lidocaine oriented mostly along the horizontal membrane plane (not the vertical transmembrane axis as in CB1), with the protonated amine group also in close proximity to the DI and DII SF region. However, the phenyl group is pointing into the fenestration region between DI and DII (see CB2 state in Fig. 6B). The other charged lidocaine at the DIVS6 F1760 site forms an interaction with F1760 in a similar manner to that in the CB1 state.
It is interesting to note that F1760 has been shown to be a key determinant for the use-dependent block, while Y1767 only has a modest effect (16). Specifically, F1760 has been shown to form cation−π interaction with class Ib antiarrhythmic drugs in a use-dependent manner (82). In addition, mutation of W1531 to Cys in NaV1.4 (W1713 in our hNaV1.5) in the DIV P2 region was shown to abolish use dependence of mexiletine and QX-222, without destabilizing fast inactivation or altering drug access (21). In our model, W1713 is part of the binding site E2 for the neutral lidocaine pathway (Fig. 4) and is the ceiling of the DIII−DIV fenestration, right above F1760. The best RosettaLigand docking models, MD simulations of both neutral and charged lidocaine, identified the DIVS6 F1760 site as a common binding site. Together, these results encourage us to propose the binding site at DIVS6 F1760, near the DIII−DIV fenestration, as the high-affinity use-dependent binding site, whereas other binding sites at the SF region (for charged lidocaine) and at central pore (for neutral lidocaine) can be considered, based on our simulations, as low-affinity binding sites. However, the low-affinity tonic drug block is beyond the scope of this study and requires comprehensive modeling of channel−drug interactions in closed state of the pore and in resting states of VSDs.
Lidocaine Binding to hNaV1.5 Attenuates Sodium Binding in the Selectivity Filter.
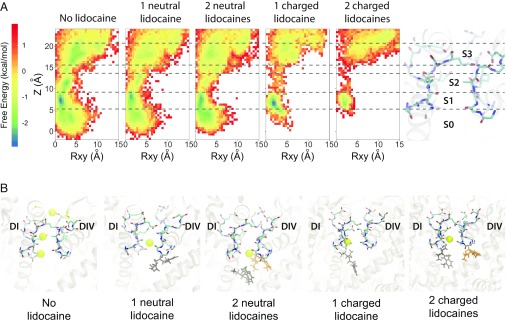
MD simulations of the hNaV1.5 channel in the absence or presence of one or two neutral or charged lidocaine molecules suggest that binding of lidocaine to its receptor site(s) within the pore lumen reduces Na+ ion binding within the SF region (Fig. 7). Free energy surfaces for a Na+ ion within the hNaV1.5 SF reveal three major Na+ binding sites within this region (see sites S1, S2, and S3 in Fig. 7) and one additional site within the pore lumen (see site S0 in Fig. 7). Site S1 is located just below the SF region and is formed by the carbonyl groups of T370 and Q371 (in DI) and C896 and G897 (in DII). Site S2 is formed by the carboxylate groups of D372 (in DI) and E898 (in DII)—residues in the classical DEKA SF motif in NaV channels. Site S3 is formed by the carboxylate groups of E375 (in DI), E901 (in DII), D1423 (in DIII), and D1714 (in DIV). In the absence of lidocaine, all three Na+ binding sites are well defined (Fig. 7). When one or two neutral lidocaine molecules are present in the pore lumen, the Na+ binding site S1 diffuses farther into the pore lumen region, while sites S2 and S3 within the SF region are occupied less frequently (Fig. 7). When one or two charged lidocaine molecules are present in the pore lumen, we observe a dramatic reduction in Na+ binding at the pore lumen site S0 and within the SF region in all three sites, especially at sites S2 and S3 (Fig. 7). This disruption of continuous ion density in those cases (Fig. 7A) may impair ion conduction through the SF.
Fig. 7.
MD simulations reveal the free energy surfaces and binding sites for sodium ion within hNaV1.5 pore. (A) Transmembrane view projection of the free energy surface for sodium ion without lidocaine and in the presence of one or two neutral or charged lidocaine molecules. Specific Na+ binding sites are labeled S0, S1, S2, and S3 in the molecular representation of the channel SF on the right. (B) Representative transmembrane views of sodium ion binding sites within the SF region of the channel observed without lidocaine and in the presence of one or two neutral or charged lidocaine molecules. Sodium ions are shown as yellow spheres. The SF region residues are shown in stick representation and labeled.
