Significance
GNIP1Aa is a protein from Chromobacterium piscinae that demonstrates specific toxicity toward Western corn rootworm, one of the most devastating corn pests in the United States. Our studies provide insight into the GNIP1Aa structure and place this protein into an insecticidal protein class, membrane attack complex/PerForin–β-tripod, different from all insect-control products of modern agricultural technology available on the market. Protein activity and uniqueness make GNIP1Aa an excellent commercial candidate for development into a transgenic product. Such a product might have a high potential to combat crop damage in corn and to delay development of resistance in insects. Our work also contributes to the general understanding of the mechanism of action of pore-forming proteins and their target specificity.
Keywords: MACPF, crystal structure, protein family, β-tripod domain, insecticidal protein
Abstract
The crystal structure of the Gram-negative insecticidal protein, GNIP1Aa, has been solved at 2.5-Å resolution. The protein consists of two structurally distinct domains, a MACPF (membrane attack complex/PerForin) and a previously uncharacterized type of domain. GNIP1Aa is unique in being a prokaryotic MACPF member to have both its structure and function identified. It was isolated from a Chromobacterium piscinae strain and is specifically toxic to Diabrotica virgifera virgifera larvae upon feeding. In members of the MACPF family, the MACPF domain has been shown to be important for protein oligomerization and formation of transmembrane pores, while accompanying domains define the specificity of the target of the toxicity. In GNIP1Aa the accompanying C-terminal domain has a unique fold composed of three pseudosymmetric subdomains with shared sequence similarity, a feature not obvious from the initial sequence examination. Our analysis places this domain into a protein family, named here β-tripod. Using mutagenesis, we identified functionally important regions in the β-tripod domain, which may be involved in target recognition.
Before the introduction of transgenic crops, corn rootworms (Diabrotica spp.) cost farmers about a billion dollars per year in corn crop damage and treatment costs (1). The Gram-positive soil bacterium Bacillus thuringiensis and its proteins are widely used in agriculture to protect plants from insect damage. More than 100 insecticidal proteins from B. thuringiensis are known, including Cry and Vip proteins (2, 3). One property of these proteins is their high specificity toward particular pests, while having no negative impact on vertebrates, the environment, and other arthropods, including beneficial insects (4, 5). Planting genetically modified crops, carrying genes for B. thuringiensis-originating insecticidal proteins, significantly reduces pest damage and increases crop yields, while reducing the use of chemical pesticides (6–8). However, continuous employment of these insecticidal products has already resulted in the evolution of significant levels of resistance in several targeted pest species (9–12). Transgenic crops expressing Cry3Bb1, a modified Cry3A, and Cry34/35Ab1, which protect corn from the Western corn rootworm (WCR or Diabrotica virgifera virgifera), exemplify commercially available products for which resistance management is needed (11, 12). To avoid or manage the development of resistance, pest-specific toxins with novel modes of action are needed.
In addition to having a different mode of action, a candidate protein for crop protection must be highly active and must be very specific with no toxicity toward nontarget organisms. The Gram-negative insecticidal protein (GNIP1Aa) described in this report fits these requirements (13). GNIP1Aa differs from the B. thuringiensis proteins in current commercial use in that it was isolated from the Gram-negative Chromobacterium piscinae species, a purple bacterium. Most of the Chromobacterium species were isolated from environmental samples, such as water, soil, and rhizosphere (14). For isolated from water Chromobacterium violaceum, a direct correlation with suspended solids was observed (15). Association with soil may suggest why some Chromobacterium species may encode toxins that kill soil-dwelling insects. Indeed, antifungal and insecticidal activity was reported for a few Chromobacterium species, including Chromobacterium Csp_P (16), Chromobacterium subtsugae (17), Chromobacterium vaccinii (18), and Chromobacterium JH7 (19). Among eight insect species against which GNIP1Aa was tested in plate-based bio-assays and in planta studies—including lepidopteran, coleopteran, and hemipteran species—it was found to be toxic exclusively to WCR. The related species Diabrotica undecimpunctata was unaffected (13).
A sequence homology search revealed that GNIP1Aa is a member of the membrane attack complex/PerForin (MACPF) superfamily. MACPF proteins are found in all kingdoms of life and have important roles in processes related to immunity, pathogenesis, and development (20). While eukaryotic MACPFs are the most abundant and the best studied representatives of the family, only a few have been functionally characterized. Some of them, such as the complement C6, C7, C8α, C8β, and C9 proteins, and perforins of mammals, are important factors in the immune system, protecting a host from infection by forming pores in target membranes of pathogens and infected cells (21–24). The reported size of MACPF pores is around 5–16 nm (22, 25), which is significantly larger than ∼1-nm pores for most of Cry proteins (26, 27) that form weakly selective cation channels in microvillar membranes. Other MACPF proteins have been shown to be involved in host development, and no pore formation was reported for them (28–30). In the prokaryotic world, GNIP1Aa is the only representative of the family for which a function has been identified, namely insecticidal activity (13). The structures for two other bacterial MACPFs have been reported and hypothetical functions were proposed. Plu-MACPF from the insecticidal bacteria Photorhabdus luminescens (PDB ID code 2QP2) (31) was shown to bind to the surface of insect cells of Trichoplusia ni, but no lytic activity was detected using various insect and mammalian cell lines. The gene for another protein, Bth-MACPF (PDB ID code 3KK7) (32), was isolated from the human gut symbiont Bacteroides thetaiotaomicron (Bth) for structural characterization. While the authors suggested an important role for this protein in the symbiotic relationship between the host and bacteria, its actual function was not identified.
Our studies provide insight into the structure of GNIP1Aa and the molecular mechanism of its insecticidal activity. The structure of the protein was determined, and it shows structural homology to perforin and other MACPF pore-forming proteins, suggesting that GNIP1Aa exerts its activity by perforating gut membranes of WCR. As in the case of perforin, the MACPF domain is accompanied by another domain, which we predict to be responsible for the specific recognition of a membrane-bound receptor. This domain has a structural fold, which constitutes a protein family we call β-tripod. Structural analysis and systematic mutagenesis of this domain was used to identify residues that are potentially important to its function.
Results and Discussion
Crystal Structure of GNIP1Aa.
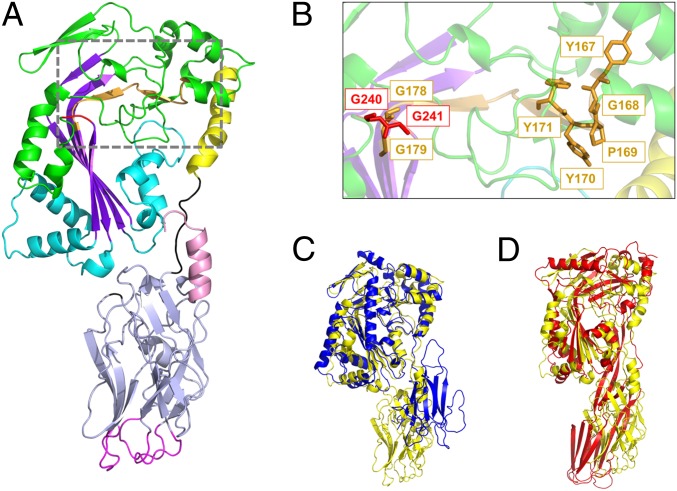
The crystal structure of GNIP1Aa was solved by single isomorphous replacement with anomalous scattering at 2.5-Å resolution and revealed an elongated shape of the protein with approximate dimensions of 100 × 40 × 30 Å (Fig. 1A). The overall structure of the 536-residue protein can be divided into two domains: an N-terminal MACPF domain (residues 9–308) and a C-terminal domain (residues 317–529) that contains mostly β-strands. The domains are connected by a short linker (residues 309–316). Several interactions at the interface suggest a tight association of the domains and limited flexibility between them. In addition, the C-terminal helix at the end of the second domain is located in a bridging position at the domain interface, adding to the overall rigidity of the structure.
Fig. 1.
