Significance
Mammals repeatedly adapted to specialized diets, including plant-based diets for herbivores and meat- or insect-based diets for carnivores. Apart from consuming diets with different nutritional compositions, obligate herbivores and carnivores differ in other aspects, such as the time spent feeding, regularity of pancreatic juice secretion, exposure to toxic plant-derived compounds, and gut microbiome diversity. To better understand how diet-related changes evolved, we performed genome-wide screens for convergent gene losses that happened preferentially in herbivores or in carnivores. We discovered repeated losses of genes involved in fat digestion, pancreatic juice secretion, glucose homeostasis, appetite regulation, detoxification, and gut microbiome diversity. Our results reveal genomic changes associated with dietary specialization and illuminate metabolic and physiological changes in herbivorous and carnivorous mammals.
Keywords: convergent gene loss, herbivorous diet, carnivorous diet, metabolism, physiology
Abstract
The repeated evolution of dietary specialization represents a hallmark of mammalian ecology. To detect genomic changes that are associated with dietary adaptations, we performed a systematic screen for convergent gene losses associated with an obligate herbivorous or carnivorous diet in 31 placental mammals. For herbivores, our screen discovered the repeated loss of the triglyceride lipase inhibitor PNLIPRP1, suggesting enhanced triglyceride digestion efficiency. Furthermore, several herbivores lost the pancreatic exocytosis factor SYCN, providing an explanation for continuous pancreatic zymogen secretion in these species. For carnivores, we discovered the repeated loss of the hormone-receptor pair INSL5–RXFP4 that regulates appetite and glucose homeostasis, which likely relates to irregular feeding patterns and constant gluconeogenesis. Furthermore, reflecting the reduced need to metabolize plant-derived xenobiotics, several carnivores lost the xenobiotic receptors NR1I3 and NR1I2. Finally, the carnivore-associated loss of the gastrointestinal host defense gene NOX1 could be related to a reduced gut microbiome diversity. By revealing convergent gene losses associated with differences in dietary composition, feeding patterns, and gut microbiomes, our study contributes to understanding how similar dietary specializations evolved repeatedly in mammals.
Adaptations to different food sources resulted in repeated dietary specializations, which constitute a cornerstone of mammalian ecology. While the exact dietary composition is probably unique to every lineage, placental mammals can be classified on a broad scale into herbivores, omnivores, and carnivores. Dietary specialization is associated with a variety of traits. To digest plant material, herbivores often possess an enlarged gastrointestinal tract, which increases the retention time of food and facilitates fermentation by specialized bacterial communities in the gut (1). Dietary specialization is associated with differences in level and activity of gut enzymes and transporters (2). An herbivorous or carnivorous diet is also a major factor that influences gut microbiome composition and diversity (3).
Comparative genomics has started to shed light on the genomic basis of metabolic and physiological differences between herbivorous and carnivorous mammals. For example, function-altering amino acid changes and positive selection on digestive enzymes and lipid-binding proteins in carnivorous cetaceans and Felidae are likely related to their fat- and protein-rich diet (4, 5). Previous candidate gene studies further revealed associations between the inactivation (loss) of protein-coding genes and dietary specializations. For example, carnivores such as cetaceans and sea lions that typically swallow their prey whole, have lost many receptors for a variety of tastes (6, 7). Cetaceans have lost the pancreatic RNASE1 gene, which is likely related to a change from an ancestral herbivorous to a carnivorous diet in this lineage (4). Carnivores also exhibit contracted families of genes involved in starch and sucrose metabolism, and detoxification of plant-derived compounds (5). Whereas the insectivorous placental mammalian ancestor possessed five chitin-digesting genes, repeated losses of these genes occurred in mammals that have few invertebrates in their diet (8). While previous studies provided valuable insights into the association between gene loss and dietary specialization, they typically investigated selected candidate genes or gene families, or were taxonomically limited to specific mammalian lineages.
To systematically detect genomic changes that are associated with an obligate herbivorous or obligate carnivorous diet, we performed an unbiased screen for convergent gene losses that are associated with such dietary specializations in 31 placental mammals. Our screen discovered a number of previously unknown gene losses in herbivores and carnivores that illuminate differences related to dietary composition, feeding patterns, and gut microbiomes.
Results
Classifying Mammals into Independent Herbivore and Carnivore Lineages.
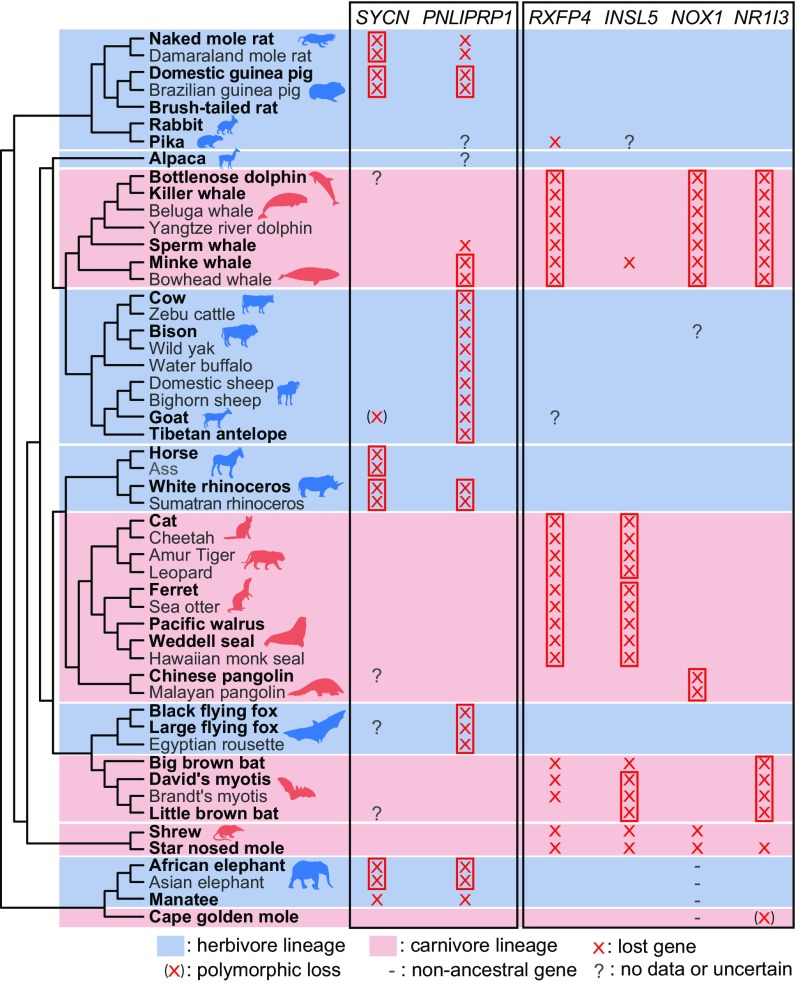
To identify convergent gene losses associated with dietary specialization into herbivory and carnivory, we classified placental mammals with a sequenced genome into 16 obligate herbivores and 15 obligate carnivores (Fig. 1 and Dataset S1). Omnivores were excluded from the analysis. Obligate insectivorous mammals were included into the carnivore group. Using a rather strict definition of herbivory and carnivory, we obtained six independent herbivore lineages and five independent carnivore lineages (Fig. 1).
