Significance
Cell surface exposure of phosphatidylserine (PtdSer) is observed not only in apoptotic cells, but also in activated lymphocytes, activated platelets, capacitated sperm, aged erythrocytes, and some cancer cells. Xkr8, a membrane protein carrying 10 putative transmembrane segments, was originally identified as a scramblase that is activated by caspase-mediated cleavage during apoptosis. Here we show that mouse Xkr8 scramblase activity can be stimulated by kinase-mediated phosphorylation independent of the caspase-mediated cleavage, suggesting that Xkr8 may play a role in various biological processes to expose PtdSer.
Keywords: scramblase, phosphorylation, phosphatidylserine, XKR, flippase
Abstract
The exposure of phosphatidylserine (PtdSer) to the cell surface is regulated by the down-regulation of flippases and the activation of scramblases. Xkr8 has been identified as a scramblase that is activated during apoptosis, but its exogenous expression in the mouse Ba/F3 pro B cell line induces constitutive PtdSer exposure. Here we found that this Xkr8-mediated PtdSer exposure occurred at 4 °C, but not at 20 °C, although its scramblase activity was observed at 20 °C. The Xkr8-mediated PtdSer exposure was inhibited by a kinase inhibitor and enhanced by phosphatase inhibitors. Phosphorylated Xkr8 was detected by Phos-tag PAGE, and a mass spectrometric and mutational analysis identified three phosphorylation sites. Their phosphomimic mutation rendered Xkr8 resistant to the kinase inhibitor for PtdSer exposure at 4 °C, but unlike phosphatase inhibitors, it did not induce constitutive PtdSer exposure at 20 °C. On the other hand, when the flippase genes were deleted, the Xkr8 induced constitutive PtdSer exposure at high temperature, indicating that the flippase activity normally counteracted Xkr8’s ability to expose PtdSer. These results indicate that PtdSer exposure can be increased by the phosphorylation-mediated activation of Xkr8 scramblase and flippase down-regulation.
Phospholipids are asymmetrically distributed between the outer and inner leaflets of the plasma membrane (1–3). Phosphatidylserine (PtdSer) and phosphatidylethanolamine (PtdEtn) are confined to the inner leaflet, while phosphatidylcholine (PtdCho) and sphingomyelin are localized mainly to the outer leaflet. This asymmetric distribution of phospholipids is maintained by the action of P4-ATPases such as ATP11A and 11C, which function as flippases in the plasma membrane to actively translocate PtdSer and PtdEtn from the outer leaflet to the inner leaflet (4, 5). In various biological processes, this asymmetric distribution of phospholipids is disrupted (6); for example, PtdSer is exposed in apoptotic cells and serves as an “eat me” signal for phagocytes (7, 8), and activated platelets expose PtdSer, which serves as a scaffold for blood-clotting factors (9). To disrupt the asymmetric phospholipid distribution, the flippases are inactivated (4, 10). In addition, scramblases, which nonspecifically and bidirectionally transport phospholipids between the leaflets in an energy-independent manner, are activated as well (2, 3, 11).
We previously identified two proteins as scramblases by isolating two different cDNAs from a subline of the mouse Ba/F3 pro B cell line (Ba/F3-PS19) that had been selected as a population exposing a high level of PtdSer (12, 13). These proteins, Transmembrane Protein 16F (TMEM16F, also called anoctamin 6) and XK-related protein 8 (Xkr8), are membrane proteins with 10 putative transmembrane regions (11). TMEM16F, a Ca2+-activated scramblase, is responsible for exposing PtdSer in activated platelets (12, 14) and for releasing microparticles in activated platelets or microvesicles carrying hydroxyapatite during bone mineralization (14, 15). Ca2+ directly binds to a TMEM16F dimer to change its conformation into an active scramblase and provides a cleft for phospholipid translocation (16–18). The second scramblase, Xkr8, is responsible for exposing PtdSer during apoptosis (13, 19, 20). Xkr8 carries a caspase 3 recognition site in its C-terminal region, and its cleavage by caspase 3 during apoptosis induces its dimerization to an active scramblase form (19). Interestingly, Ba/F3, but not WR19L, T cell transformants expressing Xkr8 constitutively expose PtdSer without an apoptotic stimulus (13); however, how Xkr8 is activated without being cleaved by caspase in a cell-specific manner is unknown.
In this study, we found that mouse (m)Xkr8-supported constitutive PtdSer exposure in Ba/F3 cells was dependent on intracellular Ca2+ and regulated by the phosphorylation of mXkr8. Mass spectrometry analysis identified three phosphorylation sites located near the caspase recognition site in the C-terminal region. Mutations preventing phosphorylation of these residues blocked the scrambling activity of mXkr8 in Ba/F3 cells, while phosphomimic mutations made mXkr8 a kinase-independent scramblase. In contrast, mutations preventing phosphorylation of these residues had no effect on Xkr8’s caspase-dependent scramblase activity, indicating that Xkr8’s scrambling activity is regulated by either caspase cleavage or phosphorylation.
Results
Constitutive Phospholipid Scrambling by Mouse Xkr8 in Ba/F3 Cells.
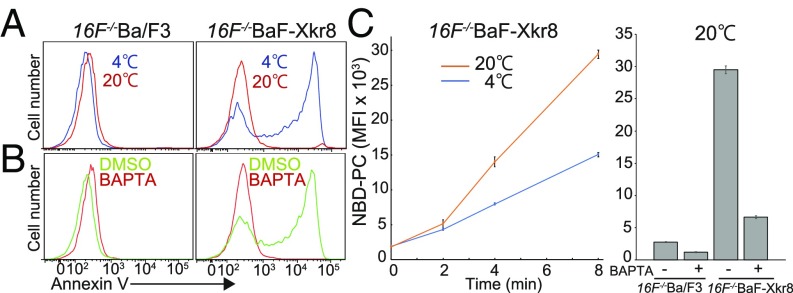
Ba/F3 transformants expressing mXkr8 (BaF-Xkr8) constitutively expose PtdSer (13). We noticed that this constitutive PtdSer exposure was strongly affected by the temperature and the Ca2+ concentration in the assay. To rule out the possibility that mXkr8 activated TMEM16F’s scramblase activity, mXkr8 cDNA was introduced into TMEM16F−/−Ba/F3 cells (16F−/−BaF), which showed no Ca2+ ionophore-induced PtdSer exposure (21). As shown in Fig. 1A, the Xkr8-supported PtdSer exposure was observed when the annexin V-binding assay was done at 4 °C, but not at 20 °C. When the cells were pretreated with 1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid (BAPTA)-AM to chelate intracellular Ca2+, PtdSer exposure was not observed even at 4 °C (Fig. 1B), indicating that Ca2+ is essential for the mXkr8-mediated PtdSer exposure.