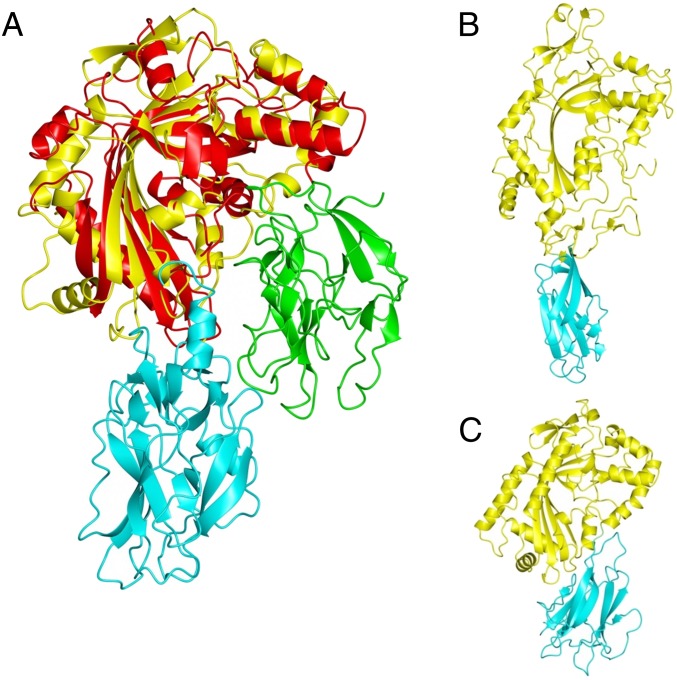
Crystal structure of GNIP1Aa in ribbon representation. The figure was made with PyMOL (33). (A) The structure of GNIP1Aa is composed of two domains. The N-terminal MACPF domain is colored green with key structural features shown in different colors: central four-stranded β-sheet in purple; two clusters of helices, CH1 and CH2, in cyan; and MACPF-conserved α-helix in yellow. Additionally, the signature MACPF motif and two conserved glycines are highlighted in orange and red, 167YGPYYX6GG179 and 240GG241, respectively. The linker between the two domains is shown in black. The C-terminal domain is colored light blue with an exception of three apical loops in magenta and the C-terminal tail in pink. (B) Close-up of the conserved MACPF residues, 167YGPYYX6GG179 and 240GG241, shown in stick presentation in orange and red, respectively. (C) Superposition of GNIP1Aa structure in yellow with bacterial MACPF protein from P. luminescens (PDB ID code 2QP2) in blue; RMSD of 2.3 Å for 298 Cα positions. (D) Superposition of GNIP1Aa structure in yellow with a representative of CDCs, perfringolysin O (PDB ID code 1M3I) in red; RMSD of 4.1 Å for 209 Cα positions.
GNIP1Aa N-Terminal Domain, MACPF Protein Family.
The N-terminal domain displays a typical MACPF fold. A characteristic feature of MACPF domains and structurally related cholesterol-dependent cytolysins (CDCs) is a central four-stranded antiparallel β-sheet with approximately a 90° bend in the middle (Fig. 1A) (31).
Similar to other MACPF and CDC proteins, GNIP1Aa carries two clusters of helices (CH1 and CH2) at the base of the β-sheet. These two helical sets undergo significant conformational changes upon protein interaction with a target membrane, turning into two β-hairpins (31, 34–36) and becoming a part of a β-barrel membrane pore. The multistep process of transmembrane pore formation starts with the protein’s initial recognition and binding to a membrane, followed by protein oligomerization, and finally pore formation via vast conformational changes (37).
The evidence for pore formation by MACPFs and CDCs derives from many biochemical, biophysical, and structural studies (22, 25, 36), including the latest cryoelectron microscopy structure determination of MACPF (35, 38) and CDC proteins (39). In the MACPF domain of GNIP1Aa, each bundle of helices spans 41–44 amino acid residues, which is similar to another bacterial MACPF from Photorhabdus luminescens (31) (40–42 residues), and somewhat shorter than in Bth-MACPF (PDB ID code 3KK7) from B. thetaiotaomicron (32) (48–58 residues), although the latter region includes a nonstandard fold with a β-hairpin. The corresponding helical sets in eukaryotic MACPFs are around 55 residues each (22, 24, 40), while structurally similar CDCs carry homologous helical sets of ∼30–34 residues each (41–43). The length of the transmembrane helical regions and the presence or absence of a conserved α-helix (Fig. 1A) have been suggested to be associated with the main difference in pore formation between CDCs and MACPFs (37). CDC-like pores undergo vertical collapse to extend their otherwise short β-barrels toward the lipid bilayer to punch a membrane pore, while MACPF proteins with longer transmembrane regions do not collapse during pore formation (37). Thus, GNIP1Aa likely forms transmembrane pores via an MACPF mechanism with no collapse, similar to the fungal MACPF pleurotolysin with 43–45 residues in the helical region (35, 37).
It is noteworthy that the residues from the helical transmembrane region (CH1 and CH2) (Fig. 1A) exclusively contribute to the contact area between the N-terminal and the C-terminal domains in GNIP1Aa. Thus, any conformational changes in the C-terminal domain, triggered by receptor recognition or any other intermolecular interaction, are directly sensed by the pore-forming region of the GNIP1Aa MACPF domain.
While sequences for various MACPF domains are quite diverse, all of them contain a unique signature motif, Y/W-G-T/S-H-F/Y-X6-GG, and two conserved glycines (31). In GNIP1Aa this motif deviates from the canonical one in positions 3 and 4 corresponding to 169 and 170 in 167YGPYYX6GG179 (Fig. 1B). Deviations in positions 3 and 4, or 3, 4, and 5, are also observed in the MACPF domains in PlyB (35) and Bth-MACPF (32), respectively, so there may be a higher tolerance to amino acid variation at these positions in fungal and bacterial MACPFs. The significance of this motif and its conservation is not fully understood. The two consecutive glycines, G240 and G241 (Fig. 1 A and B), are located immediately downstream of the last β-strand of the central β-sheet, β4, and are conserved in both MACPFs and CDCs. It has been suggested that these glycines function as a hinge during conformational changes involved in pore formation (31).
Superposition of the available MACPF and CDC structures revealed that Plu-MACPF, the bacterial MACPF protein from P. luminescens (PDB ID code 2QP2) (31), is the closest structural homolog of the N-terminal domain of GNIP1Aa, with an RMSD of 2.3 Å for 298 Cα positions (Fig. 1C). A more distant relationship to CDCs is reflected in the significantly higher RMSD numbers to the closest representatives of this protein class, perfringolysin O (PDB ID code 1M3I) (42) and suilysin (PDB ID code 3HVN) (43), with RMSD values of 4.1 Å and 4.7 Å for 209 and 230 Cα atoms, respectively (Fig. 1D).
GNIP1Aa C-Terminal Domain, a β-Tripod Protein Family.
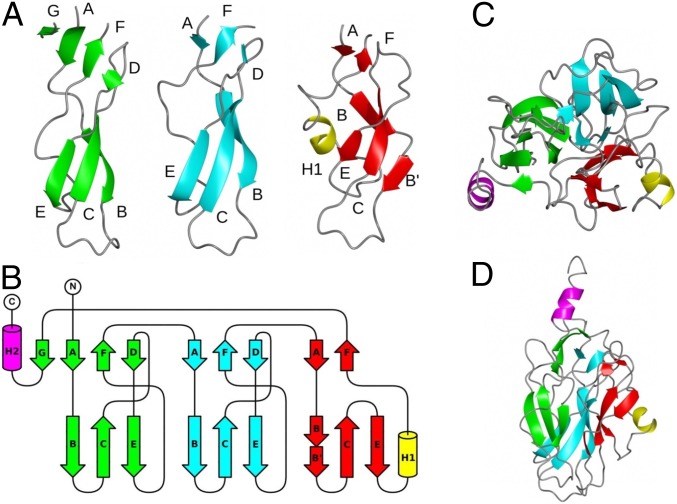
The C-terminal 220-residue domain of GNIP1Aa has dimensions of ∼50 × 30 × 30 Å and contains mostly short β-strands. The domain can be divided into three subdomains (Fig. 2A) with similar topologies (Fig. 2B). Each subdomain core contains ∼65 residues and forms two antiparallel β-sheets. In subdomains 1 and 2, the first β-sheet is located on the side that contacts the MACPF domain and contains strands A, F, and D (top part of each subdomain in Fig. 2 A and B). The second β-sheet is located in the distal half of the domain and contains strands B, C, and E (bottom part of each subdomain in Fig. 2 A and B). Subdomain 3 shows a few deviations from this regular pattern: its β-strand B is broken in two parts, the β-strand D is missing, and an extra short helix H1 is inserted downstream of strand E. The three subdomains are arranged around a threefold pseudosymmetry axis that goes through the center of the C-terminal domain (Fig. 2C). Additional structural elements of the domain are a small β-strand after subdomain 3, which becomes a part of the first β-sheet of subdomain 1, and the α-helix H2 at the extreme C terminus that bridges the two GNIP1Aa domains.
Fig. 2.
Structure and topology of the C-terminal domain of GNIP1Aa. (A) Ribbon diagrams for subdomains 1 (green), 2 (cyan), and 3 (red). The apical loops are located between strands B and C. The additional helix H1 in subdomain 3 is shown in yellow. The C-terminal α-helix H2 is omitted for clarity. (B) Topology diagram for the C-terminal domain. Structural elements are colored according to A. The C-terminal helix H2 is colored in magenta. (C) Arrangement of subdomains in the C-terminal domain. A view along the threefold pseudosymmetry axis, from the MACPF domain’s side. (D) View perpendicular to C.