The positioning of neutral or charged lidocaine molecules under the SF region in the MD simulations is notable with respect to experimental data that have identified specific mutations in the SF region that significantly affect the slow inactivation of NaV channels (65–68). Interestingly, decreasing extracellular [Na+] potentiates use-dependent block by lidocaine (69). Lidocaine binding under the SF region may induce conformational changes in the SF that may enhance slow inactivation (69, 70). However, raising extracellular [Na+] inhibits native slow inactivation of NaV channels (69).
Conclusions
Our structural modeling and simulation of antiarrhythmic and local anesthetic drugs interaction with the hNaV1.5 channel revealed the following key observations: (i) Our hNaV1.5 model does not conduct sodium ions under applied voltage, which suggests that it does not represent an open conductive state (Fig. 1C). (ii) The region above F1760 in the DIVS6 segment forms a hot spot for drug binding and extends from the fenestration region between the DIIIS6 and DIVS6 segments to the hydrophobic pockets under the SF regions in DIII and DIV, which agrees with experimental data that identified key residues forming the high-affinity drug binding to NaV channels in an inactivated state (Figs. 2, 3, 5, and 6). (iii) The amine/ammonium group of lidocaine, etidocaine, and QX-314 is positioned above and near F1760 (Fig. 2). The phenyl ring of lidocaine, etidocaine, and QX-314 is observed in multiple different orientations near F1760 (Fig. 2). (iv) Flecainide and ranolazine bind to a larger protein surface area that spans from the fenestration region between DIII and DIV to the ion conduction pathway under the SF region (Fig. 3); (v) Lidocaine enters the hNaV1.5 pore via the hydrophilic pathway through the intracellular gate and via a hydrophobic pathway through the vertical lipid–channel interface formed by the P1 helix in DIII, P2 helix in DIV, and DIII−DIV fenestration (Fig. 4 and Movie S2). (vi) Up to two lidocaine molecules can simultaneously bind within the hNaV1.5 pore lumen, which agrees with experimental data demonstrating cooperative binding of multiple lidocaine molecules to NaV channels in an inactivated state (80) (Figs. 5 and 6). (vii) Bound lidocaine molecules can interfere with the ion occupancy in the hNaV1.5 SF (Fig. 7). Based on our results, we suggest that our hNaV1.5 model is consistent with an inactivated state.
Our results provide crucial atomic-scale mechanistic insights into protein–drug interactions, necessary for the rational design of novel modulators of the cardiac NaV channel to be used for the treatment of cardiac arrhythmias. The fundamental novelty of bringing together Rosetta molecular modeling and MD simulations to study drug−channel interactions has the potential to enable automated virtual drug screening in the future. Critically, this approach can be applied to any ion channel, which might be used to predict individual patient responses to drug therapy based on which specific ion channel mutations they have. For instance, we can predict how a single mutation in ion channel-encoding gene would affect protein−drug binding and how an effect of such alteration propagates from a protein to a single cell and the cardiac rhythm of the whole organ. This work sets the stage for expansion to novel linkages by connecting mature experimental structural and functional approaches to emerging modeling approaches at the atomic and organ scales. There is potential for future simulations to be carried out to predict how functional properties of drugs can be perturbed in an emergent multiscale modeling system, and these predictions may ultimately be used to inform structural models to screen drug analogs that confer the requisite functional properties predicted critical for therapy.
In particular, this study represents a critical step for elucidating structural determinants of drug cardiac safety profiles at atomic resolution. We have observed differences in NaV1.5 binding profiles for cardiac-safe lidocaine versus flecainide, a drug with a known proclivity for deadly arrhythmia. Our previous multiscale modeling and experimental study suggested that such molecular-scale differences can propagate and emerge at the tissue and organ levels as notable proarrhythmia markers (4). We have also performed multimicrosecond molecular dynamic simulations to explore drug–channel binding pathways for charged and neutral forms of lidocaine, which provided a molecular picture consistent with previous experimental observations.