Fig. 1.
Overview of convergent gene losses in herbivorous or carnivorous mammals. A strict herbivorous or carnivorous diet evolved several times independently in mammals. The six herbivore and five carnivore lineages are indicated by red and blue backgrounds, respectively. Species in bold font were included in the initial genome-wide screen; species in dark gray font were manually inspected for the presence of shared gene-inactivating mutations. The loss patterns of diet-related genes that are preferentially lost in either herbivores or carnivores are shown by red crosses. Gene losses that already occurred in the ancestor of related species, inferred from shared inactivating mutations, are indicated by red boxes. Animal silhouettes were downloaded from phylopic.org/ and are a courtesy of Steven Traver, David Orr, Oscar Sanisidro, Yan Wong, and Michael Keesey.
Systematically Identifying Convergent Gene Losses in Herbivores and Carnivores.
We conducted a systematic screen for protein-coding genes that are preferentially lost either in independent herbivore lineages or in independent carnivore lineages. To this end, we used gene-loss data generated by a computational approach that accurately detects mutations that inactivate protein-coding genes based on genome alignments (9, 10). Specifically, this approach screens for premature stop codons, splice site-disrupting mutations, frameshifting insertions and deletions, and the deletion of entire exons or even entire genes. All steps that we performed to identify and validate gene losses are shown in SI Appendix, Fig. S1.
To robustly detect convergent gene losses in herbivores, we required that a gene is lost in at least three independent herbivore lineages. In addition, we required that the gene exhibits an intact reading frame in at least 80% of all carnivores. An analogous approach was applied to identify convergent gene losses in carnivores. These screens identified 37 genes that are preferentially lost in herbivores and 44 genes that are preferentially lost in carnivores (Datasets S2 and S3).
Serving as positive controls, our screen detected the convergent carnivore loss of two taste receptor genes, TAS1R1 and TAS1R2, and the sour taste marker gene, PKD2L1, which have previously been linked to mammals with a carnivorous diet (6, 7). We also confirmed and extended previous findings that UGT1A6, a gene encoding a xenobiotic-metabolizing enzyme, is lost in cat, Weddell seal, and walrus (11, 12) (SI Appendix, Fig. S2 and Dataset S4). However, UGT1A6 is not lost in other independent carnivore lineages; hence, it did not meet our criterion of being lost in least three independent lineages. Apart from detecting a few known gene losses, the majority of the 37 and 44 convergent gene losses have not been described before.
Identifying Convergent Gene Losses Related to Dietary Specializations.
To explore which of the 37 herbivore- and 44 carnivore-associated genes have diet-related functions, we investigated whether these genes are enriched for certain gene ontology (GO) terms. Although we did not identify significant enrichments after correcting for multiple testing, the top-ranked GO terms indicate potentially diet-related associations (Datasets S5 and S6). For herbivore-associated gene losses, these GO terms include “exocytosis” [SYCN (syncollin), MIA3]. For carnivore-associated gene losses, top-ranked GO terms include “positive regulation of feeding behavior” [RXFP4 (relaxin family peptide/INSL5 receptor 4), INSL5 (insulin like 5)], “taste receptor activity” (TAS1R1, TAS1R2), and “arachidonic acid secretion” (PLA2G2C, PLA2G2A).
Importantly, not every gene that is preferentially lost in either herbivores or carnivores has an obvious diet-related function. This is exemplified by PLCZ1, the top-ranked gene identified in the herbivore screen (Dataset S2), which is involved in oocyte activation (13). Therefore, to evaluate which of the lost genes have diet-related functions, we performed a literature search for the two sets of top-ranked 20 genes. This search revealed six genes with functions that are relevant for dietary specialization: PNLIPRP1 (pancreatic lipase related protein 1) and SYCN, which have different functions in the pancreas and are preferentially lost in herbivores; and INSL5, RXFP4, NR1I3 (nuclear receptor subfamily 1 group I member 3), and NOX1 (NADPH oxidase 1), which are involved in food intake regulation, glucose homeostasis, xenobiotics detoxification, and innate immunity and are preferentially lost in carnivores (Fig. 1).