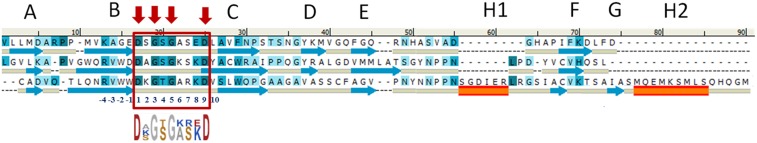
Despite the striking structural similarity of the individual subdomains in GNIP1Aa, their sequence resemblance is not obvious. Only when subdomain sequences are aligned based on their 3D structure (Fig. 3) does the sequence similarity become more noticeable. The highest sequence identity is observed between three symmetrical loops connecting strands B and C (Fig. 2 A and B) at the very distal tip of the C-terminal GNIP1Aa domain (Fig. 1A, at the bottom). Each apical loop has the same conformation and is composed of nine residues with sequence consensus “D-X-G-S/T-G-X3-D,” flanked by two aspartates #1 and #9, and containing two glycine residues #3 and #5 in each loop (Fig. 3). The same rigid loop conformation is adopted as a result of interaction between the carboxyl group of aspartate #1, the hydroxyl group of serine or threonine #4, and the main-chain amino group of a residue at position #10. The glycines at positions #3 and #5 all show combinations of main-chain angles that are located in a region of the Ramachandran plot that is allowed only for glycine. Any other amino acid at these positions would therefore destabilize the observed loop conformation. While the nature of all other conserved amino acids in the deduced motif seems consistent with a role in loop stabilization, the reason for conservation of aspartates at position #9 is less obvious. The side chains of all three aspartates point toward the solvent, and only one of them, D403 in subdomain 2, forms a single intramolecular hydrogen bond. The conservation, the solvent-exposed nature, and the location at the distal tip of the domain, suggest that the aspartates at position #9 may play a key role in target molecule recognition and will be discussed further below. The overall threefold pseudosymmetry, the rigid structure of the loops, and the nature of the conserved sequence motifs may play an essential role in receptor binding and in the subsequent steps of the pore formation.
Fig. 3.
Structure-based sequence alignment of three subdomains of the C-terminal domain. The darker shades represent higher, and lighter shades lower levels of sequence similarity. The secondary structure elements are shown underneath the sequence, where blue arrows correspond to β-stands, and orange rectangles to α-helices. Amino acids corresponding to the D-X-G-S/T-G-X3-D motif are boxed in red, with the sequence logo below. Identical residues are marked by the red arrows. Each motif is composed of nine residues numbered 1–9; the four N-terminally preceding positions are numbered from −4 to −1, accordingly.
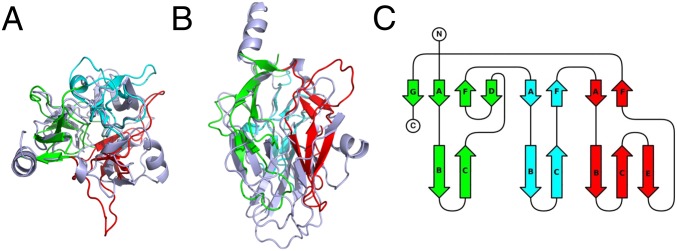
The observed topology of the C-terminal domain of GNIP1Aa has not previously been described in any protein. A sequence-based search for structurally related proteins in the Protein Data Bank (44) failed to produce any hits. However, a structure-based search using the DALI server (45) revealed that the C-terminal domain of PlyB is distantly related to the C-terminal domain of GNIP1Aa. PlyB (PDB ID code 4OEJ) (35) is part of the bicomponent toxin pleurotolysin from the edible fungus Pleurotus ostreatus. GNIP1Aa and PlyB have the same two-domain architecture, with an N-terminal pore-forming MACPF-like domain and an accompanying C-terminal domain. However, PlyB requires an additional protein component, a lipid-binding aegerolysin protein (PlyA or OlyA6), to perforate the membrane bilayer (25, 35).
While the C-terminal domains of GNIP1Aa and PlyB can be superimposed with an RMSD of 2.18 Å for 127 Cα atoms (of 213 and 173 available backbone Cα atoms, respectively), the resulting structure-based alignment shows only 11.8% identical amino acids at the equivalent positions. When structural alignment is visually presented (Fig. 4 A and B), their similarity is not obvious either, yet the topology diagram of the PlyB domain (Fig. 4C) clearly shows the same arrangement of structural elements, with a few β-strands and α-helices missing in the shorter 177-residue PlyB domain (compare with GNIP1Aa in Fig. 2B).
Fig. 4.
The C-terminal domain of PlyB, (A) Ribbon diagram for superimposed C-terminal domains of GNIP1Aa (light blue) and PlyB. Three subdomains of the PlyB β-tripod domain are shown in three different colors: 1 (green), 2 (cyan), and 3 (red). A view along the threefold pseudosymmetry axis. See Fig. 2C for comparison. (B) The same structural alignment with a view perpendicular to A. See Fig. 2D for comparison. (C) Topology diagram for the C-terminal domain of PlyB. The strands are colored and labeled according to their GNIP1Aa counterparts in Fig. 2B.
Because we were unable to identify any other structure with a similar topology, we propose that the C-terminal domains of both GNIP1Aa and PlyB have a fold different from all previously described β-proteins. The overall domain conformation with three structural subdomains or “legs” is reminiscent of a tripod, so we suggest calling this a β-tripod domain. A prototypical β-tripod domain has an internal pseudo threefold symmetry, with each of the three subdomains forming a leg of the tripod. Each leg contains two antiparallel three-stranded β-sheets, one in the upper part of the leg near an associated domain (in the case of GNIP1Aa, near the MACPF domain) and another in the lower part. The β-strands are parallel to the threefold axis.
Comparison of GNIP1Aa Architecture with Other Proteins.
The MACPF domains of GNIP1Aa and PlyB are combined with C-terminal domains that share a similar fold, but the relative orientation of the domains is fundamentally different in the two proteins (Fig. 5A). In GNIP1Aa the C-terminal domain is in contact with both CH1 and CH2 of the MACPF domain and is located in the same plane as the lower end of the central β-sheet. In PlyB, which lacks the linker between the domains, the C-terminal domain is attached only to CH1 above the plane of the central β-sheet, opposite to CH2. Because in pleurotolysin an additional component, PlyA (or OlyA6), is required for membrane recognition and association, the C-terminal domains in GNIP1Aa and PlyB may have different roles. Indeed, electron microscopy data (35) indicate that during pore formation the lipid binding component PlyA is located in a position relative to the MACPF domain that is similar to the position of the C-terminal domain in GNIP1Aa. Furthermore, the D-X-G-S/T-G-X3-D motif of GNIP1Aa C-terminal subdomains is not found in PlyB, and the residues in the BC loops of GNIP1Aa do not have direct equivalents in PlyB, due to the shorter loops and strands in this protein. This may explain that while the fold is conserved between the C-terminal domains of GNIP1Aa and PlyB, the target specificity is not.
Fig. 5.
Relative orientation of domains in MACPF proteins. (A) Superposition of MACPF domains from GNIP1Aa (yellow) and PlyB (red) results in different positioning of the C-terminal domains in GNIP1Aa (cyan) and PlyB (green). (B) Domain orientation in perforin (yellow) with C2 domain in cyan. (C) Domain orientation in Plu-MACPF (yellow) with β-prism domain in cyan.
A 3D arrangement of domains similar to that of GNIP1Aa is also found in other MACPF proteins, but the nature of the C-terminal domain varies. Perforin contains a C-terminal C2 domain in the equivalent position (Fig. 5B) (PDB ID code 3NSJ) (22), and in Plu-MACPF a β-prism domain is found (Fig. 5C) (PDB ID code 2QP2) (31). In both proteins the C-terminal domains are known to be involved in membrane binding. Similarly, a C-terminal Ig domain occupies a similar position in the more distantly related CDCs (Fig. 1D) and is critical for interacting with lipid and protein cofactors (46). Overall, the comparison of GNIP1Aa with other proteins of known structure strongly suggests a role of the C-terminal domain in membrane recognition and association. Thus, the C-terminal domain may determine the target specificity of the MACPF domain.
GNIP1Aa β-Tripod Domain as an Evolutionarily Distinct Unit.
To characterize the β-tripod domain, searches of the various protein domain databases were performed. A sequence search in Pfam (Pfam 31.0, pfam.xfam.org/family/PF06101.10) (47) placed the C-terminal domain of GNIP1Aa in the DUF946 family, which was originally described as a plant protein of unknown function. Our analysis demonstrates that the similarity between the DUF946 hidden Markov model (HMM) and β-tripod domain sequences is limited to a 185-residue region and matches the N-terminal part of the DUF946 model (SI Appendix, Fig. S1). Recently the whole DUF946 HMM sequence of 542 residues was reassigned to the Vps62 (PF06101) family. Vps62 (UniProtKB/Swiss-Prot accession no. P53285.1), composed of 467 residues, is a vacuolar protein-sorting protein required in yeast for targeting proteins from the cytoplasm to the vacuole (48). Not only does this function not make sense in the context of a bacterial protein toxin, but Vps62 protein aligns to the C-terminal part of the DUF946 model (SI Appendix, Fig. S1), rather than the β-tripod domain in the N-terminal region. In other words, the β-tripod domain and Vps62 correspond to nonoverlapping regions of DUF946.