Fig. 1.
Constitutive phospholipid scrambling by mXkr8. (A and B) Cold temperature- and Ca2+-dependent exposure of PtdSer by mXkr8 in Ba/F3 cells. (A) TMEM16F−/−Ba/F3 (16F−/−Ba/F3) and its transformants expressing mXkr8 (16F−/−BaF-Xkr8) were incubated at 4 °C (blue) or 20 °C (red) for 15 min with Cy5-annexin V and then analyzed by flow cytometry. (B) 16F−/−Ba/F3 and 16F−/−BaF-Xkr8 cells were incubated with (red) or without (green) BAPTA-AM and then stained at 4 °C with Cy5-annexin V. Annexin V-staining profiles in the PI-negative population are shown. (C) Ca2+-dependent internalization of NBD-PC by mXkr8 in Ba/F3 cells. (Left) 16F−/−BaF-Xkr8 cells were incubated in annexin V buffer with NBD-PC and SYTOX red at 4 °C or 20 °C for the indicated times, treated with BSA, and subjected to flow cytometry. The incorporated NBD-PC in the SYTOX red-negative population is expressed as mean fluorescence intensity (MFI). (Right) 16F−/−Ba/F3 and 16F−/−BaF-Xkr8 cells were incubated with or without BAPTA-AM and then incubated at 20 °C for 8 min with NBD-PC in annexin V buffer containing SYTOX red, and the incorporated NBD-PC was determined as described above. The experiments were carried out in triplicate, and average MFI values were plotted with SE (bars).
The PtdSer exposure on cells is regulated by both flippases and scramblases (11). Flippases are ATP-dependent enzymes, while scramblases do not require ATP, suggesting that flippases were more affected by the reaction temperature than the Xkr8 scramblase, and probably blocked the PtdSer exposure at 20 °C. Therefore, scramblase activity was assayed by the incorporation of NBD-PC, which should not be affected by flippase. As shown in Fig. 1C, 16F−/−BaF-Xkr8, but not the parental 16F−/−BaF cells, incorporated NBD-PC in a time-dependent manner. The incorporation of NBD-PC was accelerated by increasing the reaction temperature and inhibited by pretreating with BAPTA-AM, indicating that mXkr8 in Ba/F3 cells could constitutively scramble phospholipids in a Ca2+-dependent manner.
Regulation of the Xkr8-Mediated Phospholipid Scrambling by Phosphorylation.
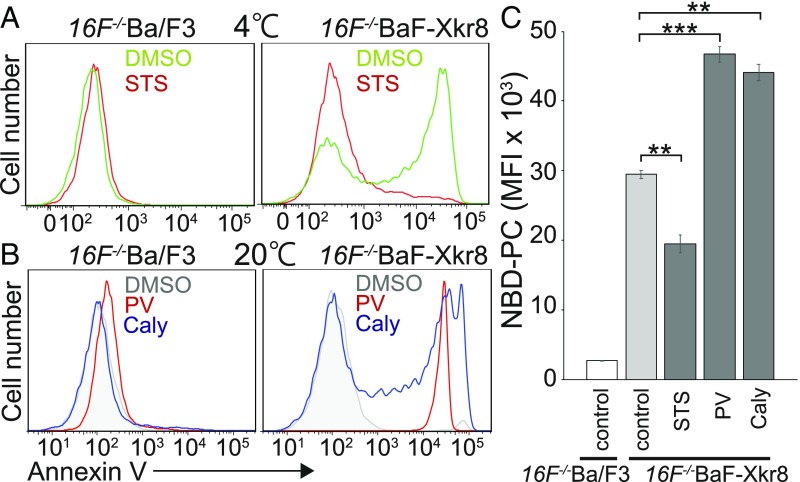
Ba/F3 is an IL-3–dependent pro-B cell line (22), and continuously receives the signal from an IL-3 receptor that activates kinases (23). To examine the possibility that mXkr8 is activated by phosphorylation, mXkr8-expressing Ba/F3 cells were treated with kinase or phosphatase inhibitors. As shown in Fig. 2A, treatment with staurosporine (STS), a broad kinase inhibitor (24), completely blocked the constitutive PtdSer exposure. In contrast, pretreatment with either pervanadate, a tyrosine-phosphatase inhibitor (25), or calyculin A, a serine/threonine phosphatase inhibitor (26), strongly enhanced the mXkr8-mediated PtdSer exposure. Furthermore, PtdSer exposure after phosphatase-inhibitor treatment could be observed when the annexin V-binding assay was carried out at 20 °C (Fig. 2B). The effects of these kinase and phosphatase inhibitors were also observed when the scramblase activity was assayed using NBD-PC; that is, pretreatment with STS significantly inhibited the NBD-PC incorporation in 16F−/−BaF-Xkr8 cells, while pervanadate and calyculin A enhanced it by approximately 50% (Fig. 2C).
Fig. 2.
