Significance
HuR is a pivotal player in binding mRNAs containing AU-rich elements and regulating their stability and decay. HuR embeds three RNA recognition motifs (RRMs). The function of RRM3 is not completely understood, and the structure of the entire Hu protein family is so far unknown. Here, we provide structural and mechanistic insights into how HuR RRM3 discriminates between U-rich and AU-rich targets. RRM3 uses additional mechanisms, like multiple-register binding and homodimerization, to fine-tune its affinity for RNA. These results highlight the multifunctional role of HuR RRM3 but also the subtle adaptability of RRMs, the most abundant RNA-binding domain in eukaryotes. Since elevated HuR levels are associated with disease, our structure may help develop new therapeutic strategies.
Keywords: NMR spectroscopy, crystal structure, RNA-binding protein, dimerization, multiple register
Abstract
Human antigen R (HuR) is a key regulator of cellular mRNAs containing adenylate/uridylate–rich elements (AU-rich elements; AREs). These are a major class of cis elements within 3′ untranslated regions, targeting these mRNAs for rapid degradation. HuR contains three RNA recognition motifs (RRMs): a tandem RRM1 and 2, followed by a flexible linker and a C-terminal RRM3. While RRM1 and 2 are structurally characterized, little is known about RRM3. Here we present a 1.9-Å-resolution crystal structure of RRM3 bound to different ARE motifs. This structure together with biophysical methods and cell-culture assays revealed the mechanism of RRM3 ARE recognition and dimerization. While multiple RNA motifs can be bound, recognition of the canonical AUUUA pentameric motif is possible by binding to two registers. Additionally, RRM3 forms homodimers to increase its RNA binding affinity. Finally, although HuR stabilizes ARE-containing RNAs, we found that RRM3 counteracts this effect, as shown in a cell-based ARE reporter assay and by qPCR with native HuR mRNA targets containing multiple AUUUA motifs, possibly by competing with RRM12.
Adenylate/uridylate–rich elements (AU-rich elements; AREs) are regulatory cis-acting elements within untranslated regions (UTRs) of short-lived mRNAs which function as a signal for rapid degradation. AREs are present in 5 to 8% of human genes involved in the regulation of many important cellular processes such as the stress response, cell-cycle regulation, inflammation, immune cell activation, apoptosis, and carcinogenesis (1), and must therefore be tightly regulated. There are three classes of AREs. Class I contains several AUUUA motifs dispersed over the 3′ UTR; class II contains multiple, overlapping copies of AUUUA; and class III is U-rich (2). Multiple RNA-binding proteins (RBPs) regulate the transport, stability, and translation of ARE-containing mRNAs. An important class of such RBPs is the embryonic lethal abnormal visual-like/human antigen (ELAVL/Hu) protein family (3). In mammals, the Hu family is composed of four highly conserved members: HuB/Hel-N1, HuC, and HuD are expressed in neurons, whereas HuR [also known as ELAV-like protein 1 (ELAVL1) or HuA] is ubiquitously expressed (3, 4). HuR positively regulates the stability and translation of ARE-containing targets (5). It is also known to destabilize a small number of mRNAs and/or to suppress their translation (6, 7).
cDNA arrays and photoactivatable-ribonucleoside–enhanced crosslinking and immunoprecipitation (PAR-CLIP) identified a rather U-rich than AU-rich motif as a HuR-binding sequence within thousands of targets (8–10). Binding sites consist of U-rich stretches, separated by adenosines or other bases (8, 11, 12). HuR is predominantly localized in the nucleus but undergoes cytoplasmic translocation under various cellular and stress conditions, where it stabilizes its targets and promotes translation (4, 13). Cellular localization of HuR as well as HuR–RNA binding are in addition regulated by various posttranslational modifications, including phosphorylation, methylation, and caspase-mediated cleavage during apoptosis (14). Mice lacking HuR show higher rates of embryonic lethality (15). Conditional HuR knockout animals revealed the essential role of HuR in organ development and tissue homeostasis (16). Elevated HuR expression levels are associated with altered physiological functions, such as promoting viral infections, neurological pathologies, and muscular disorders. Predominantly, HuR overexpression increases up-regulation of cancer-associated transcripts, correlating with tumor growth and various cancer types (e.g., breast, colon, prostate, etc.) (17). Moreover, HuR is involved in the stabilization of transcripts encoding drug-resistant proteins (18). Consequently, HuR emerges as a potential biomarker and therapeutic drug target, and deciphering its molecular function and details of its RNA target recognition will be directly relevant for informed progression.
HuR is composed of three highly conserved canonical RNA recognition motifs (RRMs). These domains adopt a βαββαβ topology, where a four-stranded antiparallel β-sheet is packed against two α-helices (19). The tandem RRM1 and 2 (RRM12) is separated from the C-terminal RRM (RRM3) by an ∼50-residue basic hinge region bearing the nucleocytoplasmic shuttling element (20). RRM12 is suggested to be critical for ARE binding by HuR (21), whereas the exact function of RRM3 is not fully understood. RRM3 was predicted to bind poly-A tails of mRNAs, as shown for HuC and HuD (22, 23). In addition, RRM3 and the hinge region have been identified to be involved in protein–protein interactions (24), dimerization (25), and multimerization in cancer cells (26). Involvement of RRM3 in RNA binding and mRNA stability is controversial (4, 21). Gel-based assays and fluorescence anisotropy measurements demonstrated that the hinge region and RRM3 play a role in cooperative binding of multiple HuR molecules to one long ARE RNA (27). The hinge region as well as RRM3 are important for counteracting miRNA-mediated repression and promoting miRNA-induced silencing complex release from target mRNAs (28). Moreover, RRM3 of HuB/Hel-N1 binds the ARE from the c-myc 3′ UTR and negatively affects HuB/Hel-N1 multimerization along the 3′ UTR of c-myc (11, 29). Multiple structural investigations of RRM12 of HuR and its family members provide information about its RNA recognition (30–32). Structural studies of the hinge region and the RRM3 domain within the whole Hu family have proven to be challenging, due to insolubility in recombinant expression systems and instability of the domains in vitro. Recently, it was shown by NMR that dimerization of RRM3 is mediated through helix 1, which is located opposite the RNA binding interface and that RRM3 prefers 5′-UUUUU-3′ over 5′-AUUUA-3′ motifs (25). Due to the low RRM3 protein solubility, structure determination by NMR has remained challenging. Thus, models of the free RRM3 and of two potential RRM3 dimer conformations were described. The models are based on chemical shifts using the CS23D server (33) and molecular dynamics (MD) simulations using the HuR RRM1 crystallographic dimer as a template, respectively. Thus, the lack of an atomic-scale analysis of the free and RNA-bound forms prevents a complete understanding of HuR RRM3 function. In this context, a mechanistic description of HuR RRM3 dimerization, RRM3–RNA recognition, and the interplay between both mechanisms is needed.
To understand the function of HuR RRM3 in ARE regulation, we determined a 1.9-Å crystal structure of an RRM3 homodimer in complex with short ARE motifs derived from the c-fos 3′ UTR. This revealed molecular insights into RNA binding by HuR RRM3 as well as RRM3 dimerization. In addition, we show the importance of RRM3–RNA binding and dimerization for ARE mRNA regulation by combining biophysical measurements in solution and functional studies using mutant HuR expression constructs in cell-based ARE reporter assays and qPCR analysis.
Results
Both HuR RRM12 as Well as RRM3 Bind ARE Motifs.
To understand the molecular basis of HuR RRM3 in gene regulation, we analyzed the RNA interactions of HuR RRM3 (residues 241 to 326; Fig. 1A) using NMR chemical shift mapping and isothermal titration calorimetry (ITC). Titrations with the c-fos ARE 11-mer (5′-AUUUUUAUUUU-3′; Fig. 1B) and 15N-labeled RRM3 or 15N-labeled RRM12 revealed chemical shift variations mostly in an intermediate exchange regime for RRM3 and slow-to-intermediate exchange regime for RRM12 on the NMR timescale (SI Appendix, Fig. S1 A and B). This is compatible with a low micromolar affinity of this RNA motif. Binding affinity measurements by ITC of the c-fos ARE 11-mer for RRM3 and RRM12 confirmed the NMR results with Kd values of 0.65 and 0.14 μM, respectively (Fig. 1 C and D). The affinity of RRM12 is consistent with the previously reported Kd of 0.20 μM (30). Interestingly, for RRM3, an ITC N value of 0.5 suggests that two RRM3 molecules bind one c-fos ARE 11-mer. RRM3 was also predicted to be involved in the binding of poly-A tails of mRNAs, as shown for HuC and HuD (22, 23), and to possess an ATP-binding pocket. This is composed of a metal ion-coordinating DxD motif, which mediates 3′-terminal adenosyl modification of nonpolyadenylated RNA substrates (34). 1H-15N HSQC titrations of 15N-labeled RRM3 with EDTA, MgCl2, or ATP showed no significant chemical shift perturbations (SI Appendix, Fig. S1C), indicating no binding to RRM3 under these conditions. Titrating an 11-nt poly-A RNA revealed smaller chemical shift perturbations at higher protein-to-RNA ratios (SI Appendix, Fig. S1D) compared with the c-fos ARE 11-mer, indicating a weaker binding to poly-A than to the c-fos 11-mer.