The β-tripod fold is created from three structurally similar subdomains that form the “legs” of the tripod (Fig. 2). Sequence analysis of each of the three subdomains using profile HMMs (HMMER) (49) against Pfam, TIGRFAM, Gene3D, Superfamily, and PIRSF databases for protein families resulted in no significant matches (https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan). Thus, our results and analysis suggest that the β-tripod fold exists as an evolutionarily distinct unit that should be separated from Vps62 in future studies, and does not overlap with any other existing protein domains or families.
To find all sequences that share the β-tripod fold, we collected protein sequences from the National Center for Biotechnology Information’s (NCBI) nonredundant database (NR) by creating a new HMM using the jackhmmer program (50) with a single subdomain as the input sequence. While the single sequence of subdomain 2 (colored cyan in Fig. 2) from the C-terminal domain of GNIP1Aa was used as the initial query, the program iteratively finds similar sequences and includes them in a new model to repeat the search. The final model includes sequences from all three subdomains of the β-tripod fold. This search resulted in 1,743 sequences retrieved from NR (sequence set 1, as described in SI Appendix, Methods). This sequence set is taxonomically distributed as follows: bacteria, 292 sequences; fungi, 72 sequences; other Eumetazoa, 26 sequences; Viridiplantae, 1,085 sequences; other Eukaryota, 140 sequences; viruses, 128 sequences.
To find the different functions that might be associated with proteins containing the β-tripod domain, we ran InterProScan against all of the representative sequences (sequence set 2; see SI Appendix, Methods for details). In total, 73 distinct domains were found in addition to Vps62 in proteins with β-tripod domains. We did not find any clear patterns in the function of associated Pfam domains (SI Appendix, Table S3). In addition to the MACPF domain found in GNIP1Aa, we found the Toxin_10 domain (PF05431) in bacterial sequences. We selected these sequences for additional analysis, as this domain is also known to be responsible for insecticidal activity (51, 52). These sequences are referred to as sequence set 3, as described in SI Appendix, Methods.
Experimental mutation analysis, as described in a later section, found several important residues (Table 1). To examine the conservation of those residues within pesticidal protein-associated sequences (sequence set 3), we looked for exact pattern matches within the bacterial and fungal proteins that also contained a Toxin_10 domain or a MACPF domain. We searched for occurrences of the single subdomain that repeats three times in the GNIP1Aa C terminus and all of the other pesticidal proteins. The presence/absence of the DX-G-S/T-G-X3-D motif is shown in concentric circles surrounding the gene phylogeny in SI Appendix, Fig. S2A. This figure also shows the possible interdomain transfer of the Toxin_10/β-tripod genes based on their phylogenetically interspersed occurrence in fungi, plants, and bacteria. The same protein sequences were used to create a sequence logo for the β-tripod protein family (SI Appendix, Fig. S2B). It is clear that the D-X-G-S/T-G-X3-D motif is well conserved across the family members, but without 100% conservation.
Table 1.
Schematic presentation of the single alanine substitutions’ effect on expression and activity of GNIP1Aa protein variants
| General description | Position of replacedresidue in GNIP1Aa | No. of affectedpositions | Proteinexpression | WCRactivity |
| Positive control | GNIP1Aa | ++ | ++ | |
| Negative control | Plu-MACPF | ++ | − | |
| Low expression | 389, 407, 415, 420, 439 | 5 | ± | ± |
| I. Complete knockouts | 393, 395, 456 | 3 | ++ | − |
| II. Reduced activity | 332, 340, 349, 351, 352, 375, 377, 381, 383, 391, 403, 413, 443, 458, 519, 523, 529 | 17 | ++ | + |
| III. Activity similar to the wild type GNIP1Aa | For the list see SI Appendix, Fig. S3 and Table S2 | 95 | ++ | ++ |
++, Expression of a target protein or WCR toxicity is similar to the wild-type GNIP1Aa, 4.00 stunt/97% mortality; stunt in the range of 3.75–4.00, 83–100% mortality; −, no activity or similar to the negative control, Plu-MACPF, 0.44 stunt/11% mortality; ±, the level of protein expression is clearly below the wild-type GNIP1Aa (SI Appendix, Fig. S3), resulting in no or low toxicity of tested samples (scores below 1.8 stunt/40% mortality); +, reduced activity (stunt in the range of 1.00–3.67, 24–92% mortality) at the level of protein expression similar to the wild-type GNIP1Aa.
Essential Amino Acids of the C-Terminal Domain.
The structural localization and conservation of the D-X-G-S/T-G-X3-D motif suggest functional significance of the composing residues. This notion was first probed by substitution of selected conserved residues with alanine. The functional effect of single- and double-position substitutions in the three D-X-G-S/T-G-X3-D loops and their flanking regions is described in Fig. 6. As expected, this protein region was found to be sensitive to modifications, with no activity detected for three single-point mutants and for four double-point mutants at the equivalent conserved positions in the three loops. The alanine-scanning approach was extended to the rest of the β-tripod domain (Table 1 and SI Appendix, Fig. S3), with more than 100 positions being probed. Still, only three single-point protein variants completely lost toxicity toward WCR: W393A, D395A, and W456A (Fig. 6 B and C, Table 1, and SI Appendix, Fig. S3 and Table S2). All three positions are part of either the conserved motif or its closest neighbors. D395A is the first N-terminal residue of the loop D-X-G-S/T-G-X3-D in subdomain 2 (#1 position) (Fig. 3); W393A and W456 sit two positions upstream of the conserved motifs in subdomains 2 and 3, at the position #−2 (Fig. 3).
Fig. 6.
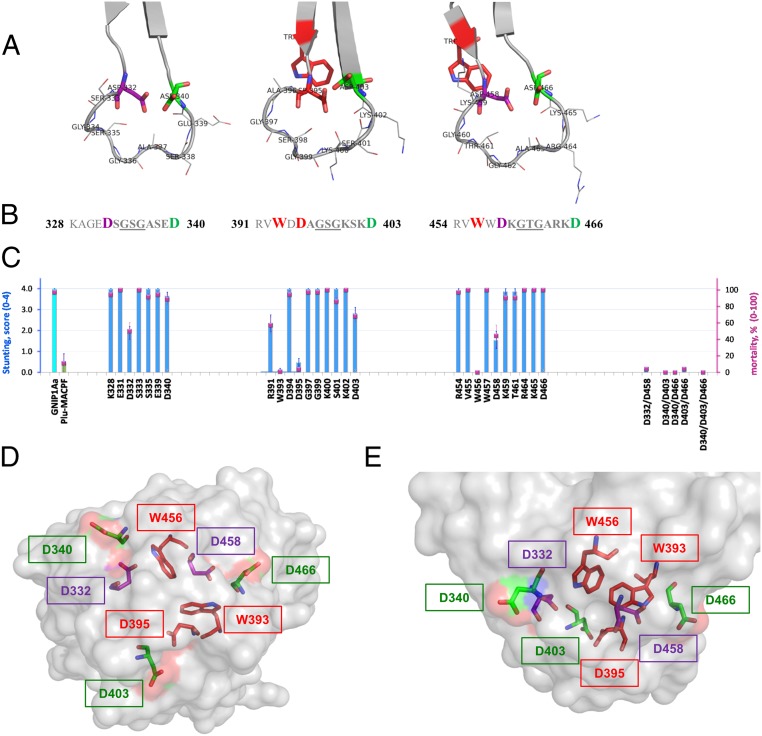
Structure–function analysis of three conserved loops of the C-terminal domain of GNIP1Aa. (A) Ribbon representation of three apical loops. Functionally important and conserved residues (Asp and Trp) are framing each loop, shown in stick representation and labeled with a corresponding residue number. The loop-composing residues are labeled and shown in lines representation. (B) Sequences of the three loop regions with the flanking regions, colored the same as in A. Red bold: residues W393 (#−2 in consensus, Fig. 3), D395 (#1), and W456 (#−2) are unique; replacement of any one completely abolishes protein toxicity. Purple bold: Asp residues in position #1 of the first and third loops, D332 and D458, are essential for GNIP1Aa activity. Green bold: Asp residues in position #9, D340, D403, and D466, for all three loops are fairly important. (C) Activity of the selected single-site Ala mutants (from the left: three groups of data), double and triple mutants (on the right side) in WCR bio-assays, shown in stunt (blue bars) and mortality (magenta squares) values. See more details in SI Appendix, Fig. S3. The GNIP1Aa is the wild-type GNIP1Aa, the positive control; its stunting activity is represented by a cyan-color bar. Plu-MACPF is the negative control with low background WCR activity, a green-colored bar for stunt. The letter and number indicate a one-letter code and position number for a residue within the wild-type GNIP1Aa1 protein, replaced by an alanine. (D) Semitransparent space-filling representation of the C-terminal domain surface with the conserved residues in positions #−2 (Trp), #1 (Asp), and #9 (Asp) colored the same as in A and B and shown in stick representation. A view along the threefold pseudosymmetry axis from the apical tip of the domain, opposite side of the view shown in Fig. 2C. (E) The side view on the apical tip of the C-terminal domain rotated 90° relative to D. The same view as in Fig. 2D.