Materials and Methods
We used the Rosetta structural modeling software (28, 56) and the cryoEM structure of the eeNaV1.4−beta1 complex (27) as a template to predict the structure of the hNaV1.5 channel. Molecular docking of antiarrhythmic and local anesthetic drugs to hNaV1.5 channel model was performed using RosettaLigand (56). Drug–membrane partitioning of charged and neutral lidocaine into a lipid membrane was assessed using the Nanoscale Molecular Dynamics (83) program. For MD simulations, the hNaV1.5 model was embedded in a bilayer using CHARMM graphical user interface (84). Further details of these methods are described in SI Appendix, SI Methods.
Supplementary Material
Acknowledgments
We thank Drs. Jon Sack, Toby Allen, Heike Wulff, Kazuharu Furutani, and Jie Zheng and members of the C.E.C., V.Y.-Y., and Sack laboratories for helpful discussions. We thank Dr. Nieng Yan (Princeton University) for sharing coordinates of electric eel and hNaV1.4 channel structures. Anton 2 computer time was provided by the Pittsburgh Supercomputing Center (PSC) through Grant R01GM116961 from the National Institutes of Health. The Anton 2 machine (74) at PSC was generously made available by D.E. Shaw Research. This research was supported by National Heart, Lung, and Blood Institute Grants U01HL126273, R01HL128537, R01HL128170, and OT2OD026580 (to C.E.C.) and American Heart Association Predoctoral Fellowship 16PRE27260295 (to K.R.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817446116/-/DCSupplemental.
References
- 1.Catterall WA, Goldin AL, Waxman SG. International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol Rev. 2005;57:397–409. doi: 10.1124/pr.57.4.4. [DOI] [PubMed] [Google Scholar]
- 2.DeMarco KR, Clancy CE. Cardiac Na channels: Structure to function. Curr Top Membr. 2016;78:287–311. doi: 10.1016/bs.ctm.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen-Izu Y, et al. Na+ channel function, regulation, structure, trafficking and sequestration. J Physiol. 2015;593:1347–1360. doi: 10.1113/jphysiol.2014.281428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moreno JD, et al. A computational model to predict the effects of class I anti-arrhythmic drugs on ventricular rhythms. Sci Transl Med. 2011;3:98ra83. doi: 10.1126/scitranslmed.3002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardiac Arrhythmia Suppression Trial (CAST) Investigators Preliminary report: Effect of encainide and flecainide on mortality in a randomized trial of arrhythmia suppression after myocardial infarction. N Engl J Med. 1989;321:406–412. doi: 10.1056/NEJM198908103210629. [DOI] [PubMed] [Google Scholar]
- 6.Waldo AL, et al. Effect of d-sotalol on mortality in patients with left ventricular dysfunction after recent and remote myocardial infarction. The SWORD Investigators. Survival with Oral d-Sotalol. Lancet. 1996;348:7–12. doi: 10.1016/s0140-6736(96)02149-6. [DOI] [PubMed] [Google Scholar]
- 7.Vassilev PM, Scheuer T, Catterall WA. Identification of an intracellular peptide segment involved in sodium channel inactivation. Science. 1988;241:1658–1661. doi: 10.1126/science.241.4873.1658. [DOI] [PubMed] [Google Scholar]
- 8.West JW, et al. A cluster of hydrophobic amino acid residues required for fast Na+-channel inactivation. Proc Natl Acad Sci USA. 1992;89:10910–10914. doi: 10.1073/pnas.89.22.10910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan X, et al. Structure of the human voltage-gated sodium channel Nav1.4 in complex with β1. Science. 2018;362:eaau2486. doi: 10.1126/science.aau2486. [DOI] [PubMed] [Google Scholar]
- 10.Hille B. Ion Channels of Excitable Membranes. 3rd Ed Sinauer Assoc; Sunderland, MA: 2001. [Google Scholar]