Fig. 1.
All three HuR RRM domains are involved in ARE binding. (A) Schematic representation of HuR domain organization, with the RRM12 and RRM3 construct lengths indicated. The RRM3 amino acid sequence including the secondary structure is highlighted. (B) Schematic representation of the c-fos mRNA with the ARE 11-mer used in this study. (C and D) ITC binding curves of RRM12 (C) or RRM3 (D) with the c-fos ARE 11-mer. Errors indicate SD of two (RRM12) or three (RRM3) measurements.
These findings imply that RRM3 preferentially binds A/U–containing RNAs and that both RRM12 as well as RRM3 are able to bind ARE motifs in a similar affinity range. This suggests that RRM3 might play a more relevant role in direct interaction with ARE-containing mRNAs than previously expected.
Crystal Structure Shows Molecular Insights into RRM3 ARE Recognition.
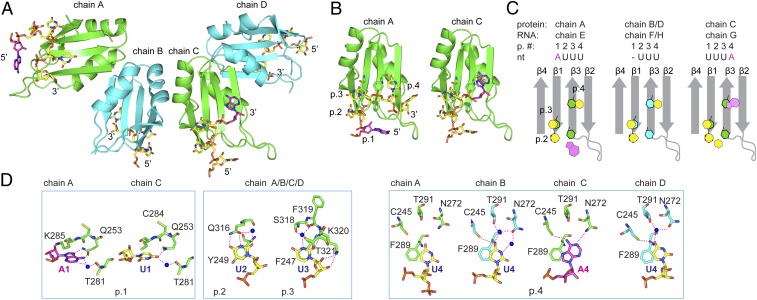
To further understand the structural basis for ARE recognition by RRM3, we crystallized RRM3 in complex with the c-fos ARE 11-mer (35). Crystals diffracted up to 1.9-Å resolution, and the crystal structure was determined by molecular replacement using the structure of HuR RRM1 [Protein Data Bank (PDB) ID code 3HI9] (Fig. 2 and SI Appendix, Fig. S2 and Table S1). The crystals belong to the C 121 space group and contain four proteins and four RNA molecules per asymmetric unit (Fig. 2A). We refer to each RRM in the asymmetric unit with the chain labels A to D, and each RNA molecule with the chain labels E to H (Fig. 2C). Each RRM adopts the expected βαββαβ fold, where four antiparallel β-strands are packed against two α-helices (Fig. 2 A and B). All four RRM molecules in the asymmetric unit adopt the same conformation, except that some electron density is missing for the β2-β3 loop for chains B and D (SI Appendix, Fig. S2 A and B). However, the RNA electron densities of chains E to H, to which we refer binding pockets according to their position, reveal variable numbers of bound nucleotides (Fig. 2C and SI Appendix, Fig. S2A). Out of the 11 nt of the c-fos ARE 11-mer, electron density for the RNA is visible for only 3, 4, or 5 nt bound to the RRM. Our crystal structure shows binding of RRM3 to pure single-stranded U-rich motifs (chains F and H), as previously suggested (25), as well as binding to AUUU and UUUA (chains E and G) (Fig. 2). These sequences are found in the c-fos ARE 11-mer. The remaining unbound nucleotides of the 11-mer are either too flexible to be observed or might have been degraded during the crystallization process. The electron density found in all four RRM3–RNA structures clearly indicates that only uracils can be bound in pockets 2 and 3. For the nucleotides in chains E and G in pockets 1 and 4, the electron density could not perfectly accommodate a uracil or an adenine (SI Appendix, Fig. S2A), which is explained by the occupancy of both adenines and uridines in these pockets.
Fig. 2.
Crystal structure of HuR RRM3 in complex with RNA shows homodimers and canonical RNA recognition. (A) Cartoon representation of the four RRM–RNA molecules (RRM chains A to D and RNA chains E to H) in one asymmetric unit. Chains A:B (green:cyan) and C:D (green:cyan) represent the same dimer interface. RNA is shown in stick representation, with carbons of uridines in yellow and carbons of adenines in magenta. (B) RRM3 of chains A and C. RNP side chains and RNA are shown in stick representation. Binding pockets are abbreviated as “p.” (C) A schematic representation of the RNP arrangement is colored in gray, uracils and adenines in yellow and magenta, respectively, and aromatic residues of the RNP in green (chains A/C) or cyan (chains B/D). Protein and RNA chains, pocket number (p. #), and nucleotides are shown (Top). (D) Protein–RNA contacts are shown as sticks for each chain. Chains are indicated (Top). Binding pockets are indicated (Bottom). (D, Left and Right) Interactions distinct to each chain in pockets 1 and 4. (D, Middle) Pocket 2 and 3 interactions representative of all four chains. Hydrogen bonds are shown as red dashed lines.
For our final structure (Fig. 2A), adenines were placed in pocket 1 of chain E and in pocket 4 of chain G, considering that in pocket 1 of chain G and in pocket 4 of chains E, F, and H we have already placed uracils. See Materials and Methods for a detailed description of the final structure determination. Overall, the structure revealed that RRM3 has a binding preference for uracil in pockets 2 and 3 and for both uracil and adenine in pockets 1 and 4. Another difference was observed by overlaying all four RNA chains (SI Appendix, Fig. S2B). Chain E shows the presence of an additional fifth pocket, containing U5, which does not exhibit any stacking contacts or hydrogen bonds to the protein. U5 is pulled away from the RRM due to interactions with a symmetry-related molecule (SI Appendix, Fig. S2D). Such a conformation is unlikely to be present in solution and therefore not biologically relevant. Three or four RNA nucleotides are bound on the surface of the RRM3 β-sheet using the conserved nucleic acid-binding ribonucleoprotein motifs RNP1, RNP2, and amino acids within the β2-β3 and β1-α1 loops (Fig. 2 B and C). In pockets 2 and 3, U2 and U3 form stacking interactions with RNP2 Y249 and F247 (both β1), respectively. In addition, U2 is sequence-specifically recognized by forming two hydrogen bonds with the Q316 side chain (β4), while U3 forms multiple hydrogen bonds. One is with the main-chain amide of T321, one occurs with the T321 side chain, and two are formed by water molecules bridging S318 and U3. Finally, the K320 side chain forms two hydrogen-bond contacts with the 2′ and 3′ oxygens of U3 (Fig. 2D) and the phosphate oxygen of U4/A4. Unlike pockets 2 and 3, which consistently bind uracils, pockets 1 and 4 are able to accommodate both adenine and uracil. A1/U1 contacts are mainly mediated by hydrophobic interactions with the aliphatic side chain of K285 (Fig. 2D). However, adenine at this position exhibits stronger stacking interactions due to its larger aromatic ring. Interestingly, HuR RRM1 (PDB ID code 4ED5) and HuR RRM3 show similar interactions with an adenine in this pocket (SI Appendix, Fig. S2C). Stacking with L61 in RRM1 is equivalent to K285 in RRM3, and Q29 in RRM1, which forms two sequence-specific hydrogen bonds with the adenine, is conserved in RRM3 as Q253. In addition, a water mediates interactions between A1/U1 and T281. A uracil at this position is only able to form one hydrogen bond with Q253. Thus, the structure clearly explains how both A and U can be accommodated in pocket 1. The uridine in pocket 4 reveals imperfect stacking with F289 (β3) but also potential water-mediated hydrogen bonds with N272 (β2) and T291 (β3) in chains B and D (Fig. 2D). Remarkably, the adenine at the same position in chain C still stacks imperfectly with F289 but is able to form two direct hydrogen bonds with N272 (β2) and C245 (β1) (Fig. 2D).
The observation of both A and U stably bound in pockets 1 and 4 within the different RRM molecules suggests multiple RNA binding modes relative to the bound sequences (AUUU, UUU, UUUA). This led us to propose (A/U)UU(A/U) as the recognition sequence for RRM3.
Molecular Dynamics Simulations Show Large-Scale Dynamics of Adenine and Uracil Binding in Pockets 1 and 4.
Considering the degenerate specificity revealed by the structure of HuR RRM3 bound to RNA, we performed MD simulations to evaluate the stability of the four binding pockets. Nucleotides in pockets 1 and 4 (which can be both uracil and adenine) were highly dynamic (SI Appendix, Table S2), forming fluctuating interactions with the protein via both direct and water-mediated hydrogen bonds (SI Appendix, Fig. S3). The simulations revealed fluctuating protein–RNA interactions also for the uracils in pockets 2 and 3, although the range and duration of the fluctuations were shorter and the average lifetimes of the interactions were significantly longer than for pockets 1 and 4 (SI Appendix, Fig. S3 and Table S3), indicating a more stable binding. We suggest the higher dynamics of the nucleotides bound in pockets 1 and 4 is due to the existence of multiple binding modes, which are separated by negligible kinetic barriers in the free-energy landscape. During the frequent transitions between these binding modes, the nucleotides can also extensively interact with the solvent, which further increases the range of their dynamics.