The tryptophan in position #−2 (Fig. 3) is fully conserved in the β-tripod family (SI Appendix, Fig. S2B) (#14 in the consensus), suggesting that Trp393 and Trp456 are important for protein folding and function. There is no an equivalent tryptophan #−2 in subdomain 1 of the C-terminal domain of GNIP1Aa1, so 2 tryptophan residues in position #−2 are sufficient for proper folding/function of a three-subdomain β-tripod domain. Because the side chains of W393 and W456 are completely buried at the interface between the subdomains, the primary role of these residues could be proper folding and stability of the soluble monomeric protein. Indeed, the purified single-site mutants, W393A and W456A, were more sensitive to protease treatment (SI Appendix, Fig. S4) than wild-type GNIP1Aa.
Another conserved position in the β-tripod family is represented by D395 of GNIP1Aa. This Asp #1 (Fig. 3) is by far the most abundant and dominates the consensus in position #16 (SI Appendix, Fig. S2B). The complete loss of activity by replacement of Asp395 with Ala explains that conservation. Alanine substitution of the other two Asp residues in position #1 of the first and the third loops of the β-tripod domain of GNIP1Aa1 also affected activity, although to a lower extent. The activity for the single-point mutants D332A and D458A dropped significantly (Fig. 6C). However, only simultaneous substitution of both aspartates completely abolished protein activity (Fig. 6C). None of the single Asp #1 mutations disrupted the folding of the protein, as shown by protease treatment experiments of purified variants (see below), suggesting a functional rather than structural importance of this position.
Position #9, the C-terminal residue within D-X-G-S/T-G-X3-D motif, was also tested by alanine substitution. Surprisingly, while Asp occupies this position in each of the three apical loops of GNIP1Aa, a small reduction of protein activity was registered for D340A and D403A only, in subdomains 1 and 2 (Fig. 6C). The activity of the D466A mutant was indistinguishable from wild-type. However, similar to the mutagenesis effect in position #1, the double mutations completely eliminated protein activity (Fig. 6C) (D340A/D403A, D340A/D466A, and D403A/D466A variants). It is noteworthy that position #9 is less conserved than positions #−2 and #1 in the β-tripod family consensus (SI Appendix, Fig. S2B). The preference for Asp in all three #9 positions of the GNIP1Aa motif, together with the lower conservation in the β-tripod family, as well as their full solvent exposure in the protein structure (Fig. 6 D and E), suggest that these residues may be important for receptor selectivity and for the specificity of the protein activity, which may vary within the family.
Thus, the results of the mutagenesis analysis suggest that among the three conserved D-X-G-S/T-G-X3-D regions, at least two residues have to be preserved in each of the equivalent positions #−2, #1, and #9 to ensure GNIP1Aa functionality (highlighted in three colors in Fig. 6 A, B, D, and E). Still, subdomain 2 and its conserved residues proved to be the most sensitive to amino acid substitutions and may play a leading role in protein activity.
It is important to stress that all aforementioned GNIP1Aa mutants were expressed at a level similar to wild-type GNIP1Aa (SI Appendix, Fig. S3). To evaluate if protein stability and overall fold was affected by a mutation, we purified selected variants and screened them for protease sensitivity. We also tested if pore formation was affected by comparing them to wild-type GNIP1Aa in liposome leakage assays.
A total of five mutants—W393A, D395A, W456A, D458A, and D332A—together with two control proteins, wild-type GNIP1Aa and Plu-MACPF, were evaluated. All tested mutants behaved similarly to wild- type GNIP1Aa. Treatment of all purified 59-kDa GNIP1Aa variants with trypsin produced protease-resistant cores or activated protein versions with an apparent molecular mass of ∼57 kDa. In fact, the digestion pattern for three mutants, D395A, D458A, and D332A, was identical to wild-type GNIP1Aa, exhibiting a single protein band for the activated protein version on an SDS/PAGE gel (SI Appendix, Fig. S4). The tryptophan mutants, W456A and W393A, appeared to be more sensitive to the protease treatment, generating three smaller fragments in addition to the main 57-kDa protein band (SI Appendix, Fig. S4). In fact, a low level of protein degradation was visible for the purified W456A variant even before the trypsin digestion, demonstrating partial protein instability. Plu-MACPF, a protein with no activity in WCR bio-assays, was found to be the most protease sensitive, showing significant protein degradation, generating multiple protein fragments with no obvious protease-resistant core (SI Appendix, Fig. S4).
The treatment of GNIP1Aa with trypsin likely resulted in a C-terminal truncation, because the N terminus of the activated wild-type GNIP1Aa stayed intact, as shown by N-terminal sequence analysis. Based on the GNIP1Aa sequence, trypsin specificity, and the size of the final activated product, the cleavage occurs after Lys520, removing the last 16 C-terminal residues, an ∼1.8-kDa fragment. Such activation removes almost the entire C-terminal helix that locks the two domains together, thus providing a higher level of flexibility for both domains. This activation seems to be crucial for GNIP1Aa to undergo significant conformational changes and to form a transmembrane pore.
All five Ala mutants of GNIP1Aa (W393A, D395A, W456A, D458A, and D332A) were tested in liposome leakage assays. They were found to behave like the wild-type protein, disrupting liposomes and releasing a fluorescent dye (SI Appendix, Fig. S4). Note that the wild-type and mutant GNIP1Aa proteins had to be proteolytically activated to trigger liposome leakage, while unactivated versions of the same proteins acted similar to the negative controls, which included buffers, BSA, and Plu-MACPF (SI Appendix, Fig. S4). Plu-MACPF was completely destroyed by protease treatment, so no activated version of the protein was available for this assay. Thus, the liposome leakage assays suggest that each of the five mutants and the wild-type GNIP1Aa are capable of binding to the lipid bilayer and adopting the proper protein conformation to form a transmembrane pore. Despite activity in the liposome assays and seemingly unaffected folding, WCR toxicity was significantly reduced or eliminated for all five GNIP1Aa mutants with a single Ala substitution in the conserved motifs of the C-terminal domain. A reasonable explanation for this could be disruption of the interaction between GNIP1Aa and a putative membrane-associated receptor in WCR (which would not be present in the liposomes).
The conserved GNIP1Aa motif DX-G-S/T-G-X3-D includes a G-X3-D motif that was described as a common carbohydrate-binding motif for jacalin-related mannose-binding lectins that have a β-prism I-fold (53, 54). Griffithsin (GRFT) is an algae-derived lectin that is structurally similar to jacaline-related lectins, but also shares some features with GNIP1Aa. GRFT is one of the most potent HIV entry inhibitors reported so far with activity against multiple viruses that have surface envelope glycoproteins with dense clusters of oligomannose N-linked glycans, which include HIV-1 envelope glycoprotein gp120. While some oligomannose N-linked glycans were also found on the surface of epithelial and peripheral blood mononuclear human cells, GRFT doesn’t affect viability of these cells and doesn’t cause significant up-regulation of markers of T cell activation. Such high specificity for virus glycoproteins makes GRFT a safe and efficacious microbicide candidate (55). Analogous to GRFT, GNIP1Aa exhibits high specific toxicity toward one insect species, WCR, with no effect on other screened insects. Presence of a specific GNIP1Aa membrane-associated receptor in WCR gut may explain that phenomenon.
Similar to jacaline-related lectins, GRFT forms a dimer, yet instead of one GXXXD motif per subunit, GRFT has three such motifs (56). Each GRFT subunit has a threefold symmetry and three GXXXD motifs that were shown to be important for carbohydrate binding (56, 57). As already described, the C-terminal domain of GNIP1Aa has threefold pseudosymmetry, with three somewhat longer motifs, D-X-G-S/T-G-X3-D. In both proteins the conserved sequence motifs are found to be in the loops connecting two strands of the β-sheet, with the conserved Asp (#9 for GNIP1Aa) at the C terminus of each loop. Moreover, all three motifs in both proteins are clustered on the same side of the corresponding domain. The crystal structures for GRFT with three mannose molecules (57) demonstrate that the side chain of Asp from each GXXXD motif is the main contributor of the specific interactions with a corresponding sugar molecule. Interestingly, mutations of a single Asp to Ala only slightly weaken the affinity of GRFT binding to HIV-1 envelope glycoprotein gp120, while mutants with all of the conserved Asp residues replaced by Ala did not bind gp120 (58). Our observations for GNIP1Aa are somewhat similar, although related to toxicity, not receptor binding. One substitution in position #9 decreased GNIP1Aa activity slightly (D3340A or D403A) or not at all (D466A), while double and triple mutants were completely inactive.