Phosphorylation-dependent Xkr8-mediated phospholipid scrambling in Ba/F3 cells. (A and B) Effect of kinase or phosphatase inhibitors on mXkr8-mediated PtdSer exposure in Ba/F3 cells. 16F−/−Ba/F3 and 16F−/−BaF-Xkr8 cells were incubated without (DMSO) or with STS and then stained at 4 °C with Cy5-annexin V (A) or were incubated with pervanadate (PV) or calyculin A (Caly) and then stained at 20 °C with Cy5-annexin V (B). Annexin V staining profiles in the PI-negative population are shown. (C) Stimulation of Xkr8-dependent phospholipid scrambling by phosphatase inhibitors. 16F−/−Ba/F3 and 16F−/−BaF-Xkr8 cells were pretreated without (control) or with STS, PV, or Caly, and incubated at 20 °C for 8 min with NBD-PC. The experiments were carried out in triplicate, and the average MFI values for the incorporated NBD-PC are plotted with SE (bars). **P < 0.01; ***P < 0.001, Welch’s t test.
Identification of Phosphorylation Sites in mXkr8.
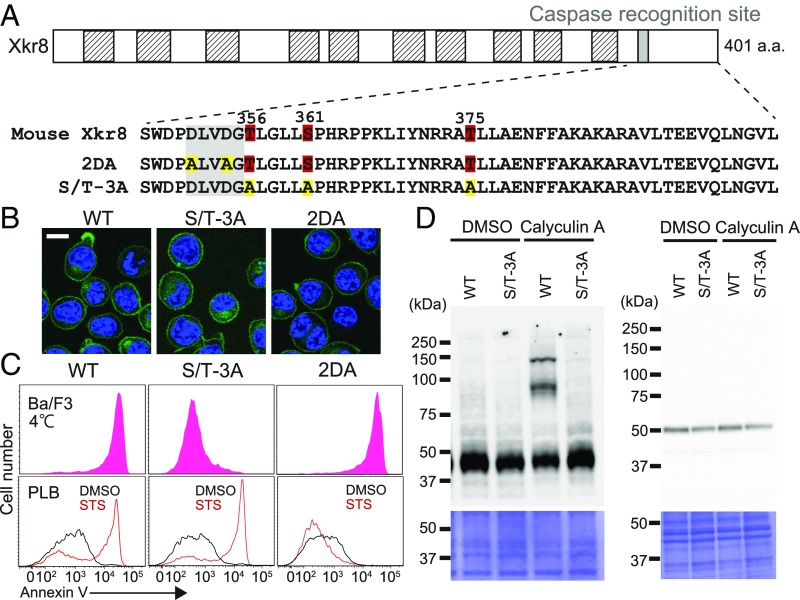
The foregoing results suggest that a kinase(s) directly phosphorylates mXkr8 to activate its scramblase activity. To identify the phosphorylation sites in mXkr8, membrane fractions were prepared from 16F−/−BaF-Xkr8 cells that had been treated with pervanadate. After solubilization with lauryl maltose neopentyl glycol, the mXkr8-EGFP chimeric protein was trapped with anti-GFP nanobody-coupled beads, digested with AspN or chymotrypsin, and analyzed by LC-MS/MS. These analyses revealed three phosphorylated residues (Thr-356, Ser-361, and Thr-375) downstream of the caspase recognition site of mXkr8 (Fig. 3A and SI Appendix, Fig. S1).
Fig. 3.
Assignment of the phosphorylation sites in mXkr8. (A) Phosphorylation sites in mXkr8. The structure of mXkr8 is shown schematically. Putative transmembrane regions are shown in hatched boxes, and the caspase 3 recognition motif is shown as a filled box. The amino acid sequence of the caspase recognition motif and its downstream region is shown. The phosphorylated residues (Thr-356, Ser-361, and Thr-375) are numbered. In the 2DA mutant, Asp-351 and Asp-354 in the caspase recognition motif were mutated to Ala, and in the S/T-3A mutant, Thr-356, Ser-361, and Thr-375 were mutated to Ala. (B) Localization of Xkr8 mutants to the plasma membrane. The WT and indicated mutants of mXkr8 were fused to EGFP and expressed in TMEM16F−/−Xkr8−/−BaF (DKO-BaF) cells. The stable transformants were stained with 5.0 μg/mL Hoechst 33342 and observed by confocal microscopy for GFP (green) and Hoechst 33342 (blue). (Scale bar: 10 μm.) (C) Kinase-dependent but caspase-independent PtdSer exposure by mXkr8. (Upper) DKO-BaF cells expressing WT, S/T-3A, or 2DA Xkr8 were stained at 4 °C for 15 min with Cy5-annexin V. The annexin V-staining profiles in the GFP-positive and PI-negative population are shown. (Lower) Human PLB cells expressing WT, S/T-3A or 2DA were treated at 37 °C for 120 min with 10 μM STS and then stained with annexin V at 4 °C. (D) Effect of the S/T-3A mutation on the phosphorylation of mXkr8. (Upper) DKO-BaF transformants (1 × 106 cells) expressing WT or S/T-3A mutant mXkr8 were incubated at 37 °C for 30 min with or without 50 nM calyculin A. After solubilization of the cells with RIPA buffer, aliquots of the lysates corresponding to 4 × 104 cells were separated by 8% Phos-tag PAGE (Left) or 10–20% SDS/PAGE (Right) and then analyzed by Western blot analysis with anti-GFP. (Lower) The membranes were stained with Coomassie brilliant blue.
To confirm that these residues were the targets for phosphorylation, they were mutated to alanine, and the mutant protein (S/T-3A) was fused to EGFP. The EGFP-fused mXkr8 mutant protein was then introduced into Xkr8-deficient 16F−/−BaF (XK−/−16F−/−BaF) cells that had been established using the CRISPR/Cas9 system (SI Appendix, Fig. S2). As shown in Fig. 3B, the S/T-3A mutant Xkr8 was localized to the plasma membrane like the wild-type (WT) mXkr8-EGFP. However, transformants expressing the S/T-3A mXkr8 mutant did not expose PtdSer (Fig. 3C). On the other hand, mXkr8 in which two aspartate residues in the caspase recognition site were mutated (2DA mutant) (13) still supported the constitutive PtdSer exposure in Ba/F3 cells. Conversely, the apoptosis-induced PtdSer exposure was blocked by the 2DA mutant, but not by mutation in the phosphorylation sites (S/T-3A) (Fig. 3C). These results indicate that the mutation in the phosphorylation site did not affect caspase-activated scramblase activity, and that the phosphorylation-induced PtdSer exposure did not require caspase.