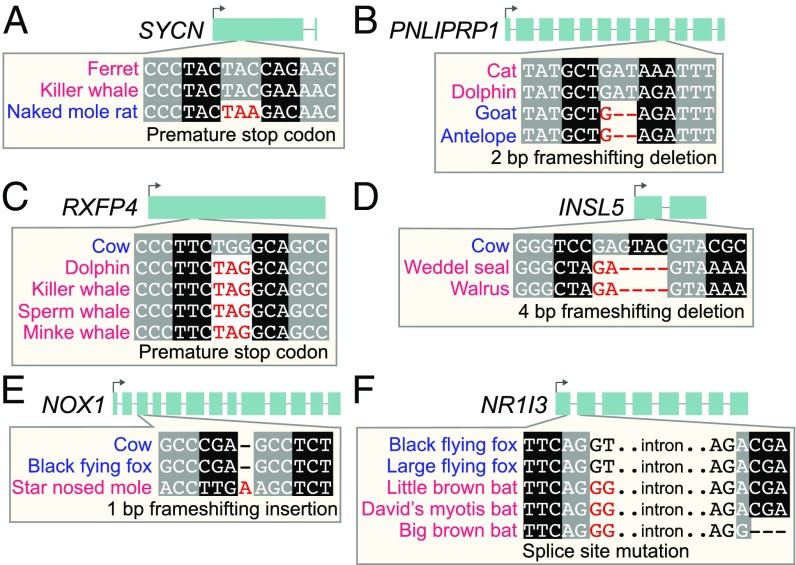
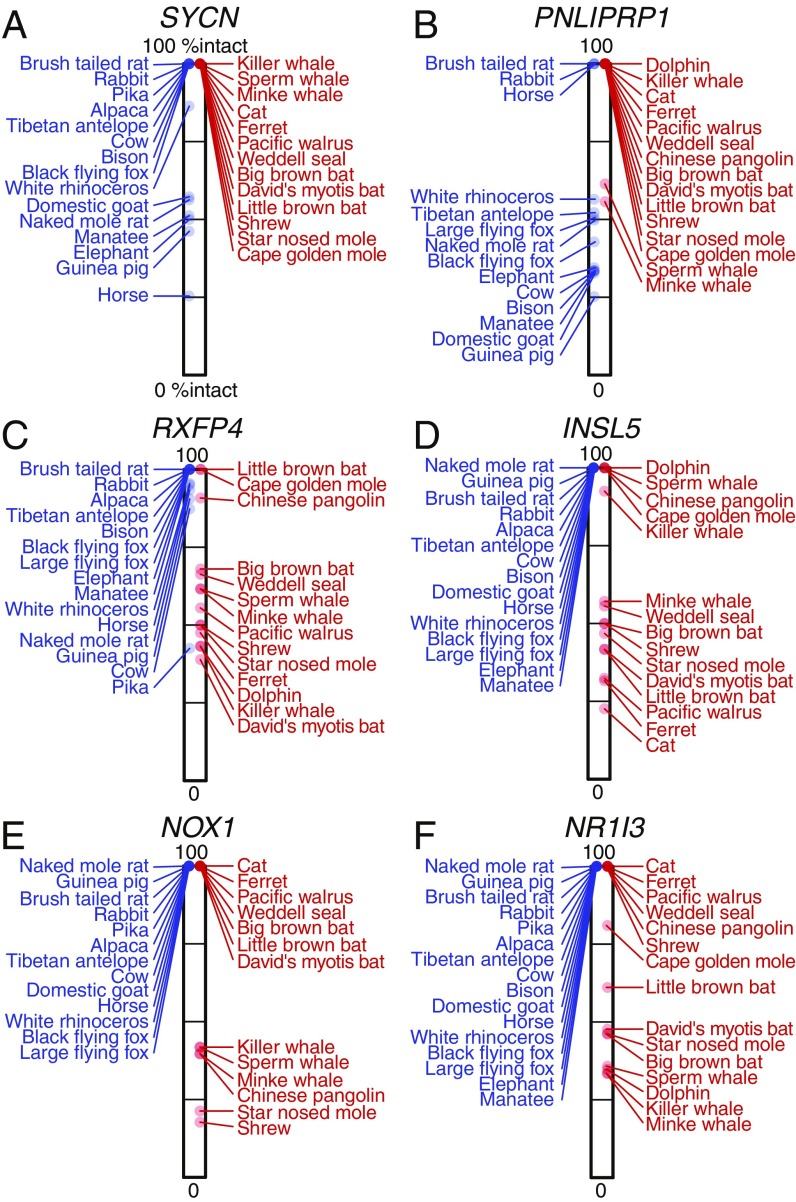
None of these six gene losses have been described before in these species. Examples of inactivating mutations in each of these genes are shown in Fig. 2. All inactivating mutations in all gene-loss species and their overlap with functional protein domains are provided in SI Appendix, Figs. S3–S8. The percentage of the reading frame that remains intact, which contrasts the pattern of gene loss versus gene conservation between herbivores and carnivores, is shown in Fig. 3.
Fig. 2.
Examples of inactivating mutations in genes that are preferentially lost in herbivores (A and B) or carnivores (C–F). The coding exon–intron structure of each diet-related gene is shown at the top. For space considerations, Insets show only one representative inactivating mutation for only one gene-loss lineage, distinguishing herbivores by blue font and carnivores by red font. Black and gray boxes indicate nucleotides that belong to the same codon. All inactivating mutations in these genes are validated by raw DNA sequencing reads and are shown in SI Appendix, Figs. S3–S8.
Fig. 3.
Maximum percentage of the intact reading frame (%intact) of genes that were preferentially lost in herbivores (A and B) and carnivores (C–F). (C) While RXFP4 is truly lost in the herbivorous pika, several other herbivores exhibit a %intact < 100% due to inactivating mutations that shorten the C terminus of the protein, but do not affect the last transmembrane domain (SI Appendix, Fig. S23). Variable C termini are relatively common in conserved genes (56) and therefore do not indicate gene loss.
Validating the Convergent Gene Losses.
To rule out that any of these gene losses were falsely detected due to artifacts, such as base errors in genome assemblies, we manually validated the correctness of all inactivating mutations in the six genes with available unassembled DNA sequencing reads. Of 415 inactivating mutations, 398 (96%) are present in a homozygous state in the sequenced individuals and 13 (3%) additional mutations are polymorphic, likely reflecting ongoing gene decay (Dataset S7). Next, we investigated whether inactivating mutations, and thus losses of these genes, are likely fixed in the respective species. Given that DNA sequencing data representing different populations of a species is often not available, we reasoned that inactivating mutations are likely fixed if they are shared with related sister species, as suggested by several examples in Fig. 2. Therefore, we manually inspected all lost genes in additional genomes of closely related species (Dataset S1). With the exception of PNLIPRP1, which appears to be independently lost in the herbivorous naked mole rat and Damaraland mole rat, and RXFP4, the shared loss of which in myotis bats is less certain, we could confirm the presence of shared inactivating mutations for all other genes where sister species genomes are available (Fig. 1 and SI Appendix, Figs. S4 and S9–S14). This suggests that these gene losses are fixed in the respective herbivore or carnivore lineages.
To further corroborate gene loss, we performed selection rate analyses using RELAX (14). We found significant evidence of relaxed selection in all lost genes, with the exception of NR1I3 (Dataset S8). Finally, we used available gene-expression data to ask whether the remnants of these genes are still expressed in tissues where expression of a functional gene would be expected. We found that PNLIPRP1 is not expressed in the pancreas of the Water buffalo (SI Appendix, Fig. S15), which is consistent with a previous study that did not detect the PNLIPRP1-encoded protein in the pancreas of cows and goats (15), two related ruminants that also lost the gene. Similarly, NR1I3 exhibits no relevant expression in the liver of cetaceans and Brandt’s myotis bat, which lost this gene (SI Appendix, Fig. S16). This indicates that the gene-loss species do not express these genes anymore in their respective tissues.
Together, validated inactivating mutations, the presence of shared inactivating mutations in closely related species, and patterns of relaxed selection show that PNLIPRP1 and SYCN are repeatedly inactivated in herbivores, and that INSL5, RXFP4, NOX1, and NR1I3 are repeatedly inactivated in carnivores. The relationship between these lost genes and dietary specializations is described in the following sections.
