Significance
The phosphatases of regenerative liver (PRL) have been shown to interact with the CNNM magnesium transport regulators. Through this protein complex, PRL controls the levels of intracellular magnesium. Our study uncovers a remarkable posttranscriptional feedback mechanism by which magnesium controls PRL expression in mammalian cells. Here we show that regulation of PRL mRNA translation by magnesium depends on a 5′UTR-located upstream ORF, which is conserved among all vertebrates, proposing an evolutionary molecular mechanism of action by a divalent ion. This magnesium-sensing mechanism, which also involves the key metabolic sensor AMPK, is thus central to maintain cellular homeostasis in mammalian cells.
Keywords: Mg2+, PRL phosphatase, uORF, PTP4A, mRNA translation
Abstract
Phosphatases of regenerating liver (PRL-1, PRL-2, and PRL-3, also known as PTP4A1, PTP4A2, and PTP4A3) control magnesium homeostasis through an association with the CNNM magnesium transport regulators. Although high PRL levels have been linked to cancer progression, regulation of their expression is poorly understood. Here we show that modulating intracellular magnesium levels correlates with a rapid change of PRL expression by a mechanism involving its 5′UTR mRNA region. Mutations or CRISPR-Cas9 targeting of the conserved upstream ORF present in the mRNA leader derepress PRL protein synthesis and attenuate the translational response to magnesium levels. Mechanistically, magnesium depletion reduces intracellular ATP but up-regulates PRL protein expression via activation of the AMPK/mTORC2 pathway, which controls cellular energy status. Hence, altered PRL-2 expression leads to metabolic reprogramming of the cells. These findings uncover a magnesium-sensitive mechanism controlling PRL expression, which plays a role in cellular bioenergetics.
The phosphatase of regenerating liver (PRL) family is composed of three members (PRL-1, -2, and -3; gene names PTP4A1, PTP4A2, and PTP4A3) of ∼20 kDa with at least 75% amino acid sequence identity shared between them that are highly expressed in a majority of cancers (1). The cyclin M (CNNM) magnesium regulators form an evolutionarily conserved complex with PRLs to regulate intracellular magnesium concentration (2, 3). The PRL/CNNM complex has been validated by structural studies characterizing the interaction between both proteins (4–6). Mutation of critical amino acid residues involved in this molecular association leads to reduced tumor growth, indicating a prooncogenic role of the protein complex (7). Furthermore, either PRL knockdown or disruption of complex formation reduces magnesium transport (2, 3, 7). The main outcome of PRL overexpression and CNNM complex formation is increased intracellular magnesium, establishing both of these as critical modulators of magnesium homeostasis. However, it is still unclear whether the CNNMs function as magnesium sensors or dual magnesium influx/efflux transporters (1, 8, 9).
Magnesium is an essential cation regulating numerous cellular functions whose intracellular concentration is tightly regulated by various transporters (10). Serum magnesium represents only ∼1% of the total body content, and thus values within the normal range (0.7 to 1 mM) might not necessarily reflect an overall somatic hypo- or hypermagnesemic state (10). Magnesium imbalance is observed in multiple diseases and is linked to alterations of several hallmarks of cancer (11). It is required for energy production either indirectly as a part of the Mg2+–ATP complex or directly as a cofactor (12). Most enzymes involved in glycolysis, the Krebs cycle, and the respiratory chain depend on magnesium as either an allosteric modulator or a cofactor. Moreover, magnesium is critical for mitochondria to carry out oxidative phosphorylation (13, 14), and low magnesium concentrations have been associated with altered uncoupled respiration (15, 16). In line with the role of PRLs in magnesium homeostasis, mitochondrial respiration was found to be similarly affected and ATP turnover decreased in cells isolated from PRL-2−/− animals (16). This is also consistent with the observed decrease in body weight of PRL-2−/− mice, suggesting that their cellular metabolism is inherently less efficient (16).
The expression of various magnesium transporters is up-regulated under conditions of magnesium deprivation, both in vitro and in vivo (17–19). The intrinsic mechanisms regulating this process are largely uncharacterized. It has been proposed that epithelial cells can sense environmental magnesium levels through transcription- and translation-dependent processes to alter magnesium transport and maintain its balance (20, 21). In bacteria, magnesium transporter expression is regulated by magnesium, both at the level of transcription by the PhoP/Q system as well as posttranscriptionally by a 5′UTR region-located riboswitch (22). In eukaryotic cells, the underlying mechanism by which magnesium regulates gene expression is largely unexplored. We previously observed that while CNNM3 expression remains unchanged following magnesium depletion, its interaction with PRL-1/2 markedly increases, indicating a pivotal regulatory role for PRL-1/2 and the PRL/CNNM complex in compensating for decreased magnesium availability (2). Furthermore, this was accompanied by an increase of PRL-1/2 protein expression by an unknown regulatory mechanism. Here we demonstrate that magnesium regulates PRL expression at the posttranscriptional level via an upstream ORF (uORF) in an AMPK/mTORC2-dependent manner.
Results
Magnesium Controls PRL Expression by an mRNA Translational Mechanism.
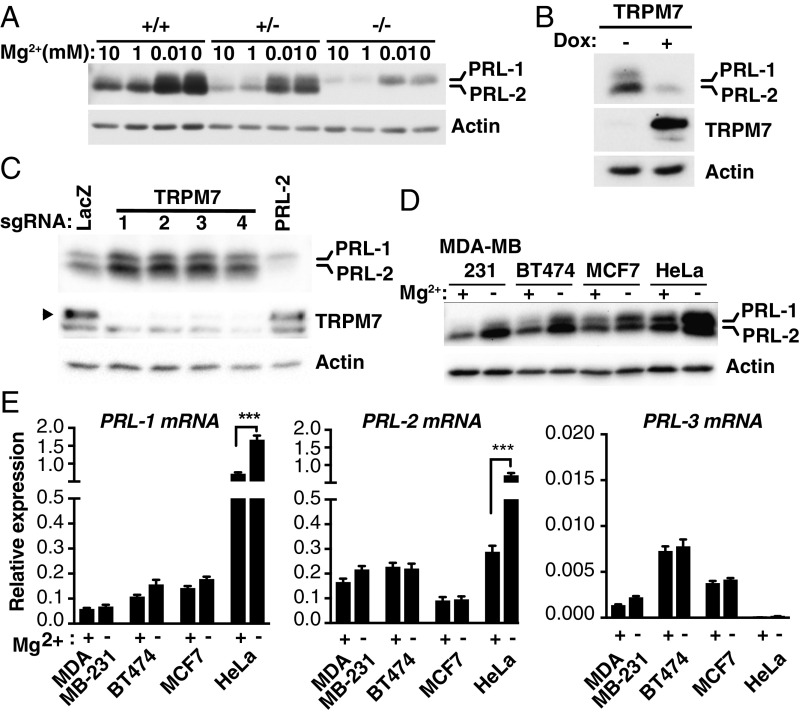
Cellular magnesium is regulated by intrinsic mechanisms which control various magnesium transporters to maintain homeostasis. Since extracellular magnesium depletion causes a decrease in intracellular magnesium levels (23, 24) (SI Appendix, Fig. S1), we examined the involvement of PRLs in the adaptive response related to changes in magnesium availability. We previously showed that extracellular magnesium levels regulate the expression of PRL-1 and -2 in various cancer cell lines by an unknown mechanism (2). Here, we further confirmed in PRL-2+/+, PRL-2+/−, and PRL-2−/− primary mouse embryonic fibroblasts that magnesium depletion induces a rise in PRL-1/2 protein levels (Fig. 1A) using a previously validated antibody that recognizes both proteins (2, 25). We next assessed the consequence of increasing intracellular magnesium instead. To this end, we overexpressed the TRPM7 magnesium transporter (26) using the well-characterized HEK293 doxycycline (dox)-inducible system to trigger an increase in intracellular magnesium levels (27, 28). We observed a decrease in PRL-1/2 expression following dox-induced TRPM7 expression (Fig. 1B). However, knockdown of TRPM7 using four independent single guide RNAs (sgRNAs) in the CRISPR-Cas9 system led to an increase in PRL-1/2 protein levels (Fig. 1C), indicating that PRL expression is regulated by intracellular magnesium levels. The knockdown of PRL-2 also confirmed the specificity of the PRL-2 signal detected by Western blot analysis. Surprisingly, while PRL protein levels were greatly induced following 24-h magnesium depletion in four different cell lines tested (Fig. 1D), no significant changes in PRL mRNA levels were detected, except in HeLa cells, which showed an increase in PRL-2 mRNA expression (Fig. 1E). Still, this change was modest compared with the increase in PRL-2 protein levels. PRL-3 mRNA and protein levels were almost undetectable in all cell lines tested. Overall, the results indicate that PRL-1/2 protein expression is regulated by magnesium.
