Recent years have seen a growing interest in generating real-time epidemic forecasts to help control infectious diseases, prompted by a succession of global and regional outbreaks. Increased availability of epidemiological data and novel digital data streams such as search engine queries and social media (1, 2), together with the rise of machine learning and sophisticated statistical approaches, have injected new blood into the science of outbreak forecasts (3, 4). In parallel, mechanistic transmission models have benefited from computational advances and extensive data on the mobility and sociodemographic structure of human populations (5, 6). In this rapidly advancing research landscape, modeling consortiums have generated systematic model comparisons of the impact of new interventions and ensemble predictions of outbreak trajectory, for use by decision makers (7–12). Despite the rapid development of disease forecasting as a discipline, however, and the interest of public health policy makers in making better use of analytics tools to control outbreaks, forecasts are rarely operational in the same way that weather forecasts, extreme events, and climate predictions are. The influenza study by Reich et al. (13) in PNAS is a unique example of multiyear infectious disease forecasts featuring a variety of modeling approaches, with consistent model formulations and forecasting targets throughout the 7-y study period (13). This is a major improvement over previous model comparison studies that used different targets and time horizons and sometimes different epidemiological datasets.
While there is considerable interest among modelers in advancing the science of disease forecasts, the level of confidence of the public health community in exploiting these predictions in real-world situations remains unclear. The disconnect is in part due to poor understanding of modeling concepts by policy experts, which is compounded by a lack of a well-established operational framework for using and interpreting model outputs. For example, the time horizon at which predictions are generally offered is in the order of 2 to 4 wk, which is generally too short for action. Prediction accuracy worsens substantially at longer time horizons, likely as a function of the modeling approach, epidemiological conditions, and type of pathogen studied, although a rigorous theoretical understanding of prediction limits is still lacking (Box 1). Further, while recent work has shown the promises of ensemble forecasts that combine outputs from different models (12–14), there is no clear understanding of best practices for this type of analysis that could stabilize operational performance in routine forecasts. In the same vein, the relationships among forecasting accuracy, data quality, and reporting rates remain elusive, due to the lack of controlled experiments and systematic analyses (Box 1).
Box 1.
Toward a principled understanding of infectious disease forecasts: Key questions
-
1.
How do prediction skills decrease with time horizon? Are there inherent limits to skilled predictions (currently 2 to 4 wk)?
-
2.
How do prediction skills scale with data accuracy and quantity? What is the value added by different datasets and disease parameters (age-specific surveillance data, digital data, social behavior, vaccination, strain composition, background population immunity)? How long and how granular does each dataset need to be?
-
3.
How should ensemble predictions be optimized? How many models? How many approaches? How should the weights of each approach be optimized?
-
4.
What is the right spatial scale for transmission, and hence for forecasts? How does it relate to the spatial scale of disease surveillance and public health interventions?
-
5.
The above questions should be studied or simulated across a variety of pathogens, populations, and data situations.
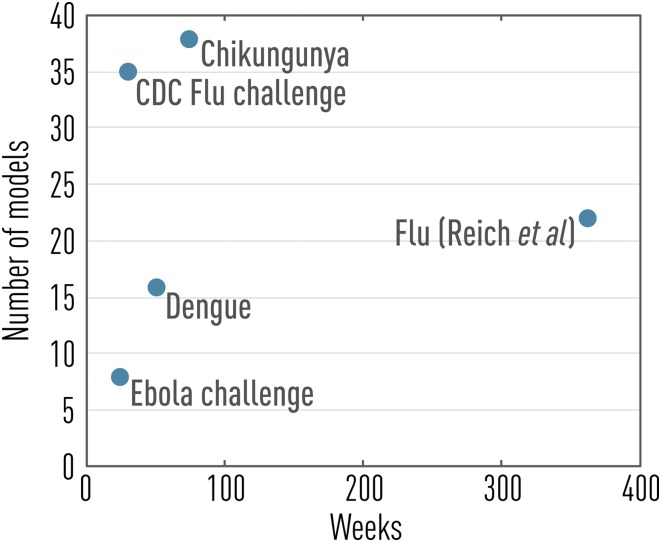
To begin to address these major issues, several infectious disease challenges have been carried out in the past few years, spanning a range of viral outbreaks such as influenza, dengue, chikungunya, and Ebola (refs. 10–14; see also Fig. 1). In particular, the influenza challenge initiated by the US Centers for Disease Control and Prevention (CDC) in the 2013/2014 winter season pioneered infectious disease forecasting in a formal way, growing from a small team of nine modelers in its inception (11) to a well-oiled machine involving 35 different approaches in the current flu season, with a recently debuted public website (14). The influenza challenge is focused on short-term prediction of the trajectory of influenza outbreaks in US regions, relying on weekly surveillance of influenza-like illness (ILI) incidence captured by a network of outpatient clinics. Seasonal influenza prediction targets are also considered, including epidemic onset week, peak size, and peak timing. Because the challenge is embedded in a public health institution, it is instrumental in bridging the gap between infectious disease modelers and end users in the public health community.
Fig. 1.