- 11.Catterall WA. Structure and function of voltage-gated sodium channels at atomic resolution. Exp Physiol. 2014;99:35–51. doi: 10.1113/expphysiol.2013.071969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hille B. Local anesthetics: Hydrophilic and hydrophobic pathways for the drug-receptor reaction. J Gen Physiol. 1977;69:497–515. doi: 10.1085/jgp.69.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boiteux C, et al. Local anesthetic and antiepileptic drug access and binding to a bacterial voltage-gated sodium channel. Proc Natl Acad Sci USA. 2014;111:13057–13062. doi: 10.1073/pnas.1408710111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeMarco KR, Bekker S, Clancy CE, Noskov SY, Vorobyov I. Digging into lipid membrane permeation for cardiac ion channel blocker d-sotalol with all-atom simulations. Front Pharmacol. 2018;9:26. doi: 10.3389/fphar.2018.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ragsdale DS, McPhee JC, Scheuer T, Catterall WA. Common molecular determinants of local anesthetic, antiarrhythmic, and anticonvulsant block of voltage-gated Na+ channels. Proc Natl Acad Sci USA. 1996;93:9270–9275. doi: 10.1073/pnas.93.17.9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ragsdale DS, McPhee JC, Scheuer T, Catterall WA. Molecular determinants of state-dependent block of Na+ channels by local anesthetics. Science. 1994;265:1724–1728. doi: 10.1126/science.8085162. [DOI] [PubMed] [Google Scholar]
- 17.Yarov-Yarovoy V, et al. Role of amino acid residues in transmembrane segments IS6 and IIS6 of the Na+ channel alpha subunit in voltage-dependent gating and drug block. J Biol Chem. 2002;277:35393–35401. doi: 10.1074/jbc.M206126200. [DOI] [PubMed] [Google Scholar]
- 18.Yarov-Yarovoy V, et al. Molecular determinants of voltage-dependent gating and binding of pore-blocking drugs in transmembrane segment IIIS6 of the Na+ channel alpha subunit. J Biol Chem. 2001;276:20–27. doi: 10.1074/jbc.M006992200. [DOI] [PubMed] [Google Scholar]
- 19.Sunami A, Dudley SC, Jr, Fozzard HA. Sodium channel selectivity filter regulates antiarrhythmic drug binding. Proc Natl Acad Sci USA. 1997;94:14126–14131. doi: 10.1073/pnas.94.25.14126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee PJ, Sunami A, Fozzard HA. Cardiac-specific external paths for lidocaine, defined by isoform-specific residues, accelerate recovery from use-dependent block. Circ Res. 2001;89:1014–1021. doi: 10.1161/hh2301.100002. [DOI] [PubMed] [Google Scholar]
- 21.Tsang SY, Tsushima RG, Tomaselli GF, Li RA, Backx PH. A multifunctional aromatic residue in the external pore vestibule of Na+ channels contributes to the local anesthetic receptor. Mol Pharmacol. 2005;67:424–434. doi: 10.1124/mol.67.2.. [DOI] [PubMed] [Google Scholar]
- 22.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bagnéris C, et al. Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism. Proc Natl Acad Sci USA. 2014;111:8428–8433. doi: 10.1073/pnas.1406855111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang L, et al. Structural basis for inhibition of a voltage-gated Ca2+ channel by Ca2+ antagonist drugs. Nature. 2016;537:117–121. doi: 10.1038/nature19102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen H, et al. Structure of a eukaryotic voltage-gated sodium channel at near-atomic resolution. Science. 2017;355:eaal4326. doi: 10.1126/science.aal4326. [DOI] [PubMed] [Google Scholar]
- 26.Shen H, et al. Structural basis for the modulation of voltage-gated sodium channels by animal toxins. Science. 2018;362:eaau2596. doi: 10.1126/science.aau2596. [DOI] [PubMed] [Google Scholar]
- 27.Yan Z, et al. Structure of the Nav1.4-β1 complex from electric eel. Cell. 2017;170:470–482.e11. doi: 10.1016/j.cell.2017.06.039. [DOI] [PubMed] [Google Scholar]
- 28.Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 29.Alford RF, et al. The Rosetta all-atom energy function for macromolecular modeling and design. J Chem Theory Comput. 2017;13:3031–3048. doi: 10.1021/acs.jctc.7b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yarov-Yarovoy V, et al. Structural basis for gating charge movement in the voltage sensor of a sodium channel. Proc Natl Acad Sci USA. 2012;109:E93–E102. doi: 10.1073/pnas.1118434109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pathak MM, et al. Closing in on the resting state of the shaker K+ channel. Neuron. 2007;56:124–140. doi: 10.1016/j.neuron.