To confirm biochemically that the assigned serine and threonine residues were phosphorylated, transformants expressing EGFP-tagged WT mXkr8 were treated with calyculin A, and protein in the cell lysates was subjected to Phos-tag SDS/PAGE (27), followed by Western blot analysis. As shown in Fig. 3D, an anti-EGFP antibody detected distinct supershifted bands in addition to the band with the expected molecular weight of approximately 50 kDa. These shifted bands were not observed in the transformants expressing the S/T-3A mutant, confirming that Thr-356, Ser-361, and Thr-375 of mXkr8 were phosphorylated in Ba/F3 cells.
Phosphorylation-Dependent Phospholipid Scrambling by mXkr8.
Aligning the amino acid sequences of Xkr8 from various animals showed that the sequence downstream of the caspase recognition site in the C-terminal tail region was well conserved among mammals (Fig. 4A). Among the three phosphorylatable serine/threonine residues, at least one residue could be found in various mammalian Xkr8s. To examine which phosphorylated residues were involved in the activation of mXkr8, the three residues were individually or together mutated into aspartate, a phosphomimic amino acid (T356D, S361D, T375D, or S/T-3D), or alanine (T356A, S361A, T375A, or S/T-3A). The EGFP-tagged WT and mutant mXkr8 were then introduced into XK−/−16F−/−BaF cells. As shown in Fig. 4B, the S/T-3D and S/T-3A mutants and WT mXkr8 were all present at the plasma membrane, indicating that the mutations did not affect the cellular localization of mXkr8. All of the phosphomimic mutants supported the constitutive PtdSer exposure at 4 °C, indicating that they did not affect scramblase activity. In the T356D or S361D mutant-transfected cells, the constitutive PtdSer exposure was efficiently inhibited by STS, while STS did not inhibit PtdSer exposure in the cells expressing the T375D or S/T-3D mutant (Fig. 4C), indicating that the phosphorylation of T375 was sufficient to activate the Xkr8 scramblase.
Fig. 4.
Phosphorylation sites of mXkr8 critical for the PtdSer exposure in Ba/F3 cells. (A, Upper) The amino acid sequences of the caspase recognition motif and its downstream region in mXkr8 of the six indicated animals were aligned. The amino acid residues that were identical in all animals are in red. The caspase recognition motifs are shadowed, while the putative phosphorylation sites are highlighted in red. (A, Lower) The three phosphorylation sites (Thr-356, Ser-361, and Thr-375) in mXkr8 were individually (T356A, S361A, T375A, T356D, S361D, or T375D) or together (S/T-3A or S/T-3D) mutated into Ala or Asp. The mutated residues are highlighted in yellow. (B) Localization of the Xkr8 mutants at the plasma membrane. The WT and the indicated mXkr8 mutants were fused to EGFP and expressed in DKO-BaF cells. The stable transformants were observed by confocal microscopy for GFP (green) and Hoechst 33342 (blue). (Scale bar: 10 μm.) (C) Kinase-independent PtdSer exposure by the phosphorylation-mimic mutations of mXkr8. DKO-BaF cells expressing WT, T356D, S361D, T375D, or S/T-3D mXkr8 were treated for 30 min at 37 °C with 10 μM STS (red) or DMSO vehicle (green) and then stained at 4 °C for 15 min with Cy5-annexin V. The annexin V staining profiles in the PI-negative and GFP-positive population are shown. (D) Effect of the Ala mutations on mXkr8-mediated constitutive PtdSer exposure in Ba/F3 cells. DKO-BaF transformants expressing the WT, T356A, S361A, T375A, or S/T-3A mXkr8 were stained at 4 °C for 15 min with Cy5-annexin V. The annexin V staining profiles in the PI-negative and GFP-positive population are shown. (E) Ca2+-dependent and temperature-sensitive PtdSer exposure by the phosphorylation-mimic mutation of mXkr8. (Upper) DKO-BaF cells expressing S/T-3D were pretreated with (red) or without (green) BAPTA-AM before being stained with Cy5-annexin V. (Lower) DKO-BaF cells expressing the WT or S/T-3D mutant mXkr8 were stained at 20 °C with Cy5-annexin V.
Accordingly, the T356A and S361A mutations had little effect on Xkr8-mediated PtdSer exposure, while the T375A mutation had a strong inhibitory effect, and the remaining activity was almost fully blocked by the S/T-3A mutation (Fig. 4D). These results suggest that the phosphorylation at T375 is essential for activating the mXkr8 scramblase, while phosphorylation of the other two residues (T356 and S361) is less important. The PtdSer exposure mediated by the S/T-3D phosphomimic mutant mXkr8 was completely inhibited by pretreating the cells with BAPTA-AM (Fig. 4E), indicating that Ca2+ is essential for the scrambling activity of phosphorylated mXkr8.
Regulation of PtdSer Exposure by Flippase.
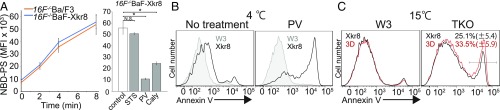
As described above, the strong constitutive PtdSer exposure on 16F−/−BaF-Xkr8 cells could be detected at 20 °C after treatment with phosphatase inhibitors (Fig. 2B). On the other hand, the PtdSer exposure detected at 20 °C in the transformants expressing the S/T-3D mutant was relatively weak (Fig. 4E). Since human and yeast flippases are down-regulated by phosphorylation (28–30), we considered the possibility that flippases in the Ba/F3 cells were also inhibited by phosphorylation. To examine this possibility, we assayed the flippase activity by measuring the incorporation of a fluorescently labeled PtdSer (NBD-PS). We found that the 16F−/−BaF and 16F−/−BaF-Xkr8 cells incorporated NBD-PS with similar efficiency (Fig. 5A), confirming that Xkr8 did not affect flippase activity. Pretreatment with STS had little effect on flippase activity, but pervanadate or calyculin A strongly inhibited the incorporation of NBD-PS, indicating that flippases present in Ba/F3 cells could be inhibited by phosphorylation.