Fig. 1.
Magnesium regulates PRL expression. (A) Western blot analysis of primary mouse embryonic fibroblasts isolated from PRL-2 knockout (−/−), heterozygote (+/−), and WT (+/+) mouse embryos incubated for 24 h in either the presence or absence of magnesium. (B) TRPM7 expression was induced with dox in HEK293 cells for 30 h and analyzed by Western blot. (C) Knockdown of TRPM7 using four independent sgRNAs by the CRISPR-Cas9 system was performed in HEK293-TRPM7–expressing cells and analyzed by Western blot. (D and E) The indicated cell lines were cultured for 24 h in either the presence or absence of magnesium and analyzed by Western blot (D) or qPCR (E). The plots in E represent data as mean ± SD (n = 3); ***P < 0.001 by two-way ANOVA. All Western blots are representative of three independent experiments.
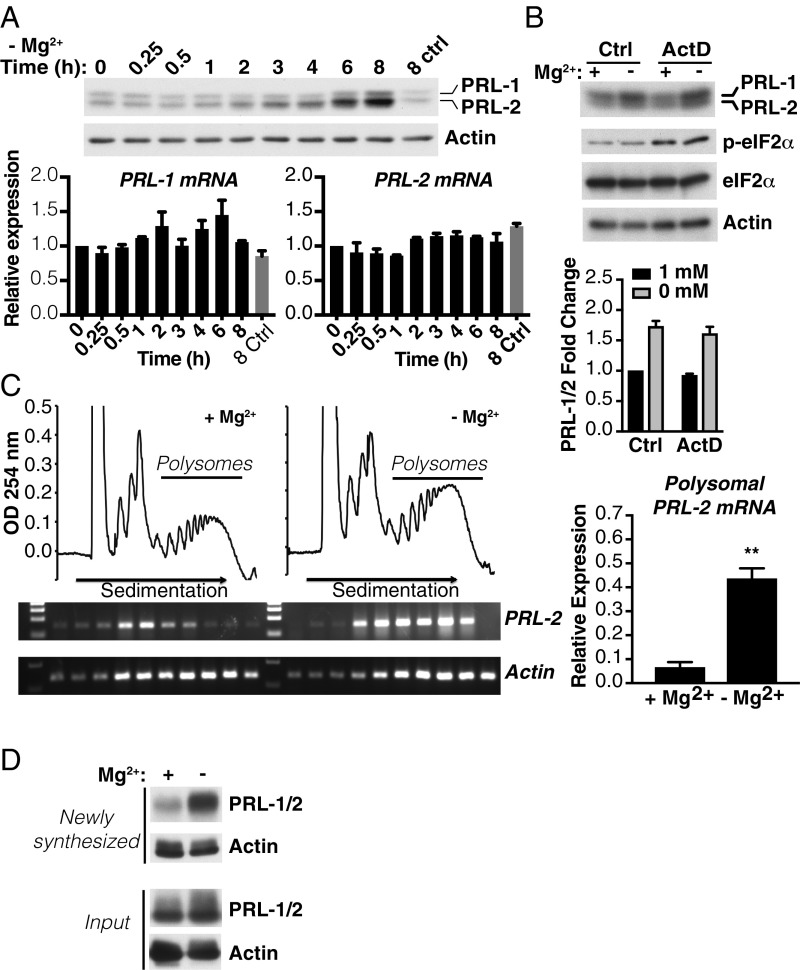
Since the data described in Fig. 1 suggested that a posttranscriptional mechanism might be responsible for the effect of magnesium on PRL expression, we performed an experiment to parallelly determine both mRNA and protein expression changes under hypomagnesemic conditions at different time points. Interestingly, we could detect an initial rise in PRL-1/2 levels by Western blot after 3 to 4 h following magnesium depletion, which increased over time (Fig. 2A). In contrast, no difference was observed up to 8 h following magnesium depletion when we examined PRL-1 and -2 mRNA levels by qPCR. We noted that PRL expression was still induced following magnesium depletion when transcription was inhibited using actinomycin D, a commonly used transcription inhibitor (Fig. 2B). eIF2α Ser-51 phosphorylation, which has been implicated in the cellular stress response, remained unaffected by magnesium depletion but was induced by actinomycin D, as previously described (29). The up-regulation of PRL-1/2 was also not caused by magnesium-related inhibition of the proteasome since treatment with the proteasome inhibitor MG132 did not interfere with the increase in PRL-1/2 proteins observed in the absence of magnesium (SI Appendix, Fig. S2). We next explored whether PRL-2 was being more efficiently translated upon magnesium depletion. To this end, we analyzed the polysome distribution of PRL-2 mRNA in HeLa cells cultured for 2 h in either the absence or presence of magnesium (Fig. 2C). There was a clear recruitment of PRL-2 mRNA into heavy polysomes upon acute magnesium depletion compared with cells cultured under normal magnesium conditions (Fig. 2C). Moreover, de novo protein synthesis of PRL-1/2 was also induced (Fig. 2D) and this was further confirmed in the MDA-MB-231 breast cancer cell line (SI Appendix, Fig. S3). Since a recent study proposed that magnesium-dependent regulation of PRL expression occurs at the transcriptional level in HeLa cells via the STAT pathway (30), we performed experiments using the same STATs inhibitor, S3I-201. Induction of PRL-1/2 protein expression was unaffected in the 8 h following magnesium depletion and STAT inhibition (SI Appendix, Fig. S4). We were only able to detect a reduction in PRL-1/2 at 24 h following treatment. Collectively, these data indicate that PRL expression is rapidly regulated by an mRNA translational mechanism in response to changes in intracellular magnesium.
Fig. 2.
Magnesium depletion promotes PRL mRNA translation. (A) HeLa cells were incubated in the absence of magnesium for the indicated times and analyzed by Western blot (Top) and qPCR (Bottom). The 8 ctrl indicates 8-h control with magnesium. (B) Western blot analysis of HeLa cells incubated for 6 h in either the presence or absence of magnesium with or without actinomycin D (ActD). Quantification is represented as values expressed as fold change relative to 1 mM magnesium control (Ctrl). Data are means ± SEM (n = 3). (C) Polysome profiles of HeLa cells treated for 2 h in either the presence or absence of magnesium. PRL-2 and actin mRNA distribution across the gradient was evaluated in each fraction by semiquantitative RT-PCR and on the polysome fractions by qPCR. Data are means ± SD (n = 3); **P < 0.01 by one-way ANOVA. (D) HeLa cells were metabolically labeled with the methionine analog l-azidohomoalanine for 2 h in either the presence or absence of magnesium. Newly synthesized proteins were covalently bound to biotin, pulled down with streptavidin beads, and analyzed by Western blotting. Input represents total protein extract before biotin pulldown. Representative of three independent experiments.
A Conserved uORF Regulates PRL Protein Synthesis in a Magnesium-Sensitive Manner.