Loss of Pancreatic Genes in Herbivores.
The exocrine pancreas plays an important role in digestion by secreting enzymes into the intestine. Adaptation to different diets might therefore influence physiological processes involving pancreatic genes. Our screen discovered that herbivores have repeatedly lost two pancreatic genes, SYCN and PNLIPRP1, which are expressed in pancreatic acinar cells (15, 16).
SYCN is exclusively lost in six independent herbivore lineages (Fig. 1). SYCN localizes to granules that contain inactive digestive enzyme precursors (zymogens) (16). Pancreatic acinar cells secrete these zymogens into the pancreatic duct that empties into the small intestine, where the enzymes are activated. To achieve a high secretory capacity, pancreatic acinar cells rely on compound exocytosis, a mechanism that involves the fusion of a zymogen granule with the apical plasma membrane (primary fusion) followed by fusion of other granules to this primary granule (secondary fusion event) (17). In SYCN knockout mice, the secretory capacity is reduced by half due to drastic reduction of secondary fusion events (18). Hence, SYCN is required for efficient compound exocytosis and necessary to achieve high rates of pancreatic secretion.
The reduced secretory capacity of the exocrine pancreas as a result of SYCN loss is likely related to the feeding habits of herbivores. Many herbivores are continuous grazers or browsers that spend many hours a day feeding (12–20 h for horse, 12–18 h for elephants, 6–12 h for goats). In contrast to carnivores that generally feed at irregular intervals and secrete large amounts of pancreatic juice during a meal, herbivores like horses, goats, and guinea pigs secrete pancreatic juice continuously, with no or little increase during feeding (19–21). Thus, the high secretory capacity mediated by compound exocytosis is not necessary for a broad range of herbivores, which may have permitted the loss of SYCN.
The second pancreatic gene, PNLIPRP1, is lost in eight herbivore lineages but only in two carnivore lineages (sperm whale and minke/bowhead whales, discussed below) (Fig. 1). Notably, while PNLIPRP1 in horse does not exhibit inactivating mutations, this pancreas-specific gene is not expressed in the pancreas of horses anymore (22).
While the PNLIPRP1-encoded pancreatic lipase related protein 1 (PLRP1) exhibits sequence similarity to the pancreatic triglyceride lipase (PTL), the key enzyme for efficient dietary triglyceride digestion (23), PLRP1 does not have any detectable enzymatic lipase activity (24). Instead, PLRP1 acts as a competitive inhibitor for PTL activity by binding colipase with a similar affinity as PTL (24). Because PTL requires colipase for stabilizing the active enzyme conformation, PLRP1-mediated competition for colipase binding reduces the activity of PTL (23). Consequently, the loss of the inhibitor gene PNLIPRP1 should result in an increased PTL activity, which was demonstrated in a mouse knockout. Compared with wild-type mice, the pancreatic juice of knockout mice has a higher lipase activity, resulting in larger adipocytes and a twofold higher fat mass (25). Hence, the loss of the competitive PTL inhibitor gene PNLIPRP1 leads to an increased capacity for digesting dietary triglycerides, which may be beneficial for herbivores that typically consume a rather fat-poor diet. In addition to an increased fat mass, PNLIPRP1 knockout also impairs insulin sensitivity in these mice, especially when fed a high-fat diet (25), which likely explains the presence of this gene in most carnivores. Interestingly, the only two carnivore lineages that lost PNLIPRP1, sperm whale and minke/bowhead whales, mostly feed on cephalopods and krill, which have a substantially lower triglyceride content (26, 27), similar to plant-based diets. This could explain why sperm, minke, and bowhead whale, but not the fish- and meat-eating bottlenose dolphin and killer whale or other carnivores lost PNLIPRP1.
Loss of Food Intake and Glucose Homeostasis Regulating Genes in Carnivores.
In general, herbivorous diets are rich in carbohydrates and carnivorous diets are rich in proteins and fatty acids. These differences in dietary composition may be reflected by changes in signaling through gastrointestinal hormones that regulate various processes ranging from nutrient absorption, lipid, and glucose homeostasis to food intake (28). Our genomic screen discovered a hormone-receptor pair that has pleiotropic roles in appetite regulation and glucose homeostasis, INSL5 and RXFP4. INSL5 and RXFP4 are lost in a number of independent carnivores (Fig. 1). INSL5 is a hormone that specifically binds to the G protein-coupled receptor encoded by RXFP4 (29). Both genes are highly expressed in the colon, where INSL5 is secreted by enteroendocrine L-cells (29, 30). First, consistent with a role in appetite regulation, INSL5 plasma levels depend on food intake. Specifically, INSL5 levels in wild-type mice increase during fasting or during prolonged calorie restriction, and decline after feeding (31). Furthermore, administration of INSL5 after fasting increased food intake in wild-type mice, but not in mice in which the INSL5 receptor RXFP4 is knocked out. These experiments suggested that INSL5 regulates food intake as an orexigenic (appetite stimulating) hormone through binding to its specific receptor RXFP4 (31). The loss of the appetite-stimulating hormone-receptor pair INSL5–RXFP4 in a number of carnivores could be related to differences in feeding patterns, given that carnivores generally feed at more irregular intervals compared with most herbivores.
Second, in addition to regulating appetite, INSL5 and RXFP4 have also been linked to glucose homeostasis. Cell line-based and mouse studies have shown that RXFP4 activation by INSL5 plays a regulatory role in glucose-stimulated insulin secretion in pancreatic β-cells, which express RXFP4 (32–34). Furthermore, the INSL5 hormone regulates glucose production (gluconeogenesis) in the liver (35). Thus, the repeated loss of both genes in carnivores may additionally be related to the lower carbohydrate content of their diet. To maintain sufficient glucose levels, carnivores such as cats and minks exhibit constant gluconeogenesis (36, 37). Thus, changes in glucose metabolism of carnivores may have rendered the need for INSL5–RXFP4-mediated regulation of glucose homeostasis obsolete. Interestingly, because gluconeogenesis is even active in cats and minks after feeding, where insulin is normally expected to suppress gluconeogenesis (36, 37), it is possible that there is a direct connection between the loss of the feeding-regulated INSL5 hormone that affects insulin secretion and the presence of constant gluconeogenesis. In summary, the loss of INSL5 and RXFP4 in carnivores may be related to both irregular feeding patterns and constant gluconeogenesis.
Loss of the Innate Immunity Gene NOX1 in Carnivores.