Fig. 5.
Regulation of the Xkr8-mediated PtdSer exposure by flippase activity. (A) Down-regulation of flippase activity by phosphatase inhibitors in Ba/F3 cells. (Left) 16F−/−Ba/F3 and 16F−/−Ba/F-Xkr8 cells were incubated for the indicated time at 20 °C with NBD-PS in Hepes-buffered saline. The BSA-nonextractable NBD-PS was determined by flow cytometry and plotted as the MFI. (Right) 16F−/−BaF-Xkr8 cells were preincubated with STS, PV, or calyculin A (Caly) as described in Fig. 2. The cells were then incubated at 20 °C for 8 min with NBD-PS, and the incorporated lipids were determined by flow cytometry. The experiments were carried out in triplicate, and the average MFI was plotted with SE (bars). *P < 0.05; NS, not significant (P > 0.05), Welch’s t test. (B) Phosphorylation-dependent mXkr8-mediated PtdSer exposure in W3 cells. W3 and its transformants expressing mXkr8-EGFP and Basigin were preincubated with 100 μM PV and then stained at 4 °C for 30 min with Cy5-annexin V. The annexin V staining profiles in the PI-negative population are shown. For the Xkr8 transformants, the GFP-positive population was gated for the annexin V profile. (C) Enhanced PtdSer exposure by the loss of ATP11A and ATP11C. WT (W3) and ATP11A−/−ATP11C−/−TMEM16F−/−W3 (TKO) cells were transformed with Basigin together with mXkr8-EGFP or its S/T-3D mutant and then stained at 15 °C for 30 min with Cy5-annexin V. The annexin V staining profiles in the PI-negative and GFP-positive population are shown. (Right) The experiments were performed five times, and the mean percentage of the annexin V-positive population is indicated with SE. There is no significant difference between the WT and the S/T-3D mutant mXkr8 (P > 0.05, Welch’s t test).
We previously observed that transforming Ba/F3, but not W3, cells with mXkr8 causes constitutive PtdSer exposure (13). However, when W3 transformants expressing mXkr8 were pretreated with pervanadate, they exposed PtdSer at 4 °C (Fig. 5B), suggesting the presence of a kinase that phosphorylates mXkr8 in W3 cells. As found with Ba/F3 cells, the S/T-3D mutant mXkr8 did not support PtdSer exposure at high temperature (Fig. 5C), suggesting that flippases antagonize the effect of Xkr8’s scramblase activity in W3 cells. W3 cells express two flippases (ATP11A and 11C) at the plasma membrane (5), which were previously knocked out by the CRISPR/Cas9 system together with TMEM16F, to generate ATP11A−/−ATP11C−/−16F−/−W3 (TKO-W3) cells (18). To confirm the effect of the flippase on Xkr8-mediated PtdSer exposure, the WT and phosphomimic mutant Xkr8s were introduced into TKO-W3 cells. As shown in Fig. 5C, the transformants expressing the phosphomimic mutant of mXkr8 as well as the WT mXkr8 exposed PtdSer. These results confirm that PtdSer exposure is determined by the balance between scramblase and flippase activities.
Discussion
In this study, we have shown that mXkr8, previously identified as a caspase-dependent phospholipid scramblase (13), can be activated by phosphorylation. The phosphorylation sites were identified downstream of the caspase recognition site in a region well conserved in mammalian Xkr8. The phosphorylation of caspase substrates at or near the caspase recognition site often affects the efficiency of caspase cleavage (31–33); however, here we found that mutations to nonphosphorylatable amino acids in mXkr8 did not affect its ability to promote apoptotic PtdSer exposure, and that mutating the caspase recognition site did not block phosphorylation-mediated PtdSer exposure. These results indicate that mXkr8’s scramblase can be activated independently by caspase-mediated cleavage or by kinase-mediated phosphorylation. Removing the 47 C-terminal amino acids by caspase induces the dimerization of Xkr8 (19), suggesting that the tail region masks the domains necessary for its dimerization. Phosphorylation at a regulatory domain controls the activity of various enzymes by inhibiting or promoting interaction with the enzymatic active site (34, 35). It is tempting to speculate that phosphorylation at the C-terminal region releases the dimerization or scrambling domain of mXkr8 from its inhibited form.
Treating Ba/F3 cells with pervanadate, a tyrosine phosphatase inhibitor, stimulated mXkr8 phosphorylation at three sites (Thr-356, Ser-361, and Thr-375) and activated its scrambling activity. Among these sites, the phosphorylation at Thr-375 was found to contribute most strongly to the activation of mXkr8. The motif around Thr-375 (RRXpTL) fully agrees with the consensus motif for cAMP-dependent protein kinase A (PKA), which is known to be activated in Ba/F3 cells (36). Ba/F3 is an IL-3–dependent pro-B cell line (22) that expresses IL-3 receptors and B cell receptors. These receptors activate JAK and SYK tyrosine kinases, respectively, leading to the activation of numerous signaling molecules, including PKA (23, 37, 38). Whether PKA is actually responsible for phosphorylating mXkr8, and what kind of kinase cascade leads to this activation, remain to be studied.
The flippase activity in Ba/F3 cells was inhibited by treatment with phosphatase inhibitors, indicating that the flippase can be regulated by phosphorylation, as has been reported previously for P-type ATPases, including flippases (28–30, 39). Among the three P4-type ATPases that function as flippases at the plasma membrane, real-time RT-PCR analysis indicated that Ba/F3 cells express ATP11A and ATP11C (SI Appendix, Fig. S3). Takatsu et al. (29) recently reported that treating Ba/F3 cells with phorbol 12-myristate 13-acetate (PMA) induces the endocytosis of ATP11C, but not of ATP11A, via protein kinase C-mediated phosphorylation at its C-terminal region. The strong reduction of flippase activity that we observed in pervanadate-treated Ba/F3 cells suggests that not only ATP11C, but also ATP11A, were down-regulated by phosphorylation. Quantitative phosphoproteomics analysis has shown that human ATP11A can be phosphorylated at two evolutionarily conserved positions (Ser-738 and Ser-740) during mitosis or by stimulation with angiotensin (40, 41) (PhosphoSitePlus; https://www.phosphosite.org/homeAction). It will be interesting to determine whether the same kinase cascade that leads to the phosphorylation of mXkr8 is responsible for phosphorylating ATP11A and ATP11C to down-regulate their flippase activity.