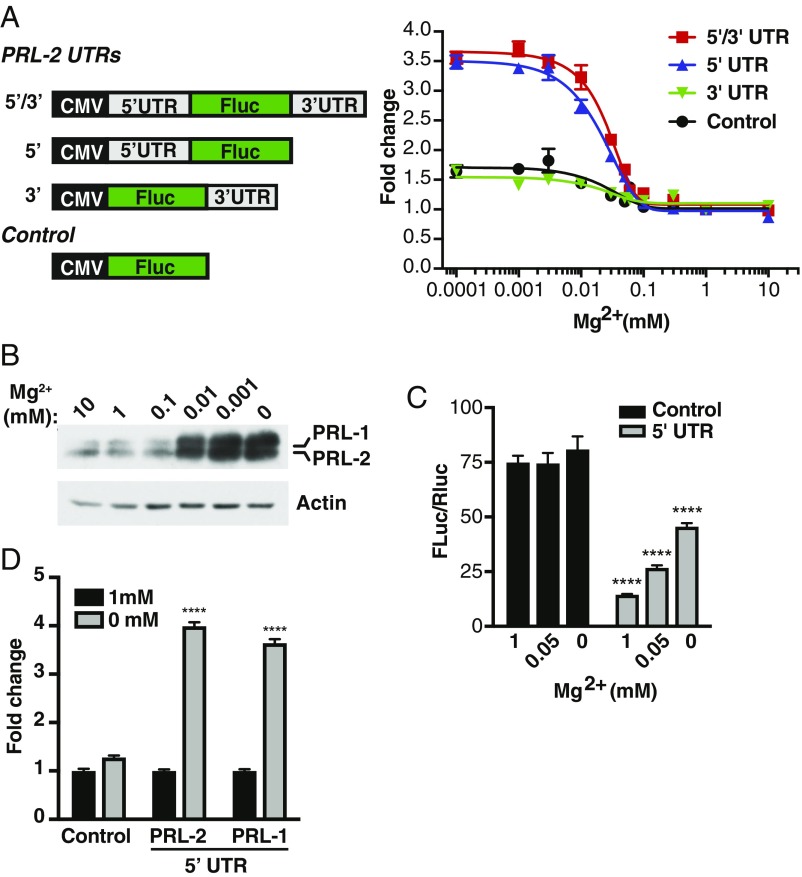
Given the importance of the UTRs in the control of mRNA translation (31), we first determined which region of the PRL mRNA could be involved in regulating its expression following magnesium depletion. We generated reporter constructs by fusing the PRL-2 5′UTR, the 3′UTR, or both regions to the luciferase gene as described in Fig. 3A. We then transfected these constructs into HeLa cells, treated them with various concentrations of magnesium, and measured luciferase activity. The results indicate that decreasing magnesium levels does not significantly affect luciferase activity in either the absence of both UTRs or the presence of only the 3′UTR (Fig. 3A). Interestingly, the PRL-2 5′UTR was sufficient to elicit a magnesium dose–response effect, indicating that this region is critical for magnesium sensing and regulation of PRL-2 protein expression. In parallel, we observed a similar dosage effect on endogenous PRL-1/2 protein expression in HeLa cells (Fig. 3B), validating the results obtained using the luciferase assay. We found that the presence of the 5′UTR drastically repressed luciferase activity under normal conditions (Fig. 3C). These various reporter constructs showed no difference in mRNA level of the luciferase gene (SI Appendix, Fig. S5A), indicating a translational mode of regulation. Finally, we confirmed the importance of the PRL-1 5′UTR using the luciferase assay, which had a response to low magnesium similar to the PRL-2 5′UTR (Fig. 3D). Overall, these results suggest that the 5′UTR of both PRL-1 and PRL-2 mRNA may harbor some cis-regulatory elements affecting their translation and response to magnesium.
Fig. 3.
The 5′UTR region of the PRL mRNA controls PRL expression in response to magnesium levels. (A) PRL-2 UTR luciferase reporter constructs used to detect translation in response to magnesium levels. HeLa cells were transfected with the indicated constructs and treated with various concentrations of magnesium for 24 h and luciferase activity was measured. Values are expressed as fold change relative to 1 mM magnesium for each construct. (B) Western blot analysis of HeLa cells incubated for 24 h in the presence of the indicated magnesium concentration. Representative of three independent experiments. (C) HeLa cells were transfected with the indicated constructs, treated with varying magnesium concentrations for 24 h, and luciferase activity was measured. Plot is representative of three independent experiments. Data are means ± SD (n = 4); ****P < 0.0001 vs. control for each magnesium concentration by two-way ANOVA. (D) HeLa cells were transfected with the indicated constructs and incubated for 24 h in either the presence or absence of magnesium followed by luciferase activity measurements. Values are expressed as fold change over 1 mM magnesium for each construct. Plot is representative of two independent experiments. Data are mean ± SD (n = 4); ****P < 0.0001 vs. 1 mM magnesium by two-way ANOVA.
mRNA translation can be regulated by various factors, including RNA secondary structures, upstream AUG (uAUG), and near-cognate non-AUG codons that may be used to initiate translation of uORFs. In addition, some mRNAs bypass cap-dependent translation (see refs. 32 and 33 for review). For example, it was recently shown that m6A modifications allow cap-independent recruitment of initiating ribosome complexes (34). To assess the possibility of cap-independent initiation we used a bicistronic reporter vector encoding Renilla (RLuc) and firefly (FLuc) luciferase on a single mRNA transcript, in which we cloned the PRL-2 5′UTR between the two reporters (SI Appendix, Fig. S5B). Production of Renilla reports on cap-dependent translation, whereas production of firefly luciferase reports on cap-independent translational activity, as assessed by the encephalomyocarditis virus control in this experiment. There is a slight increase in FLuc production when the PRL-2 5′UTR is placed between FLuc and RLuc, indicating that this region might possesses elements involved in cap-independent translation (SI Appendix, Fig. S5B), although the use of a bicistronic reporter alone is insufficient evidence (see ref. 35 for discussion). Even if cap-independent initiation is involved, we did not detect an increased signal upon magnesium withdrawal. This excludes this mode of regulation involving this cation.
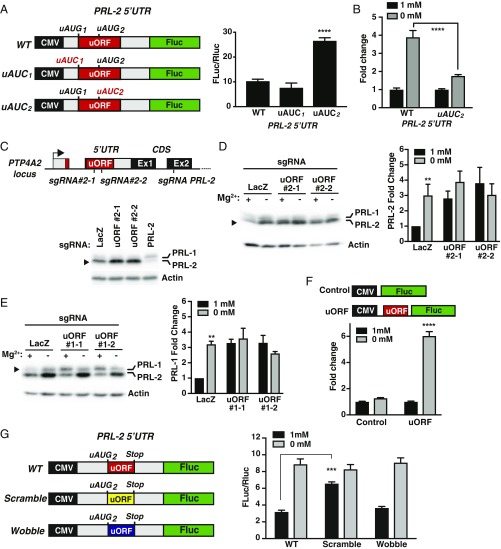
Using GWIPS-viz (36, 37), which provides access to the genomic alignments of publicly available ribosome profiling data that maps the position of translating ribosomes over the entire transcriptome, we were able to identify a translated short uORF generating ribosome footprints located in the 5′UTR of both PRL-1 and -2 mRNAs (SI Appendix, Figs. S6 and S7). Most importantly, this small uORF is extremely well conserved among vertebrates and between PRL-1 and PRL-2, suggesting a potential function in regulating PRL expression. Of note, this uORF has been proposed to be associated with the resistance of PRL mRNA translation to global down-regulation during integrated stress response (38). In addition, these ribosome-profiling data, which were obtained under normal magnesium conditions, suggested that there was an accumulation of ribosomes stalled at the conserved uORFs in the 5′UTR of both PRL-1 and -2 mRNAs. Since both conserved uORFs contain two AUGs (SI Appendix, Figs. S6–S8), we first wanted to assess if they are used for translation initiation and whether this type of cis-acting mode of regulation controls the expression of the main coding sequence (CDS). Therefore, we individually mutated both AUG codons in the 5′UTR of the PRL-2 mRNA to AUC (AUG1 and AUG2) and performed the luciferase assay described above (Fig. 4A). In contrast to the AUG1 mutant, we detected a stronger luciferase signal in the AUG2 mutant, indicating that this codon is used to initiate translation of the regulatory uORF controlling the expression of the main CDS (Fig. 4A). Importantly, the mutation reduced the reporter signal induced by magnesium depletion, suggesting that this uORF2 affects mRNA translation in a magnesium-sensitive manner (Fig. 4B). We next targeted the endogenous uORF2 located in the 5′UTR of the PRL-2 mRNA using two different CRISPR-Cas9 sgRNAs, which were directed to the corresponding genomic region in the PRL-2 locus. Also, an sgRNA targeting the coding sequence of PRL-2 was used as a control. As expected, we observed an increase in PRL-2 protein expression in MDA-MB-231 cells expressing the two sgRNAs targeting the uORF2 (Fig. 4C). Furthermore, magnesium depletion, which increased PRL-2 levels in lacZ control cells, did not have an additive effect on PRL-2 expression in the uORF-targeted cells (Fig. 4D). Similar results were obtained on targeted MDA-MB-231 clones in which we confirmed the presence of indels near the C-terminal region of the uORF-encoded peptide (SI Appendix, Fig. S9). Consistent with this observation, when we targeted the uORF2 of PRL-1 in a similar fashion near the C-terminal region of the uORF-encoded peptide, its expression was not further affected in hypomagnesemic conditions, although we detected an increase in PRL-1 expression using two different sgRNAs in normal magnesium (Fig. 4E). Importantly, we did not detect any changes in PRL-1 and -2 mRNA levels in the targeted cells (SI Appendix, Fig. S10). Using a reporter construct containing only the uORF cloned directly upstream of the luciferase gene (SI Appendix, Fig. S8), we showed that presence of the uORF was sufficient to provoke a magnesium-dependent response of the luciferase signal (Fig. 4F). Finally, we scrambled the amino sequence corresponding to the uORF2 in the 5′UTR of the PRL-2 mRNA without changing the start and stop codons (SI Appendix, Fig. S11) and showed that it abrogates translational control using the luciferase assay (Fig. 4G), thus indicating that the uORF amino acid sequence is critical for the regulation of the main coding sequence. In addition, we modified the nucleotide sequence without changing the amino acid sequence (wobble construct) to show that the nucleotide sequence itself is not important for the observed regulation. Taken together, these data identify a role for a cis-regulatory element present on the 5′UTR affecting PRL mRNA translation in a magnesium-sensitive manner.