In all simulations, a nucleotide located at binding pocket 1 was continuously fluctuating, with the notable exception of an adenine base adopting a syn conformation. When uracil is bound in this pocket, mainly two different binding modes were observed. In one, the protein–RNA hydrogen bonds were formed with the K285, K313, and Q253 side chains. In another, a water was mediating an interaction between the U1 imino and the carbonyl oxygen of C284 (SI Appendix, Fig. S2E). When an adenine was bound in pocket 1, the nucleotide interaction with the protein was dependent on the orientation of the base. A base in the anti conformation was highly dynamic, comparable to the uracil (SI Appendix, Table S2). The anti-adenine formed interactions with the Q253 side chain and a water-mediated interaction with the C284 carbonyl oxygen. Occasional water-mediated interaction with the carbonyl of L251 was also observed. Interestingly, when adenine was present in a syn conformation, it was more stably bound, with hydrogen bonds formed with the L251, C284, and K313 main chains. Occasionally, the direct hydrogen bond with C284 became temporarily water-mediated. Lastly, an interaction between the K285 carbonyl oxygen and the 2′ hydroxyl group of both uracil and adenine nucleotides was observed in all simulations (SI Appendix, Fig. S2E). We then checked if we could observe a syn conformation in solution by NMR, which would be obvious with a strong H8-to-H1′ NOE. In the NOESY spectra of RRM3 in complex with AUUAUU, we could detect a syn conformation (SI Appendix, Fig. S2G). However, when we tried to build a syn-adenine conformation in the electron density, it differed from the syn-adenine seen in MD, and its proposed protein–RNA interactions were unstable in MD simulations. Thus, during refinement, the syn-adenine flipped immediately to the anti or the syn conformation as seen in the MD; however, this syn conformation did not fit into the electron density. Therefore, an anti-adenine was placed in pocket 1 in our final crystal structure. In binding pocket 4, depending on whether uracil or adenine was present, different protein residues would become involved in binding. Specifically, the simulations showed the adenine forming a greater number of direct hydrogen bonds with the protein than the uracil. However, additional water-mediated interactions around the uracil were compensating for this difference, resulting in similar dynamics. When simulations were performed with an adenine at this pocket, direct hydrogen bonds were formed with the side chains of both N272 and C245, as described previously, but also with the side chains of K320 and K274 (SI Appendix, Fig. S2F). However, the fairly large dynamics in binding pocket 4 caused these interactions to constantly fluctuate in simulations, and they would rarely all coexist at the same time (SI Appendix, Fig. S3). For instance, the K320 side chain would sometimes interact with the RNA sugar–phosphate backbone between binding pockets 3 and 4 instead of with A4 N7. The K274 and N272 side chains would sometimes temporarily flip away, forming other protein–protein interactions or interacting with the solvent. Lastly, the entire adenine base flipped temporarily away numerous times in the course of simulations, breaking even the stacking interaction with F289. However, in most cases, the base did not drift too far from its initial position during these short flips, resulting in slightly lesser overall dynamics and a quicker restoration of its native interactions compared with bases in pocket 1 (SI Appendix, Fig. S3 and Table S3). Simulations with uracil bound at this pocket showed only the K320 and K274 forming direct hydrogen-bond interactions, supplemented by water-mediated hydrogen bonds to the backbone of N272. Since the K320 would sometimes also interact with the RNA sugar–phosphate backbone instead of the base, the uracil at this position was often directly recognized only by the K274 (SI Appendix, Fig. S2E).
In conclusion, the MD simulations showed a highly dynamic RNA recognition and hydrogen-bond network in pockets 1 and 4. This is in perfect agreement with our crystal structure, where a degenerate specificity is detected for these pockets. Furthermore, MD simulations revealed that adenine in pocket 1 is able to bind in both anti and syn conformations, and that a syn-adenine would be better stabilized by several direct interactions with the protein.
Solution Studies of HuR RRM3–ARE Recognition.
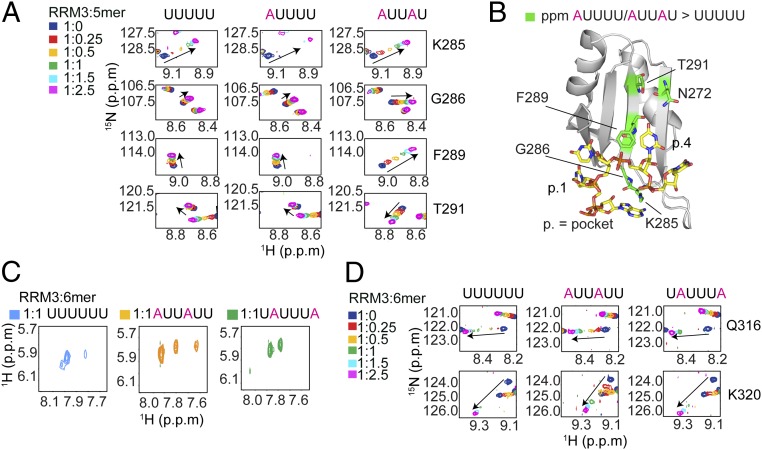
To investigate further the (A/U)UU(A/U) recognition sequence suggested by the crystal structure, we studied the binding of RRM3 to several 5- and 6-nt-long U-rich RNA molecules using NMR titrations (Fig. 3) and ITC (SI Appendix, Table S4). RRM3 binding to 5′-AUUUU-3′ or 5′-AUUAU-3′ led to different chemical shift perturbations compared with binding to 5′-UUUUU-3′ (Fig. 3A and SI Appendix, Figs. S4 and S5). K285 as well as G286 show larger 1H-15N perturbations upon binding to 5′-AUUUU-3′, whereas G288, F289, and T291 resonances exhibit larger perturbations for 5′-AUUAU-3′ (Fig. 3 A and B). All these residues are located in pocket 1 or pocket 4 (Fig. 3B), in agreement with the crystal structure and MD simulations, where both pockets can accommodate an adenine. Placing the A in the middle, as in 5′-UUAUU-3′, led to a similar chemical shift pattern as 5′-AUUAU-3′, indicating the preference for an A in pocket 4. An A at the end, as in 5′-UUUUA-3′, shows the least differences compared with 5′-UUUUU-3′ (SI Appendix, Fig. S4). With ITC titrations, we obtained Kds for 5′-UUUUUU-3′, 5′-AUUUUU-3′, 5′-UUUAUU-3′, and 5′-AUUAUU-3′ of 3.58 ± 0.21, 4.03 ± 0.25, 5.60 ± 0.40, and 5.75 ± 0.27 μM, respectively (SI Appendix, Fig. S5D and Table S4). The slight increase in affinity for the first two oligonucleotides could be explained by the fact that RRM3 can bind these oligonucleotides at more registers than in the last two, as previously seen for several other RRMs (36, 37). To validate this hypothesis, we recorded 1H-1H TOCSY experiments of RRM3 in complex with various 5- and 6-mer oligonucleotides (Fig. 3C and SI Appendix, Fig. S5B). In the 5′-UUUUU-3′–RRM3 TOCSY spectrum, five H5–H6 cross-peaks would be expected (one for each uridine) if the RRM binds a single register, but only one sharp and one broad correlation are observable (SI Appendix, Fig. S5B). Broadening or absence of a cross-peak results from chemical exchange that could be due to multiple-register binding. In the 5′-AUUUU-3′–RRM3 complex, exchange is still present, as only three peaks are visible, whereas three sharp signals are observable for the complex with 5′-AUUAU-3′, indicating a single binding register. The same observation can be seen for the 6-mer oligonucleotides (Fig. 3C). Only 5′-AUUAUU-3′ shows the four expected signals (two being overlapped), indicative of a single register.
Fig. 3.
RRM3 recognizes various motifs in solution and binds multiple registers. (A) Close-up view of the 1H-15N HSQC spectrum for key residues showing different chemical shift perturbations for AUUUU or AUUAU compared with UUUUU [RRM3:RNA ratios: 1:0 (blue), 1:0.25 (red), 1:0.5 (yellow), 1:1 (green), 1:1.5 (cyan), and 1:2.5 (magenta)]. (B) Residues with chemical shift differences larger for AUUUU and AUUAU than for UUUUU are represented as sticks and highlighted in green on chain A. (C) Comparison of H5–H6 correlations in 2D TOCSY spectra at 750 MHz of RRM3 in complex with various 6-mer RNAs (blue, 5′-UUUUUU-3′; orange, 5′-AUUAUU-3′; green, 5′-UAUUUA-3′). (D) Close-up view of the 1H-15N HSQC spectrum for key residues showing a different exchange regime for AUUAUU compared with UUUUU or UAUUUA. RRM3:RNA ratios: 1:0 (blue), 1:0.25 (red), 1:0.5 (yellow), 1:1 (green), 1:1.5 (cyan), and 1:2.5 (magenta).
Overall, these solution studies confirm the (A/U)UU(A/U) consensus sequence seen in the crystal structure.
Binding to the Prototypical AUUUA ARE.