Ca2+ ionophore treatment does not activate Xkr8 for scrambling (13). However, mXkr8 phosphomimic mutant-mediated phospholipid scrambling was inhibited by an intracellular Ca2+ chelator. Calcium ion is an important signal for various cellular processes, including muscle contraction, neurotransmitter release, and cell proliferation (42). TMEM16F is responsible for the PtdSer exposure in activated platelets, in which stimulation with thrombin and collagen can increase the local Ca2+ concentration near the plasma membrane to several hundred micromolars. In contrast, mXkr8 requires a resting level of calcium ion of approximately 100 nM (42, 43). Whether Ca2+ directly binds to Xkr8 or indirectly regulates Xkr8 scramblase activity remains to be studied.
In addition to apoptotic cells and activated platelets, PtdSer exposure is observed in activated CD4 and CD8 T cells (44, 45), activated B cells (46), capacitated sperm (47), aged erythrocytes (48), some cancer cells (49), and tumor-associated endothelial cells (50). The extremely high expression of Xkr8 in the testis (13) and the central role of PKA in sperm capacitation (51) suggest that phosphorylation-mediated Xkr8 activation may be involved in this process. Since a kinase cascade is strongly activated in cancer cells (52), it is tempting to speculate that the PtdSer exposure in transformed cancer cells is regulated by phosphorylated Xkr8.
Finally, Xkr8 belongs to the Xkr family, which consists of eight to nine members in human and mouse (53). In addition to Xkr8, Xkr4 and Xkr9 are cleaved by caspase and act as scramblases, while the functions of the other members remain unknown. It will be interesting to study whether other Xkr family members are phosphorylated to function as scramblases. In this regard, it may be noteworthy that human XKR5 was recently shown to be phosphorylated by the KIT receptor tyrosine kinase (54).
Materials and Methods
Cell Lines and Expression Plasmids.
Mouse Ba/F3 cells were grown in RPMI 1640 containing 10% FCS and mouse IL-3. Mouse WR19L cells expressing Fas (W3) and human PLB985 cells (PLB) were cultured in RPMI 1640 containing 10% FCS and 50 μM β-mercaptoethanol. The TMEM16F−/−Ba/F3 and ATP11A−/−ATP11C−/−TMEM16F−/−W3 (TKO-W3) cell lines have been described previously (18, 21). The mXKR8 gene in Ba/F3 cells was edited with the CRISPR-Cas9 system.
To establish Ba/F3 cell transformants expressing mutant mXkr8, cells were transfected with Ahd1-cleaved plasmid DNA by electroporation using NEPA21 and then selected with 0.5–1.0 mg/mL G418 or 1.0 μg/mL puromycin. PLB and W3 cells were transformed by infection with a pantropic retrovirus, and the transformants were selected with 1 μg/mL puromycin or 10 μg/mL blasticidin. Expression plasmids for mXkr8, mXkr8-2DA, and mouse Basigin have been described previously (13, 19). The cDNAs coding for the mXkr8 mutants T356A, S361A, T375A, T356A/S361A/T375A (S/T-3A), T356D, S361D, T375D, and T356D/S361D/T375D (S/T-3D) were generated by PCR.
Assay for Scramblase and Flippase Activity, Induction of Apoptosis, and Treatment with Inhibitors.
Phospholipid scrambling activity was measured by PtdSer exposure or internalization of NBD-PC. Flippase activity was assayed by internalization of NBD-PS. Apoptosis was induced by incubating cells with 10 μM STS for 2 h at 37 °C. For BAPTA-AM treatment, cells were incubated at 37 °C for 15 min with 25 μM BAPTA-AM. Pervanadate was prepared as described previously (25). To examine the effect of phosphatase inhibitors, cells were incubated at 37 °C for 30 min with 100 μM pervanadate or 50 nM calyculin A. The effect of kinase inhibitor was examined by incubating the cells at 37 °C for 30 min with 10 μM STS.
Zn2+ Phos-Tag SDS/PAGE and Identification of Phosphorylation Sites.
For Zn2+ Phos-tag SDS/PAGE (27), proteins were separated by electrophoresis on an 8% polyacrylamide gel containing 30 μM Phos-tag acrylamide (Wako). To determine the phosphorylation sites, GFP-Flag–tagged mXkr8 was immunoprecipitated from the membrane fractions of Ba/F3 transformants expressing mXkr8-GFP-Flag, with anti-GFP nanobody-coupled magnetic agarose beads (GFP-Trap_MA; ChromoTek). Proteins on the beads were digested with Asp-N or chymotrypsin, and the resultant peptides were analyzed by LC-MS/MS on an EASY-nLC 1200 ultra-high-performance liquid chromatograph connected to a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific). Raw data were analyzed against the SwissProt database using Proteome Discoverer version 2.2 (Thermo Fisher Scientific) with Mascot version 2.5 (Matrix Science).
Statistical Analysis.
All data are expressed as mean with SE. Differences between groups were examined for statistical significance using Welch’s t-test. Full details are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank S. Omura (Kitasato Institute for Life Science, Kitasato University) for the STS, M. Kawano for technical assistance, and M. Fujii for secretarial assistance. This work was supported by Grant-in-Aid for Scientific Research (S) for the Promotion of Science 15H05785 from the Japan Society for the Promotion of Science and Grant JPMJCR14M4 from the Core Research for Evolutional Science and Technology of the Japanese Science and Technology Agency (to S.N.). T.S. is the recipient of a Tadamitsu Kishimoto Fellowship from the Japan Society of Immunology.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820499116/-/DCSupplemental.