Fig. 4.
A conserved uORF in the PRL 5′UTR senses magnesium levels to regulate its expression. (A) PRL-2 5′UTR luciferase reporter constructs used to test uORFs identified by ribosome profiling data. HeLa cells were transfected with the indicated constructs and luciferase activity was measured. Plot is representative of three independent experiments. Data are means ± SD (n = 4); ****P < 0.0001 vs. WT by two-way ANOVA. (B) HeLa cells were transfected with the indicated constructs and incubated for 24 h in either the presence or absence of magnesium followed by luciferase activity measurements. Values are expressed as fold change relative to 1 mM magnesium for each construct. Plot is representative of three independent experiments. Data are means ± SD (n = 4); ****P < 0.0001 by two-way ANOVA. (C) Targeting of the PTP4A2 locus using two independent sgRNAs against the uORF and a specific sgRNA against the main ORF encoding PRL-2 by the CRISPR-Cas9 system was performed in MDA-MB-231 cells and analyzed by Western blot. LacZ sgRNA was used as a control. (D and E) PRL-2 and PRL-1 uORF-targeted MDA-MB-231 cells were incubated for 24 h in the absence of magnesium and analyzed by Western blot. Quantification is presented on each panel. Values are expressed as fold change relative to 1 mM magnesium control (Ctrl). Data are means ± SEM (n = 3). **P < 0.01 vs. 1 mM by two-way ANOVA. (F and G) PRL-2 5′UTR luciferase reporter constructs to test the uORF response to magnesium. HeLa cells were transfected with the indicated constructs and luciferase activity was measured. Values are expressed as fold change relative to 1 mM magnesium for each construct (F) or as luminescence ratio signal (G). Plot is representative of three independent experiments. Data are means ± SD (n = 4); ****P < 0.0001 vs. uORF 1 mM and ***P < 0.001 vs. WT 1 mM by two-way ANOVA.
Magnesium Regulates PRL Expression by a Mechanism Linked to the AMPK/mTORC2 Pathway.
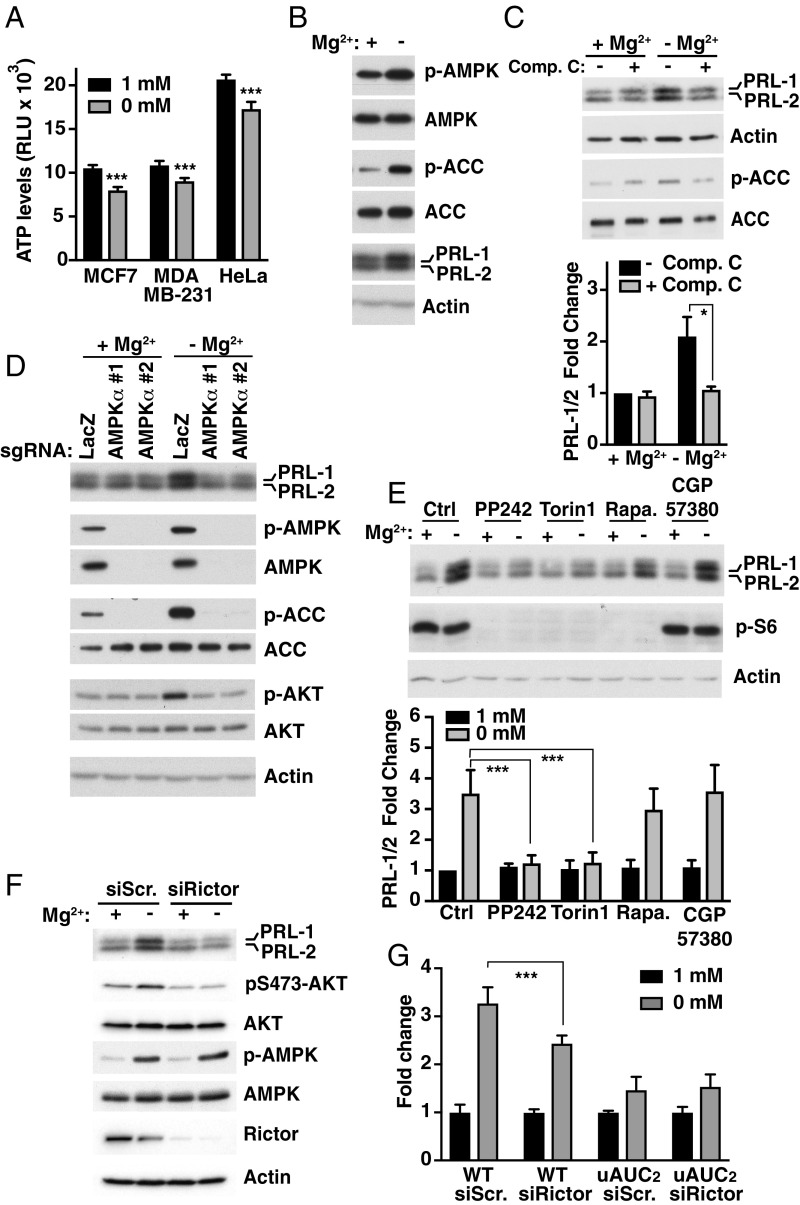
The main source of energy in cells is ATP, which must be bound to magnesium to be biologically active and is therefore commonly referred to as Mg-ATP (39). A decrease in its intracellular levels leads to an increase in AMPK activity, which is a major regulator of metabolism and mRNA translation (40). We detected a decrease in ATP levels following magnesium depletion in various cell lines (Fig. 5A). This correlated with an increase in AMPK activity, as measured by the phosphorylation of acetyl-CoA carboxylase (ACC), a direct downstream target of AMPK (Fig. 5B). Surprisingly, inhibiting AMPK using compound C inhibited PRL-1/2 expression induced by the absence of magnesium in MCF-7 cells (Fig. 5C) and other cell lines (SI Appendix, Fig. S12). Furthermore, we used CRISPR-Cas9 to target the catalytic alpha subunit of AMPK using two different sgRNAs and confirmed that this enzyme is involved in the regulation of PRL-1/2 protein expression, but not mRNA abundance (SI Appendix, Fig. S13), in hypomagnesemic conditions (Fig. 5D). This result was confirmed in MDA-MB-231 cells (SI Appendix, Fig. S14). Interestingly, reducing magnesium levels stimulated mTORC2 activity as measured by increased pS473-AKT, its direct downstream target, and this rise was abrogated in AMPK-targeted cells (Fig. 5D). One of the major downstream signaling pathways regulated by AMPK is the mammalian target-of-rapamycin (mTOR) pathway, which includes the rapamycin-sensitive mTORC1 complex and rapamycin-insensitive mTORC2 complex. Activation of AMPK represses mTORC1, but not mTORC2, which in turn suppresses translation of select mRNAs (41, 42). Exposing cells to rapamycin or an inhibitor of MNK1/2 (CGP57380), an upstream regulator of the cap-binding protein eukaryotic initiation factor 4E (eIF4E), did not affect PRL-1/2 protein levels following magnesium depletion in MCF-7 cells (Fig. 5E). In contrast, inhibiting mTORC2 in addition to mTORC1 using PP242 or Torin1 resulted in reduced expression of PRL-1/2. Similar results were obtained in two other cell lines with PP242 (SI Appendix, Fig. S15), further indicating that mTORC2 might be more important than mTORC1 for the control of PRL-1/2 levels in response to low magnesium. Consistent with this, we observed reduced magnesium-dependent up-regulation of PRL-1/2 proteins, without change in their mRNA levels, when Rictor, an essential component of the mTORC2 complex, was knocked down (Fig. 5F and SI Appendix, Fig. S13). This was accompanied by a block in pS473-AKT and no change in AMPK activity, suggesting that the latter acts upstream of mTORC2. Finally, we targeted Rictor in WT and uAUC2 5′UTR reporter-expressing cells (described in Fig. 4A) and show that targeting mTORC2 activity reduces the luciferase signal following magnesium depletion when the uORF is functional (Fig. 5G). These findings demonstrate that restriction of magnesium availability leads to mTORC2 activation in an AMPK-dependent manner to increase PRL expression by a mechanism involving the uORF-regulatory functions.
Fig. 5.
