Abstract
Background
The importance of gut bacteria in human physiology, immune regulation, and disease pathogenesis is well established. In contrast, the composition and dynamics of the gut virome are largely unknown; particularly lacking are studies in pregnancy. We used comprehensive virome capture sequencing to characterize the gut virome of pregnant women with and without type 1 diabetes (T1D), longitudinally followed in the Environmental Determinants of Islet Autoimmunity study.
Methods
In total, 61 pregnant women (35 with T1D and 26 without) from Australia were examined. Nucleic acid was extracted from serial fecal specimens obtained at prenatal visits, and viral genomes were sequenced by virome capture enrichment. The frequency, richness, and abundance of viruses were compared between women with and without T1D.
Results
Two viruses were more prevalent in pregnant women with T1D: picobirnaviruses (odds ratio [OR], 4.2; 95% confidence interval [CI], 1.0–17.1; P = .046) and tobamoviruses (OR, 3.2; 95% CI, 1.1–9.3; P = .037). The abundance of 77 viruses significantly differed between the 2 maternal groups (≥2-fold difference; P < .02), including 8 Enterovirus B types present at a higher abundance in women with T1D.
Conclusions
These findings provide novel insight into the composition of the gut virome during pregnancy and demonstrate a distinct profile of viruses in women with T1D.
Keywords: enterovirus, pregnancy, type 1 diabetes, virome capture sequencing
Pregnancy is a complex immunological state in which the balance between an inimical alloimmune response and an environment of maternal tolerance may be perturbed by virus infections [1], potentially resulting in significant perinatal morbidity [2]. Helper T-lymphocyte (Th)-associated cytokines shift from Th1 towards Th2 to protect the fetus; however, this Th2 bias can diminish cell-mediated immunity and increase vulnerability to intracellular infections, including viruses [3–5]. In pregnant women with type 1 diabetes (T1D), hyperglycemia may impede pathogen clearance, increasing the duration of gestational infections [6] and heightening the risk of adverse outcomes in the fetus, including subsequent development of T1D during childhood [7]. This is supported by our recent systematic review and meta-analysis of 2992 women and children, which demonstrated a significant association between maternal virus infection in pregnancy and T1D in offspring (odds ratio [OR], 2.2; 95% confidence interval [CI], 1.2–3.8; P = .008) [8]. To date, no study has examined the gut virome during pregnancy and how it may be altered by T1D.
METHODS
Study Population and Sample Selection
The gut virome of 61 pregnant women (35 with T1D and 26 without) in the ENDIA (Environmental Determinants of Islet Autoimmunity) study (Australian New Zealand Clinical Trials Registry registration number ACTRN12613000794707) was examined (Supplementary Table 1). ENDIA is a prospective cohort study of children at risk of T1D (have ≥1 first-degree relative with T1D), followed longitudinally from pregnancy to 3 years of life [9]. Women recruited between 2012 and 2016 were included in this analysis. Overall, 59 pregnancies were singleton and 2 were twin pregnancies. Fecal samples were collected during the first (n = 18), second (n = 47), and/or third trimester of pregnancy (n = 59), which were defined as gestational age of 1–14, 15–26, and 27–42 weeks, respectively. All samples were stored at −80oC in aliquots before analysis. For every participant, all available samples were examined. In total, 49 of 61 women had samples for multiple timepoints and 12 of 61 had samples for all 3 trimesters.
The study was reviewed and approved (July 13, 2016) by the study’s lead Human Research Ethics Committee at the Women’s and Children’s Health Network under the National Mutual Acceptance Scheme (HREC/16/WCHN/66) and at all participating study sites in Australia. All participants provided written informed consent and were free to withdraw from the study at any time. Families were excluded if the mother could not comprehend her participation in the study and therefore was unable to provide informed consent.