Carnivores and herbivores do not only face different challenges related to digestion and metabolism, they also host different bacteria in their gut (3). Furthermore, carnivory is generally associated with less-diverse gut microbiomes than herbivory (3). These gut microbiome differences may be reflected by alterations in the innate immune system, which in turn can also influence gut microbiome composition (38). Our genomic screen detected the loss of the innate immune gene NOX1 in four carnivore lineages (Fig. 1).
NOX1 encodes a transmembrane protein that is highly expressed in the colon epithelium (39). Like other NADPH oxidases, NOX1 facilitates the generation of reactive oxygen species (ROS) (39), and this activity has been linked to antimicrobial immune defense and restitution of the colon mucosa (innermost layer of cells in the colon that is exposed to the gut microbiome). Supporting a function in antimicrobial immune response, ROS production by NOX1 can be induced by bacterial lipopolysaccharides and flagellin or by immune system cytokines, such as IFN-γ (40–42). NOX1 knockout mice further revealed a function of NOX1 in wound healing and restitution of the colon mucosa (43, 44). The antimicrobial defense and mucosal restitution functions of NOX1 are likely related. Pathogenic bacteria can damage the colon mucosa, which causes inflammation. The inflammatory response would then trigger ROS generation by NOX1 to kill pathogenic bacteria and activate wound-healing pathways. Hence, NOX1 plays important roles at the intersection between the host and its microbiome.
The loss of NOX1 might be related to differences between herbivorous and carnivorous gut microbiomes. For example, carnivores typically possess half the bacterial species diversity than herbivores (3). Exposure to less diverse bacterial communities in the carnivore gut may have led to a decreased selection pressure to preserve NOX1 in carnivores.
Loss of Detoxification Genes in Carnivores.
Herbivorous mammals possess a variety of receptors and enzymes to cope with potentially toxic, plant-derived xenobiotics in their diet (45). Because a carnivorous diet does not provide large amounts of plant-derived xenobiotics, detoxification genes are less important for carnivores. Our genomic screen detected the convergent loss of NR1I3 in three independent carnivore lineages (Fig. 1). NR1I3 encodes one of the major xenobiotic receptors. Interestingly, while not meeting our strict screening criteria, the functionally related NR1I2 gene is also lost in two carnivore lineages (cetaceans and shrew) (SI Appendix, Fig. S17 and Dataset S4). NR1I3 and NR1I2 are primarily expressed in the liver and intestine, and activate a large number of cytochrome P450 and other xenobiotic metabolizing enzymes, including UGT1A6 (46). Similar to NR1I3, the remnants of NR1I2 are not expressed in the liver of cetaceans (SI Appendix, Fig. S18). The loss of NR1I3 and NR1I2 suggests that key components of the hepatic and intestinal xenobiotic pathway are lost in several carnivores, which could be related to a lower exposure to plant-derived xenobiotics in a carnivorous diet.
Discussion
By performing a systematic genomic screen for convergent gene losses in independent herbivore and carnivore lineages, we identified several previously unknown gene losses that are related to three aspects of dietary specialization: (i) dietary composition, (ii) feeding pattern, and (iii) gut microbiome diversity. First, given that herbivorous diets are generally rich in carbohydrates and plant-derived xenobiotics while carnivorous diets are generally rich in proteins and fats, the loss of PNLIPRP1, INSL5, RXFP4, and NR1I3 is likely associated with key differences in dietary composition. The triglyceride digestion inhibitor gene PNLIPRP1 protects from adverse consequences of excessive digestion and absorption of fatty acids, a function that is likely less important for herbivores than for carnivores. In contrast, the glucose homeostasis-regulating function of INSL5 and RXFP4 may not be important for carnivores, especially for the species that exhibit constant gluconeogenesis. Similarly, the loss of the xenobiotic receptors NR1I3, and also NR1I2, in carnivores reflects reduced amounts of plant-derived xenobiotics in their diets. Second, the loss of INSL5, RXFP4, and SYCN could be related to differences in feeding patterns. In general, carnivores feed at more irregular intervals than herbivores. These differences may have contributed to the repeated carnivore-associated loss of INSL5 and RXFP4 given that both genes are also involved in regulating appetite. In contrast, while SYCN, a gene required for efficient pancreatic zymogen secretion, is important for carnivores that feed at irregular intervals, this gene is less relevant for herbivores with regular feeding patterns and continuous zymogen secretion. Third, the carnivore-associated loss of the gastrointestinal host-defense gene NOX1 is likely related to a different composition and lower diversity of the gut microbiome of carnivores. Given that microbiome research has been recognized as an important field of study, not only in evolution and ecology but also in medicine (3, 47), the loss of NOX1 in carnivores may offer new insights into how the genes encoded by the mammalian host interact with and shape its gut microbiome.
Relaxed or no selection to maintain the respective gene function is likely a major factor that explains the gene losses detected in this study. For example, similar to the previously reported loss of taste receptor genes in mammals that swallow their prey whole (6, 7), the “use it or lose it” principle probably also applies to the loss of detoxification genes in carnivores. However, during evolution, gene loss can also be beneficial under certain circumstances (9, 48), although after longer evolutionary time periods it is not possible to infer which mutation was initially adaptive. Gene loss can proceed via different trajectories. For example, a regulatory mutation can initially abolish gene expression, after which neutral, reading frame-inactivating mutations can accumulate over time. One such case is a promoter mutation in human populations in the DARC gene, which protects from infection with the malaria parasite Plasmodium (49). PNLIPRP1 in the herbivorous horse might represent another such case in its early stage, as this pancreas-specific gene is not expressed in the pancreas anymore (22), but presently exhibits an intact reading frame in horse. Another possibility is that a reading frame-inactivating mutation initially causes gene loss, after which the promoter and regulatory regions would be free to accumulate neutral mutations. This mechanism is exemplified by a frameshift mutation in human CCR5 that protects from HIV infection (50). Regardless of the different mutational trajectories that lead to the elimination of a functional gene product, gene loss as an evolutionary event can confer an advantage. Regarding the gene losses detected here, it is possible that a reduced secretory capacity due to the loss of SYCN could be beneficial for herbivores by attenuating secretory spikes after a meal. This could allow herbivores to preserve a sufficiently large zymogen granule pool for continuous zymogen secretion, as observed in several herbivores. Similarly, the loss of the competitive triglyceride lipase inhibitor gene PNLIPRP1 improves the efficiency of digesting dietary triglycerides, which could be an advantage for herbivores whose diet contains a lower amount of triglycerides. Finally, Nox1 deficiency in mouse protects against liver damage induced by a diet rich in fat and cholesterol (51), which might also be an advantage for carnivores that naturally consume a similar diet.