References
- 1.Kobayashi T, Menon AK. Transbilayer lipid asymmetry. Curr Biol. 2018;28:R386–R391. doi: 10.1016/j.cub.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Bevers EM, Williamson PL. Getting to the outer leaflet: Physiology of phosphatidylserine exposure at the plasma membrane. Physiol Rev. 2016;96:605–645. doi: 10.1152/physrev.00020.2015. [DOI] [PubMed] [Google Scholar]
- 3.Hankins HM, Baldridge RD, Xu P, Graham TR. Role of flippases, scramblases and transfer proteins in phosphatidylserine subcellular distribution. Traffic. 2015;16:35–47. doi: 10.1111/tra.12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Segawa K, et al. Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science. 2014;344:1164–1168. doi: 10.1126/science.1252809. [DOI] [PubMed] [Google Scholar]
- 5.Segawa K, et al. Phospholipid flippases enable precursor B cells to flee engulfment by macrophages. Proc Natl Acad Sci USA. 2018;115:12212–12217. doi: 10.1073/pnas.1814323115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birge RB, et al. Phosphatidylserine is a global immunosuppressive signal in efferocytosis, infectious disease, and cancer. Cell Death Differ. 2016;23:962–978. doi: 10.1038/cdd.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henson PM. Cell removal: Efferocytosis. Annu Rev Cell Dev Biol. 2017;33:127–144. doi: 10.1146/annurev-cellbio-111315-125315. [DOI] [PubMed] [Google Scholar]
- 8.Nagata S, Hanayama R, Kawane K. Autoimmunity and the clearance of dead cells. Cell. 2010;140:619–630. doi: 10.1016/j.cell.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 9.Zwaal RF, Comfurius P, Bevers EM. Lipid-protein interactions in blood coagulation. Biochim Biophys Acta. 1998;1376:433–453. doi: 10.1016/s0304-4157(98)00018-5. [DOI] [PubMed] [Google Scholar]
- 10.Segawa K, Kurata S, Nagata S. Human type IV P-type ATPases that work as plasma membrane phospholipid flippases, and their regulation by caspase and calcium. J Biol Chem. 2016;291:762–772. doi: 10.1074/jbc.M115.690727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagata S, Suzuki J, Segawa K, Fujii T. Exposure of phosphatidylserine on the cell surface. Cell Death Differ. 2016;23:952–961. doi: 10.1038/cdd.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki J, Umeda M, Sims PJ, Nagata S. Calcium-dependent phospholipid scrambling by TMEM16F. Nature. 2010;468:834–838. doi: 10.1038/nature09583. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki J, Denning DP, Imanishi E, Horvitz HR, Nagata S. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science. 2013;341:403–406. doi: 10.1126/science.1236758. [DOI] [PubMed] [Google Scholar]
- 14.Fujii T, Sakata A, Nishimura S, Eto K, Nagata S. TMEM16F is required for phosphatidylserine exposure and microparticle release in activated mouse platelets. Proc Natl Acad Sci USA. 2015;112:12800–12805. doi: 10.1073/pnas.1516594112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehlen HW, et al. Inactivation of anoctamin-6/TMEM16f, a regulator of phosphatidylserine scrambling in osteoblasts, leads to decreased mineral deposition in skeletal tissues. J Bone Miner Res. 2013;28:246–259. doi: 10.1002/jbmr.1751. [DOI] [PubMed] [Google Scholar]
- 16.Ishihara K, Suzuki J, Nagata S. Role of Ca2+ in the stability and function of TMEM16F and 16K. Biochemistry. 2016;55:3180–3188. doi: 10.1021/acs.biochem.6b00176. [DOI] [PubMed] [Google Scholar]
- 17.Bethel NP, Grabe M. Atomistic insight into lipid translocation by a TMEM16 scramblase. Proc Natl Acad Sci USA. 2016;113:14049–14054. doi: 10.1073/pnas.1607574113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gyobu S, Ishihara K, Suzuki J, Segawa K, Nagata S. Characterization of the scrambling domain of the TMEM16 family. Proc Natl Acad Sci USA. 2017;114:6274–6279. doi: 10.1073/pnas.1703391114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki J, Imanishi E, Nagata S. Xkr8 phospholipid scrambling complex in apoptotic phosphatidylserine exposure. Proc Natl Acad Sci USA. 2016;113:9509–9514. doi: 10.1073/pnas.1610403113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawano M, Nagata S. Lupus-like autoimmune disease caused by a lack of Xkr8, a caspase-dependent phospholipid scramblase. Proc Natl Acad Sci USA. 2018;115:2132–2137. doi: 10.1073/pnas.1720732115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gyobu S, et al. A role of TMEM16E carrying a scrambling domain in sperm motility. Mol Cell Biol. 2015;36:645–659. doi: 10.1128/MCB.00919-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palacios R, Steinmetz M. Il-3-dependent mouse clones that express B-220 surface antigen, contain Ig genes in germ-line configuration, and generate B lymphocytes in vivo. Cell. 1985;41:727–734. doi: 10.1016/s0092-8674(85)80053-2. [DOI] [PubMed] [Google Scholar]
- 23.Reddy EP, Korapati A, Chaturvedi P, Rane S. IL-3 signaling and the role of Src kinases, JAKs and STATs: A covert liaison unveiled. Oncogene. 2000;19:2532–2547. doi: 10.1038/sj.onc.1203594. [DOI] [PubMed] [Google Scholar]
- 24.Ōmura S, Asami Y, Crump A. Staurosporine: New lease of life for parent compound of today’s novel and highly successful anti-cancer drugs. J Antibiot (Tokyo) 2018;71:688–701. doi: 10.1038/s41429-018-0029-z. [DOI] [PubMed] [Google Scholar]
- 25.Huyer G, et al. Mechanism of inhibition of protein-tyrosine phosphatases by vanadate and pervanadate. J Biol Chem. 1997;272:843–851. doi: 10.1074/jbc.272.2.843. [DOI] [PubMed] [Google Scholar]
- 26.Wakimoto T, Egami Y, Abe I. Calyculin: Nature’s way of making the sponge-derived cytotoxin. Nat Prod Rep. 2016;33:751–760. doi: 10.1039/c5np00123d. [DOI] [PubMed] [Google Scholar]