Nucleic Acid Extraction
Total nucleic acid (NA) was extracted using the MagMAX Total NA Isolation Kit (Thermo Fisher Scientific) on the semi-automated KingFisher FLEX Purification System (Thermo Fisher Scientific), following manufacturer’s guidelines with minor modifications. Thirty percent (w/v) of fecal suspensions were prepared in 1× phosphate-buffered saline and centrifuged for 5 minutes. All spin steps were performed at 13 000 ×g at room temperature. After centrifugation, 175 μL supernatant was transferred to zirconium bead tubes containing 235 μL Lysis/Binding Solution. Bead tubes were shaken at 2400 rpm on the Bioshake iQ (Quantifoil Instruments, Jena, Germany) for 15 minutes, then centrifuged 3 minutes. Into new tubes, 300 μL lysate was transferred and further centrifuged for 6 minutes. Total NA was purified from 200 μL lysate and stored at −80oC.
Sequence-Independent Amplification
Total NA was subjected to complementary deoxyribonucleic acid (cDNA) synthesis and sequence-independent preamplification (SIP) using the Transplex Complete Whole Transcriptome Amplification Kit (WTA1; Sigma-Aldrich, St. Louis, MO), following a published protocol [10]. In brief, 3.5 μL total NA was denatured at 95oC for 5 minutes instead of 70oC before cDNA synthesis to ensure amplification of both DNA and ribonucleic acid (RNA) molecules. Denatured NA was cooled to 18oC, and cDNA was synthesized using the following thermocycling conditions: 18oC 10 minutes, 25oC 10 minutes, 37oC 30 minutes, 42oC 10 minutes, 70oC 20 minutes, and 4oC holding. The entire cDNA library was used as template for SIP using the following cycling conditions repeated 22 times: 94oC 30 seconds and 70oC 5 minutes. After amplification, polymerase chain reaction (PCR) products were visualized on an agarose gel before purification using the ChargeSwitch-Pro PCR CleanUp Kit (Thermo Fisher Scientific).
Virome Capture Sequencing
One microgram of double-stranded DNA (dsDNA) was used for library synthesis using the KAPA Hyperplus kit (KAPA Biosystems, Wilmington, MA) with single-index adapters. In brief, dsDNA was enzymatically fragmented to an average of 200 base pairs. Fragments were purified using AmpureXP beads (Beckman Coulter, Brea, CA). Libraries were amplified for 6–9 cycles, quality checked on the LabChip GX Touch 24 Bioanalyzer (PerkinElmer, Waltham, MA), and quantified using the picogreen assay (Thermo Fisher Scientific) on the Victor X2 Fluorescent Microplate Reader (PerkinElmer). Completed libraries were pooled by equal mass for sequence capture. VirCapSeq-VERT was performed according to the Nimblegen SeqCap protocol (Roche, Basel, Switzerland) as described previously [11]. Postcapture libraries were purified and amplified before sequencing. To ensure sufficient depth of coverage (approximately 10 million unique sequence reads/sample), uniquely barcoded samples were pooled at a maximum of 20 libraries per pool (20-plex) and sequenced on a lane of HiSeq 4000 (Illumina, San Diego, CA).
Genome Sequence Analysis
Genome assembly, contig generation, and taxonomic classification of reads were performed as previously described [11]. Demultiplexed and quality-trimmed sequence reads were aligned against host reference databases from GenBank (National Center for Biotechnology Information) using the Bowtie2 mapping algorithm (version 2.1.0) [12] to remove the host background. Filtered reads were assembled de novo using either SOAPdenovo2 [13], MEGAHIT [14], or MIRA assemblers [15], then contigs and unique singletons were subjected to homology search at the nucleotide level using MegaBLAST. Sequences that exhibited poor or no homology at the nucleotide level were screened further by BLASTX against the viral GenBank protein database. Viral sequences detected from BLASTX analysis were subjected to another round of BLASTX homology search against the entire GenBank protein database to correct for biased E values and inaccurate taxonomic classifications. For reference-based alignments, to visualize depth and spread of coverage for individual viruses, both Integrated Genomics Viewer [16] and Geneious (version 9.0.5) [17] were used. After taxonomic classification, read counts were corrected to account for sample bleeding due to Illumina index cross-talk, where sequences with single index barcodes are erroneously sorted, resulting in approximately 0.1% of total reads being distributed to the incorrect sample Fastq file. Cutoffs were applied across each pool separately. For each virus, 0.1% of the highest read count in that pool was calculated and subtracted from the number of reads of that virus in each sample. All resulting read counts below 1 were corrected to zero. This process minimized the risk of false-“positive” identification of viruses in samples. To evaluate virus positivity, a threshold of 100 viral reads matched by Basic Local Alignment Search Tool (BLAST) at the species level, randomly distributed over the target genome, was applied. This threshold was selected for its proximity to the typical limit of detection of targeted quantitative PCR (~100 viral copies/mL), determined based on previous VirCapSeq-VERT experiments using whole blood [11] and feces (unpublished data).