Our genomic screen not only highlights the need for further studies to address how the loss of the above-described genes affects physiology and metabolism in these natural knockout species, but it also revealed additional less well-characterized genes whose preferential loss in herbivores or carnivores could be related to dietary changes. For example, the PHGR1 gene ranks third in our screen for carnivore-associated gene losses (SI Appendix, Fig. S19). The PHGR1 protein is specifically expressed in secretory goblet cells and absorptive enterocytes of the intestines, where it might be involved in vesicle-mediated transport (52); however, its exact function has not been characterized yet. Other examples are the two phospholipase A2 genes, PLA2G2A and PLA2G2C, that are preferentially lost in carnivores (SI Appendix, Figs. S20 and S21). While automatic pathway annotations suggest that both genes are involved in the metabolism of arachidonic acid, a fatty acid that is abundant in meat, this function has not been experimentally demonstrated, and instead a role in inflammatory responses has been suggested for PLA2G2A (53). Finally, we detected the convergent loss of the RNA virus receptor gene DDX58 in all cetaceans, the star-nosed mole, and cape golden mole (SI Appendix, Fig. S22). While DDX58 is known to detect rotaviruses in the intestine (54), it is not obvious how this gene loss relates to a carnivorous diet. Therefore, additional functional studies are required to understand if and how the evolutionary loss of these genes is related to dietary specialization.
In conclusion, convergent gene losses provide new insights into a classic ecological question from a molecular perspective. More generally, our study highlights how comparative genomics can shed light on biological processes that changed in obligate herbivores or carnivores, which extends our understanding of how similar dietary specializations evolved repeatedly in placental mammals.
Materials and Methods
Detailed descriptions of GO enrichment, selection rate, and expression data analyses are provided in SI Appendix.
Detection of Gene Loss Events.
We used a previously developed pipeline to systematically detect gene-inactivating mutations (9). To overcome assembly and alignment issues and address evolutionary exon–intron structure changes, this pipeline: (i) distinguishes assembly gaps from real deletions (55), (ii) realigns coding exons with CESAR to consider reading frame and splice site information (56, 57), (iii) excludes paralog or processed pseudogene alignments, and (iv) considers all principal or alternative APPRIS isoforms of a gene (58). As input, we used a whole-genome alignment between the human hg38 genome assembly (reference) and the genome assemblies of other (query) placental mammals (10), and the human Ensembl v90 gene annotation (59). We excluded olfactory receptors whose relationship to dietary specializations have been investigated previously (60), and thus considered a total of 16,135 genes.
We used the positions of inactivating mutations to compute the maximum percentage of the reading frame that remains intact (%intact). For example, a 90% intact reading frame can arise by an inactivating mutation that is close to the N or C terminus. Given that conserved genes can have inactivating mutations close to the N or C terminus (56), we considered genes with %intact ≥ 90% as conserved. Genes with %intact ≤ 60% have one or more mutations in the middle, which indicates that these genes cannot encode a functional protein anymore, and are considered as lost.
Screening for Diet-Related Convergent Gene Losses.
We adopted the Forward Genomics approach (61, 62) and screened for genes where %intact is generally lower in one group compared with the other group (9). To identify herbivore-associated gene losses, we searched for genes where at least 10% of the herbivore species have lost the gene (%intact ≤ 60%), while at least 80% of the carnivore species have an intact gene (%intact ≥ 90%). We required that the gene-loss herbivore species belong to at least three independent lineages. An analogous approach was applied to identify convergent gene losses in carnivores. We excluded genes that were only lost in carnivores with poor vision (cape golden mole, star-nosed mole, pangolin, and insectivorous bats) as such gene losses may be related to vision but not diet.
Gene Loss Validation.
First, we validated the correctness of stop codon and frameshift mutations with unassembled DNA reads provided by the National Center for Biotechnology Information Sequence Read Archive or Trace Archive (63), as previously described (64, 65). Second, we manually verified that each lost gene is located in the ancestral synteny context by inspecting the pair-wise alignment chains in the University of California, Santa Cruz Genome Browser (66) for the presence of up- and downstream genes. This examination of the genomic locus harboring the lost gene was also used to rule out that an intact copy of the lost gene exists elsewhere in the genome. Third, in addition to the genomes provided by the whole-genome alignment (10), we manually investigated for the six genes whether the same inactivating mutations occur in genomes or DNA read data of other mammals that are sister species to those that lost the gene.
Supplementary Material
Acknowledgments
We thank Sylvia Ortmann and Irina Ruf for helpful comments on the manuscript, and the Computer Service Facilities of the Max Planck Institute of Molecular Cell Biology and Genetics and Max Planck Institute for the Physics of Complex Systems for their support. This work was supported by the Max Planck Society, the German Research Foundation (Grant HI1423/3-1), and the Leibniz Association (Grant SAW-2016-SGN-2).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818504116/-/DCSupplemental.