Another interesting feature was discovered for GRFT upon its interaction with oligosaccharides: even though the protein has three symmetrical and identical GXXXD binding sites per subunit, only two of them were found occupied by two arms of a three-armed nonamannoside, while the third mannose-binding site was occupied by the D2 arm of another nonamannoside molecule (58, 59). GNIP1Aa, which also has three identical and nearly symmetrical D-X-G-S/T-G-X3-D motifs (Fig. 6 D and E), could be expected, similar to GRFT, to have three putative receptor-binding sites. The three sites of GNIP1Aa also may not necessarily be equally involved in receptor recognition; they may provide independent contributions and differentially bind one or more glycan molecules. Thus, a unique receptor that explains the high specificity of GNIP1Aa toward WCR could be a glycosylated protein or glycosylated lipid. While the GXXXD binding site seems to be a part of the D-X-G-S/T-G-X3-D motif, the identity of the receptor for GNIP1Aa could be quite different from the receptor for GRFT.
The identification of the receptor for GNIP1Aa or other proteins from the β-tripod family would help us understand the nature of GNIP1Aa specificity. Subsequent studies of protein–receptor interactions might reveal the contribution of each binding site to receptor recognition and to protein activity.
Alanine Scanning Analysis of the C-Terminal Domain.
A total of 120 positions of the 220-residue long C-terminal domain (V317-L536) were probed by the single-position alanine scan (Table 1 and SI Appendix, Fig. S3 and Table S2). Five Ala substitutions (4%) led to no or low protein expression, and as a result, undetectable or low WCR toxicity (Table 1). For all of the other 115 variants with a single alanine substitution, protein expression was similar to wild-type GNIP1Aa (SI Appendix, Fig. S3). These mutant proteins were split into three groups based on their WCR toxicity, with the stunting activity being chosen as a more accurate measure of protein toxicity than mortality (Table 1 and SI Appendix, Fig. S3 and Table S2).
The first group was described above, with no detectable protein toxicity because of the essential nature of amino acids within or near the conserved loop D-X-G-S/T-G-X3-D motif, and includes only three variants: W393A, D395A, and W456A. Their activity was at the level of the buffer control or the negative protein control, Plu-MACPF (0.44 stunt/11% mortality) (Table 1 and SI Appendix, Fig. S3 and Table S2).
The second group contains 95 variants and represents the majority of the studied variants (79% of all single site alanine mutants). Activity for them was not visibly negatively affected by the mutation, with the toxicity being very similar to the wild-type GNIP1Aa protein (stunting activity of 3.75 and above and mortality between 83% and 100%).
A reduced level of protein activity was detected for 17 GNIP1Aa Ala-substituted versions (14% of all screened positions) (Table 1 and SI Appendix, Fig. S3), comprising group 3. Only mutants that consistently resulted in lower than the wild- type GNIP1Aa activity are included in this list. The stunting activity for these 17 mutants ranged between 1.00 and 3.67, while mortality varied from 24 to 92%. Mutants Q413A and Y352A are examples of variants with small yet detectable reduction of protein toxicity, 3.67 stunt/89% mortality and 3.56 stunt/92% mortality, respectively. The variant L523A, on the other hand, is an example of mutant proteins with the lowest yet detectable WCR toxicity within this group, 1.00 stunt/24% mortality.
It is noteworthy that 3 of 17 negatively affected positions are not a part of the core β-tripod domain, but are localized at the very C-terminal end instead. M519 and L523 are the part of the second helix H2 of GNIP1Aa (Fig. 7); M529 is the last C-terminal amino acid with interpretable electron density that was included in the protein structure. L523A and M529A mutations resulted in significant reduction of protein toxicity, demonstrating 1.00 and 1.22 stunt, and 24% and 29% mortality, respectively. M519 was also sensitive to alanine substitution, demonstrating a noticeable and reproducible decrease of protein activity down to 3.11 stunt/72% mortality.
Fig. 7.
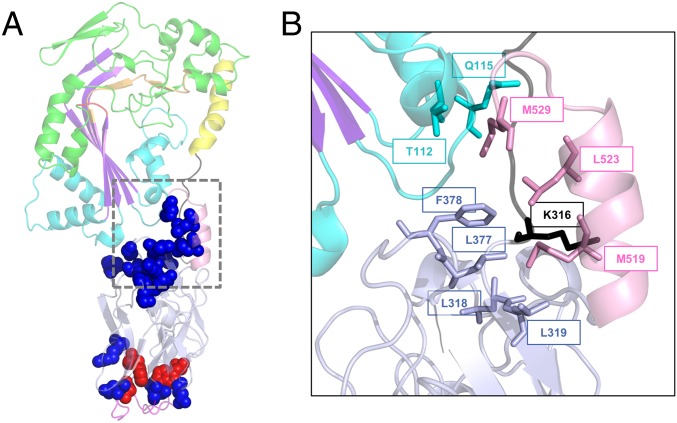
Functionally important positions within the C-terminal domain of GNIP1Aa. (A) Ribbon representation of the GNIP1Aa protein. The same colors were used as in Fig. 1A, displayed at 50% transparency. All positions shown in sphere representation are sensitive to a single substitution with an alanine: replacement of residues in red leads to complete knockout of protein WCR toxicity, and in blue to the reduced protein activity (total of 17). See Table 1 and SI Appendix, Fig. S3 for details. (B) Close-up of the extensive interaction of the selected C-terminal residues with different protein modules, shown in ribbon representation. The M519, L523, and M529 of the C-terminal helix are in contact with four protein regions. The aforementioned residues and their interacting partners are shown in stick representation, labeled with a corresponding position number, and colored the same as in Fig. 1A.
All three residues, M519, L523, and M529, are spatially close to each other (Fig. 7B). Their side chains are stacked together on the inner side of the C-terminal helix, facing and interacting with four different regions of the protein: CH1 from MACPF domain (T112 and Q115), the N- and the C-terminal ends of subdomain 1 of the β-tripod domain (L318, L319, L377, and F378), and the linker connecting protein domains (K316). Thus, the C-terminal helix fastens all three protein parts together like a safety pin (Fig. 7B).
When mapped on the structure of GNIP1Aa (Fig. 7A), all 20 mutants (complete knockouts and variants with reduced activity) (Table 1) negatively affected by the alanine substitutions form two clusters. One group of 10 residues (shown as a cluster of blue spheres in Fig. 7A) is localized at the interface between two protein domains, while another group is located on the apical surface of the C-terminal domain of GNIP1Aa (a cluster of seven blue and three red spheres in Fig. 7A). This clustering of the positions that affect activity, combined with the overall GNIP1Aa resemblance to other β-pore–forming proteins like perforin and CDCs (22, 60, 61), suggests that the apical residues are the most critical for primary target recognition and are responsible for the specific interaction with a receptor or a unique molecule on a membrane surface. It is also possible that some apical residues play a direct role in protein oligomerization or pore formation (SI Appendix, Fig. S4). The other functionally important residues (blue cluster) reflect the significance of communication between the two protein domains. This group of residues transduces the primary target-recognition signal from the C-terminal domain to the N-terminal MACPF domain, triggering enormous conformational changes in the MACPF domain, leading to the transformation of two helical CH sets into two transmembrane β-hairpins, and ultimately to pore formation (35).
Overall our mutational analysis proposes that the β-tripod domain plays the role of an accessory target-recognition domain, whereas the MACPF domain serves as a pore-forming protein component.
Our alanine analysis also touched on the importance of the interdomain linker. Five single positions (307, 309, 311, 313, and 315) were individually replaced with alanine, and two were found to be crucial for protein expression. Both Ile311A and Lys313A substitutions resulted in low level of protein expression. That suggests that the linker between the two GNIP1Aa domains is not just a flexible hinge, but is an integral part of the full-length protein that affects protein folding and stability.
Conclusions
Our studies provide insight into the GNIP1Aa structure and its mode of action, which appear to be different from all other known insecticidal proteins. The activity and the mode of action of this protein make it an excellent candidate for commercial development into a transgenic agricultural product. Such a product might have a high potential to combat crop damage in corn and to delay development of resistance in WCR insects.
GNIP1Aa is also a representative of a growing group of dual-nature pore-forming proteins that undergo significant conformational changes to exert their activity, transforming from water-soluble to integral membrane form. This group includes defense proteins involved in the innate immune response, anticancer proteins, protein toxins, pesticidal proteins, and many more. The mechanism of pore formation by MACPFs is an actively studied field, yet the first step of this process, specific target recognition, is poorly understood. A crucial part of the present study is the discovery of a previously uncharacterized type of MACPF accessory domain. Structural–functional analysis of this domain allowed us to pinpoint functionally important amino acid residues, and to suggest an explanation for the protein’s highly specific toxicity. Moreover, analysis of the structure and sequence of this domain led us to identify it as a member of a protein family that we dub β-tripod. This may lead to the discovery of other family representatives with the same or different target specificities. Our results, together with the accumulated knowledge about perforin (61) and structurally similar CDCs (60) and pesticidal proteins (62–64), suggest that the β-tripod domain of GNIP1Aa specifically recognizes a receptor in the target pest’s gut and triggers the lethal pore-formation process. The high specificity of this accessory domain for a receptor in WCR may make this member of the MACPF family particularly useful for controlling this pest and managing resistance to other commercial insecticidal proteins, and suggest the possibility of rationally designing insecticidal proteins specific for other targets.