Statistical Analysis
The STROBE reporting guidelines for observational cohort studies were followed [18]. Continuous demographic variables are reported as a mean ± standard deviation (SD) for parametric data and median (interquartile range) for skewed data, and categorical data as number (%). Participant characteristics, including demographic variables, lifestyle factors, and comorbidities, are reported according to T1D status and were compared using independent t tests and Fisher’s Exact tests for continuous and categorical data, respectively. The socioeconomic index for areas (index for relative socioeconomic disadvantage) percentile for the postal area in which each patient resided was used as an indicator of socioeconomic status (SES) [19]. High SES was defined as >75th percentile [20]. Virus positivity was determined by a positivity threshold of 100 viral reads matched by BLAST at the species level.
The differential abundance of viruses between mothers with and without T1D was examined using the edgeR package (version 3.14.0) [21] in R (version 3.3.0). A matrix of read counts, corrected for index cross-talk, was generated encompassing all samples and detected viruses. Each matrix entry had a count of 1 added to avoid issues with division by or log function of zero [22] before conversion to counts per million. Data were normalized using the Relative Log Expression method with respect to library size [23]. Two methods, common and tag-wise [24], were used to estimate the biological coefficient of variation. Samples were divided into case and control groups, and the “exact” test was used to perform hypothesis testing [25]. P values were adjusted to control false-discovery rates with the Benjamini-Hochberg multiple testing correction procedure [26]. Viruses with an adjusted P < .05 were identified as displaying statistically significant differential abundance between case and control groups. Heatmaps were created to visualize the data using iheatmapr package in R [27].
The richness of vertebrate-infecting viruses in feces was estimated using EstimateS software (version 9.1.0) [28]. Estimates used to calculate richness at the genus level and sample-based rarefaction curves were computed using prepublished analytical formulas [29]. In total, 100 randomizations (runs) were completed, extrapolating by a factor of 1.0 with estimates (knots) at every sample. Estimates of vertebrate-infecting virus richness in cases were compared with those of controls using a non-parametric test, assuming the estimates in cases and controls are statistically independent of each other. Rarefaction curves were plotted in Microsoft Excel, version 15.3 (Redmond, WA).
The association between maternal T1D and positivity for each virus detected in mothers during pregnancy was examined using generalized estimating equation (GEE) models. The GEE method for the binary outcome of virus positivity at the genus level (logit link function) was applied to account for the correlation among longitudinal observations of the same participant. In the GEE model, virus infection at the genus level was the independent variable and the major dependent variable was maternal T1D. Other explanatory variables examined were maternal age at conception, parity, pet ownership, SES, maternal body mass index (BMI), education, maternal smoking, and age at sample in the model.
Univariate and multivariable analyses were undertaken on both study groups. These covariates and factors in the GEE model were selected from a larger set of potential covariates by forwards and backwards regression. An individual model was used for each genus of virus, with independent variables that exhibited association with the outcome variable being included in multivariable analysis. The GEE models were compared using the quasi-likelihood under independence model criterion, and the lowest scoring and most parsimonious models are reported. An exchange correlation structure was used, and the results were expressed as OR determined by the regression coefficient expβ, with 95% CIs. Frequency of virus positivity limited multivariable analysis to the 5 most frequent genera of virus detected in both study groups. Statistical analyses were performed using IBM SPSS Statistics, version 24.0 (Chicago, IL). Statistical significance was defined as P ≤ .05.
