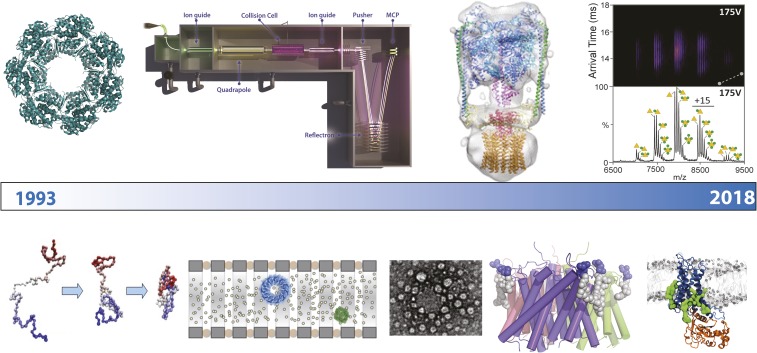
Significance
Following initial discoveries of noncovalent associations surviving in the gas phase, only a few practitioners pursued this research area. Today scientists around the world are using these approaches to ascertain the heterogeneity and stoichiometry of proteins within complexes. Recent developments further highlight opportunities for studying the effects of protein glycosylation on antibody–antigen interactions and drug binding, as well as site-directed mutagenesis and posttranslational modification on membrane protein interfaces. As a result of many developments over the last two decades, mass spectrometry of protein complexes has exploded and is now undertaken not just in dedicated research laboratories in academia, but also in pharmaceutical and biotechnology companies. It is therefore timely to trace the history of these developments in this personal perspective.
Keywords: membrane proteins, mass spectrometry, biophysics
Abstract
In this Inaugural Article, I trace some key steps that have enabled the development of mass spectrometry for the study of intact protein complexes from a variety of cellular environments. Beginning with the preservation of the first soluble complexes from plasma, I describe our early experiments that capitalize on the heterogeneity of subunit composition during assembly and exchange reactions. During these investigations, we observed many assemblies and intermediates with different subunit stoichiometries, and were keen to ascertain whether or not their overall topology was preserved in the mass spectrometer. Adapting ion mobility and soft-landing methodologies, we showed how ring-shaped complexes could survive the phase transition. The next logical progression from soluble complexes was to membrane protein assemblies but this was not straightforward. We encountered many pitfalls along the way, largely due to the use of detergent micelles to protect and stabilize complexes. Further obstacles presented when we attempted to distinguish lipids that copurify from those that are important for function. Developing new experimental protocols, we have subsequently defined lipids that change protein conformation, mediate oligomeric states, and facilitate downstream coupling of G protein-coupled receptors. Very recently, using a radical method—ejecting protein complexes directly from native membranes into mass spectrometers—we provided insights into associations within membranes and mitochondria. Together, these developments suggest the beginnings of mass spectrometry meeting with cell biology.
Set against the early days of electrospray, when the removal of water was considered deleterious to the folded structure of proteins (1), the development of mass spectrometry (MS) to study protein complexes was not intuitive (Fig. 1). The concept of protein assemblies maintaining their subunit interactions was received with considerable skepticism for some years (2). For membrane proteins, issues were further compounded because, without the support of the lipid bilayer, how could we expect such assemblies to remain folded? Their predominantly hydrophobic associations had long been considered unfavorable for the survival of subunit interactions, while their requirement for membrane mimetics or high concentrations of detergent were predicted to suppress protein signals and overwhelm electrospray mass spectra (3). The fact that eventually we were able to find conditions wherein our first membrane complex did survive, with interactions between cytoplasmic and transmembrane proteins retained (4), has opened up new opportunities for deducing the stoichiometry of membrane proteins and, in particular, the study of their fine-tuning by lipid interactions.
Fig. 1.
A timeline for milestones along the path from protein folding to GPCRs. (Left to Right) In the 1990s our research was focused on developing hydrogen deuterium exchange methodologies to monitor the folding of proteins, capturing folding intermediates (7) and probing their interactions with molecular chaperones (8). Transmission of an intact GroEL14-mer using instrumentation modified in our laboratory was an exciting milestone for us because it demonstrated the potential for MS to maintain intact macromolecular complexes (10, 48). Subsequently, we collaborated with others on prototype ion mobility spectrometers to demonstrate preservation of ring-shaped assemblies, and to produce early images of complexes on electron microscopy grids (15, 17, 18, 49). Our first membrane protein complexes were ejected from micelles in 2008 (4), with intact rotary ATPases surviving the phase transition in 2011 (24). In 2014 we began our quest to uncover the many roles of lipids, starting with those that modulate the structure of membrane proteins, including the ammonia channel (34). In 2016, we recorded our first mass spectra of a folded GPCR with both endogenous ligand and drug retained (41).
The interplay between proteins and lipids in cellular membranes, and their inherent dynamic nature, poses significant challenges for all structural biologists. Crystallography and electron microscopy require the appropriate choice of detergent micelles, lipid cubic phase, or membrane mimetic, to extract proteins from the heterogeneous membrane environment (5). Capturing interactions without perturbing the native lipid environment is a common concern. Beyond the protection of the membrane-embedded regions, some structural studies require the flexible termini and dynamic loops to be removed or the receptor to be stabilized with fusion proteins and mutagenesis is often employed to increase the overall stability of the native fold (6).