References
- 1.Mackie RI. Mutualistic fermentative digestion in the gastrointestinal tract: Diversity and evolution. Integr Comp Biol. 2002;42:319–326. doi: 10.1093/icb/42.2.319. [DOI] [PubMed] [Google Scholar]
- 2.Karasov WH, Douglas AE. Comparative digestive physiology. Compr Physiol. 2013;3:741–783. doi: 10.1002/cphy.c110054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z, et al. Evolution of digestive enzymes and RNASE1 provides insights into dietary switch of cetaceans. Mol Biol Evol. 2016;33:3144–3157. doi: 10.1093/molbev/msw191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim S, et al. Comparison of carnivore, omnivore, and herbivore mammalian genomes with a new leopard assembly. Genome Biol. 2016;17:211. doi: 10.1186/s13059-016-1071-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng P, Zheng J, Rossiter SJ, Wang D, Zhao H. Massive losses of taste receptor genes in toothed and baleen whales. Genome Biol Evol. 2014;6:1254–1265. doi: 10.1093/gbe/evu095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang P, et al. Major taste loss in carnivorous mammals. Proc Natl Acad Sci USA. 2012;109:4956–4961. doi: 10.1073/pnas.1118360109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emerling CA, Delsuc F, Nachman MW. Chitinase genes (CHIAs) provide genomic footprints of a post-Cretaceous dietary radiation in placental mammals. Sci Adv. 2018;4:eaar6478. doi: 10.1126/sciadv.aar6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma V, et al. A genomics approach reveals insights into the importance of gene losses for mammalian adaptations. Nat Commun. 2018;9:1215. doi: 10.1038/s41467-018-03667-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma V, Hiller M. Increased alignment sensitivity improves the usage of genome alignments for comparative gene annotation. Nucleic Acids Res. 2017;45:8369–8377. doi: 10.1093/nar/gkx554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Court MH, Greenblatt DJ. Molecular genetic basis for deficient acetaminophen glucuronidation by cats: UGT1A6 is a pseudogene, and evidence for reduced diversity of expressed hepatic UGT1A isoforms. Pharmacogenetics. 2000;10:355–369. doi: 10.1097/00008571-200006000-00009. [DOI] [PubMed] [Google Scholar]
- 12.Kakehi M, et al. Uridine diphosphate-glucuronosyltransferase (UGT) xenobiotic metabolizing activity and genetic evolution in pinniped species. Toxicol Sci. 2015;147:360–369. doi: 10.1093/toxsci/kfv144. [DOI] [PubMed] [Google Scholar]
- 13.Escoffier J, et al. Homozygous mutation of PLCZ1 leads to defective human oocyte activation and infertility that is not rescued by the WW-binding protein PAWP. Hum Mol Genet. 2016;25:878–891. doi: 10.1093/hmg/ddv617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K. RELAX: Detecting relaxed selection in a phylogenetic framework. Mol Biol Evol. 2015;32:820–832. doi: 10.1093/molbev/msu400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Caro J, et al. Occurrence of pancreatic lipase-related protein-2 in various species and its relationship with herbivore diet. Comp Biochem Physiol B Biochem Mol Biol. 2008;150:1–9. doi: 10.1016/j.cbpb.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Edwardson JM, An S, Jahn R. The secretory granule protein syncollin binds to syntaxin in a Ca2(+)-sensitive manner. Cell. 1997;90:325–333. doi: 10.1016/s0092-8674(00)80340-2. [DOI] [PubMed] [Google Scholar]
- 17.Pickett JA, Edwardson JM. Compound exocytosis: Mechanisms and functional significance. Traffic. 2006;7:109–116. doi: 10.1111/j.1600-0854.2005.00372.x. [DOI] [PubMed] [Google Scholar]
- 18.Wäsle B, et al. Syncollin is required for efficient zymogen granule exocytosis. Biochem J. 2005;385:721–727. doi: 10.1042/BJ20041064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim CD, Lee KY, Chang TM, Chey WY. Negative feedback regulation of pancreatic exocrine secretion in guinea pigs. Pancreas. 1995;10:173–179. doi: 10.1097/00006676-199503000-00011. [DOI] [PubMed] [Google Scholar]
- 20.Kurilov NV, Obukhov BM. Study of pancreatic and biliary secretion in horses by means of a chronic fistula. Bull Exp Biol Med. 1958;46:877–880. [PubMed] [Google Scholar]
- 21.Xu M, et al. Influence of rumen escape starch on pancreatic exocrine secretion of goats. J Anim Physiol Anim Nutr (Berl) 2009;93:122–129. doi: 10.1111/j.1439-0396.2007.00792.x. [DOI] [PubMed] [Google Scholar]
- 22.Jayne S, Kerfelec B, Foglizzo E, Chapus C, Crenon I. High expression in adult horse of PLRP2 displaying a low phospholipase activity. Biochim Biophys Acta. 2002;1594:255–265. doi: 10.1016/s0167-4838(01)00309-0. [DOI] [PubMed] [Google Scholar]
- 23.Lowe ME. The triglyceride lipases of the pancreas. J Lipid Res. 2002;43:2007–2016. doi: 10.1194/jlr.r200012-jlr200. [DOI] [PubMed] [Google Scholar]
- 24.Crenon I, et al. Pancreatic lipase-related protein type I: A specialized lipase or an inactive enzyme. Protein Eng. 1998;11:135–142. doi: 10.1093/protein/11.2.135. [DOI] [PubMed] [Google Scholar]
- 25.Ren J, et al. Increased fat mass and insulin resistance in mice lacking pancreatic lipase-related protein 1. J Nutr Biochem. 2011;22:691–698. doi: 10.1016/j.jnutbio.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Tou JC, Jaczynski J, Chen YC. Krill for human consumption: Nutritional value and potential health benefits. Nutr Rev. 2007;65:63–77. doi: 10.1111/j.1753-4887.2007.tb00283.x. [DOI] [PubMed] [Google Scholar]
- 27.Ozogul Y, Duysak O, Ozogul F, Özkütük AS, Türeli C. Seasonal effects in the nutritional quality of the body structural tissue of cephalopods. Food Chem. 2008;108:847–852. doi: 10.1016/j.foodchem.2007.11.048. [DOI] [PubMed] [Google Scholar]
- 28.Chaudhri OB, Salem V, Murphy KG, Bloom SR. Gastrointestinal satiety signals. Annu Rev Physiol. 2008;70:239–255. doi: 10.1146/annurev.physiol.70.113006.100506. [DOI] [PubMed] [Google Scholar]
- 29.Liu C, et al. INSL5 is a high affinity specific agonist for GPCR142 (GPR100) J Biol Chem. 2005;280:292–300. doi: 10.1074/jbc.M409916200. [DOI] [PubMed] [Google Scholar]
- 30.Thanasupawat T, et al. INSL5 is a novel marker for human enteroendocrine cells of the large intestine and neuroendocrine tumours. Oncol Rep. 2013;29:149–154. doi: 10.3892/or.2012.2119. [DOI] [PubMed] [Google Scholar]
- 31.Grosse J, et al. Insulin-like peptide 5 is an orexigenic gastrointestinal hormone. Proc Natl Acad Sci USA. 2014;111:11133–11138. doi: 10.1073/pnas.1411413111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burnicka-Turek O, et al. INSL5-deficient mice display an alteration in glucose homeostasis and an impaired fertility. Endocrinology. 2012;153:4655–4665. doi: 10.1210/en.2012-1161. [DOI] [PubMed] [Google Scholar]
- 33.Luo X, et al. The insulinotrophic effect of insulin-like peptide 5 in vitro and in vivo. Biochem J. 2015;466:467–473. doi: 10.1042/BJ20141113. [DOI] [PubMed] [Google Scholar]