RESULTS
The pregnancy gut virome of women with and without T1D was characterized using VirCapSeq-VERT on 124 fecal specimens collected from 61 mothers (n = 35 with T1D, n = 26 without T1D) in the ENDIA study (Supplementary Table 1). The mean (±SD) age of mothers at conception was 32 ± 4 years and BMI was 27 ± 6 kg/m2. Compared to women without T1D, women with T1D gave birth at a significantly younger age, after a shorter gestational length, and had fewer children (Supplementary Table 1).
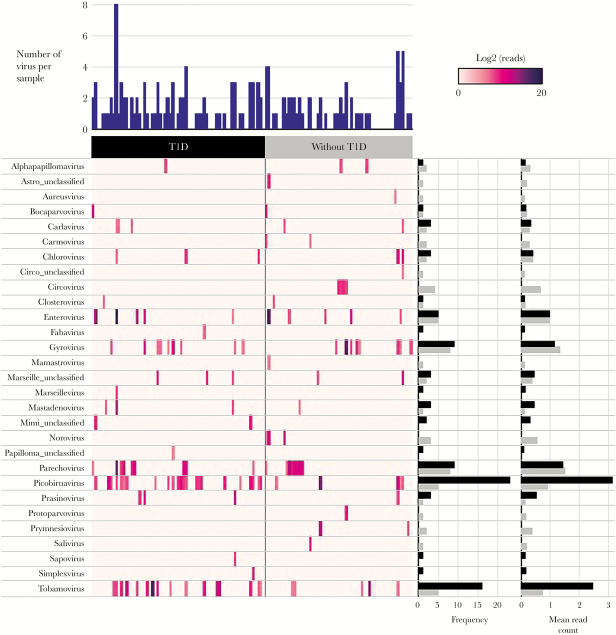
High-throughput sequencing generated ~2 billion raw reads, which reduced to 1.6 billion reads after filtration of host and primer sequences. This equated to 12.7 ± 4.2 million filtered reads per sample. In total, 29 genera of eukaryotic viruses were detected, and 63% of samples (78 of 124) tested positive for at least 1 virus (Figure 1). Members of the Picobirnavirus, Parechovirus, and Enterovirus (EV) genera were among the most frequent vertebrate-infecting viruses sequenced. Although nonvertebrate-infecting viruses were excluded from VirCapSeq-VERT enrichment, tobamoviruses were frequently detected, suggesting that plant viruses are highly abundant in the gut during pregnancy and prevalent in human feces [30]. Although not reaching statistical significance, there was a trend to higher virus positivity in mothers with T1D versus without (64% vs 50%; P = .14). Rarefaction analysis revealed no difference in the richness of vertebrate-infecting viruses between women with and without T1D (Supplementary Figure 1), suggesting that all participants were exposed to a comparable community of viruses, independent of their T1D status.
Figure 1.
Viruses detected using VirCapSeq-VERT. Heatmap of viral reads (log2 scale) sequenced in 124 fecal samples collected from 35 women with type 1 diabetes (T1D) (n = 69 samples) and 26 without (n = 55 samples) during pregnancy. Only viruses with ≥100 reads matched by Basic Local Alignment Search Tool (BLAST) at the species level were included and represented at the genus level. Number of viruses detected per specimen, frequency of each virus within the case or control group, and the mean log read counts are summarized by bar charts.
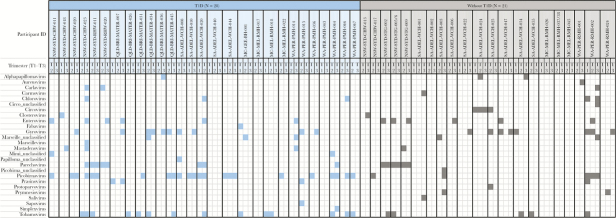
Examination of longitudinal samples from 49 of 61 participants (n = 28 with T1D, n = 21 without T1D) identified alphapapillomaviruses, circoviruses, parechoviruses, and picobirnaviruses in multiple trimesters of pregnancy within individuals (Figure 2). This may indicate persistent or recurring infection by closely related strains. There was no difference in the proportion of virus-positive samples across the 3 trimesters (P = .95). Chicken anaemia virus, genus Gyrovirus, and plant virus, pepper mild mottle virus, genus Tobamovirus were also detected across multiple trimesters, but these most likely originated from dietary intake [30].Two viruses were more prevalent in women with T1D: picobirnaviruses (33% vs 9%; OR = 4.2; 95% CI, 1.0–17.1; P = .046) and tobamoviruses (22% vs 9%; OR = 3.2; 95% CI, 1.1–9.3; P = .037). In multivariable GEE models, the higher odds of having picobirnaviruses and tobamoviruses in women with T1D remained significant after adjustment for maternal age. In addition, there was a trend towards higher rates of gyroviruses, chloroviruses, and carlaviruses in women with T1D, but differences did not reach statistical significance. The frequency of EV did not differ between the 2 maternal groups; however, significant differences in EV types were observed. Coxsackievirus A2 (CVA2), CVB4, CVB5, Rhinovirus B, and ECHOviruses were detected exclusively in women with T1D, whereas CVA6, CVA10, CVA14, and EV71 were present only in mothers without T1D (Supplementary Table 2).