Characterizing wild-type membrane proteins as close to their native-like bilayer environments as possible has been a long-term goal for my research group over the past decade. We anticipated that, if this could be achieved, we would uncover many new roles for lipids. Moreover we would be able to highlight differences between detergent-extracted complexes versus those ejected directly from their native membrane environments. The many developments that have led us to this point are the focus of this Inaugural Article.
Historical Perspective
My earliest interests in the development of MS for the study of folded proteins date back to the formation of intermediates on protein-folding pathways and their characterization through the incorporation of deuterium labeling (7). A natural progression from these studies was to try to recreate cellular folding environments wherein the presence of chaperones or ribosomes are known to be important. Projecting the GroEL molecular chaperone with substrates intact required us to adapt the electrospray interface to enable lower temperatures than were previously possible, and hence preserve these interactions during electrospray (8). At this stage, however, we could not observe the intact GroEL14-mer. Further adaptation of instrumentation was necessary. Working with prototype electrospray time-of-flight mass spectrometers, we showed that the composition and subunit stoichiometry of complexes could be retained, even from relatively crude extracts such as plasma (9), and observed our first 800-kDa complex of GroEL in 1999 (10). This led to opportunities to monitor changes in subunit composition, either through spontaneous exchange (11) or induced through thermal activation following construction of a thermally controlled nanoflow device (12, 13). The precision with which masses could be assigned to heterogeneous populations was such that incorporation of different subunits could be uncovered and monitored as a function of time.
A turning point came with the application of ion mobility MS to the study of native complexes. Following many excellent earlier developments for clusters and biomolecules (14), we adapted modeling strategies and demonstrated that the shape of a protein complex could be preserved to maintain a defined ring-shaped structure within the mass spectrometer (15). Alternatively, by increasing the internal energy of the ions, these structures could be induced to collapse to form spherical structures, which could then be separated from the larger ring-shaped structures. Collision cross sections can be obtained from these measurements, which in turn can be modeled and compared with theoretical values, calculated from known structures where available (16). In effect, this adds a new dimension to the experiment: that of topology of the complex. These experiments, together with early attempts to soft-land protein complexes and subsequent imaging following negative stain using electron microscopy (17, 18), helped convince remaining skeptics that the shapes of protein assemblies could be maintained in the gas phase and, for the large part, corresponded to those anticipated from X-ray structures.
From Membranes to Micellated Complexes
A long-term goal for us has been to achieve the same insights for membrane proteins that we were beginning to amass for soluble proteins. Overcoming the high concentrations of detergent necessary to retain solubility was a significant challenge, with early attempts yielding mainly aggregates of both detergents and proteins (19, 20). It was only when we realized that to effect efficient delivery we needed to increase detergent concentrations, above the critical micelle concentration, that we managed to retain transmembrane and cytoplasmic subunits. Following further instrument modification and application of bespoke parameters, we observed an ABC transporter in a well-defined subunit stoichiometry (4) and knew that we had finally achieved conditions whereby we could begin a systematic study of membrane protein–lipid interactions with a high chance of success. From our very first mass spectra of membrane proteins, however, it was clear that associated lipids would remain, despite extensive delipidation protocols (21). A major challenge for us, therefore, was to consider how to use the lipid-binding we readily observed to inform a more complete picture of the structure and function of membrane proteins. Specifically, we wanted to develop experiments that would enable identification of lipids that were important for fine-tuning functions and for modulating oligomeric states.
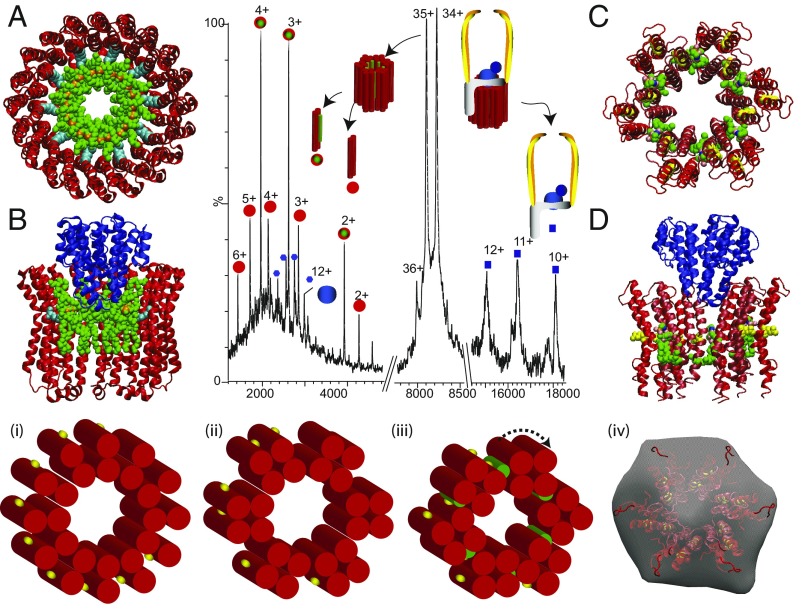