Materials and Methods
GNIP1Aa Overexpression and Purification.
The construction of the MBP-gnip1Aa plasmid was described previously (13). Details are provided in SI Appendix.
For crystallization, the wild-type GNIP1Aa was overexpressed as the MBP-GNIP1Aa fusion and purified as described previously (13), with slight modifications as described in SI Appendix.
Crystallization.
Crystallization was performed with Crystallization Screens from Hampton Research, as detailed in SI Appendix.
Structure Determination.
Crystallographic data were collected on an Xcalibur Nova system from Oxford Diffraction. The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org [PDB ID codes 6FBM (65)]. Details are provided in SI Appendix.
Plasmid Constructions to Test Protein Activity in Bioassays.
To analyze activity of GNIP1Aa and its variants, a construct expressing untagged full-length wild-type protein was used: gnip1Aa plasmid. Details are provided in SI Appendix.
Protein Expression for Bioassays.
GNIP1Aa protein, its mutants, and the negative control Plu-MACPF were expressed in BL21 Star (DE3), as detailed in SI Appendix.
Protein Analysis.
Protein concentration for structural studies was determined according to the method of Bradford (66) or by BCA protein assay (67) with BSA as a standard as described in SI Appendix.
WCR Bioassays.
WCR eggs were received from Crop Characteristics. Bioassays were performed as described in SI Appendix.
Sequence Discovery and Domain Search.
To find the largest set of sequences containing homology to the β-tripod fold, a local copy of the NCBI NR database was searched using a single repeat from the β-tripod fold as the seed sequence with the jackhmmer program (50). The protocol is described in detail in SI Appendix.
Phylogeny.
The insecticidal-associated domain phylogeny shown in SI Appendix, Fig. S2 is created from the alignment of the full sequences in sequence set 3 (all MACPF and Toxin_10 domain containing sequences), as described in SI Appendix.
Sequence Logo.
The sequence logo was generated from the parts of sequence set 3 to the single repeat HMM generated by jackhmmer. The alignment was used to create the sequence logo using the Geneious software package (68).
Supplementary Material
Acknowledgments
We thank Volker Heinrichs for design of the library of Ala mutants; Jayme Williams for her help with screening and mutagenesis; Dan Tomso for the design of the original construct gnip1Aa plasmid; Rong Guo for design and construct of MBP-gnip1Aa plasmid; Xia Cao for her assistance in performing some of the initial exploratory bioinformatics analysis; Leo Magalhaes, Amanda Morrison, Dianne Cantoni, Sharon Bagasol, Kellie Lucas, and Emily Boutaud for optimization, screening, and bio-assay support; and Ted Kahn and Cris Oppert for constructive criticism of the manuscript.
Footnotes
Conflict of interest statement: J.Z., D.V., and S.L.M. are current employees of BASF; J.F., J.D., C.K., and D.B. are current employees of Bayer; N.B.D. was formerly employed by Bayer. GNIP1Aa is protected under US Patent 8575425 and family members, and US Patent 9862965 and family members.
This article is a PNAS Direct Submission. C.B. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and the structure factors reported in this paper have been deposited in the RCSB Protein Data Bank www.rcsb.org (PDB ID code 6FBM).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815547116/-/DCSupplemental.
References
- 1.Metcalf RL. Foreword. In: Krysan JL, Miller TA, editors. Methods for the Study of Pest Diabrotica. Springer; New York: 1986. pp. vii–xv. [Google Scholar]
- 2.Crickmore N, et al. 2017 Bacillus thuringiensis toxin nomenclature. Available at www.btnomenclature.info/. Accessed September 1, 2018.
- 3.Crickmore N, et al. Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998;62:807–813. doi: 10.1128/mmbr.62.3.807-813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Comas C, Lumbierres B, Pons X, Albajes R. No effects of Bacillus thuringiensis maize on nontarget organisms in the field in southern Europe: A meta-analysis of 26 arthropod taxa. Transgenic Res. 2014;23:135–143. doi: 10.1007/s11248-013-9737-0. [DOI] [PubMed] [Google Scholar]
- 5.Yu HL, Li YH, Wu KM. Risk assessment and ecological effects of transgenic Bacillus thuringiensis crops on non-target organisms. J Integr Plant Biol. 2011;53:520–538. doi: 10.1111/j.1744-7909.2011.01047.x. [DOI] [PubMed] [Google Scholar]
- 6.Brookes G, Barfoot P. Key environmental impacts of global genetically modified (GM) crop use 1996-2011. GM Crops Food. 2013;4:109–119. doi: 10.4161/gmcr.24459. [DOI] [PubMed] [Google Scholar]
- 7.Brookes G, Barfoot P. The global income and production effects of genetically modified (GM) crops 1996-2011. GM Crops Food. 2013;4:74–83. doi: 10.4161/gmcr.24176. [DOI] [PubMed] [Google Scholar]
- 8.Kathage J, Qaim M. Economic impacts and impact dynamics of Bt (Bacillus thuringiensis) cotton in India. Proc Natl Acad Sci USA. 2012;109:11652–11656. doi: 10.1073/pnas.1203647109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabashnik BE, Carrière Y. Surge in insect resistance to transgenic crops and prospects for sustainability. Nat Biotechnol. 2017;35:926–935. doi: 10.1038/nbt.3974. [DOI] [PubMed] [Google Scholar]
- 10.Tabashnik BE, Brévault T, Carrière Y. Insect resistance to Bt crops: Lessons from the first billion acres. Nat Biotechnol. 2013;31:510–521. doi: 10.1038/nbt.2597. [DOI] [PubMed] [Google Scholar]
- 11.Gassmann AJ, et al. Field-evolved resistance by Western corn rootworm to multiple Bacillus thuringiensis toxins in transgenic maize. Proc Natl Acad Sci USA. 2014;111:5141–5146. doi: 10.1073/pnas.1317179111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gassmann AJ, et al. Evidence of resistance to Cry34/35Ab1 corn by Western corn rootworm (Coleoptera: Chrysomelidae): Root injury in the field and larval survival in plant-based bioassays. J Econ Entomol. 2016;109:1872–1880. doi: 10.1093/jee/tow110. [DOI] [PubMed] [Google Scholar]
- 13.Sampson K, et al. Discovery of a novel insecticidal protein from Chromobacterium piscinae, with activity against Western corn rootworm, Diabrotica virgifera virgifera. J Invertebr Pathol. 2017;142:34–43. doi: 10.1016/j.jip.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Batista JH, da Silva Neto JF. Chromobacterium violaceum pathogenicity: Updates and insights from genome sequencing of novel Chromobacterium species. Front Microbiol. 2017;8:2213. doi: 10.3389/fmicb.2017.02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Byamukama D, et al. Contrasting occurrence of Chromobacterium violaceum in tropical drinking water springs of Uganda. J Water Health. 2005;3:229–238. doi: 10.2166/wh.2005.028. [DOI] [PubMed] [Google Scholar]
- 16.Ramirez JL, et al. Chromobacterium Csp_P reduces malaria and dengue infection in vector mosquitoes and has entomopathogenic and in vitro anti-pathogen activities. PLoS Pathog. 2014;10:e1004398. doi: 10.1371/journal.ppat.1004398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin PAW, Gundersen-Rindal D, Blackburn M, Buyer J. Chromobacterium subtsugae sp. nov., a betaproteobacterium toxic to Colorado potato beetle and other insect pests. Int J Syst Evol Microbiol. 2007;57:993–999. doi: 10.1099/ijs.0.64611-0. [DOI] [PubMed] [Google Scholar]
- 18.Soby SD, Gadagkar SR, Contreras C, Caruso FL. Chromobacterium vaccinii sp. nov., isolated from native and cultivated cranberry (Vaccinium macrocarpon Ait.) bogs and irrigation ponds. Int J Syst Evol Microbiol. 2013;63:1840–1846. doi: 10.1099/ijs.0.045161-0. [DOI] [PubMed] [Google Scholar]
- 19.Han J-H, Park G-C, Kim KS. Antagonistic evaluation of Chromobacterium sp. JH7 for biological control of ginseng root rot caused by Cylindrocarpon destructans. Mycobiology. 2017;45:370–378. doi: 10.5941/MYCO.2017.45.4.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderluh G, Kisovec M, Kraševec N, Gilbert RJC. Distribution of MACPF/CDC proteins. In: Anderluh G, Gilbert R, editors. MACPF/CDC Proteins—Agents of Defence, Attack and Invasion. Springer; Dordrecht, The Netherlands: 2014. pp. 7–30. [Google Scholar]
- 21.Hadders MA, et al. Assembly and regulation of the membrane attack complex based on structures of C5b6 and sC5b9. Cell Rep. 2012;1:200–207. doi: 10.1016/j.celrep.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Law RH, et al. The structural basis for membrane binding and pore formation by lymphocyte perforin. Nature. 2010;468:447–451. doi: 10.1038/nature09518. [DOI] [PubMed] [Google Scholar]
- 23.Lopez JA, et al. Perforin forms transient pores on the target cell plasma membrane to facilitate rapid access of granzymes during killer cell attack. Blood. 2013;121:2659–2668. doi: 10.1182/blood-2012-07-446146. [DOI] [PubMed] [Google Scholar]