- 27.Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka K, Fujimura-Kamada K, Yamamoto T. Functions of phospholipid flippases. J Biochem. 2011;149:131–143. doi: 10.1093/jb/mvq140. [DOI] [PubMed] [Google Scholar]
- 29.Takatsu H, et al. Phospholipid flippase ATP11C is endocytosed and downregulated following Ca2+-mediated protein kinase C activation. Nat Commun. 2017;8:1423. doi: 10.1038/s41467-017-01338-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roelants FM, et al. Protein kinase Gin4 negatively regulates flippase function and controls plasma membrane asymmetry. J Cell Biol. 2015;208:299–311. doi: 10.1083/jcb.201410076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Desagher S, et al. Phosphorylation of bid by casein kinases I and II regulates its cleavage by caspase 8. Mol Cell. 2001;8:601–611. doi: 10.1016/s1097-2765(01)00335-5. [DOI] [PubMed] [Google Scholar]
- 32.McDonnell MA, et al. Phosphorylation of murine caspase-9 by the protein kinase casein kinase 2 regulates its cleavage by caspase-8. J Biol Chem. 2008;283:20149–20158. doi: 10.1074/jbc.M802846200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turowec JP, et al. An unbiased proteomic screen reveals caspase cleavage is positively and negatively regulated by substrate phosphorylation. Mol Cell Proteomics. 2014;13:1184–1197. doi: 10.1074/mcp.M113.037374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 35.Wallach D, Kovalenko A. Phosphorylation and dephosphorylation of the RIG-I-like receptors: A safety latch on a fateful pathway. Immunity. 2013;38:402–403. doi: 10.1016/j.immuni.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 36.Harada H, et al. Phosphorylation and inactivation of BAD by mitochondria-anchored protein kinase A. Mol Cell. 1999;3:413–422. doi: 10.1016/s1097-2765(00)80469-4. [DOI] [PubMed] [Google Scholar]
- 37.Mócsai A, Ruland J, Tybulewicz VLJ. The SYK tyrosine kinase: A crucial player in diverse biological functions. Nat Rev Immunol. 2010;10:387–402. doi: 10.1038/nri2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morris R, Kershaw NJ, Babon JJ. The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. 2018;27:1984–2009. doi: 10.1002/pro.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poulsen H, Morth P, Egebjerg J, Nissen P. Phosphorylation of the Na+,K+-ATPase and the H+,K+-ATPase. FEBS Lett. 2010;584:2589–2595. doi: 10.1016/j.febslet.2010.04.035. [DOI] [PubMed] [Google Scholar]
- 40.Christensen GL, et al. Quantitative phosphoproteomics dissection of seven-transmembrane receptor signaling using full and biased agonists. Mol Cell Proteomics. 2010;9:1540–1553. doi: 10.1074/mcp.M900550-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olsen JV, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 42.Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 43.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: Dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 44.Elliott JI, et al. Membrane phosphatidylserine distribution as a non-apoptotic signalling mechanism in lymphocytes. Nat Cell Biol. 2005;7:808–816. doi: 10.1038/ncb1279. [DOI] [PubMed] [Google Scholar]
- 45.Fischer K, et al. Antigen recognition induces phosphatidylserine exposure on the cell surface of human CD8+ T cells. Blood. 2006;108:4094–4101. doi: 10.1182/blood-2006-03-011742. [DOI] [PubMed] [Google Scholar]
- 46.Elliott JI, et al. Phosphatidylserine exposure in B lymphocytes: A role for lipid packing. Blood. 2006;108:1611–1617. doi: 10.1182/blood-2005-11-012328. [DOI] [PubMed] [Google Scholar]
- 47.de Vries KJ, Wiedmer T, Sims PJ, Gadella BM. Caspase-independent exposure of aminophospholipids and tyrosine phosphorylation in bicarbonate responsive human sperm cells. Biol Reprod. 2003;68:2122–2134. doi: 10.1095/biolreprod.102.012500. [DOI] [PubMed] [Google Scholar]
- 48.Qadri SM, Bissinger R, Solh Z, Oldenborg P-A. Eryptosis in health and disease: A paradigm shift towards understanding the (patho)physiological implications of programmed cell death of erythrocytes. Blood Rev. 2017;31:349–361. doi: 10.1016/j.blre.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 49.Riedl S, et al. In search of a novel target—phosphatidylserine exposed by non-apoptotic tumor cells and metastases of malignancies with poor treatment efficacy. Biochim Biophys Acta. 2011;1808:2638–2645. doi: 10.1016/j.bbamem.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stafford JH, Thorpe PE. Increased exposure of phosphatidylethanolamine on the surface of tumor vascular endothelium. Neoplasia. 2011;13:299–308. doi: 10.1593/neo.101366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buffone MG, Wertheimer EV, Visconti PE, Krapf D. Central role of soluble adenylyl cyclase and cAMP in sperm physiology. Biochim Biophys Acta. 2014;1842:2610–2620. doi: 10.1016/j.bbadis.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.López-Otín C, Hunter T. The regulatory crosstalk between kinases and proteases in cancer. Nat Rev Cancer. 2010;10:278–292. doi: 10.1038/nrc2823. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki J, Imanishi E, Nagata S. Exposure of phosphatidylserine by Xk-related protein family members during apoptosis. J Biol Chem. 2014;289:30257–30267. doi: 10.1074/jbc.M114.583419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun J, Thingholm T, Højrup P, Rönnstrand L. XK-related protein 5 (XKR5) is a novel negative regulator of KIT/D816V-mediated transformation. Oncogenesis. 2018;7:48. doi: 10.1038/s41389-018-0057-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.