We reasoned that the rotary ATPase, with a large proportion of subunits rotating within membranes, would have close interactions with surrounding lipids (22). Our earliest mass spectra had been limited to subcomplexes arising from the soluble head; without the detergent micelle, the membrane-embedded components were not observed (23). We concluded at that time that the loss of membrane subunits was most likely due to their inability to ionize sufficiently. Returning to these targets with our new knowledge that we need to retain a high concentration of detergent to form protective micelles, we obtained dramatically different results. Using this protective “bubble” it was now possible to project an intact rotary ATPase, comprising ∼30 subunits, into the gas phase (24). Initially, we mistook the precise cohorts of lipids that came with the ATPase for additional subunits. Designing new software to assign these spectra (25), applying quantitative proteomics and lipidomics, and confirming these interactions by tandem MS, we revealed lipids bound directly to c-subunits within the rotor (Fig. 2). From these data we concluded that large lipid plugs, consisting of 10 cardiolipins (Fig. 2 A and B) or 6 phosphatidylethanolamine (PE) lipids (Fig. 2 C and D), reside within the rings of two V-type ATPases from Enterococcus hirae and Thermus thermophilus, respectively. These lipid plugs, tailored to fit the different species, act to seal and adapt the rings to yield closely similar orifices, which accommodate the central stalk of the ATPase. Intriguingly, extraction with detergent appears to stabilize the plug, because recently we found that the lipid plug was absent in assemblies ejected directly from membranes (26). After fitting these lipids within the rotary rings, our models imply that the central orifices of both rotors (48 Å and 54 Å) are of similar dimensions, such that they can rotate their respective central stalk of comparable size.
Fig. 2.
Tandem MS of a subcomplex from T. thermophilus ATPase (ICL12E2G2F) leads to disruption of the L12 ring, releasing proteolipids L ± PE (red/green circle, 8,539 Da; red circles, 7,849 Da) and a stripped-complex ICE2G2F (blue squares, 184,242 Da). Atomic structure of the K10 ring of E. hirae ATPase (50) with docking of 10 cardiolipins to show reduction in the inner diameter (A) and after docking subunit C (B). Models for sixfold symmetry of the L12 ring with six PE molecules (green) (C) and with subunit C (blue) (51) docked into the ring (D). (Lower) Schematics of the rotor ring with 12-L subunits each having two transmembrane helices (red cylinders) and one conserved glu-63 (yellow) as seen in electron microscopy of 2D crystals (52) (i). Transformation into a sixfold symmetric ring (ii and iii). (iv) Comparison of the sixfold symmetrical model with electron microscopy data reported previously for the T. thermophilus ATPase (iv) (53).
Uncovering Lipids That Fine-Tune Membrane Protein Function
Having defined the critical lipid plug within the rotary ATPases, we became intrigued by the likelihood that lipids would be implicated in the function of other membrane protein complexes. Returning to ABC transporters, with which we first developed our approach (4), we were particularly interested in the roles played by lipids in the conduit of drugs or small molecules, and in the transition between the many different conformational states. Combining the MS of membrane proteins with ion mobility we were able to demonstrate the synergy between lipid and drug binding to P-glycoprotein (27), the importance of annular lipids in the ATPase activity of TmrAB (28), and the preference for negatively charged phosphatidyl glycerol for MsbA (29).
This ability to define preferential lipid binding, from bulk lipid association, suggested that we could settle a long-standing controversy over the identity of the bacterial lipid II flippase (30). Two possible lipid II flippases had been proposed: MurJ and FtsW (31, 32). Following expression of both proteins and study by MS, we revealed only low levels of lipid II binding to FtsW compared with MurJ, consistent with MurJ having a higher affinity (33). We also demonstrated that the antibiotic ramoplanin dissociates lipid II from MurJ, whereas vancomycin binds to form a stable ternary complex. Furthermore, we showed that cardiolipin associates with MurJ, but not FtsW, and that exogenous cardiolipins reduce lipid II binding to MurJ. These observations identify MurJ as the primary lipid II flippase and allowed us to suggest roles for endogenous lipids in fine-tuning lipid II binding.
While it was possible to observe lipid binding, and to demonstrate synergistic interactions with other small molecules, we wanted to define those lipids that had a structural impact on the membrane protein itself. We hypothesized that mechanosensitive channels of large conductance (MscL) would likely be influenced significantly by lipid binding, given that the complex responds to tension in the bilayer. Using MscL from Mycobacterium tuberculosis, and resolving lipid-bound states in mass spectra, we ranked bound lipids on the basis of their ability to resist gas-phase unfolding. We found that lipids bind with high affinity and all impart comparable stability (34). The highest-ranking lipid was found to be phosphatidylinositol phosphate, in line with its proposed functional role in mechanosensation. Turning to aquaporin Z (AqpZ) from Escherichia coli, we found that many lipids enhanced stability; however, we found that only cardiolipin affected AqpZ function. For the ammonia channel (AmtB) from E. coli, we found that it was highly selective for phosphatidylglycerol, prompting us to obtain an X-ray structure of this protein in this lipid membrane-like environment. The resulting 2.3-Å resolution structure, which we compared with others obtained without lipids bound, showed distinct conformational changes that reposition AmtB residues to interact with the lipid bilayer (34). These experiments highlighted to us the importance of the lipid cohort, not only for structure but also for function.