Magnesium regulates PRL expression by an AMPK/mTORC2-dependent pathway. (A) ATP measurements in various cell lines following 2-h magnesium depletion. Data are means data ± SEM (n = 4); ***P < 0.001 by two-way ANOVA. RLU, relative luminescent unit. (B) Western blot analysis of MCF-7 cells incubated for 8 h in either the presence or absence of magnesium. Representative of three independent experiments. (C) MCF-7 cells were incubated for 24 h in the presence or absence of magnesium with or without compound C (Comp. C) and analyzed by Western blot. Quantification is shown as values expressed as fold change relative to 1 mM magnesium without Comp. C. Data are means ± SEM (n = 3); *P < 0.05 by two-way ANOVA. (D) Knockdown of AMPKα using two sgRNAs by the CRISPR-Cas9 system was performed in MCF-7 cells. Targeted cells were incubated for 24 h in the presence or absence of magnesium and analyzed by Western blot. LacZ sgRNA was used as a control. Representative of three independent experiments. (E) MCF-7 cells were incubated for 24 h in the presence or absence of magnesium with or without the indicated inhibitors and analyzed by Western blot. Quantification is shown as values expressed as fold change relative to Ctrl 1 mM magnesium. Data are means ± SEM (n = 3); ***P < 0.001 by two-way ANOVA. Ctrl, vehicle control with DMSO; Rapa, rapamycin. (F) MCF-7 cells were transfected for 30 h with the indicated siRNAs and incubated for 12 h in the presence or absence of magnesium before being analyzed by Western blot. Scr., scramble. (G) HeLa cells were transfected with the indicated constructs and siRNAs followed by luciferase activity measurement. Plot is representative of three independent experiments. Data are means ± SD (n = 4); ***P < 0.001 by two-way ANOVA.
PRL-2 Modulates Cell Metabolism.
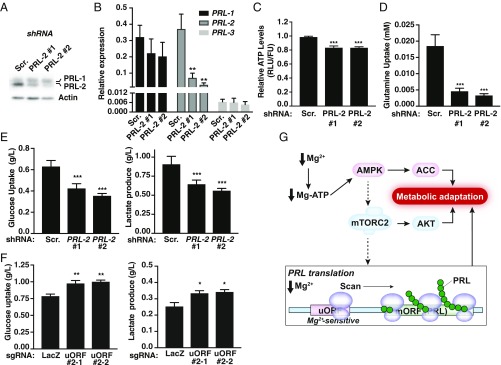
Since magnesium is a critical regulator of various metabolic enzymes (10), and given the role of the AMPK/mTOR pathway in metabolism (42, 43), we next assessed the downstream bioenergetic consequences of altering PRL-2 expression. Of note, we initially generated PRL-2 knockout MDA-MB-231 cells using CRISPR-Cas9, but we observed compensatory PRL-1 up-regulation (SI Appendix, Fig. S16). Since long-term deletion of PRL-2 may cause metabolic cell adaptation or compensation by the other PRLs, we established MDA-MB-231 cells expressing dox-inducible PRL-2 shRNAs. After 48 h in the presence of dox, we observed ∼80% knockdown efficiency using two shRNAs compared with the scramble control (Fig. 6A). This acute treatment did not result in increased PRL-1 protein levels. At the transcriptional level, although we observed a decrease in PRL-2 mRNA as expected, PRL-1 and -3 transcription profiles were not altered (Fig. 6B). Given the importance of magnesium in metabolism, we observed that knockdown of PRL-2 leads to a decrease in intracellular ATP levels (Fig. 6C) and glutamine uptake (Fig. 6D). Moreover, this correlated with a reduction of glucose uptake by the PRL-2 targeted cells, which also led to lower lactate production (Fig. 6E), suggesting a lower glycolytic flux in these cells. Surprisingly, since glutamine uptake was also reduced, this indicated that other catabolic pathways might compensate for the reduced utilization of these two major nutrients. Finally, to evaluate the role of the uORF-mediated translational control in cell metabolism, MDA-MB-231 cells expressing the two sgRNAs targeting the uORF (described in Fig. 4C) were analyzed. As these cells showed increased PRL-2 expression, higher glucose uptake and increased lactate production was observed (Fig. 6F). Overall, these data demonstrate metabolic reprogramming in response to PRL-2, indicating that PRL-2 overexpression may provide a survival advantage by allowing cancer cells to meet the high bioenergetic demands required for rapid proliferation.
Fig. 6.
PRL-2 mediates cellular metabolic reprogramming. (A) Western blot analysis of MDA-MB-231 cells expressing dox-inducible PRL-2 or scramble (Scr.) shRNAs treated with dox for 48 h. (B) Quantification of PRL-1, -2, and -3 mRNAs was performed on dox-inducible PRL-2 or Scr. shRNA-expressing cells treated with dox for 48 h. Data are mean ± SD (n = 3); **P < 0.01 vs. Scr. by two-way ANOVA. (C–E) MDA-MB-231 cells expressing dox-inducible PRL-2 or Scr. shRNAs were treated with dox for 48 h and the indicated metabolites measured. All plots are representative of at least three independent experiments. Data are means ± SD (n = 4–5); ***P < 0.001 vs. Scr. by two-way ANOVA. (F) Metabolite measurement of MDA-MB-231 cells expressing the two sgRNAs targeting the uORF cultured in 2.5 mM glucose. All plots are representative of at least three independent experiments. Data are means ± SD (n = 4); **P < 0.01 or *P < 0.05 vs. LacZ by two-way ANOVA. (G) Schematic depicting how low intracellular magnesium leads to an increase in PRL protein levels. When magnesium is reduced, the AMPK/mTORC2-dependent pathways, which are generally involved in regulating cellular bioenergetics, get activated to promote a rise in PRL protein synthesis to contribute to the observed metabolic switch. Concomitantly, a uORF contributes to the regulation of PRL mRNA translation by a “magnesium-sensing” mechanism involving scanning ribosomes that will be stalled at the uORF in the presence of magnesium but become competent for reinitiation at the downstream start codon of the main ORF (mORF) under conditions of low magnesium, leading to PRL protein synthesis.
Discussion
We identified a mode of control of PRL expression involving the 5′UTR in which the presence of a conserved uORF is critical for its regulation. Furthermore, this cis-regulatory element acts as a magnesium sensor by a mechanism involving in part the AMPK/mTORC2 pathway to regulate PRL expression, which will lead to a cellular metabolic response (Fig. 6G). As we previously showed, PRL-2 can modulate bioenergetic functions in vivo (16) and a magnesium-deficient diet can stimulate PRL-2 expression in various mouse tissues (44), all of which establish the PRLs as important metabolic mediators which respond to magnesium to maintain homeostasis.
The lower ATP levels that we observed following magnesium depletion correlate well with the cellular function of magnesium. ATP must be bound to this cation to be biologically active (39) and is maintained within a tight concentration range by AMPK, which senses the energy state of the cell largely as a ratio of AMP or ADP to ATP (40). Consistent with this, we also observed AMPK activation in low-magnesium conditions, thus highlighting the essential function of magnesium as a cofactor of various metabolic pathways (10, 13). During energetic stress, the role of AMPK is to rapidly replenish intracellular ATP by shutting down major anabolic pathways and activating the catabolic ones to promote metabolic adaptation leading to cell survival. Given the role of the PRLs in cell survival, proliferation, and metabolism, our results suggest that the rapid increase in PRL protein levels following magnesium depletion via the AMPK pathway is part of this adaptive process. One likely possibility is that the binding of PRL to the CNNM magnesium transporter leads to an increase in intracellular magnesium levels, as we and others have previously shown (2, 3), to replenish intracellular ATP-Mg. This correlates well with the reported up-regulation of various magnesium transporters in response to low magnesium to maintain its intracellular balance (17–19, 21).
A rise in intracellular magnesium via TRPM7-mediated influx is essential for sustained mTORC1 activation (45). We show that overexpression of this magnesium transporter leads to a reduction in PRL levels. Interestingly, activated AMPK directly suppresses the function of mTORC1 (41, 42) but potentially up-regulates mTORC2 activity (46, 47). Here, blocking mTORC1 with rapamycin did not affect PRL expression under hypomagnesemic conditions. Instead, we establish that mTORC2 activation in response to low magnesium acts as an upstream modulator of PRL expression. This suggests that when intracellular magnesium levels become limiting, AMPK is activated and suppresses mTORC1, thus favoring the mTORC2 pathway to promote PRL protein synthesis. The exact role of mTORC2 is still obscure but it has been shown to affect cell metabolism, as loss of Rictor in mice, a critical component of the mTORC2 complex, affects glucose homeostasis (43). This is in line with the metabolic function of PRL-2 previously identified in PRL-2−/− mice (16) and our observation that the acute knockdown of PRL-2 leads to a decrease in glucose uptake.