The 5′-UAUUUA-3′ sequence, which is the prototypical element enriched in class I and II AREs, does not fit the consensus for RRM3 since three uracils separate the two adenines. However, if one would consider binding to AUUUA, we would predict binding to two registers, AUUU and UUUA, in exchange. In such a binding mode, the affinity would increase compared with a single binding mode (AUUA). In addition, the protein would be bound at a specific locus, compared with a long poly-U sequence, where the multiple registers would lead to a constant movement along the sequence (SI Appendix, Fig. S5C). We therefore investigated RRM3 binding to 5′-UAUUUA-3′ by NMR and ITC. TOCSY revealed only two cross-peaks, confirming the exchange between the two registers AUUU and UUUA (Fig. 3C). In addition, the Kd measured by ITC is between the poly-U– and AUUA-containing oligo (SI Appendix, Fig. S5D and Table S4), and the NMR titration experiments reveal a different exchange regime for AUUAUU compared to UUUUUU and UAUUUA for several residues (Fig. 3D), indicating a weaker binding of AUUAUU.
This unusual dynamic mode of recognition which is composed of sequence-specific binding registers in exchange could explain why AUUUA pentamers are found in many mRNAs regulated by HuR.
The Crystal Structure Reveals a Dimerization Interface in RRM3.
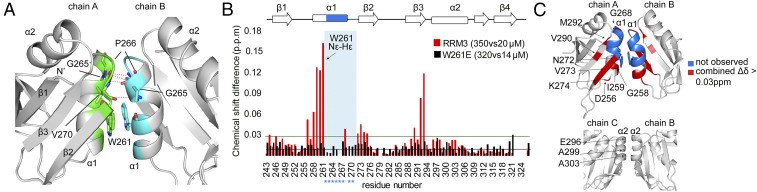
Two potential dimerization interfaces are visible within one asymmetric unit (Fig. 2A): one between chains A and B, mediated by several hydrogen bonds and hydrophobic interactions, and a second one, mainly hydrophobic, formed between chains B and C (Fig. 4A). Interestingly, the dimer between chains A and B is stabilized by stacking interactions of a conserved tryptophan side chain, W261 (Fig. 4A and SI Appendix, Fig. S6A). The surface area buried within this dimer is ∼347 Å, calculated with PISA (38). The involvement of the W261 residue in dimerization was reported previously by NMR and analytical ultracentrifugation studies (25). A model generated in this study showed two potential orientations of the domains with respect to each other but did not provide an in-depth molecular mechanism of dimerization. Our crystal structure clearly highlights the involvement of the conserved interface in dimerization. Interactions are mainly mediated by hydrophobic residues within α1. In particular, the aromatic rings of W261 are stacking between the two monomers. In addition, backbone atoms of G265/P266 and V270 of the first monomer form hydrogen bonds with V270 and G265/P266 of the second monomer, respectively (Fig. 4A). The dimer between chains B and C, mediated by α2, does not occur in solution (Fig. 4B).
Fig. 4.
RRM3 dimerizes in solution via the conserved Trp261. (A) Cartoon representation of the RRM3 homodimer interface between RRM chains A and B. Amino acids involved in hydrophobic interactions are colored green (chain A) and cyan (chain B). The conserved W261 involved in stacking and other amino acids involved in hydrogen bonds are shown in stick representation. Hydrogen bonds are represented as dashed lines. (B) Comparison of the combined chemical shift perturbations of RRM3 and W261E, 350 vs. 20 μM (red) and 320 vs. 14 μM (black), respectively. Blue asterisks indicate amino acids located within dimerization helix 1 of which signals are missing in the 1H-15N HSQC of RRM3. The W261 side chain Nε-Hε was also included in the analysis (indicated by the arrow). The green and black horizontal lines represent the SD of all chemical shift differences for RRM3 and W261E, respectively. (C) Cartoon representation of the RRM3 homodimer (Top) (chains A and B) and the crystallographic dimer (chains B and C) (Bottom). Residues missing in the 1H-15N HSQC of the RRM3 spectrum are highlighted in blue, and residues showing ∆δ > 0.03 ppm are in red.
RRM3 Dimerizes in Solution via the Conserved W261 and upon Increasing Concentrations.
To characterize the mechanism of dimerization in detail, we performed titrations by NMR and ITC. 1H-15N HSQC spectra of RRM3 were recorded over a range of concentrations, showing chemical shift perturbations for several residues with increasing protein concentration (Fig. 4B and SI Appendix, Fig. S6 B and C). This observation suggests that increasing RRM3 concentration shifts the equilibrium toward RRM3 dimerization. The chemical shift differences are mainly localized within α1 and β2, consistent with the location of the hydrophobic dimerization interface observed in the crystal structure, but changes are additionally seen in the residues of the RNP1 motif within β3 (Fig. 4 B and C), suggesting that dimerization and RNA binding could be allosterically coupled. In addition, residues 260 to 270 were missing in the 1H-15N HSQC spectra, possibly due to exchange broadening of the resonances caused by the equilibrium between monomer and dimer. Mutating the conserved W261 to E261 abolished the concentration-dependent chemical shift changes of RRM3 (Fig. 4B and SI Appendix, Fig. S6 B and C). A comparison of the 1H-15N HSQC spectra of the wild type and the W261E mutant showed a similar pattern, indicating that the mutant preserved the fold of RRM3. Consistent with this, no significant differences in 13Cα chemical shift deviations from random coil values were observed when comparing wild-type RRM3 and W261E, indicating that the secondary structure is unchanged by the mutation (SI Appendix, Fig. S6D). Strikingly, the resonances of residues 260 to 270 could be detected in the W261E mutant, consistent with the absence of dimer–monomer exchange. To directly probe the oligomerization state, we carried out longitudinal (T1) and transverse (T2) NMR relaxation experiments at various protein concentrations and estimated the overall rotational correlation time τc of RRM3, from which the approximate molecular mass can be deduced (SI Appendix, Fig. S6E). τc was calculated from the ratio of 15N T1 and T2 relaxation times of each residue. Indeed, at high concentrations of about 300 μM, we observed a mean τc of 13.59 ± 0.76 ns, which corresponds to an ∼22.65 ± 1.27-kDa dimer, assuming a spherical particle at 298 K (39). In contrast, W261E has a mean τc of 7.31 ± 0.45 ns under the same conditions, which corresponds to a monomer with a molecular mass of 12.18 ± 0.75 kDa. Interestingly, the wild-type RRM3 showed a reduction of τc to 9.05 ± 0.94 ns (∼15.08 ± 1.57 kDa) at 80 μM, indicating that even at this concentration RRM3 is still in equilibrium between monomer and dimer. T1 and T2 NMR relaxation experiments of HuR RRM12 resulted in a mean τc of 12.80 ± 0.92 ns for residues 20 to 185 (SI Appendix, Fig. S6E). Residues 2 to 19 are flexible, and were not included in the τc determination. This correlation time, which is close to the value obtained for RRM3 at high concentration, suggests that RRM1 and RRM2 tumble as a single particle. This indicates that the RRM12 particles are mainly in the monomeric state under our reducing conditions. At the beginning of our studies, we identified that ITC titrations of RRM3 into RNA showed two events, one endothermic and one exothermic, the latter event representing the RNA binding. The endothermic event was explained by the presence of dimers at high concentrations, and diluting them during the titration process induces dissociation into monomers.
We then performed ITC experiments to quantify the dissociation constant for dimer–monomer equilibrium of wild-type RRM3. A highly concentrated RRM3 sample was injected into buffer. Measurements revealed a dimerization constant of 31.7 ± 0.47 μM (SI Appendix, Fig. S6F), which is twofold lower than the Kddimer obtained by analytical ultracentrifugation (25). The difference can be explained by different experimental conditions, such as buffers, especially the salt dependence for dimer formation, described previously (25).
Together, our data suggest that RRM3 indeed forms dimers in solution at higher concentrations, with a Kddimer of 31.7 ± 0.47 μM. Mutation of the conserved W261 abolishes the dimerization. Furthermore, dimerization of free RRM3 shows chemical shift changes on the RNA binding surface, suggesting the possibility of an interplay between dimerization and RNA binding.
Mutation of RRM3 W261 Affects RNA Binding.