That strong interactions between lipids and proteins occur primarily through association of charged headgroups and amino acid side chains is well established. In accord with this, we found that binding to OmpF of anionic phosphatidylglycerol (POPG) or zwitterionic phosphatidylcholine (POPC) is sensitive to changes in the polarity of the mass spectrometer, and thereby the charge on the amino acid headgroup. The effects of polarity are less pronounced for other proteins in outer or mitochondrial membranes: the ferripyoverdine receptor (FpvA) or the voltage-dependent anion channel (VDAC), for example. Only marginal charge-induced differences were observed for inner membrane proteins: the ammonia channel or MscL.
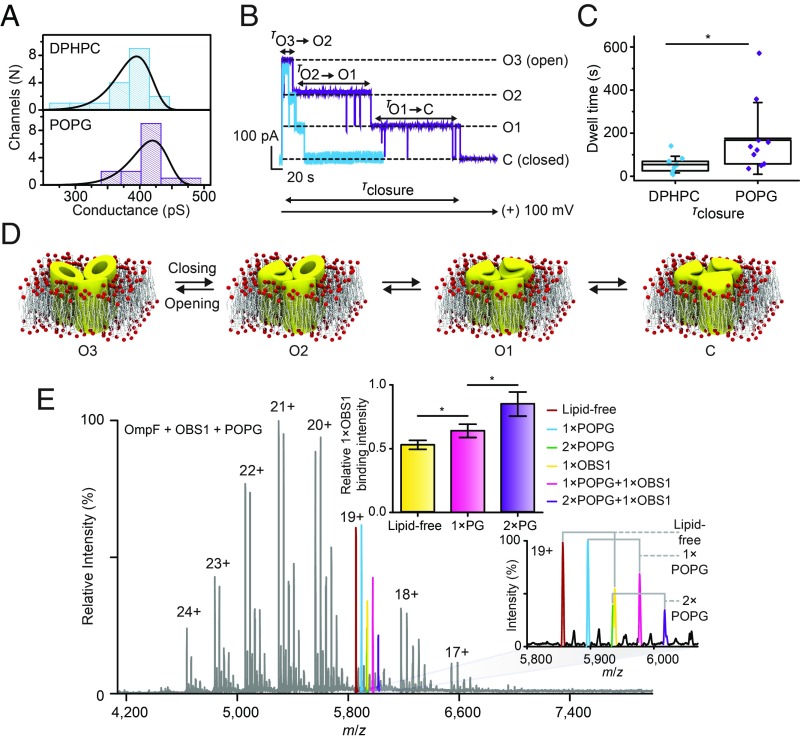
To understand these different sensitivities, we performed an extensive bioinformatics analysis of membrane protein structures and found that OmpF, and to a lesser extent FpvA and VDAC, have atypically high local densities of basic and acidic residues in their lipid headgroup-binding regions. We performed channel-recording experiments, at low pH, to show that POPG can maintain OmpF channels in open conformations for extended time periods (Fig. 3 A–C). Subsequently, we reasoned that if OmpF channels were held by POPG in open conformations for extended time periods, an intrinsically disordered peptide OBS1 (OmpF-binding site 1) would gain increased access to the inside of the channels (35). Using a high-resolution Orbitrap instrument, adapted for native MS (36) and further developed for membrane proteins (37), we showed increased binding of the OBS1 peptide in the presence of POPG. Because the outer membrane is composed almost entirely of anionic lipopolysaccharide, with similar headgroup properties to POPG, such anionic lipid binding could prevent closure of OmpF channels, thereby increasing access of antibiotics that use porin-mediated pathways. That these lipids can maintain open channels was discovered almost entirely by accident, through our attempts to rationalize changes in lipid binding in response to changes in electrospray polarity (38).
Fig. 3.
Negatively charged lipid (POPG) influences OmpF porin gating at low pH. (A) OmpF channel conductance values (all three pores open) were obtained in DPhPC planar bilayers (blue) and in DPhPC/POPG (3:1 ratio) bilayers (purple) with 19 and 15 independent OmpF porins, respectively. (B) Representative current versus time traces for single OmpF porins in a DPhPC bilayer (purple) and in a DPhPC/POPG (3:1) bilayer (blue). (C) Plot of closure times (box and whisker) showing that they are statistically different. (D) Schematic depicting stepwise OmpF gating with three, two, one, and no pores open (O3, O2, O1, C). (E) High-resolution mass spectrum of OmpF in the presence of the peptide OBS1 and POPG. Expansion of the 19+ charge state reveals a greater intensity of peptide binding in the presence of POPG than when bound in the absence of this lipid. *P < 0.05.