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 32.Tuluc P, Yarov-Yarovoy V, Benedetti B, Flucher BE. Molecular interactions in the voltage sensor controlling gating properties of CaV calcium channels. Structure. 2016;24:261–271. doi: 10.1016/j.str.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang F, et al. The conformational wave in capsaicin activation of transient receptor potential vanilloid 1 ion channel. Nat Commun. 2018;9:2879. doi: 10.1038/s41467-018-05339-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang S, et al. A pain-inducing centipede toxin targets the heat activation machinery of nociceptor TRPV1. Nat Commun. 2015;6:8297. doi: 10.1038/ncomms9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta K, et al. Tarantula toxins use common surfaces for interacting with Kv and ASIC ion channels. eLife. 2015;4:e06774. doi: 10.7554/eLife.06774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang JZ, et al. Mapping the interaction site for a β-scorpion toxin in the pore module of domain III of voltage-gated Na+ channels. J Biol Chem. 2012;287:30719–30728. doi: 10.1074/jbc.M112.370742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeLuca S, Khar K, Meiler J. Fully flexible docking of medium sized ligand libraries with RosettaLigand. PLoS One. 2015;10:e0132508. doi: 10.1371/journal.pone.0132508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen HM, et al. Structural insights into the atomistic mechanisms of action of small molecule inhibitors targeting the KCa3.1 channel pore. Mol Pharmacol. 2017;91:392–402. doi: 10.1124/mol.116.108068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang F, Vu S, Yarov-Yarovoy V, Zheng J. Rational design and validation of a vanilloid-sensitive TRPV2 ion channel. Proc Natl Acad Sci USA. 2016;113:E3657–E3666. doi: 10.1073/pnas.1604180113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang F, et al. Structural mechanism underlying capsaicin binding and activation of the TRPV1 ion channel. Nat Chem Biol. 2015;11:518–524. doi: 10.1038/nchembio.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen PT, DeMarco KR, Vorobyov I, Clancy CE, Yarov-Yarovoy V. Structural modeling of local anesthetic and antiarrhythmic drug binding to the human cardiac voltage gated sodium channel. Biophys J. 2018;114:39a. [Google Scholar]
- 42.Martin LJ, Chao R, Corry B. Molecular dynamics simulation of the partitioning of benzocaine and phenytoin into a lipid bilayer. Biophys Chem. 2014;185:98–107. doi: 10.1016/j.bpc.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 43.Corry B, Lee S, Ahern CA. Pharmacological insights and quirks of bacterial sodium channels. Handb Exp Pharmacol. 2014;221:251–267. doi: 10.1007/978-3-642-41588-3_12. [DOI] [PubMed] [Google Scholar]
- 44.Barber AF, Carnevale V, Klein ML, Eckenhoff RG, Covarrubias M. Modulation of a voltage-gated Na+ channel by sevoflurane involves multiple sites and distinct mechanisms. Proc Natl Acad Sci USA. 2014;111:6726–6731. doi: 10.1073/pnas.1405768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tikhonov DB, Zhorov BS. Mechanism of sodium channel block by local anesthetics, antiarrhythmics, and anticonvulsants. J Gen Physiol. 2017;149:465–481. doi: 10.1085/jgp.201611668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buyan A, Sun D, Corry B. Protonation state of inhibitors determines interaction sites within voltage-gated sodium channels. Proc Natl Acad Sci USA. 2018;115:E3135–E3144. doi: 10.1073/pnas.1714131115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carnevale V. Protonation underlies tonic vs. use-dependent block. Proc Natl Acad Sci USA. 2018;115:3512–3514. doi: 10.1073/pnas.1802178115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martí-Renom MA, et al. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 49.Muroi Y, Chanda B. Local anesthetics disrupt energetic coupling between the voltage-sensing segments of a sodium channel. J Gen Physiol. 2009;133:1–15. doi: 10.1085/jgp.200810103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Capes DL, Goldschen-Ohm MP, Arcisio-Miranda M, Bezanilla F, Chanda B. Domain IV voltage-sensor movement is both sufficient and rate limiting for fast inactivation in sodium channels. J Gen Physiol. 