- 24.Lovelace LL, Cooper CL, Sodetz JM, Lebioda L. Structure of human C8 protein provides mechanistic insight into membrane pore formation by complement. J Biol Chem. 2011;286:17585–17592. doi: 10.1074/jbc.M111.219766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ota K, et al. Membrane cholesterol and sphingomyelin, and ostreolysin A are obligatory for pore-formation by a MACPF/CDC-like pore-forming protein, pleurotolysin B. Biochimie. 2013;95:1855–1864. doi: 10.1016/j.biochi.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 26.Peyronnet O, et al. Estimation of the radius of the pores formed by the Bacillus thuringiensis Cry1C delta-endotoxin in planar lipid bilayers. Biochim Biophys Acta. 2002;1567:113–122. doi: 10.1016/s0005-2736(02)00605-3. [DOI] [PubMed] [Google Scholar]
- 27.Kao C-Y, et al. Global functional analyses of cellular responses to pore-forming toxins. PLoS Pathog. 2011;7:e1001314. doi: 10.1371/journal.ppat.1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haag ES, Sly BJ, Andrews ME, Raff RA. Apextrin, a novel extracellular protein associated with larval ectoderm evolution in Heliocidaris erythrogramma. Dev Biol. 1999;211:77–87. doi: 10.1006/dbio.1999.9283. [DOI] [PubMed] [Google Scholar]
- 29.Johnson TK, et al. Torso-like functions independently of Torso to regulate Drosophila growth and developmental timing. Proc Natl Acad Sci USA. 2013;110:14688–14692. doi: 10.1073/pnas.1309780110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng C, Heintz N, Hatten ME. CNS gene encoding astrotactin, which supports neuronal migration along glial fibers. Science. 1996;272:417–419. doi: 10.1126/science.272.5260.417. [DOI] [PubMed] [Google Scholar]
- 31.Rosado CJ, et al. A common fold mediates vertebrate defense and bacterial attack. Science. 2007;317:1548–1551. doi: 10.1126/science.1144706. [DOI] [PubMed] [Google Scholar]
- 32.Xu Q, et al. Structure of a membrane-attack complex/perforin (MACPF) family protein from the human gut symbiont Bacteroides thetaiotaomicron. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:1297–1305. doi: 10.1107/S1744309110023055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeLano WL. 2002. The PyMOL Molecular Graphics System (Schrödinger, LLC, New York), Version 1.5.0.4.
- 34.Hadders MA, Beringer DX, Gros P. Structure of C8alpha-MACPF reveals mechanism of membrane attack in complement immune defense. Science. 2007;317:1552–1554. doi: 10.1126/science.1147103. [DOI] [PubMed] [Google Scholar]
- 35.Lukoyanova N, et al. Conformational changes during pore formation by the perforin-related protein pleurotolysin. PLoS Biol. 2015;13:e1002049. doi: 10.1371/journal.pbio.1002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shatursky O, et al. The mechanism of membrane insertion for a cholesterol-dependent cytolysin: A novel paradigm for pore-forming toxins. Cell. 1999;99:293–299. doi: 10.1016/s0092-8674(00)81660-8. [DOI] [PubMed] [Google Scholar]
- 37.Reboul CF, Whisstock JC, Dunstone MA. Giant MACPF/CDC pore forming toxins: A class of their own. Biochim Biophys Acta. 2016;1858:475–486. doi: 10.1016/j.bbamem.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 38.Serna M, Giles JL, Morgan BP, Bubeck D. Structural basis of complement membrane attack complex formation. Nat Commun. 2016;7:10587. doi: 10.1038/ncomms10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Pee K, et al. CryoEM structures of membrane pore and prepore complex reveal cytolytic mechanism of Pneumolysin. eLife. 2017;6:e23644. doi: 10.7554/eLife.23644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aleshin AE, et al. Structure of complement C6 suggests a mechanism for initiation and unidirectional, sequential assembly of membrane attack complex (MAC) J Biol Chem. 2012;287:10210–10222. doi: 10.1074/jbc.M111.327809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polekhina G, Giddings KS, Tweten RK, Parker MW. Insights into the action of the superfamily of cholesterol-dependent cytolysins from studies of intermedilysin. Proc Natl Acad Sci USA. 2005;102:600–605. doi: 10.1073/pnas.0403229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossjohn J, Feil SC, McKinstry WJ, Tweten RK, Parker MW. Structure of a cholesterol-binding, thiol-activated cytolysin and a model of its membrane form. Cell. 1997;89:685–692. doi: 10.1016/s0092-8674(00)80251-2. [DOI] [PubMed] [Google Scholar]
- 43.Xu L, et al. Crystal structure of cytotoxin protein suilysin from Streptococcus suis. Protein Cell. 2010;1:96–105. doi: 10.1007/s13238-010-0012-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosado CJ, et al. The MACPF/CDC family of pore-forming toxins. Cell Microbiol. 2008;10:1765–1774. doi: 10.1111/j.1462-5822.2008.01191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bonangelino CJ, Chavez EM, Bonifacino JS. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2486–2501. doi: 10.1091/mbc.02-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finn RD, Clements J, Eddy SR. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 51.Hire RS, Hadapad AB, Dongre TK, Kumar V. Purification and characterization of mosquitocidal Bacillus sphaericus BinA protein. J Invertebr Pathol. 2009;101:106–111. doi: 10.1016/j.jip.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 52.Humphreys MJ, Berry C. Variants of the Bacillus sphaericus binary toxins: Implications for differential toxicity of strains. J Invertebr Pathol. 1998;71:184–185. doi: 10.1006/jipa.1997.4711. [DOI] [PubMed] [Google Scholar]
- 53.Kanagawa M, et al. Structural basis for multiple sugar recognition of Jacalin-related human ZG16p lectin. J Biol Chem. 2014;289:16954–16965. doi: 10.1074/jbc.M113.539114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meagher JL, Winter HC, Ezell P, Goldstein IJ, Stuckey JA. Crystal structure of banana lectin reveals a novel second sugar binding site. Glycobiology. 2005;15:1033–1042. doi: 10.1093/glycob/cwi088. [DOI] [PubMed] [Google Scholar]
- 55.Kouokam JC, et al. Investigation of griffithsin’s interactions with human cells confirms its outstanding safety and efficacy profile as a microbicide candidate. PLoS One. 2011;6:e22635. doi: 10.1371/journal.pone.0022635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lusvarghi S, Bewley CA. Griffithsin: An antiviral lectin with outstanding therapeutic potential. Viruses. 2016;8:E296. doi: 10.3390/v8100296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ziółkowska NE, et al. Domain-swapped structure of the potent antiviral protein griffithsin and its mode of carbohydrate binding. Structure. 2006;14:1127–1135. doi: 10.1016/j.str.2006.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xue J, et al. The role of individual carbohydrate-binding sites in the function of the potent anti-HIV lectin griffithsin. Mol Pharm. 2012;9:2613–2625. doi: 10.1021/mp300194b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moulaei T, et al. Monomerization of viral entry inhibitor griffithsin elucidates the relationship between multivalent binding to carbohydrates and anti-HIV activity. Structure. 2010;18:1104–1115. doi: 10.1016/j.str.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hotze EM, Tweten RK. Membrane assembly of the cholesterol-dependent cytolysin pore complex. Biochim Biophys Acta. 2012;1818:1028–1038. doi: 10.1016/j.bbamem.2011.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Traore DA, et al. Defining the interaction of perforin with calcium and the phospholipid membrane. Biochem J. 2013;456:323–335. doi: 10.1042/BJ20130999. [DOI] [PubMed] [Google Scholar]
- 62.Palma L, Berry C. Understanding the structure and function of Bacillus thuringiensis toxins. Toxicon. 2016;109:1–3. doi: 10.1016/j.toxicon.2015.10.020. [DOI] [PubMed] [Google Scholar]
- 63.Jurat-Fuentes JL, Crickmore N. Specificity determinants for Cry insecticidal proteins: Insights from their mode of action. J Invertebr Pathol. 2017;142:5–10. doi: 10.1016/j.jip.2016.07.018. [DOI] [PubMed] [Google Scholar]
- 64.Berry C, Crickmore N. Structural classification of insecticidal proteins—Towards an in silico characterisation of novel toxins. J Invertebr Pathol. 2017;142:16–22. doi: 10.1016/j.jip.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 65.Freigang J, Zaitseva J. 2019 Crystal structure of GNIP1Aa from Chromobacterium piscinae. RCSB Protein Data Bank. Available at https://www.rcsb.org/structure/6FBM. Deposited December 19, 2017.
- 66.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 67.Smith PK, et al. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 68.Kearse M, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.