Because MS can monitor simultaneously the oligomeric state and lipid-binding properties, it should be ideally suited to uncover lipid-mediated oligomerization. To investigate if this were the case, we first developed a high-energy MS platform to disrupt the micelle and then enable tandem MS of protein–lipid complexes. We then considered the mass spectra of 12 membrane proteins and found that they were stable without lipids present. This was surprising because many of these assemblies, for example MscL, might be expected to have an intimate relationship with the lipid membrane because it is known to respond to tension in the bilayer (39). In contrast, for the bacterial homolog of the eukaryotic biogenic transporter (LeuT), we observed a precise cohort of lipids bound to the dimer, and importantly, removal of these lipids abrogated dimer formation. Combining this observation with molecular dynamics simulations revealed that cardiolipin acts as a bidentate ligand, bridging across subunits. Subsequently, we showed that for the Vibrio splendidus sugar transporter (SemiSWEET), cardiolipin shifts the equilibrium from monomer to functional dimer. We therefore hypothesized that lipids might be essential for dimerization of the Na+/H+ antiporter NhaA from E. coli, but not for the substantially more stable homologous T. thermophilus protein NapA. We found that lipid binding is obligatory for dimerization of NhaA, whereas NapA has adapted to form an interface that is stable without lipids. Correlating the interface strength of a series of dimers with the presence or absence of interfacial lipids, we proposed roles for lipids in both transient and stable interactions (39). We realized that this not only explained our observations for the LeuT, semiSWEET, NhaA, and NapA, but also could explain associations in other α-helical membrane proteins, including G protein-coupled receptors (GPCRs).
We set out to discover whether or not lipids played a role in the anticipated dimerization of GPCRs (40). Our first major obstacle, however, was to maintain the folded state of a GPCR in the gas phase. After conducting many trials, we found conditions whereby we could detect drug binding to a GPCR, implying that folded structure was, at least to some extent, preserved (41). Building on from this, we developed an approach to uncover lipids that might stabilize GPCRs. Although detailed structural information is available for GPCRs, the effect of lipids on these receptors is largely unknown. We were able to maintain the trimeric Gαsβγ protein complex bound to the adenosine A2A receptor (A2AR) and stabilized by a nanobody with all five components present. Interestingly, when we released the receptor from this complex in the gas phase, we observed binding of two lipids: phosphatidylserine (PS) and phosphatidylinositol (PI) (42).
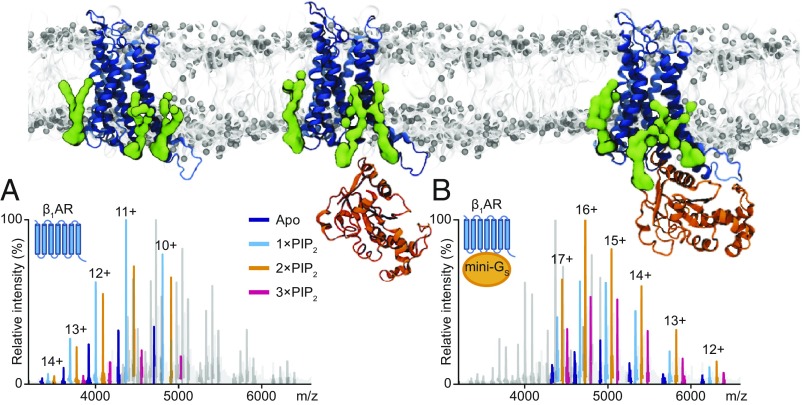
To investigate this further, we added PS and PI phosphates (PIPs) exogenously to three class A GPCRs. We found preferential binding of PIP2 over related lipids and, using engineered Gα subunits, showed that binding of two PIP2 molecules stabilizes the complex of mini-Gαs with the β1-adrenergic receptor (β1AR) (Fig. 4). We did not observe this stabilizing effect for other Gα subunits (mini-Gαi or mini-Gα12) or a high-affinity nanobody. Other endogenous lipids that we found to bind to these receptors had no effect on coupling, highlighting the specificity of PIP2. Increased Guanosine-5′-triphosphate (GTP) turnover by the activated neurotensin receptor when coupled to trimeric Gαiβγ complex in the presence of PIP2 provided us with further evidence for a specific effect of this lipid on coupling. These modulating effects of lipids on receptors suggest possibilities for understanding function, G protein selectivity, and drug targeting of class A GPCRs.
Fig. 4.
Molecular dynamics simulations identify hotspots for binding of PIP2 (green) on the intracellular side of class A receptors (blue), enhancing binding and docking of mini Gs (orange). (A) Mass spectrum showing the binding of PIP2 to the apo receptor and (B) after coupling of mini- Gs, wherein the intensity of the species binding to two PIP2 molecules is significantly higher than all others.
Moving Away from Membrane Mimetics
Over the past decade our MS studies of membrane protein complexes have been largely performed in detergent micelles. During this period we observed equilibrium of MscL complexes of differing stoichiometries in response to various detergents, lipids, and temperatures (37 °C), and propensity for lipids to bind in the presence of detergents (43, 44). These results, as well as other controversies in the literature, suggested to us a need to move away from detergent, ideally producing complexes from native membranes without recourse to detergents. While many approaches, when combined with MS are promising in this area (45)—including the styrene-maleic acid lipid particle technology (SMALPs), which requires a low pH step—subsequent release of protein assemblies in the gas phase is not straightforward (46). Approaches that could proceed directly from the membrane to the mass spectrometer, without chemical intervention, would undoubtedly be ideal.