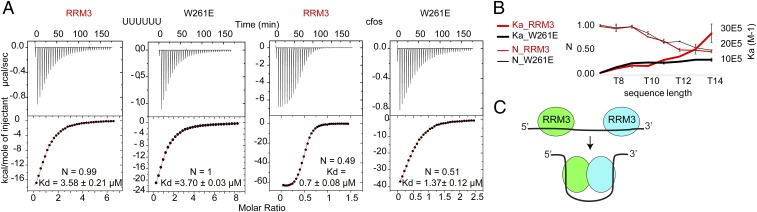
Previous studies reported that both the Drosophila ELAV protein and HuR dimerize through short amino acid patterns within RRM3, which are conserved in the Hu family, including the tryptophan within α1 (3, 25, 40). Drosophila ELAV–ELAV interaction requires the presence of RNA (40). HuD–HuD interaction is reduced after RNase treatment (41). HuR RRM3 is required for cooperative assembly of HuR oligomers on RNA (27). All these findings lead to the possibility that dimerization might affect RNA binding affinity. The c-fos ARE 11-mer sequence, which was used for crystallization, is significantly longer than the usual RNA length bound by one RRM [2 to 8 nt per RRM (42)]. In addition, our ITC experiments showed a 1:2 RNA:protein stoichiometry (Fig. 1D). Therefore, we investigated if the c-fos ARE 11-mer could be bound by a dimer or two separated RRMs. Looking at our crystal structure, the distance between the U5 of chain E and the U2 of chain F is 42 Å. This is too long to accommodate the 3-nt spacer between two chains if one RRM would bind the first 5 nt and the second one would bind the last 3 nt of the c-fos ARE (with an extended single-stranded RNA, the distance between two phosphate atoms is around 7 Å). However, due to the crystal packing, the orientation of the monomers within the crystal structure might slightly differ from a dimer in solution. In addition, a dimer in solution might be more dynamic and flexible to rearrange upon RNA binding. To test whether RNA binding has an effect on the dimerization of RRM3, we performed ITC experiments with the W261E mutant and the 11-mer c-fos ARE motif. Interestingly, the affinity is decreased twofold for the mutant compared with the RRM3 WT (Kd of 1.4 μM compared with 0.65 μM for WT) (Fig. 5A, Right). ITC experiments with the shorter 5′-UUUUUU-3′ motif, on the other hand, showed no change in affinity between the two proteins (Fig. 5A, Left). This demonstrates that the ability to dimerize increases the RNA binding affinity. We performed further ITC experiments with the two proteins and DNA analogs of increasing size (from 6 to 21 nt). Similar to U6, T6 showed no difference in Kd for RRM3 or W261E (SI Appendix, Table S5). With a length equivalent to c-fos (T11), RRM3 and W261E show no difference in Kd. This indicates that the sequence, and not only the nucleotide length, is a factor affecting dimerization. Only at a length of 12 nt or above can an increase in affinity be detected for the RRM3 WT compared with the W261E mutant RRM. This coincides with the N value decreasing to 0.5 (indicating a 1:2 stoichiometry of RNA:protein) for the WT, while for the mutant the N value decreases only to 0.68 when binding T12 (SI Appendix, Fig. S5B and Table S5). For T14 or for T6C10T6, the affinity of the RRM3 WT is almost three times higher than for the mutant. The Kd for this last DNA sequence of 0.26 μM approaches the value measured for RRM12 and c-fos (Fig. 1C), suggesting that an RRM3 dimer and RRM12 might have a comparable affinity for AREs. NMR titrations confirmed qualitatively the data obtained by ITC. Binding of c-fos by the WT RRM3 showed a fast-to-intermediate exchange regime, while binding to short oligos for both proteins or for the mutant W261E to c-fos shows mostly a fast exchange regime (SI Appendix, Fig. S6G).
Fig. 5.
Dimerization and RNA binding of RRM3 directly affect each other. (A) Comparison of ITC profiles of RRM3 or W261E titrated with UUUUUU or c-fos. Errors indicate the SD of at least two measurements. (B) Ka in M−1 (thick line, Right axis) and N value (thin line, Left axis) measured by ITC for RRM3 (red) and W261E (black). Errors indicate the SD of two or three measurements. (C) Model of RNA binding-induced dimerization. Binding to long RNAs, comprising two motifs separated by a long linker, induces dimerization.
Overall, we show evidence that the ability of RRM3 to dimerize increases RNA binding affinity for RNAs above a length of 11 or 12 nt. At the length of 11 or 12 nt, as for c-fos, the different exchange regime and the affinity increase could originate from an increase in local concentration of the RRMs when one of the RRMs is bound to the RNA. The dimer might not persist, since the RNA is too short to span the two RRM binding surfaces. However, when the RNA is longer (over 14 nt), the dimerization can be maintained and the two RRM surfaces can be bound by the same RNA (Fig. 5C). The apparent gain of affinity is rather small (threefold maximum), which is in agreement with the high RRM3 dimerization Kd of 31.7 μM, indicating a weak affinity to dimerize.
Dimerization and RNA Binding of RRM3 Regulate an AUUUA Repeat-Containing Reporter in a Destabilizing Manner in Living Cells.
The high dimerization Kd of 31.7 ± 0.2 μM and the weaker affinity of RRM3 for RNA compared with RRM12 raised the question of whether these interactions are functionally important in vivo. To address this, we compared the efficiency of the wild-type HuR and HuR mutants defective in either RRM3 dimerization, RRM3 RNA binding, or both in expression regulation of an ARE-containing reporter mRNA (Fig. 6). Wild-type or mutant Flag-HuR expression constructs as well as a dual-luciferase reporter bearing the first 60 nt of the COX-2 3′ UTR in the 3′ UTR of the Renilla coding sequence (CDS) (Fig. 6 A and B) were cotransfected in Huh7 or C3H/10T1/2 cells (multipotent cells isolated from C3H mouse embryos). Renilla luciferase measurements were normalized to the firefly luciferase signal within each sample. We observed a significant increase in luciferase activity and hence translation up-regulation when transiently overexpressing HuR (Fig. 6 C and D) compared with the empty control vector, similar to previous reports for different ARE-containing mRNAs (e.g., ref. 4). The dimerization-inactive mutant (W261E) caused a statistically significant increase in luciferase activity in Huh7 and C3H/10T1/2 cells (Fig. 6C). This is surprising, since one would have rather expected that inhibition of HuR dimerization would lower HuR’s affinity for the ARE motif and therefore have reduced ability to stabilize COX-2 ARE reporter mRNA. The increase in luciferase activity was further seen both in the presence of endogenous HuR (Fig. 6C, Right) as well as after siRNA knockdown of endogenous HuR in C3H/10T1/2 cells (SI Appendix, Fig. S7C). Interestingly, the luciferase activity in Huh7 cells was also elevated when overexpressing HuR with the RRM3-containing RNP mutations F247A to Y249A (abbreviated FY; Fig. 6C, Left). The RRM3 FY mutant is still folded but defective in RRM3–RNA binding (SI Appendix, Fig. S7A). The triple-HuR W261E FY mutant, deficient in both dimerization and RNA binding, showed no changes in Renilla up-regulation compared with the RRM3–RNA binding-inactive HuR FY mutant (Fig. 6C). This suggests that in Huh7 cells, RRM3 dimerization depends on RNA binding. If RRM3 cannot bind to the RNA, dimerization plays a minimal role or cannot even take place. The effect of HuR FY seems to be cell line-dependent, as the same experiments in C3H/10T1/2 cells showed a slightly decreased effect of HuR FY (Fig. 6D, Right).
Fig. 6.
Dimerization and RNA binding of HuR RRM3 negatively contribute to RNA target regulation in cells. (A) Schematic description of the pSICHECK-2 dual-luciferase reporter, containing the first 60 nt of the COX-2 3′ UTR, fused to Renilla luciferase (RL). (B) Schematic description of HuR and the position of the W261E, FY, and RR mutations. (C) Dual-luciferase reporter assay for evaluating the effect of HuR mutants on up-regulation of the Cox-2 ARE reporter. Proteins were coexpressed with the Cox-2 1–60 luciferase reporter in Huh7 cells (Left) or C3H/10T1/2 cells in the presence of Scr-RNA (Right). Firefly luciferase (FL) was used for normalization. (D) Dual-luciferase reporter assay for evaluating the effect of HuR mutants on different reporter constructs. Proteins were coexpressed with either the empty luciferase reporter (red; hyphen), luciferase reporter harboring Cox-2 1–60 (blue), or Cox-2 1–60 AtoUmut, where all adenines are mutated to uracils (green) (SI Appendix, Fig. S7B) in Huh7 cells. (E) mRNA levels of TNFα were quantified by quantitative reverse transcription–PCR using the RPLP0 housekeeping gene as a normalization control. Fold changes in mRNA expression were normalized according to the ΔΔCt method. For all assays, the mean values ± SD from at least three independent experiments are shown. P values were determined by the Student’s t test. *P < 0.05, **P < 0.001.
The destabilizing effect mediated by RRM3 is not observed anymore when using a luciferase reporter where all adenines of the COX-2 3′ UTR are mutated to uridines (abbreviated COX-2 AtoUmut; SI Appendix, Fig. S7B), in agreement with a sequence-specific recognition of AUUUA. Similar to the COX-2 AtoUmut, an empty luciferase reporter without the ARE sequence showed a less than 1.5-fold increase in luciferase activity upon HuR overexpression compared with the luciferase reporter with the COX-2 ARE (2.5-fold), indicating unspecific binding of HuR to the psiCHECK-2 vector. This can be explained by the prolonged HuR overexpression, leading to secondary effects on the cell RNA metabolism. Here, RRM3 mutants also show no effect on this empty luciferase reporter (Fig. 6D).
To test if the increase in luciferase activity was RRM12-dependent, we introduced mutations into RRM12 of HuR to reduce the RNA binding affinity. Two mutations, R97A and R136A (abbreviated RR; Fig. 6B), which were shown in a previous study to decrease RNA binding without impacting the fold and stability of the tandem RRM12 (30), were additionally introduced into our HuR FY double or HuR FY W261E triple mutant. Other key RNA-binding residues were also mutated in that study; however, the secondary structures of these mutants were changed (30). Therefore, additional mutations to abolish RRM12 RNA binding were not tested. All HuR RR mutants showed as expected a significant decrease in luciferase activity compared with the HuR wild type (SI Appendix, Fig. S7D). The elevated luciferase activity of the dimerization-inactive HuR W261E and the HuR FY, which is incapable of RNA binding, was abolished when including the RR mutations (SI Appendix, Fig. S7D). This indicates that the effect seen for RRM3 dimerization and RNA binding depends on RNA binding by RRM12 of HuR. HuR knockdown as well as expression levels of all mutants were characterized by Western blotting (SI Appendix, Fig. S7 E–G). These data suggest that the mRNA stabilization induced by HuR RRM12 RNA binding can be counteracted by RRM3 RNA binding and dimerization. These results could be rationalized by a competition between RRM3 and HuR RRM12 or other AU-rich binding proteins for binding to the AUUUA repeats in the COX-2 ARE sequence (SI Appendix, Fig. S8A).