2013;142:101–112. doi: 10.1085/jgp.201310998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith MR, Goldin AL. Interaction between the sodium channel inactivation linker and domain III S4-S5. Biophys J. 1997;73:1885–1895. doi: 10.1016/S0006-3495(97)78219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McPhee JC, Ragsdale DS, Scheuer T, Catterall WA. A critical role for the S4-S5 intracellular loop in domain IV of the sodium channel alpha-subunit in fast inactivation. J Biol Chem. 1998;273:1121–1129. doi: 10.1074/jbc.273.2.1121. [DOI] [PubMed] [Google Scholar]
- 53.Tang L, Kallen RG, Horn R. Role of an S4-S5 linker in sodium channel inactivation probed by mutagenesis and a peptide blocker. J Gen Physiol. 1996;108:89–104. doi: 10.1085/jgp.108.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McPhee JC, Ragsdale DS, Scheuer T, Catterall WA. A mutation in segment IVS6 disrupts fast inactivation of sodium channels. Proc Natl Acad Sci USA. 1994;91:12346–12350. doi: 10.1073/pnas.91.25.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qu Y, Rogers J, Tanada T, Scheuer T, Catterall WA. Molecular determinants of drug access to the receptor site for antiarrhythmic drugs in the cardiac Na+ channel. Proc Natl Acad Sci USA. 1995;92:11839–11843. doi: 10.1073/pnas.92.25.11839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bender BJ, et al. Protocols for molecular modeling with Rosetta3 and RosettaScripts. Biochemistry. 2016;55:4748–4763. doi: 10.1021/acs.biochem.6b00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meiler J, Baker D. ROSETTALIGAND: Protein-small molecule docking with full side-chain flexibility. Proteins. 2006;65:538–548. doi: 10.1002/prot.21086. [DOI] [PubMed] [Google Scholar]
- 58.Singh BN. Acute management of ventricular arrhythmias: Role of antiarrhythmic agents. Pharmacotherapy. 1997;17:56S–64S, and discussion (1997) 17:89S–91S. [PubMed] [Google Scholar]
- 59.Nau C, Wang SY, Wang GK. Point mutations at L1280 in Nav1.4 channel D3-S6 modulate binding affinity and stereoselectivity of bupivacaine enantiomers. Mol Pharmacol. 2003;63:1398–1406. doi: 10.1124/mol.63.6.1398. [DOI] [PubMed] [Google Scholar]
- 60.Liu H, Atkins J, Kass RS. Common molecular determinants of flecainide and lidocaine block of heart Na+ channels: Evidence from experiments with neutral and quaternary flecainide analogues. J Gen Physiol. 2003;121:199–214. doi: 10.1085/jgp.20028723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benhorin J, et al. Effects of flecainide in patients with new SCN5A mutation: Mutation-specific therapy for long-QT syndrome? Circulation. 2000;101:1698–1706. doi: 10.1161/01.cir.101.14.1698. [DOI] [PubMed] [Google Scholar]
- 62.Wang GK, Russell C, Wang SY. State-dependent block of wild-type and inactivation-deficient Na+ channels by flecainide. J Gen Physiol. 2003;122:365–374. doi: 10.1085/jgp.200308857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang GK, Calderon J, Wang SY. State- and use-dependent block of muscle Nav1.4 and neuronal Nav1.7 voltage-gated Na+ channel isoforms by ranolazine. Mol Pharmacol. 2008;73:940–948. doi: 10.1124/mol.107.041541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fredj S, Lindegger N, Sampson KJ, Carmeliet P, Kass RS. Altered Na+ channels promote pause-induced spontaneous diastolic activity in long QT syndrome type 3 myocytes. Circ Res. 2006;99:1225–1232. doi: 10.1161/01.RES.0000251305.25604.b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Balser JR, et al. External pore residue mediates slow inactivation in mu 1 rat skeletal muscle sodium channels. J Physiol. 1996;494:431–442. doi: 10.1113/jphysiol.1996.sp021503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kambouris NG, et al. Mechanistic link between lidocaine block and inactivation probed by outer pore mutations in the rat micro1 skeletal muscle sodium channel. J Physiol. 1998;512:693–705. doi: 10.1111/j.1469-7793.1998.693bd.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ong BH, Tomaselli GF, Balser JR. A structural rearrangement in the sodium channel pore linked to slow inactivation and use dependence. J Gen Physiol. 