Very recently we showed that we could project complexes directly from lipid vesicles following sonication. After further development of the Orbitrap platform, and in the first report of this technology (26) known as SOLVE (sonicated lipid vesicles), we demonstrated release of proteins directly from native membranes into the gas phase of the mass spectrometer. Assigning the spectra was, however, a formidable task. Many strategies were employed, including proteomics and lipidomics, assignment of split peaks to cofactor or protein binding, identification of diffuse peaks to the heterogeneity of lipid chain length, accurate mass measurement, and correlation with reported posttranslational modifications. New interactions were uncovered. From E. coli outer membranes we identified interactions including an OmpA dimer bound to the chaperone DnaK. We also observed an additional subunit in the Bam complex, and in its absence, we found cardiolipin-binding interactions, prompting us to suggest a membrane-targeting mechanism. For inner membranes from E. coli we observed cytochromes and the F1Fo ATP synthase, surprisingly with a c12 ring stoichiometry, in association with the SecYEG translocon. For bovine mitochondrial membranes, one of the primary complexes, the adenine nucleotide translocator (ANT1), was ejected predominantly as a dimer with palmitate caught in the act of transport. Complexes in the inner membrane also shed light on the mechanism of SOLVE because ATP synthase in inner membranes was sheared during sonication but protected when the outer mitochondrial membrane was present (Fig. 5), enabling a well-resolved spectrum of complex V with the previously established c8 ring stoichiometry.
Fig. 5.
High m/z region of the mass spectrum recorded for complexes ejected directly from the sonicated lipid vesicles, formed directly from native bovine mitochondrial membranes. Complexes I, III, IV, and V are expelled from intact mitochondria. Complex V with associated nucleotides and partial assemblies of complex I are observed in the absence of the catalytic core. (Right ) A depiction of the protein assemblies ejected from sonicated intact mitochondrial membranes color-coded according to the labels on the peaks. Gray subunits were not observed.
Overall, therefore, for mitochondrial membranes we were able to observe many complexes in the oxphos chain, together with their lipid and cofactor binding properties. It is clear that the full potential of this approach is yet to be explored. As we attempt to advance the method, we will explore further the range of membranes accessible to the technique, as well as the types of complexes that are ejected preferentially from the membrane and the mechanism of their expulsion. With these developments we hope that SOLVE will become universal as a procedure for the study of membrane proteins and will mitigate the problems that can arise from chemical intervention.
Future Perspectives
In parallel with developments in MS, other techniques are becoming ever more powerful: for example, cryoelectron microscopy and tomography. Breath-taking images of complexes in action are now emerging. As the complexity of assemblies amenable to imaging increases dramatically, molecular details of their subunit interfaces, posttranslational modification status, and small molecule-binding attributes will need to be defined. Further developments that combine imaging at the single-molecule level with MS offer great promise (47) and, with further development, will provide molecular detail of the structure and regulation of proteins and their complexes.
As we begin to study membrane protein complexes in the context of their native bilayers, opportunities arise to compare analogous complexes from different membranes and to learn how specific lipids modulate their properties. This will further advance our knowledge of membrane proteins, their native lipid environments, and enable better drug targeting. With further developments in MS we will be in a position to resolve complexes from heterogeneous matrices, with modifications and binding patterns intact, and truly conquer all aspects of their structure and function.
Acknowledgments
I thank all group members, past and present, for their many varied and creative contributions. Without their constant enthusiasm for this research, our collective understanding of protein complexes in the gas phase would be considerably weaker and my enjoyment and enthusiasm for their study would be significantly less. I also acknowledge, with great appreciation, contributions of all colleagues and collaborators in this exciting field. This work was supported by the European Research Council Grant 69551-ENABLE, a Wellcome Trust Investigator Award (104633/Z/14/Z), and a Medical Research Council Programme Grant MRC (MR/N020413/1).
Footnotes
The author declares no conflict of interest.