TRPM7 is regulated by magnesium levels via a posttranscriptional mechanism involving two uORFs located within its mRNA (48). The proposed model is one where the first uORF inhibits the overall translation of the main coding sequence, whereas the second confers specific magnesium sensitivity of translation leading to TRPM7 up-regulation at decreased magnesium concentrations. In TRPM7, the first uORF overlaps the main ORF, thus preventing proper scanning by the translation machinery past the main ORF, thus dampening TRPM7 translation (48). In contrast, the main PRL ORF is located downstream of the conserved uORF, and the accumulation of ribosomes within this uORF indicates a strong inhibitory function under normal conditions. It is when magnesium becomes limiting that this ribosome stalling is likely relieved. There are at least three nonmutually exclusive ways that this could enhance translation of the downstream ORF. First, it is possible that the ribosome pause makes ribosomes incompetent for reinitiation downstream as they lose translation initiation factors associated with the ribosome. When the pause is removed, the ribosome would become competent for reinitiation at the downstream main start codon, leading to the synthesis of PRL protein. The other possibility is that the pause may work as a roadblock to the leaky scanning 43S preinitiation complex (PIC). This possibility is supported by deep phylogenetic conservation of uAUG2 Kozak context for both PRL-1 and PRL-2, which is very weak (UUUAUGG). Finally, it is also possible that the pausing is involved in modulating the efficiency of translation initiation itself; paused ribosomes could lead to the generation of ribosome queues at the beginning of the uORF, resulting in a prolonged time spent by the 43S PIC at uAUG2, increasing the chance of initiation. Such a mode of regulation has been demonstrated recently in polyamine-dependent regulation of CUG-initiated uORFs in antzime inhibitor mRNA (49). While this pause-dependent initiation has been shown for a non-AUG start, it is likely that initiation at an AUG codon in a very weak Kozak context may also be highly sensitive to the time that 43S PIC spends at this start.
What might be responsible for the pause? A very likely possibility is that the uORF encodes a cis-acting regulatory short peptide that responds to specific small molecules in the cellular environment (49, 50). The translation of the arg-2 gene of Neurospora crassa and CPA1 in fungi is negatively controlled at the translational level in response to the level of arginine (51, 52). Such a mechanism is supported by the exceptional cross-species conservation of the uORF amino acid sequence, which indicates that the sequence has an important functional role since it is evolutionally conserved. This is reinforced by our data showing that amino acid sequence of the uORF is critical for the translational control of PRL-2. Magnesium could interact with the nascent peptide either directly or through an intermediary to stall ribosomes at the uORF, which would attenuate PRL translation. While it is likely that the conservation of the amino acid sequence of this uORF is due to its properties associated with ribosome pausing and magnesium-dependent regulation of the main ORF translation, we cannot exclude a possibility that its product has its own function. Indeed, products of many uORFs have been detected in proteomics studies, and those of SLC35A4 and MIEF1 uORFs are predicted to be functional based on their conservation (38) and function as a component of mitochondrial membrane (53) and a mitochondrial ribosome assembly factor (54), respectively.
We provide strong evidence that magnesium regulates PRL expression by a posttranscriptional mechanism. However, it was recently proposed that PRL is transcriptionally regulated by magnesium in a STAT1-dependent manner in HeLa cells (30). With the STAT inhibitor used in that study, we observed a slight decrease in PRL expression at both basal conditions and following magnesium depletion only after 24 h. As the rise in PRL protein expression in HeLa cells is detected much earlier in magnesium-depleted media, we strongly believe that the main regulation occurs at the mRNA translation level to provide a rapid adaptive response by the cell and that transcription changes might occur as a secondary event. Except for HeLa cells, we did not see major changes in PRL mRNA levels under low-magnesium conditions at late time points in all cell lines tested, implying that posttranscriptional mechanisms are important for the regulation of this PTP family. In line with this, the existence of a posttranscriptional mechanism involving the binding of poly(C)-binding protein 1 (PCBP1) to the 5′UTR of PRL-3 mRNA was shown to down-regulate its translation (55). Moreover, an inverse correlation between the protein levels of PRL-3 and PCBP1 was shown in different human primary cancers, providing a clinical significance to this mechanism. Therefore, the rapid translational response to hypomagnesemia preceding the transcriptional response could be a general mode of cellular regulation involving PRL expression in response to various stimuli. Fast translational induction has been described in response to various stresses, including acute metabolic stress induced by oxygen and glucose deprivation, which provokes rapid widespread uORF-mediated alterations in mRNA translation with minimal transcription effects (56). As we saw under low-magnesium conditions, AMPK is also activated by hypoxia, low glucose, and impaired mitochondrial function (40), thus contributing to selective translational reprograming via the AMPK/mTOR pathway, leading to the survival of cells exposed to these adverse environmental conditions. Similar to magnesium deprivation, these conditions mimic the stringent tumor microenvironment which imposes a dramatic stress on cellular bioenergetics. Thus, we believe that an acute change in PRL expression contributes to the metabolic rewiring of cells to rapidly meet the bioenergetic requirements for cell survival.
Overall, our work establishes a mode of posttranscriptional regulation of gene expression involving a magnesium-sensing mechanism controlled by a conserved cis-regulatory element present in the 5′ UTR of PRL-1 and PRL-2 mRNAs. The findings presented here provide insight on how PRL up-regulation regulates magnesium homeostasis, promotes cancer progression, and offers a metabolic advantage to cells in harsh growth environments.
Materials and Methods
Cell Lines and Cell Culture.
HeLa, MCF-7, HEK293, BT474, and MDA-MB-231 cells were initially obtained from the American Type Culture Collection, and the DB-7 cell line has been described previously (57). Primary murine embryonic fibroblasts were isolated from E14.5 embryos according to the standard protocols. All cell lines were cultured in DMEM (Thermo Fisher Scientific) supplemented with 10% FBS (Thermo Fisher Scientific). Where indicated, magnesium-free DMEM (Wisent) and dialyzed FBS (Thermo Fisher Scientific) were used to vary the concentrations of magnesium by the addition of MgSO4, which is about 1 mM in regular DMEM. Where stated, the following concentrations of inhibitors were used: actinomycin D (1 μg/mL), compound C (10 μM), rapamycin (50 nM), PP242 (2.5 μM), CGP57380 (10 μM), Torin1 (100 nM), and MG132 (5 μM). Cells were tested for mycoplasma contamination by PCR. All cell lines were maintained at 37 °C in a 5% CO2 incubator.
Western Blot Analysis.
Analysis was performed as previously described (58). Antibodies used in this study are from Sigma (alpha-actin), EMD Millipore (PRL-2), and Cell Signaling Technology (AMPK, pT172-AMPK, ACC, pS79-ACC, pS240/244-S6, pS51-eIF2α, eIF2α, AKT, pS473-AKT, and Rictor). All Western blots performed in this study are representative of at least three independent experiments.
RNA Isolation and Real-Time PCR.
RNA isolation, cDNA synthesis, and qPCR were performed as described using the primers described in SI Appendix, Fig. S17. NormFinder analysis (59) determined that actin beta isoform (ACTB) and protein-phosphatase PP1-beta (PP1B) were the most stable genes upon magnesium modulation and were thus used as reference genes to calculate the relative abundance of the indicated genes of interest.
Intracellular Magnesium Measurement.
MDA-MB-231 cells were seeded in 100-mm plates 1 d before treatment. Cells were washed with PBS and scraped into ICP-grade polypropylene tubes (SCP Science) and digested sequentially with 500 μL of both trace element-grade HNO3 (Macron Fine Chemicals) and 30% H2O2 (BioShop) for 1 h each at 100 °C. Samples were then diluted to 5% HNO3 and analyzed with inductively coupled plasma optical emission spectroscopy (iCAP 6000; Thermo Fisher Scientific). Magnesium concentration was normalized with total protein content measured from each sample before digestion.
Polysome Profiling.
Polysome profiles were generated as previously described (60). The primers used for semiquantitative PCR are as follows: PRL-2: ACGCGCAGTGTCCATCAGTAT and GGAGGGCTAATGTTTCATGTTGC; beta-actin: CTCTTCCAGCCTTCCTTCCT and AGCACTGTGTTGGCGTACAG. qPCR was performed on the pooled polysomal fractions using the PRL-2 and actin primers described in SI Appendix, Fig. S17.
Metabolic Labeling of Newly Synthesized Proteins.
Newly synthesized proteins were determined as previously described (61) with minor modifications. See SI Appendix, Materials and Methods for detailed labeling protocols.
Plasmid Constructs, Transfection, and Luciferase Assay.