2000;116:653–662. doi: 10.1085/jgp.116.5.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Todt H, Dudley SC, Jr, Kyle JW, French RJ, Fozzard HA. Ultra-slow inactivation in mu1 Na+ channels is produced by a structural rearrangement of the outer vestibule. Biophys J. 1999;76:1335–1345. doi: 10.1016/S0006-3495(99)77296-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen Z, et al. Lidocaine induces a slow inactivated state in rat skeletal muscle sodium channels. J Physiol. 2000;524:37–49. doi: 10.1111/j.1469-7793.2000.t01-1-00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fukuda K, Nakajima T, Viswanathan PC, Balser JR. Compound-specific Na+ channel pore conformational changes induced by local anaesthetics. J Physiol. 2005;564:21–31. doi: 10.1113/jphysiol.2004.081646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pless SA, Galpin JD, Frankel A, Ahern CA. Molecular basis for class Ib anti-arrhythmic inhibition of cardiac sodium channels. Nat Commun. 2011;2:351. doi: 10.1038/ncomms1351. [DOI] [PubMed] [Google Scholar]
- 72.Avdeef A, Box KJ, Comer JE, Hibbert C, Tam KY. pH-metric logP 10. Determination of liposomal membrane-water partition coefficients of ionizable drugs. Pharm Res. 1998;15:209–215. doi: 10.1023/a:1011954332221. [DOI] [PubMed] [Google Scholar]
- 73.Weizenmann N, Huster D, Scheidt HA. Interaction of local anesthetics with lipid bilayers investigated by 1H MAS NMR spectroscopy. Biochim Biophys Acta. 2012;1818:3010–3018. doi: 10.1016/j.bbamem.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 74.Shaw DE, et al. SC’14: International Conference for High Performance Computing, Networking, Storage and Analysis. Inst Electr Electron Eng; New York: 2014. Anton 2: Raising the bar for performance and programmability in a special-purpose molecular dynamics supercomputer; pp. 41–53. [Google Scholar]
- 75.Frazier DT, Narahashi T, Yamada M. The site of action and active form of local anesthetics. II. Experiments with quaternary compounds. J Pharmacol Exp Ther. 1970;171:45–51. [PubMed] [Google Scholar]
- 76.Sunami A, Glaaser IW, Fozzard HA. A critical residue for isoform difference in tetrodotoxin affinity is a molecular determinant of the external access path for local anesthetics in the cardiac sodium channel. Proc Natl Acad Sci USA. 2000;97:2326–2331. doi: 10.1073/pnas.030438797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sunami A, Glaaser IW, Fozzard HA. Structural and gating changes of the sodium channel induced by mutation of a residue in the upper third of IVS6, creating an external access path for local anesthetics. Mol Pharmacol. 2001;59:684–691. doi: 10.1124/mol.59.4.684. [DOI] [PubMed] [Google Scholar]
- 78.Pérez-García MT, et al. Mechanisms of sodium/calcium selectivity in sodium channels probed by cysteine mutagenesis and sulfhydryl modification. Biophys J. 1997;72:989–996. doi: 10.1016/S0006-3495(97)78751-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hilber K, et al. Selectivity filter residues contribute unequally to pore stabilization in voltage-gated sodium channels. Biochemistry. 2005;44:13874–13882. doi: 10.1021/bi0511944. [DOI] [PubMed] [Google Scholar]
- 80.Leuwer M, et al. An improved model for the binding of lidocaine and structurally related local anaesthetics to fast-inactivated voltage-operated sodium channels, showing evidence of cooperativity. Br J Pharmacol. 2004;141:47–54. doi: 10.1038/sj.bjp.0705594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smith JA, et al. Evidence for a multivalent interaction of symmetrical, N-linked, lidocaine dimers with voltage-gated Na+ channels. Mol Pharmacol. 2006;69:921–931. doi: 10.1124/mol.105.019257. [DOI] [PubMed] [Google Scholar]
- 82.Ahern CA, Eastwood AL, Dougherty DA, Horn R. Electrostatic contributions of aromatic residues in the local anesthetic receptor of voltage-gated sodium channels. Circ Res. 2008;102:86–94. doi: 10.1161/CIRCRESAHA.107.160663. [DOI] [PubMed] [Google Scholar]
- 83.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.