References
- 1.Wood TD, et al. Gas-phase folding and unfolding of cytochrome c cations. Proc Natl Acad Sci USA. 1995;92:2451–2454. doi: 10.1073/pnas.92.7.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McLafferty FW. Mass spectrometry across the sciences. Proc Natl Acad Sci USA. 2008;105:18088–18089. doi: 10.1073/pnas.0800784105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeung YG, Nieves E, Angeletti RH, Stanley ER. Removal of detergents from protein digests for mass spectrometry analysis. Anal Biochem. 2008;382:135–137. doi: 10.1016/j.ab.2008.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrera NP, Di Bartolo N, Booth PJ, Robinson CV. Micelles protect membrane complexes from solution to vacuum. Science. 2008;321:243–246. doi: 10.1126/science.1159292. [DOI] [PubMed] [Google Scholar]
- 5.Hendrickson WA. Atomic-level analysis of membrane-protein structure. Nat Struct Mol Biol. 2016;23:464–467. doi: 10.1038/nsmb.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milić D, Veprintsev DB. Large-scale production and protein engineering of G protein-coupled receptors for structural studies. Front Pharmacol. 2015;6:66. doi: 10.3389/fphar.2015.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miranker A, Robinson CV, Radford SE, Aplin RT, Dobson CM. Detection of transient protein folding populations by mass spectrometry. Science. 1993;262:896–900. doi: 10.1126/science.8235611. [DOI] [PubMed] [Google Scholar]
- 8.Robinson CV, et al. Conformation of GroEL-bound alpha-lactalbumin probed by mass spectrometry. Nature. 1994;372:646–651. doi: 10.1038/372646a0. [DOI] [PubMed] [Google Scholar]
- 9.Rostom AA, et al. Dissection of multi-protein complexes using mass spectrometry: Subunit interactions in transthyretin and retinol-binding protein complexes. Proteins. 1998;33(Suppl 2):3–11. doi: 10.1002/(sici)1097-0134(1998)33:2+<3::aid-prot2>3.3.co;2-8. [DOI] [PubMed] [Google Scholar]
- 10.Rostom AA, Robinson CV. Detection of the intact GroEL chaperonin assembly by mass spectrometry. J Am Chem Soc. 1999;121:4718–4719. [Google Scholar]
- 11.Sobott F, Benesch JL, Vierling E, Robinson CV. Subunit exchange of multimeric protein complexes. Real-time monitoring of subunit exchange between small heat shock proteins by using electrospray mass spectrometry. J Biol Chem. 2002;277:38921–38929. doi: 10.1074/jbc.M206060200. [DOI] [PubMed] [Google Scholar]
- 12.Fändrich M, et al. Observation of the noncovalent assembly and disassembly pathways of the chaperone complex MtGimC by mass spectrometry. Proc Natl Acad Sci USA. 2000;97:14151–14155. doi: 10.1073/pnas.240326597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benesch JL, Sobott F, Robinson CV. Thermal dissociation of multimeric protein complexes by using nanoelectrospray mass spectrometry. Anal Chem. 2003;75:2208–2214. doi: 10.1021/ac034132x. [DOI] [PubMed] [Google Scholar]
- 14.Clemmer DE, Jarrold MF. Ion mobility measurements and their applications to clusters and biomolecules. J Mass Spectrom. 1997;32:577–592. [Google Scholar]
- 15.Ruotolo BT, et al. Evidence for macromolecular protein rings in the absence of bulk water. Science. 2005;310:1658–1661. doi: 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]
- 16.Ruotolo BT, et al. Ion mobility-mass spectrometry reveals long-lived, unfolded intermediates in the dissociation of protein complexes. Angew Chem Int Ed Engl. 2007;46:8001–8004. doi: 10.1002/anie.200702161. [DOI] [PubMed] [Google Scholar]
- 17.Benesch JL, et al. Separating and visualising protein assemblies by means of preparative mass spectrometry and microscopy. J Struct Biol. 2010;172:161–168. doi: 10.1016/j.jsb.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Mikhailov VA, Mize TH, Benesch JL, Robinson CV. Mass-selective soft-landing of protein assemblies with controlled landing energies. Anal Chem. 2014;86:8321–8328. doi: 10.1021/ac5018327. [DOI] [PubMed] [Google Scholar]
- 19.Ilag LL, Ubarretxena-Belandia I, Tate CG, Robinson CV. Drug binding revealed by tandem mass spectrometry of a protein-micelle complex. J Am Chem Soc. 2004;126:14362–14363. doi: 10.1021/ja0450307. [DOI] [PubMed] [Google Scholar]
- 20.Lengqvist J, Svensson R, Evergren E, Morgenstern R, Griffiths WJ. Observation of an intact noncovalent homotrimer of detergent-solubilized rat microsomal glutathione transferase-1 by electrospray mass spectrometry. J Biol Chem. 2004;279:13311–13316. doi: 10.1074/jbc.M310958200. [DOI] [PubMed] [Google Scholar]
- 21.Barrera NP, et al. Mass spectrometry of membrane transporters reveals subunit stoichiometry and interactions. Nat Methods. 2009;6:585–587. doi: 10.1038/nmeth.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stock D, Gibbons C, Arechaga I, Leslie AG, Walker JE. The rotary mechanism of ATP synthase. Curr Opin Struct Biol. 2000;10:672–679. doi: 10.1016/s0959-440x(00)00147-0. [DOI] [PubMed] [Google Scholar]
- 23.Esteban O, et al. Stoichiometry and localization of the stator subunits E and G in Thermus thermophilus H+-ATPase/synthase. J Biol Chem. 2008;283:2595–2603. doi: 10.1074/jbc.M704941200. [DOI] [PubMed] [Google Scholar]
- 24.Zhou M, et al. Mass spectrometry of intact V-type ATPases reveals bound lipids and the effects of nucleotide binding. Science. 2011;334:380–385. doi: 10.1126/science.1210148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgner N, Robinson CV. Massign: An assignment strategy for maximizing information from the mass spectra of heterogeneous protein assemblies. Anal Chem. 2012;84:2939–2948. doi: 10.1021/ac300056a. [DOI] [PubMed] [Google Scholar]