Generation of all DNA constructs for the luciferase assays is described in SI Appendix, Materials and Methods. Also, see SI Appendix, Materials and Methods for detailed information on transfection and luciferase assays.
Analysis of Publicly Available Data.
For the analysis of publicly available data we used GWIPS-viz browser (36, 37). For the analysis of nucleotide conservation, we used 100-way vertebrate alignment that was visually explored with phyloP (62) and CodAlignView (https://data.broadinstitute.org/compbio1/CodAlignViewUsersGuide.html).
CRISPR-Cas9 Genome-Editing Technology and Lentivirus Infection.
sgRNAs were designed using the online tool (zlab.bio/guide-design-resources) and are listed in SI Appendix, Fig. S17. sgRNAs were inserted into the BsmBI sites of the following lentiviral constructs: pL-CRISPR.SFFV.GFP vector (Addgene plasmid no. 57827) for TRPM7 and PRL-2; lentiCRISPR v2 vector (Addgene plasmid no. 52961) for PRL-2, AMPKα, and the uORF of the PRL-1 gene; and LentiCRISPRv2GFP (Addgene plasmid no. 82416) for the uORF of the PRL-2 gene. LacZ sgRNA control was cloned in all these lentiviral vectors. Lentivirus production and infection were described previously (58). Cells were used as a pool of targeted cells for the assays described in this study and targeting efficiency was determined by Western blot analysis.
Generation of shRNA-Inducible Cell Lines.
A stable reverse tetracycline-controlled transactivator (rtTA) MDA-MB-231 cell line was generated using CRISPR-Cas9 technology by targeting rtTA to the AAVS1 safe harbor human locus. sgRNA targeting the AAVS1 locus was cloned into pSpCas9(BB)-2A-GFP (PX458) (Addgene plasmid no. 48138). A DNA fragment consisting of the CAG promoter located in front of an rtTA-T2A-Fluc transgene was generated by overlapping PCR and cloned into the AgeI/EcoRI sites of pZDonor-AAVS1 Puromycin targeting vector (Sigma). This targeting vector was then cotransfected along with the CRISPR-Cas9 AAVS1 sgRNA construct in MDA-MB-231 using Fugene HD (Roche) and clones were selected for puromycin resistance (5 μg/mL). Efficient targeting of a selected clone was validated by PCR on the genomic DNA as previously described (63). The newly generated rtTA MDA-MB-231 cells were then infected with two individual shRNAs targeting PRL-2 and a scramble control (58), which were cloned into the lentiviral vector pLentiCMVTRE3GNeo_Dest (Addgene plasmid no. 27566) using the gateway system (Invitrogen). Cells were selected with G418 (800 μg/mL) and used as a pool for the various assays described in this study. shRNA expression was induced using 1 μg/mL of dox (Sigma).
ATP and Metabolite Measurement.
ATP levels were measured using the CellTiter-Glo Luminescent Cell Viability Assay (Promega), which directly measures cellular ATP. Glucose and glutamine consumption and lactate production were measured using a Bioprofile Analyzer (NOVA Biomedical). Data were normalized to cell number using Cyquant (Thermo Fisher Scientific).
Supplementary Material
Acknowledgments
We thank Drs. Carsten Schmitz and Anne-Laure Perraud (Integrated Department of Immunology, National Jewish Health and University of Colorado) for HEK293-inducible TRPM7, N. Uetani for the schematic summary, and Irwin Jungreis and Manolis Kellis (Massachusetts Institute of Technology) for the permission to use CodAlignView. This work was supported by Canadian Institutes of Health Research Grants MOP-142497 and FDN-159923 (to M.L.T.) and FDN-14366 (to J.P.) and Science Foundation Ireland–Health Research Board–Wellcome Trust Biomedical Research Partnership Investigator Award in Science 210692/Z/18/Z (to P.V.B.). M.L.T. is a Jeanne and Jean-Louis Lévesque Chair in Cancer Research. E.K. is a recipient of a doctoral training award from the Fonds de Recherche Santé Quebec. T.H. is a recipient of the Canada graduate scholarship master award from the Canadian Institutes of Health Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815361116/-/DCSupplemental.
References
- 1.Hardy S, et al. Physiological and oncogenic roles of the PRL phosphatases. FEBS J. 2018;285:3886–3908. doi: 10.1111/febs.14503. [DOI] [PubMed] [Google Scholar]
- 2.Hardy S, et al. The protein tyrosine phosphatase PRL-2 interacts with the magnesium transporter CNNM3 to promote oncogenesis. Oncogene. 2015;34:986–995. doi: 10.1038/onc.2014.33. [DOI] [PubMed] [Google Scholar]
- 3.Funato Y, et al. Membrane protein CNNM4-dependent Mg2+ efflux suppresses tumor progression. J Clin Invest. 2014;124:5398–5410. doi: 10.1172/JCI76614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giménez-Mascarell P, et al. Structural basis of the oncogenic interaction of phosphatase PRL-1 with the magnesium transporter CNNM2. J Biol Chem. 2017;292:786–801. doi: 10.1074/jbc.M116.759944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gulerez I, et al. Phosphocysteine in the PRL-CNNM pathway mediates magnesium homeostasis. EMBO Rep. 2016;17:1890–1900. doi: 10.15252/embr.201643393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H, et al. PRL3 phosphatase active site is required for binding the putative magnesium transporter CNNM3. Sci Rep. 2017;7:48. doi: 10.1038/s41598-017-00147-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kostantin E, et al. Inhibition of PRL-2·CNNM3 protein complex formation decreases breast cancer proliferation and tumor growth. J Biol Chem. 2016;291:10716–10725. doi: 10.1074/jbc.M115.705863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arjona FJ, de Baaij JHF. CrossTalk opposing view: CNNM proteins are not Na+/Mg2+ exchangers but Mg2+ transport regulators playing a central role in transepithelial Mg2+ (re)absorption. J Physiol. 2018;596:747–750. doi: 10.1113/JP275249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Funato Y, Furutani K, Kurachi Y, Miki H. CrossTalk proposal: CNNM proteins are Na+/Mg2+ exchangers playing a central role in transepithelial Mg2+ (re)absorption. J Physiol. 2018;596:743–746. doi: 10.1113/JP275248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Baaij JH, Hoenderop JG, Bindels RJ. Magnesium in man: Implications for health and disease. Physiol Rev. 2015;95:1–46. doi: 10.1152/physrev.00012.2014. [DOI] [PubMed] [Google Scholar]
- 11.Wolf FI, Trapani V. Magnesium and its transporters in cancer: A novel paradigm in tumour development. Clin Sci (Lond) 2012;123:417–427. doi: 10.1042/CS20120086. [DOI] [PubMed] [Google Scholar]
- 12.Romani AMP. Intracellular magnesium homeostasis. In: Vink R, Nechifor M, editors. Magnesium in the Central Nervous System. Univ of Adelaide Press; Adelaide, SA, Australia: 2011. [PubMed] [Google Scholar]
- 13.Garfinkel L, Garfinkel D. Magnesium regulation of the glycolytic pathway and the enzymes involved. Magnesium. 1985;4:60–72. [PubMed] [Google Scholar]
- 14.Yamanaka R, et al. Mitochondrial Mg(2+) homeostasis decides cellular energy metabolism and vulnerability to stress. Sci Rep. 2016;6:30027. doi: 10.1038/srep30027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cadenas S, Brand MD. Effects of magnesium and nucleotides on the proton conductance of rat skeletal-muscle mitochondria. Biochem J. 2000;348:209–213. [PMC free article] [PubMed] [Google Scholar]
- 16.Uetani N, et al. PRL2 links magnesium flux and sex-dependent circadian metabolic rhythms. JCI Insight. 2017;2:91722. doi: 10.1172/jci.insight.91722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Baaij JH, et al. Elucidation of the distal convoluted tubule transcriptome identifies new candidate genes involved in renal Mg(2+) handling. Am J Physiol Renal Physiol. 2013;305:F1563–F1573. doi: 10.1152/ajprenal.00322.2013. [DOI] [PubMed] [Google Scholar]
- 18.Dai LJ, Quamme GA. Intracellular Mg2+ and magnesium depletion in isolated renal thick ascending limb cells. J Clin Invest. 1991;88:1255–1264. doi: 10.1172/JCI115429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai LJ, Raymond L, Friedman PA, Quamme GA. Mechanisms of amiloride stimulation of Mg2+ uptake in immortalized mouse distal convoluted tubule cells. Am J Physiol. 1997;272:F249–F256. doi: 10.1152/ajprenal.1997.272.2.F249. [DOI] [PubMed] [Google Scholar]
- 20.Goytain A, Quamme GA. Identification and characterization of a novel mammalian Mg2+ transporter with channel-like properties. BMC Genomics. 2005;6:48. doi: 10.1186/1471-2164-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quamme GA. Molecular identification of ancient and modern mammalian magnesium transporters. Am J Physiol Cell Physiol. 2010;298:C407–C429. doi: 10.1152/ajpcell.00124.2009. [DOI] [PubMed] [Google Scholar]