- 34.Ang SY, et al. Signal transduction pathways activated by insulin-like peptide 5 at the relaxin family peptide RXFP4 receptor. Br J Pharmacol. 2017;174:1077–1089. doi: 10.1111/bph.13522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee YS, et al. Insulin-like peptide 5 is a microbially regulated peptide that promotes hepatic glucose production. Mol Metab. 2016;5:263–270. doi: 10.1016/j.molmet.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rogers QR, Morris JG, Freedland RA. Lack of hepatic enzymatic adaptation to low and high levels of dietary protein in the adult cat. Enzyme. 1977;22:348–356. doi: 10.1159/000458816. [DOI] [PubMed] [Google Scholar]
- 37.Schermerhorn T. Normal glucose metabolism in carnivores overlaps with diabetes pathology in non-carnivores. Front Endocrinol (Lausanne) 2013;4:188. doi: 10.3389/fendo.2013.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vijay-Kumar M, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suh YA, et al. Cell transformation by the superoxide-generating oxidase Mox1. Nature. 1999;401:79–82. doi: 10.1038/43459. [DOI] [PubMed] [Google Scholar]
- 40.Kawahara T, et al. Type I Helicobacter pylori lipopolysaccharide stimulates Toll-like receptor 4 and activates mitogen oxidase 1 in gastric pit cells. Infect Immun. 2001;69:4382–4389. doi: 10.1128/IAI.69.7.4382-4389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawahara T, et al. Role of nicotinamide adenine dinucleotide phosphate oxidase 1 in oxidative burst response to Toll-like receptor 5 signaling in large intestinal epithelial cells. J Immunol. 2004;172:3051–3058. doi: 10.4049/jimmunol.172.5.3051. [DOI] [PubMed] [Google Scholar]
- 42.Kuwano Y, et al. Interferon-gamma activates transcription of NADPH oxidase 1 gene and upregulates production of superoxide anion by human large intestinal epithelial cells. Am J Physiol Cell Physiol. 2006;290:C433–C443. doi: 10.1152/ajpcell.00135.2005. [DOI] [PubMed] [Google Scholar]
- 43.Kato M, et al. The ROS-generating oxidase Nox1 is required for epithelial restitution following colitis. Exp Anim. 2016;65:197–205. doi: 10.1538/expanim.15-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leoni G, et al. Annexin A1, formyl peptide receptor, and NOX1 orchestrate epithelial repair. J Clin Invest. 2013;123:443–454. doi: 10.1172/JCI65831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dearing MD, Foley WJ, McLean S. The influence of plant secondary metabolites on the nutritional ecology of herbivorous terrestrial vertebrates. Annu Rev Ecol Evol Syst. 2005;36:169–189. [Google Scholar]
- 46.Tolson AH, Wang H. Regulation of drug-metabolizing enzymes by xenobiotic receptors: PXR and CAR. Adv Drug Deliv Rev. 2010;62:1238–1249. doi: 10.1016/j.addr.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Albalat R, Cañestro C. Evolution by gene loss. Nat Rev Genet. 2016;17:379–391. doi: 10.1038/nrg.2016.39. [DOI] [PubMed] [Google Scholar]
- 49.Tournamille C, Colin Y, Cartron JP, Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet. 1995;10:224–228. doi: 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]
- 50.Liu R, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 51.Matsumoto M, et al. The NOX1 isoform of NADPH oxidase is involved in dysfunction of liver sinusoids in nonalcoholic fatty liver disease. Free Radic Biol Med. 2018;115:412–420. doi: 10.1016/j.freeradbiomed.2017.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oltedal S, et al. Expression profiling and intracellular localization studies of the novel proline-, histidine-, and glycine-rich protein 1 suggest an essential role in gastro-intestinal epithelium and a potential clinical application in colorectal cancer diagnostics. BMC Gastroenterol. 2018;18:26. doi: 10.1186/s12876-018-0752-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Birts CN, Barton CH, Wilton DC. Catalytic and non-catalytic functions of human IIA phospholipase A2. Trends Biochem Sci. 2010;35:28–35. doi: 10.1016/j.tibs.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 54.Broquet AH, Hirata Y, McAllister CS, Kagnoff MF. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J Immunol. 2011;186:1618–1626. doi: 10.4049/jimmunol.1002862. [DOI] [PubMed] [Google Scholar]
- 55.Hiller M, Schaar BT, Bejerano G. Hundreds of conserved non-coding genomic regions are independently lost in mammals. Nucleic Acids Res. 2012;40:11463–11476. doi: 10.1093/nar/gks905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma V, Elghafari A, Hiller M. Coding exon-structure aware realigner (CESAR) utilizes genome alignments for accurate comparative gene annotation. Nucleic Acids Res. 2016;44:e103. doi: 10.1093/nar/gkw210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharma V, Schwede P, Hiller M. CESAR 2.0 substantially improves speed and accuracy of comparative gene annotation. Bioinformatics. 2017;33:3985–3987. doi: 10.1093/bioinformatics/btx527. [DOI] [PubMed] [Google Scholar]
- 58.Rodriguez JM, et al. APPRIS 2017: Principal isoforms for multiple gene sets. Nucleic Acids Res. 2017;46:D213–D217. doi: 10.1093/nar/gkx997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zerbino DR, et al. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayden S, et al. Ecological adaptation determines functional mammalian olfactory subgenomes. Genome Res. 2010;20:1–9. doi: 10.1101/gr.099416.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hiller M, et al. A “forward genomics” approach links genotype to phenotype using independent phenotypic losses among related species. Cell Rep. 2012;2:817–823. doi: 10.1016/j.celrep.2012.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prudent X, Parra G, Schwede P, Roscito JG, Hiller M. Controlling for phylogenetic relatedness and evolutionary rates improves the discovery of associations between species’ phenotypic and genomic differences. Mol Biol Evol. 2016;33:2135–2150. doi: 10.1093/molbev/msw098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kodama Y, Shumway M, Leinonen R. International Nucleotide Sequence Database Collaboration The sequence read archive: Explosive growth of sequencing data. Nucleic Acids Res. 2012;40:D54–D56. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hecker N, Sharma V, Hiller M. Transition to an aquatic habitat permitted the repeated loss of the pleiotropic KLK8 gene in mammals. Genome Biol Evol. 2017;9:3179–3188. doi: 10.1093/gbe/evx239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sharma V, Lehmann T, Stuckas H, Funke L, Hiller M. Loss of RXFP2 and INSL3 genes in Afrotheria shows that testicular descent is the ancestral condition in placental mammals. PLoS Biol. 2018;16:e2005293. doi: 10.1371/journal.pbio.2005293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Casper J, et al. The UCSC genome browser database: 2018 update. Nucleic Acids Res. 2018;46:D762–D769. doi: 10.1093/nar/gkx1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.