- 26.Chorev DS, et al. Protein assemblies ejected directly from native membranes yield complexes for mass spectrometry. Science. 2018;362:829–834. doi: 10.1126/science.aau0976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcoux J, et al. Mass spectrometry reveals synergistic effects of nucleotides, lipids, and drugs binding to a multidrug resistance efflux pump. Proc Natl Acad Sci USA. 2013;110:9704–9709. doi: 10.1073/pnas.1303888110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bechara C, et al. A subset of annular lipids is linked to the flippase activity of an ABC transporter. Nat Chem. 2015;7:255–262. doi: 10.1038/nchem.2172. [DOI] [PubMed] [Google Scholar]
- 29.Gupta K, et al. Identifying key membrane protein lipid interactions using mass spectrometry. Nat Protoc. 2018;13:1106–1120. doi: 10.1038/nprot.2018.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruiz N. Lipid flippases for bacterial peptidoglycan biosynthesis. Lipid Insights. 2016;8:21–31. doi: 10.4137/LPI.S31783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sham LT, et al. Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science. 2014;345:220–222. doi: 10.1126/science.1254522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohammadi T, et al. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 2011;30:1425–1432. doi: 10.1038/emboj.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolla JR, et al. Direct observation of the influence of cardiolipin and antibiotics on lipid II binding to MurJ. Nat Chem. 2018;10:363–371. doi: 10.1038/nchem.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laganowsky A, et al. Membrane proteins bind lipids selectively to modulate their structure and function. Nature. 2014;510:172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Housden NG, et al. Intrinsically disordered protein threads through the bacterial outer-membrane porin OmpF. Science. 2013;340:1570–1574. doi: 10.1126/science.1237864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rose RJ, Damoc E, Denisov E, Makarov A, Heck AJ. High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nat Methods. 2012;9:1084–1086. doi: 10.1038/nmeth.2208. [DOI] [PubMed] [Google Scholar]
- 37.Gault J, et al. High-resolution mass spectrometry of small molecules bound to membrane proteins. Nat Methods. 2016;13:333–336. doi: 10.1038/nmeth.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liko I, et al. Lipid binding attenuates channel closure of the outer membrane protein OmpF. Proc Natl Acad Sci USA. 2018;115:6691–6696. doi: 10.1073/pnas.1721152115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupta K, et al. The role of interfacial lipids in stabilizing membrane protein oligomers. Nature. 2017;541:421–424. doi: 10.1038/nature20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dijkman PM, et al. Dynamic tuneable G protein-coupled receptor monomer-dimer populations. Nat Commun. 2018;9:1710. doi: 10.1038/s41467-018-03727-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yen HY, et al. Ligand binding to a G protein-coupled receptor captured in a mass spectrometer. Sci Adv. 2017;3:e1701016. doi: 10.1126/sciadv.1701016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yen HY, et al. PtdIns(4,5)P2 stabilizes active states of GPCRs and enhances selectivity of G-protein coupling. Nature. 2018;559:423–427. doi: 10.1038/s41586-018-0325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reading E, et al. The effect of detergent, temperature, and lipid on the oligomeric state of MscL constructs: Insights from mass spectrometry. Chem Biol. 2015;22:593–603. doi: 10.1016/j.chembiol.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Landreh M, Costeira-Paulo J, Gault J, Marklund EG, Robinson CV. Effects of detergent micelles on lipid binding to proteins in electrospray ionization mass spectrometry. Anal Chem. 2017;89:7425–7430. doi: 10.1021/acs.analchem.7b00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reading E, et al. Interrogating membrane protein conformational dynamics within native lipid compositions. Angew Chem Int Ed Engl. 2017;56:15654–15657. doi: 10.1002/anie.201709657. [DOI] [PubMed] [Google Scholar]
- 46.Hellwig N, et al. Native mass spectrometry goes more native: Investigation of membrane protein complexes directly from SMALPs. Chem Commun (Camb) 2018;54:13702–13705. doi: 10.1039/c8cc06284f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Longchamp JN, et al. Imaging proteins at the single-molecule level. Proc Natl Acad Sci USA. 2017;114:1474–1479. doi: 10.1073/pnas.1614519114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sobott F, Hernández H, McCammon MG, Tito MA, Robinson CV. A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal Chem. 2002;74:1402–1407. doi: 10.1021/ac0110552. [DOI] [PubMed] [Google Scholar]
- 49.Giles K, et al. Applications of a travelling wave-based radio-frequency-only stacked ring ion guide. Rapid Commun Mass Spectrom. 2004;18:2401–2414. doi: 10.1002/rcm.1641. [DOI] [PubMed] [Google Scholar]
- 50.Murata T, Yamato I, Kakinuma Y, Leslie AG, Walker JE. Structure of the rotor of the V-Type Na+-ATPase from Enterococcus hirae. Science. 2005;308:654–659. doi: 10.1126/science.1110064. [DOI] [PubMed] [Google Scholar]
- 51.Iwata M, et al. Crystal structure of a central stalk subunit C and reversible association/dissociation of vacuole-type ATPase. Proc Natl Acad Sci USA. 2004;101:59–64. doi: 10.1073/pnas.0305165101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toei M, et al. Dodecamer rotor ring defines H+/ATP ratio for ATP synthesis of prokaryotic V-ATPase from Thermus thermophilus. Proc Natl Acad Sci USA. 2007;104:20256–20261. doi: 10.1073/pnas.0706914105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bernal RA, Stock D. Three-dimensional structure of the intact Thermus thermophilus H+-ATPase/synthase by electron microscopy. Structure. 2004;12:1789–1798. doi: 10.1016/j.str.2004.07.017. [DOI] [PubMed] [Google Scholar]