Past and present infectious disease forecasting challenges as a function of prediction horizon and number of models considered (data from refs. 10–14).
While there have been prior evaluations of the performances of the influenza challenge on yearly basis (11, 15), the study by Reich et al. (13) is the first multiyear comparison of a sizeable number of models. This extensive evaluation allows for a careful analysis of prediction performances not subject to a single specific season or model implementation. A multiyear perspective is particularly important because no two flu seasons look alike due to constant changes in the genetic makeup of circulating strains, combined with fluctuations in population immunity from natural infection and vaccination.
Reich et al. (13) consider a portfolio of statistical and mechanistic models and a range of datasets beyond the primary incidence targets, including digital surveillance and social media such as Google queries and Twitter. The study thus provides a broad perspective on most of the current approaches to influenza time series forecasting and is highly relevant to other infectious diseases. The study shows that the top-performing models in the statistical and mechanistic categories achieve similar performances. The study also stresses the importance of data quality, particularly because weekly flu data are regularly retroactively adjusted, even in nonpandemic seasons. The issue of retroactive data adjustment is even more pervasive during large-scale health emergencies such as the West African Ebola epidemic (6).
Reich et al. (13) present a thorough comparison of prediction skills across seasons and US regions, shedding light on how forecasting performances vary with geography. Some regions appear more difficult to predict than others (e.g., a null model based on historical averages does poorly, indicating large interannual variability in ILI incidences). In contrast, other regions are easier to predict due to greater stability in observed historical patterns and substantial improvement of predictive models over historical averages. Moving forward, it will be important to understand whether regional differences in predictive skills are a reporting artifact or whether they reflect heterogeneities in influenza transmission dynamics. Demographic and environmental differences among regions, connectivity, and spatial extent could all affect predictive skills. This question could have practical implications because regions displaying consistently high predictive power could be used as sentinels for influenza surveillance.
In line with other infectious disease challenges (10, 12), the study by Reich et al. (13) shows that statistical approaches perform particularly well, especially given the short time horizon. It is important to keep in mind that existing prediction targets rely on noisy epidemiological data that are proxies of disease activity (here, weekly ILI incidence), rather than the true accumulation of patients with influenza virus infection, which is unobservable directly. ILI is a sensible prediction target because it is used by health authorities to set epidemic alerts and gauge the impact of interventions and because it is monitored globally (16). As a result, however, all forecasting approaches are optimized to forecast a composite signal, which includes reporting biases, healthcare-seeking behavior, incompleteness, and the contribution of nonflu pathogens. It is thus perhaps not surprising that statistical models can perform as well as they currently do. Conceivably, alternative prediction targets closer to the true disease process may result in more skilled predictions, especially for mechanistic models and over longer time horizons. Reliance on composite signals like ILI may also explain why models that contain key features of influenza biology and epidemiology, such as information on population immunity, viral evolution, or vaccine coverage, have not outperformed other approaches so far.
Ultimately, one would hope that mechanistic models could be used to their full capacity and provide valuable information to policy makers on key disease parameters (e.g., reproduction number, residual immunity) and the benefits of interventions, which is generally not accessible to statistical approaches solely based on time series analysis. The mechanistic models considered so far often ignore population heterogeneities, such as age and contact patterns, that might improve forecasting skills in the future. It is also useful to recall that weather forecasting went through a similar transition 60 y ago, from primarily statistical time series predictions to mechanistic models numerically solving the hydrodynamic and thermodynamic equations ruling atmospheric evolution. This transition took a careful understanding of the performances of different models, optimization of the spatial scales most appropriate for different components of the forecasts, and improvement in the quality and quantity of weather observations available to calibrate models (17, 18).
The study by Reich et al. (13) points at an overall degradation of disease forecasts at the 4-wk time horizon, beyond which the average of historical incidences (i.e., a null model) becomes the most reliable option for prediction. From a public health standpoint, a longer horizon, in the order of 2 mo or more, would be particularly useful to ramp up interventions and adjust hospital surge capacity. Ideally, even longer timescales should be considered so that the prediction of epidemic intensity (epidemic size) and severity (total number of hospitalizations and deaths) aligns with the vaccine manufacturing process. In other words, in the future, the influenza forecasting community will need to offer weather forecasts as well as climate predictions.
To see substantial progress in infectious disease forecasts to the level of other forecasting disciplines, it will be crucial to achieve a principled and theoretically informed understanding of the predictive skills of different models, the inherent limits to prediction horizon, and the minimum quantity and quality of data needed (Box 1). For instance, for a wide range of diseases, the inclusion of longer time series and a statistical signal does not seem to improve predictions, but rather deteriorates them (19). Additional case studies of empirical outbreaks featuring different transmission modes and measurement processes would be particularly informative (Box 1). We believe that synthetic challenges (12), for which the epidemiological data are generated in full or in part by a model in which the ground truth of transmission and measurement processes are fully known (20), are important tools to test the inherent limits of infectious disease forecasts. Synthetic challenges could also be particularly useful to prepare for pandemic and emerging infectious disease threats, which are more complicated by lack of historical data and coordinated modeling efforts of the kind presented by Reich et al. (13).