To test whether the stabilizing effect of HuR W261E in the luciferase assay is due to increased mRNA stabilization, we performed quantitative PCR of known HuR targets in Huh7 and in C3H/10T1/2 cells. In Huh7 cells, we could not detect any amplification of Cox-2 mRNA due to a low Cox-2 cDNA abundance after reverse transcription (four different primer pairs were tested). Various different mRNAs were tested in Huh7 cells, with representative examples shown in SI Appendix, Fig. S7H; however, stabilization by HuR was not detected due to the missing stress signal to induce HuR translocation to the cytoplasm (5). However, we found that TNFα mRNA is up-regulated by HuR even without a stimulation signal. Here, HuR W261E shows a significantly higher increase in TNFα mRNA levels compared with the HuR wild type (Fig. 6E). Remarkably, TNFα mRNA contains an AUUUA triple motif, a classical type II ARE, like Cox-2 (SI Appendix, Fig. S7J). As for Huh7 cells, in unstressed C3H/10T1/2 cells, no changes are observable. However, upon tamoxifen treatment, various mRNAs were significantly increased upon HuR overexpression (SI Appendix, Fig. S7I). Interestingly, HuR W261E overexpression showed different effects for various mRNAs. Autotaxin mRNA levels are unaffected. c-fos even showed a trend toward lower mRNA levels. Both mRNAs are lacking the triple-AUUUA motif found in Cox-2 and TNF mRNA (SI Appendix, Fig. S7J). These data are in agreement with our luciferase assay using a non-AUUUA sequence. Cox-2, on the contrary, showed a trend toward an increase in mRNA levels upon HuR W261E overexpression, supporting the qPCR data in Huh7 cells. Thus, the qPCR analysis of two natural HuR targets, at least for type II AREs, validates the destabilizing effect of HuR RRM3 observed in the luciferase assay.
Overall, these data suggest that both the dimerization and the AUUUA sequence-specific binding of RRM3 do not positively contribute to the activity of HuR in ARE up-regulation. Rather, RRM3 might autoregulate the protein function by limiting productive complex formation with mRNA harboring AUUUA motifs in their AREs, while not affecting those containing only U-rich sequences.
Discussion
Our study shows that HuR RRM3 binds a c-fos ARE 11-mer RNA with a Kd in the low micromolar range in vitro (Fig. 1D). This demonstrates that all three HuR RRM domains are able to bind AREs, indicating that RRM3 might have a potential contribution in direct interaction with mRNA targets. RRM3 is also able to bind poly-A, but with a much lower affinity (SI Appendix, Fig. S1D). This does not support a primary role of RRM3 in binding to poly-A tails of mRNAs (22, 23). Binding studies with other ligands, such as EDTA, MgCl2, or ATP, showed no significant chemical shift perturbations (SI Appendix, Fig. S1C), indicating no binding of these ligands to RRM3. However, we cannot exclude that in the context of the full-length protein as well as in the presence of cellular cofactors a direct interaction of RRM3 with the poly-A tail or other factors might be possible.
While structural investigation of the full-length HuR or its relatives HuB, HuC, and HuD remains challenging due to their poor stability and solubility, a study of the separate RRMs enables us to obtain functional insights. The crystal structures of the free HuR RRM12 and HuR RRM12 in complex with an 11-nt ARE from c-fos reveal a conformational change upon RNA binding, a preference for pyrimidine-rich sequences, and a low degree of sequence specificity (30). HuR RRM3 was previously reported to bind poly-U (25). In addition, cDNA arrays and PAR-CLIP experiments identified a U-rich rather than an AU-rich motif for HuR binding in thousands of targets (9, 10). While building our model of RRM3 in complex with c-fos AREs and taking into account this information, we realized that placing uracils in some electron density pockets proved challenging, and we soon realized that pockets 1 and 4 showed occupancy for both adenine and uracil. Combining NMR, ITC, and MD simulations, we could validate that RRM3 can recognize sequence-specifically both nucleotides in these pockets (Fig. 3 and SI Appendix, Figs. S2 and S3). RRM3 can therefore recognize multiple sequences, UUUUU, AUUUU, UUUAU, AUUUA, or AUUAU, with comparable affinities but experiences a slight increase in affinity for U-rich sequences due to multiple-register binding or avidity (SI Appendix, Fig. S3 and Table S4). A similar lack of sequence specificity was reported for RRM12 of HuR and HuD (30, 31). This could enable Hu proteins to bind motifs from all three classes of AREs, and explains how this versatile protein family can regulate thousands of targets found in pre-mRNAs, mature mRNAs, miRNAs, and long noncoding RNAs (9, 10, 43–45).
When encountering poly-U sequences, RRM3 binding might be highly dynamic, with a constant binding and unbinding along U-rich sequences within 3′ UTRs. We found that the possibility to accommodate adenines in pockets 1 and 4 reduces or even prevents multiple-register binding when bound to AUUA, for example. This results in RRM3 being locked and localized to a precise position within the 3′ UTRs. However, loss of multiple registers leads also to a decrease in affinity, making the RRM3 more prone to be displaced by other AU-rich binding proteins. When binding the AUUUA pentamer motif, which is enriched in class I and II AREs, we predict two binding registers in exchange, AUUU and UUUA, on the basis of our structure. Our data confirmed the register change and the increase in affinity compared with AUUAU. In binding AUUUA motifs, RRM3 localizes at these motifs and still binds with substantial affinity due to the two-register binding. Thus, we provide an original hypothesis on why AUUUA is a prototypical motif in AREs and how RRM3 recognizes it (SI Appendix, Fig. S5C). This could be a tuning mechanism for HuR to find its target sequences on 3′ UTRs. This is reminiscent of what we previously observed for the poly-U–binding protein heterogeneous nuclear ribonucleoprotein C (hnRNP C) in terms of structure and affinity enhancement (36).
In addition to new insights into HuR RRM3–ARE recognition, our structure provides an additional example of the various strategies by RRM-containing proteins to regulate gene expression. HuR RRM3 is a multifunctional domain containing an RNA binding platform but also a protein interaction surface to further tune its functions. Concentration-dependent dimerization of RRM3 is mediated primarily by W261 within helix α1, which is conserved in all Hu proteins, suggesting that an analogous dimerization mechanism via RRM3 is likely to exist for HuB, HuC, and HuD. Considering the high dimerization Kd of 30 μM measured for RRM3 in vitro (SI Appendix, Fig. S6F), we questioned whether RRM3 of HuR would be able to dimerize under physiological concentrations. It should be considered, though, that in our in vitro studies, characterization of dimerization was performed in the context of the isolated RRM3 domain, while the full-length HuR dimerization affinity might be different due to the additional hinge region. Furthermore, we could show that the capacity of RRM3 to dimerize increases the RNA binding affinity to oligonucleotide sequences above 12 nt in length. In turn, RNA binding also affects HuR dimerization. AREs are mostly 30- to 100-nt-long and typically in the context of 3′ UTRs harboring additional HuR binding sites scattered across the sequence. Upon oligomerization of HuR along RNA sequences, as reported before (27), the local HuR concentration is increased, which would certainly shift the equilibrium toward dimerization. Under certain stress signals, HuR is highly accumulated in stress granules (46). In cancerous cells or tumors, HuR concentration is also elevated and was recently shown to form multimers in glioma cells (26). Given the micromolar affinity of RRM3 dimers, such concentration changes could become relevant to directly affect RRM3-mediated HuR dimerization and thereby affect its function in ARE-based gene regulation.
Many RRM-containing proteins contain multiple RRMs to fine-tune their specificity and affinity. Tandem RRMs can interact with each other, sometimes involving their interdomain linkers, to create either extended RNA binding surfaces or deep clefts for interaction with RNA, as shown for HuD and HuR RRM12 (30, 31). Other RRMs interact through their helices opposite the RNA binding surface, leading to conformations inducing looping of bound RNAs. This is exemplified by the RRM34 of PTB or RRM12 of hnRNPA1 (47, 48). Interestingly, while PTB interacts with its RRM4 α2 and RRM3 α1, hnRNPA1 RRM1 uses its β4-strands to bind α2 of RRM2. Homodimerization via α1 as in HuR RRM3 results also in the β-sheet surfaces being in opposite directions. Dimer formation could therefore induce RNA looping and bring together distant motifs within one mRNA target, like in PTB, or it could bring close together different mRNAs, which could induce liquid–liquid phase transition and formation of cellular granules (49).