- 22.Groisman EA, et al. Bacterial Mg2+ homeostasis, transport, and virulence. Annu Rev Genet. 2013;47:625–646. doi: 10.1146/annurev-genet-051313-051025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowen-Pope DF, Vidair C, Sanui H, Rubin AH. Separate roles for calcium and magnesium in their synergistic effect on uridine uptake by cultured cells: Significance for growth control. Proc Natl Acad Sci USA. 1979;76:1308–1312. doi: 10.1073/pnas.76.3.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Covacci V, et al. Magnesium restriction induces granulocytic differentiation and expression of p27Kip1 in human leukemic HL-60 cells. J Cell Biochem. 1998;70:313–322. doi: 10.1002/(sici)1097-4644(19980901)70:3<313::aid-jcb4>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 25.Bai Y, et al. Role of phosphatase of regenerating liver 1 (PRL1) in spermatogenesis. Sci Rep. 2016;6:34211. doi: 10.1038/srep34211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chubanov V, Mittermeier L, Gudermann T. Role of kinase-coupled TRP channels in mineral homeostasis. Pharmacol Ther. 2018;184:159–176. doi: 10.1016/j.pharmthera.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Valinsky WC, Jolly A, Miquel P, Touyz RM, Shrier A. Aldosterone upregulates transient receptor potential melastatin 7 (TRPM7) J Biol Chem. 2016;291:20163–20172. doi: 10.1074/jbc.M116.735175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmitz C, et al. Regulation of vertebrate cellular Mg2+ homeostasis by TRPM7. Cell. 2003;114:191–200. doi: 10.1016/s0092-8674(03)00556-7. [DOI] [PubMed] [Google Scholar]
- 29.Jiang HY, et al. Phosphorylation of the alpha subunit of eukaryotic initiation factor 2 is required for activation of NF-kappaB in response to diverse cellular stresses. Mol Cell Biol. 2003;23:5651–5663. doi: 10.1128/MCB.23.16.5651-5663.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida A, Funato Y, Miki H. Phosphatase of regenerating liver maintains cellular magnesium homeostasis. Biochem J. 2018;475:1129–1139. doi: 10.1042/BCJ20170756. [DOI] [PubMed] [Google Scholar]
- 31.Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science. 2016;352:1413–1416. doi: 10.1126/science.aad9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilbert WV. Alternative ways to think about cellular internal ribosome entry. J Biol Chem. 2010;285:29033–29038. doi: 10.1074/jbc.R110.150532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shatsky IN, Terenin IM, Smirnova VV, Andreev DE. Cap-independent translation: What’s in a name? Trends Biochem Sci. 2018;43:882–895. doi: 10.1016/j.tibs.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 34.Meyer KD, et al. 5′ UTR m(6)A promotes cap-independent translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terenin IM, Smirnova VV, Andreev DE, Dmitriev SE, Shatsky IN. A researcher’s guide to the galaxy of IRESs. Cell Mol Life Sci. 2017;74:1431–1455. doi: 10.1007/s00018-016-2409-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michel AM, et al. GWIPS-viz: Development of a ribo-seq genome browser. Nucleic Acids Res. 2014;42:D859–D864. doi: 10.1093/nar/gkt1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michel AM, Kiniry SJ, O’Connor PBF, Mullan JP, Baranov PV. GWIPS-viz: 2018 update. Nucleic Acids Res. 2018;46:D823–D830. doi: 10.1093/nar/gkx790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andreev DE, et al. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. eLife. 2015;4:e03971. doi: 10.7554/eLife.03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gout E, Rébeillé F, Douce R, Bligny R. Interplay of Mg2+, ADP, and ATP in the cytosol and mitochondria: Unravelling the role of Mg2+ in cell respiration. Proc Natl Acad Sci USA. 2014;111:E4560–E4567. doi: 10.1073/pnas.1406251111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herzig S, Shaw RJ. AMPK: Guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol. 2018;19:121–135. doi: 10.1038/nrm.2017.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thoreen CC, et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169:361–371. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 43.Hagiwara A, et al. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab. 2012;15:725–738. doi: 10.1016/j.cmet.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 44.Gungabeesoon J, Tremblay ML, Uetani N. Localizing PRL-2 expression and determining the effects of dietary Mg(2+) on expression levels. Histochem Cell Biol. 2016;146:99–111. doi: 10.1007/s00418-016-1427-6. [DOI] [PubMed] [Google Scholar]
- 45.Sahni J, Scharenberg AM. TRPM7 ion channels are required for sustained phosphoinositide 3-kinase signaling in lymphocytes. Cell Metab. 2008;8:84–93. doi: 10.1016/j.cmet.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao M, et al. AMPK-mediated up-regulation of mTORC2 and MCL-1 compromises the anti-cancer effects of aspirin. Oncotarget. 2016;7:16349–16361. doi: 10.18632/oncotarget.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y, et al. AMPK blunts chronic heart failure by inhibiting autophagy. Biosci Rep. 2018;38:BSR20170982. doi: 10.1042/BSR20170982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nikonorova IA, Kornakov NV, Dmitriev SE, Vassilenko KS, Ryazanov AG. Identification of a Mg2+-sensitive ORF in the 5′-leader of TRPM7 magnesium channel mRNA. Nucleic Acids Res. 2014;42:12779–12788. doi: 10.1093/nar/gku951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ivanov IP, et al. Polyamine control of translation elongation regulates start site selection on antizyme inhibitor mRNA via ribosome queuing. Mol Cell. 2018;70:254–264. doi: 10.1016/j.molcel.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jorgensen RA, Dorantes-Acosta AE. Conserved peptide upstream open reading frames are associated with regulatory genes in angiosperms. Front Plant Sci. 2012;3:191. doi: 10.3389/fpls.2012.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Z, Sachs MS. Ribosome stalling is responsible for arginine-specific translational attenuation in Neurospora crassa. Mol Cell Biol. 1997;17:4904–4913. doi: 10.1128/mcb.17.9.4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Werner M, Feller A, Messenguy F, Piérard A. The leader peptide of yeast gene CPA1 is essential for the translational repression of its expression. Cell. 1987;49:805–813. doi: 10.1016/0092-8674(87)90618-0. [DOI] [PubMed] [Google Scholar]
- 53.Ma J. 2016. Discovery and characterization of novel smORF-encoded polypeptides (SEPs). PhD dissertation (Harvard Univ, Cambridge, MA)
- 54.Brown A, et al. Structures of the human mitochondrial ribosome in native states of assembly. Nat Struct Mol Biol. 2017;24:866–869. doi: 10.1038/nsmb.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang H, et al. PCBP1 suppresses the translation of metastasis-associated PRL-3 phosphatase. Cancer Cell. 2010;18:52–62. doi: 10.1016/j.ccr.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 56.Andreev DE, et al. Oxygen and glucose deprivation induces widespread alterations in mRNA translation within 20 minutes. Genome Biol. 2015;16:90. doi: 10.1186/s13059-015-0651-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Borowsky AD, et al. Syngeneic mouse mammary carcinoma cell lines: Two closely related cell lines with divergent metastatic behavior. Clin Exp Metastasis. 2005;22:47–59. doi: 10.1007/s10585-005-2908-5. [DOI] [PubMed] [Google Scholar]
- 58.Hardy S, Wong NN, Muller WJ, Park M, Tremblay ML. Overexpression of the protein tyrosine phosphatase PRL-2 correlates with breast tumor formation and progression. Cancer Res. 2010;70:8959–8967. doi: 10.1158/0008-5472.CAN-10-2041. [DOI] [PubMed] [Google Scholar]
- 59.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 60.Akimitsu N, Tanaka J, Pelletier J. Translation of nonSTOP mRNA is repressed post-initiation in mammalian cells. EMBO J. 2007;26:2327–2338. doi: 10.1038/sj.emboj.7601679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kar AN, MacGibeny MA, Gervasi NM, Gioio AE, Kaplan BB. Intra-axonal synthesis of eukaryotic translation initiation factors regulates local protein synthesis and axon growth in rat sympathetic neurons. J Neurosci. 2013;33:7165–7174. doi: 10.1523/JNEUROSCI.2040-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosenbloom KR, et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015;43:D670–D681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.