In conclusion, the study by Reich et al. (13) is an exemplary and much-needed study that should serve as a template for future forecasting work. The multiyear CDC seasonal influenza challenge has been remarkably successful in maintaining momentum for a coordinated and large-scale network of modeling teams focused on operationalization of disease forecasting, while strengthening the link with decision making. This coordinated effort has been useful beyond the immediate output of seasonal flu forecasts; as a case in point, several teams involved in the flu challenge were also involved in forecasts of emerging threats such as Zika and Ebola. Expanding this type of long-term coordinated effort to emerging infections in low- and middle-income regions with less-robust surveillance data and a greater need to optimize interventions will be an important next frontier.
Acknowledgments
This article does not necessarily represent the views of the NIH or the US government.
Footnotes
The authors declare no conflict of interest.
See companion article on page 3146.
References
- 1.Althouse BM, et al. Enhancing disease surveillance with novel data streams: Challenges and opportunities. EPJ Data Sci. 2015;4:17. doi: 10.1140/epjds/s13688-015-0054-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bansal S, Chowell G, Simonsen L, Vespignani A, Viboud C. Big data for infectious disease surveillance and modeling. J Infect Dis. 2016;214:S375–S379. doi: 10.1093/infdis/jiw400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ginsberg J, et al. Detecting influenza epidemics using search engine query data. Nature. 2009;457:1012–1014. doi: 10.1038/nature07634. [DOI] [PubMed] [Google Scholar]
- 4.Shaman J, Karspeck A. Forecasting seasonal outbreaks of influenza. Proc Natl Acad Sci USA. 2012;109:20425–20430. doi: 10.1073/pnas.1208772109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tizzoni M, et al. Real-time numerical forecast of global epidemic spreading: Case study of 2009 A/H1N1pdm. BMC Med. 2012;10:165. doi: 10.1186/1741-7015-10-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chretien JP, Riley S, George DB. Mathematical modeling of the West Africa Ebola epidemic. eLife. 2015;4:e09186. doi: 10.7554/eLife.09186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pitzer VE, et al. Direct and indirect effects of rotavirus vaccination: Comparing predictions from transmission dynamic models. PLoS One. 2012;7:e42320. doi: 10.1371/journal.pone.0042320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flasche S, et al. The long-term safety, public health impact, and cost-effectiveness of routine vaccination with a recombinant, live-attenuated dengue vaccine (Dengvaxia): A model comparison study. PLoS Med. 2016;13:e1002181. doi: 10.1371/journal.pmed.1002181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eaton JW, et al. HIV treatment as prevention: Systematic comparison of mathematical models of the potential impact of antiretroviral therapy on HIV incidence in South Africa. PLoS Med. 2012;9:e1001245. doi: 10.1371/journal.pmed.1001245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Valle SY, et al. Summary results of the 2014-2015 DARPA Chikungunya challenge. BMC Infect Dis. 2018;18:245. doi: 10.1186/s12879-018-3124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biggerstaff M, et al. Influenza Forecasting Contest Working Group Results from the Centers for Disease Control and Prevention’s Predict the 2013-2014 Influenza Season Challenge. BMC Infect Dis. 2016;16:357. doi: 10.1186/s12879-016-1669-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Viboud C, et al. RAPIDD Ebola Forecasting Challenge group The RAPIDD ebola forecasting challenge: Synthesis and lessons learnt. Epidemics. 2018;22:13–21. doi: 10.1016/j.epidem.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reich NG,etal. A collaborative multiyear, multimodel assessment of seasonal influenza forecasting in the United States. Proc Natl Acad Sci USA. 2019;116:3146–3154. doi: 10.1073/pnas.1812594116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Centers for Disease Control and Prevention 2019 FluSight forecasting. Available at https://www.cdc.gov/flu/weekly/flusight/index.html. Accessed January 20, 2019.
- 15.Biggerstaff M, et al. Results from the second year of a collaborative effort to forecast influenza seasons in the United States. Epidemics. 2018;24:26–33. doi: 10.1016/j.epidem.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.World Health Organization 2019 FluID—A global influenza epidemiological data sharing platform. Available at https://www.who.int/influenza/surveillance_monitoring/fluid/en/. Accessed January 20, 2019.
- 17.Lynch P. The origins of computer weather prediction and climate modeling. J Comput Phys. 2008;227:3431–3444. [Google Scholar]
- 18.Bauer P, Thorpe A, Brunet G. The quiet revolution of numerical weather prediction. Nature. 2015;525:47–55. doi: 10.1038/nature14956. [DOI] [PubMed] [Google Scholar]
- 19.Scarpino SV, Petri G. 2019. On the predictability of infectious disease outbreaks. Nat Commun, 10.1038/s41467-019-08616-0. [DOI] [PMC free article] [PubMed]
- 20.Ajelli M, et al. The RAPIDD Ebola forecasting challenge: Model description and synthetic data generation. Epidemics. 2018;22:3–12. doi: 10.1016/j.epidem.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]