In fact, members of the RNA-binding protein with multiple splicing (RBPMS) family are also able to form homodimers, via residues in the first α-helix and adjacent loop regions (50). However, the dimer interface of the RBPMS family is formed by multiple salt bridges, additional hydrophobic contacts, and hydrogen bonds, resulting in a stable and permanent dimer (50–52), whereas HuR RRM3 is in an equilibrium between monomer and dimer. Interaction studies of RBPMS family members with an RNA containing two binding sites and a sufficiently long linker to overcome the distance led to a two- to ninefold increase in affinity compared with a short RNA containing a single binding site (52). This is in good agreement with our measurements under conditions where the HuR RRM3 is not quantitatively in the dimer state (Fig. 5 and SI Appendix, Table S5). In addition, dimerization mutations of RBPMS members result in 2.5- to 4-fold lower binding affinities to RNA comprising two binding sites (51), similar to what we measured for HuR RRM3 for binding c-fos (Fig. 5) or other long DNA sequences (SI Appendix, Table S5).
We could show quite unexpectedly that in Huh7 cells, overexpression of HuR mutants, which are inactive in RRM3 dimerization or RNA binding, shows a higher increase in translation of the reporter construct harboring multiple AUUUA motifs compared to HuR wild type (Fig. 6C, Left). Moreover, HuR W261E shows an increase in endogenous TNFα mRNA levels in Huh7 cells and a trend toward an increase of endogenous Cox-2 mRNA levels in C3H/10T1/2 cells (Fig. 6E and SI Appendix, Fig. S7I), validating the destabilizing effect of HuR RRM3. These findings may suggest that the ARE binding capability of HuR RRM3 might exert an autoinhibitory role by competing with HuR RRM12 for ARE binding and thereby limiting its role in up-regulation of the COX-2 ARE reporter. The COX-2 as well as the TNFα sequences contain multiple AUUUA motifs (type II AREs), which are also known targets of HuR RRM12, and mutating all adenines abolished the positive effect of the RRM3 mutants (Fig. 6D). This could be due to the locked confirmation of RRM3 on the two AUUUA registers versus a dynamic binding and unbinding on longer U-rich sequences. As both types of binding could lead to different effects, it is thus conceivable that mRNAs containing different classes of AREs could be differently fine-tuned by such an autoinhibitory function of RRM3 (Fig. 6 and SI Appendix, Fig. S7).
In addition, the negative effect of RRM3 could also be due to protein–protein interactions mediated by this domain via interaction with other proteins, possibly independent of ARE binding and/or dimerization. Interestingly, the HuR RRM3/KH-type splicing regulatory protein (KSRP) complex destabilizes nucleophosmin (NPM) mRNA to regulate muscle fiber formation (6). Depletion of HuR by siRNAs resulted in elevation of NPM mRNA. During muscle cell differentiation, HuR is cleaved into two products, HuR-CP1 (24 kDa) and HuR-CP2 (8 kDa), the latter containing the C-terminal RRM3. HuR-CP2 associates with KSRP and recruits two ribonucleases to induce destabilization of the NPM mRNA. The destabilizing effect observed in our assays could also act via a similar mechanism, the recruitment of ribonucleases via KSRP–RRM3 dimer binding (SI Appendix, Fig. S8B). In addition, RBPMS2, an RBPMS paralog which forms a stable dimer, is able to interact with the translation elongation factor eEF2. This interaction is abolished in the dimerization-inactive mutant (50). Similarly, RRM3 monomers could also influence binding to other proteins which play a role in translation (SI Appendix, Fig. S8B). Future studies are needed to address whether dimerization- and RNA binding-dependent RRM3 interactions with other proteins play a role in HuR ARE interaction to up- or down-regulate gene expression.
Since elevated HuR levels are associated with several chronic and deadly diseases, understanding its molecular mechanism is of high importance to develop new therapeutic strategies to target and down-regulate it. Our model of an autoinhibitory role of HuR RRM3 opens new perspectives on how to control HuR function by therapeutic agents.
Materials and Methods
Cloning and Site-Directed Mutagenesis.
The DNA regions coding for HuR or HuR RRM3 (UniProtKB Q15717) were used to generate His-tagged RRM3 (residues 241 to 326), mammalian expression constructs, and mutants by PCR and QuikChange site-directed mutagenesis (see SI Appendix for more details). Other constructs used are described in SI Appendix.
Expression and Purification of Recombinant Proteins.
Recombinant human 6-His-tagged GB1-fusion proteins (RRM3, RRM3 W261E, RRM3 FY) were overexpressed in BL21 (DE3) cells (Novagen) and purified by immobilized metal affinity chromatography (IMAC). The His tag was removed by TEV digestion. The protein was further purified by an additional IMAC step to remove the 6-His-GB1 tag, followed by size-exclusion chromatography. A detailed expression and purification protocol can be found in SI Appendix.
RNA and DNA Synthesis.
The c-fos ARE 11-mer (5′-AUUUUUAUUUU-3′) and the 11-nt A-rich RNA were provided by J. Hunziger, Novartis, and synthesized as described (53). Short single-stranded RNAs (5′-UUUUU-, -AUUUU-, -UUAUU-, -AUUAU-, -UUUUA-, -UUUUUU-, -AUUUUU-, -UUUAUU-, -AUUAUU-3′) were purchased from Thermo Fisher Scientific and deprotected as described in the manufacturer’s instructions, lyophilized, and dissolved in NMR or ITC buffer. Single-stranded DNAs were purchased from Microsynth.
Sample Preparation and Crystallization.
The protein was concentrated to 7 mg/mL in 20 mM Tris (pH 8), 100 mM NaCl, 10% (wt/vol) glycerol, 1 mM Tris(2-carboxyethyl)phosphine. The RRM3–RNA complex was prepared by adding 1.2 molar excess c-fos ARE RNA. Crystals were obtained by sitting drop vapor diffusion at 298 K, in which 200 nL protein–RNA complex was mixed with 200 nL precipitant and suspended over 80 μL precipitant. Crystals were obtained in a 2 M ammonium sulfate, 0.1 M Bis-Tris well (Index HT Screen; Hampton Research) and flash-frozen in liquid nitrogen (without cryoprotection).
Data Collection and Structure Determination.
The crystals were measured under an N2 cryostream at the PXII-X10SA beamline (equipped with PILATUS detector) of the Swiss Light Source synchrotron in Villigen, Switzerland. The wavelength of data collection was 1 Å. The data were processed, and the structure was solved and refined as described in SI Appendix.
NMR Spectroscopy.
All NMR spectroscopy measurements were done in 20 mM Na2HPO4 (pH 7), 100 mM NaCl, 1 mM DTT, 10% D2O at 298 K using Bruker AVIII-500 MHz, AVIII-600 MHz (equipped with cryoprobes), and AVIII-750 MHz. NMR data were processed with TopSpin 3.1 (Bruker), and the analysis was performed using SPARKY 3 (T. D. Goddard and D. G. Kneller, University of California, San Francisco). For more details, see SI Appendix.
Isothermal Titration Calorimetry.
ITC measurements were performed on a VP-ITC instrument (MicroCal) at 25 °C. Raw RNA titration data were integrated and analyzed according to a one–binding-site model provided in Origin 7.0 software. We used the simple dimer dissociation model to determine Kddimer and ΔHdimer. Reported errors correspond to the SD of at least two replicates. Sample preparation, buffer conditions, and experimental setup are described in SI Appendix.
Cell Culture, Luciferase Assay, qPCR, and Western Blot.
Huh7 hepatoma cells (a generous gift from Wilhelm Krek, ETH Zürich) were cultured in Dulbecco’s modified Eagle’s medium + GlutaMAX (Thermo Fisher Scientific) supplemented with 10% FBS, 1% penicillin/streptomycin at 37 °C and 5% CO2. All plasmids were transiently transfected with Lipofectamine 2000 (Invitrogen) as described by the manufacturer’s instructions. Cells were harvested after 48 h by adding passive lysis buffer (Promega). Renilla and firefly activities were measured using the Dual-Luciferase Assay Kit (Promega) as described in the manufacturer’s instructions. Renilla measurements were normalized to the firefly luciferase signal within each sample. Endogenous mRNA levels were quantified by qPCR. Western blot analysis and detailed qPCR procedure, reagents, and normalization, as well as the experiments in C3H/10T1/2 cells, are described in SI Appendix, Materials and Methods.
Molecular Dynamics Simulations.
The X-ray structure of the HuR RRM3 in complex with c-fos ARE was used as the starting structure of all MD simulations. Each simulated system contained only a single, monomeric copy of the complex. Different chains from the X-ray structure were utilized to obtain starting structures where either uracil or adenine was positioned in binding pockets 1 and 4, respectively. See SI Appendix for the detailed simulated systems, simulation length, and force fields.
Supplementary Material
Acknowledgments
We thank G. Stier and A. Geerlof [European Molecular Biology Laboratory (EMBL)] for providing the petGB1-1a plasmid, Prof. Dr. Wilhelm Krek (ETH Zürich) for the Huh7 hepatoma cells, and Dr. Jürg Hunziger (Novartis) for the c-fos ARE RNA. In addition, we thank the MX group at the Swiss Light Source Villigen for their excellent support in data collection, and F. F. Damberger (ETH Zürich) for help with the setup, analysis of the NMR experiments, and proofreading of the manuscript. This work was supported by EU FP7 ITN Project RNPnet (Contract 289007) and NCCR RNA and Disease. M.K. and J.Š. acknowledge support by the Czech Science Foundation (Grant P305/12/G034) and by Project LO1305 of the Ministry of Education, Youth and Sports of the Czech Republic under National Sustainability Programme II. This project is part of the dissertation of N.R.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6GC5).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808696116/-/DCSupplemental.
References
- 1.Bakheet T, Williams BRG, Khabar KSA. ARED 3.0: The large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–D114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benjamin D, Moroni C. mRNA stability and cancer: An emerging link? Expert Opin Biol Ther. 2007;7:1515–1529. doi: 10.1517/14712598.7.10.1515. [DOI] [PubMed] [Google Scholar]
- 3.Good PJ. A conserved family of elav-like genes in vertebrates. Proc Natl Acad Sci USA. 1995;92:4557–4561. doi: 10.1073/pnas.92.10.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, et al. Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis. Int J Mol Sci. 2013;14:10015–10041. doi: 10.3390/ijms140510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cammas A, et al. Destabilization of nucleophosmin mRNA by the HuR/KSRP complex is required for muscle fibre formation. Nat Commun. 2014;5:4190. doi: 10.1038/ncomms5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kullmann M, Göpfert U, Siewe B, Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev. 2002;16:3087–3099. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc Natl Acad Sci USA. 2000;97:14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.López de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lebedeva S, et al. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 11.Levine TD, Gao F, King PH, Andrews LG, Keene JD. Hel-N1: An autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol Cell Biol. 1993;13:3494–3504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meisner N-C, et al. mRNA openers and closers: Modulating AU-rich element-controlled mRNA stability by a molecular switch in mRNA secondary structure. ChemBioChem. 2004;5:1432–1447. doi: 10.1002/cbic.200400219. [DOI] [PubMed] [Google Scholar]
- 13.Gallouzi IE, et al. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc Natl Acad Sci USA. 2000;97:3073–3078. doi: 10.1073/pnas.97.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip Rev RNA. 2010;1:214–229. doi: 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katsanou V, et al. The RNA-binding protein Elavl1/HuR is essential for placental branching morphogenesis and embryonic development. Mol Cell Biol. 2009;29:2762–2776. doi: 10.1128/MCB.01393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zucal C, et al. Targeting the multifaceted HuR protein, benefits and caveats. Curr Drug Targets. 2015;16:499–515. doi: 10.2174/1389450116666150223163632. [DOI] [PubMed] [Google Scholar]
- 17.Srikantan S, Gorospe M. HuR function in disease. Front Biosci. 2012;17:189–205. doi: 10.2741/3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filippova N, et al. The RNA-binding protein HuR promotes glioma growth and treatment resistance. Mol Cancer Res. 2011;9:648–659. doi: 10.1158/1541-7786.MCR-10-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maris C, Dominguez C, Allain FH-T. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 20.Fan XC, Steitz JA. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc Natl Acad Sci USA. 1998;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C-YA, Xu N, Shyu A-B. Highly selective actions of HuR in antagonizing AU-rich element-mediated mRNA destabilization. Mol Cell Biol. 2002;22:7268–7278. doi: 10.1128/MCB.22.20.7268-7278.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma WJ, Chung S, Furneaux H. The Elav-like proteins bind to AU-rich elements and to the poly(A) tail of mRNA. Nucleic Acids Res. 1997;25:3564–3569. doi: 10.1093/nar/25.18.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abe R, Sakashita E, Yamamoto K, Sakamoto H. Two different RNA binding activities for the AU-rich element and the poly(A) sequence of the mouse neuronal protein mHuC. Nucleic Acids Res. 1996;24:4895–4901. doi: 10.1093/nar/24.24.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brennan CM, Gallouzi IE, Steitz JA. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J Cell Biol. 2000;151:1–14. doi: 10.1083/jcb.151.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheiba RM, et al. The C-terminal RNA binding motif of HuR is a multi-functional domain leading to HuR oligomerization and binding to U-rich RNA targets. RNA Biol. 2014;11:1250–1261. doi: 10.1080/15476286.2014.996069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Filippova N, et al. Hu antigen R (HuR) multimerization contributes to glioma disease progression. J Biol Chem. 2017;292:16999–17010. doi: 10.1074/jbc.M117.797878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fialcowitz-White EJ, et al. Specific protein domains mediate cooperative assembly of HuR oligomers on AU-rich mRNA-destabilizing sequences. J Biol Chem. 2007;282:20948–20959. doi: 10.1074/jbc.M701751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kundu P, Fabian MR, Sonenberg N, Bhattacharyya SN, Filipowicz W. HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res. 2012;40:5088–5100. doi: 10.1093/nar/gks148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao FB, Keene JD. Hel-N1/Hel-N2 proteins are bound to poly(A)+ mRNA in granular RNP structures and are implicated in neuronal differentiation. J Cell Sci. 1996;109:579–589. doi: 10.1242/jcs.109.3.579. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, et al. The structure of the ARE-binding domains of Hu antigen R (HuR) undergoes conformational changes during RNA binding. Acta Crystallogr D Biol Crystallogr. 2013;69:373–380. doi: 10.1107/S0907444912047828. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Tanaka Hall TM. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat Struct Biol. 2001;8:141–145. doi: 10.1038/84131. [DOI] [PubMed] [Google Scholar]
- 32.Inoue M, Muto Y, Sakamoto H, Yokoyama S. NMR studies on functional structures of the AU-rich element-binding domains of Hu antigen C. Nucleic Acids Res. 2000;28:1743–1750. doi: 10.1093/nar/28.8.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wishart DS, et al. CS23D: A web server for rapid protein structure generation using NMR chemical shifts and sequence data. Nucleic Acids Res. 2008;36:W496–W502. doi: 10.1093/nar/gkn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meisner N-C, et al. Terminal adenosyl transferase activity of posttranscriptional regulator HuR revealed by confocal on-bead screening. J Mol Biol. 2009;386:435–450. doi: 10.1016/j.jmb.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 35.Ripin N, et al. 2019 Molecular basis for AU-rich element recognition and dimerization by the HuR C-terminal RRM. Protein Data Bank. Available at https://www.rcsb.org/structure/6GC5. Deposited April 17, 2018.
- 36.Cieniková Z, Damberger FF, Hall J, Allain FHT, Maris C. Structural and mechanistic insights into poly(uridine) tract recognition by the hnRNP C RNA recognition motif. J Am Chem Soc. 2014;136:14536–14544. doi: 10.1021/ja507690d. [DOI] [PubMed] [Google Scholar]
- 37.Mackereth CD, et al. Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature. 2011;475:408–411. doi: 10.1038/nature10171. [DOI] [PubMed] [Google Scholar]
- 38.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 39.Rossi P, et al. A microscale protein NMR sample screening pipeline. J Biomol NMR. 2010;46:11–22. doi: 10.1007/s10858-009-9386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toba G, White K. The third RNA recognition motif of Drosophila ELAV protein has a role in multimerization. Nucleic Acids Res. 2008;36:1390–1399. doi: 10.1093/nar/gkm1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kasashima K, Sakashita E, Saito K, Sakamoto H. Complex formation of the neuron-specific ELAV-like Hu RNA-binding proteins. Nucleic Acids Res. 2002;30:4519–4526. doi: 10.1093/nar/gkf567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Auweter SD, Oberstrass FC, Allain FH-T. Sequence-specific binding of single-stranded RNA: Is there a code for recognition? Nucleic Acids Res. 2006;34:4943–4959. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukherjee N, et al. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mukherjee K, et al. Reversible HuR-microRNA binding controls extracellular export of miR-122 and augments stress response. EMBO Rep. 2016;17:1184–1203. doi: 10.15252/embr.201541930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Legnini I, Morlando M, Mangiavacchi A, Fatica A, Bozzoni I. A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol Cell. 2014;53:506–514. doi: 10.1016/j.molcel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gallouzi IE, Brennan CM, Steitz JA. Protein ligands mediate the CRM1-dependent export of HuR in response to heat shock. RNA. 2001;7:1348–1361. doi: 10.1017/s1355838201016089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oberstrass FC, et al. Structure of PTB bound to RNA: Specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 48.Beusch I, Barraud P, Moursy A, Cléry A, Allain FHT. Tandem hnRNP A1 RNA recognition motifs act in concert to repress the splicing of survival motor neuron exon 7. eLife. 2017;6:1–35. doi: 10.7554/eLife.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Protter DSW, Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;26:668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sagnol S, et al. Homodimerization of RBPMS2 through a new RRM-interaction motif is necessary to control smooth muscle plasticity. Nucleic Acids Res. 2014;42:10173–10184. doi: 10.1093/nar/gku692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teplova M, Farazi TA, Tuschl T, Patel DJ. Structural basis underlying CAC RNA recognition by the RRM domain of dimeric RNA-binding protein RBPMS. Q Rev Biophys. 2016;49:e1. doi: 10.1017/S0033583515000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soufari H, Mackereth CD. Conserved binding of GCAC motifs by MEC-8, couch potato, and the RBPMS protein family. RNA. 2017;23:308–316. doi: 10.1261/rna.059733.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Masliah G, et al. Structural basis of siRNA recognition by TRBP double-stranded RNA binding domains. EMBO J. 2018;37:e97089. doi: 10.15252/embj.201797089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.