Figure 2.
Longitudinal changes in the gut virome during pregnancy. Presence-absence heatmap of viruses detected over multiple trimesters of pregnancy (T1, T2, and T3) in women with type 1 diabetes (n = 28 individuals) and without (n = 21 individuals).
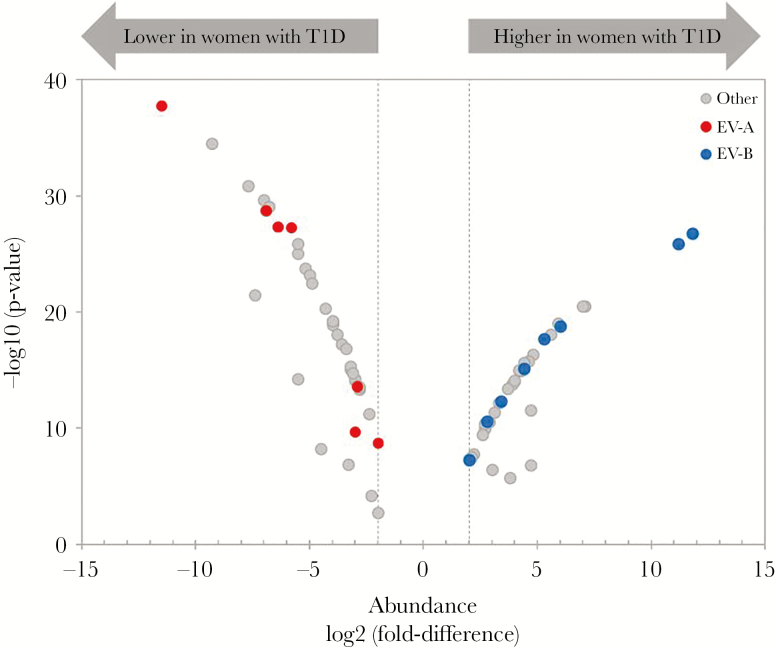
Differential abundance analysis identified 77 virus types with ≥2-fold significant difference (P < .02) between pregnant women with T1D versus those without, with a false discovery rate <5% (Figure 3 and Supplementary Data). Among the top 15 differentially abundant viruses were 3 EV-B types (CVB4, CVB3, and ECHOvirus E18), all present at higher abundance in women with T1D (Table 1). In contrast, 4 EV-A types (CVA10, CVA16, CVA5, and CVA14) were more abundant during pregnancy in women without T1D.
Figure 3.
Viruses differentially abundant between the gut of women with and without type 1 diabetes during pregnancy. Volcano plot of viruses with ≥2-fold difference (marked by vertical dotted lines) in abundance between pregnant women with and without type 1 diabetes. Only differences with false discovery rate below 5% (P < .05) as determined by edgeR are represented. Species A (EV-A) and B enteroviruses (EV-B) are marked in red and blue, respectively. All other viruses represented in gray.
Table 1.
Top 15 Differentially Abundant Species of Viruses between the Gut of Pregnant Women With Type 1 Diabetes Versus Without
| Virus | LogFD | P Value | FDR | Rank (Magnitude of FD) |
|---|---|---|---|---|
| Higher in Women with T1D | ||||
| Human coxsackievirus B4 | 11.8 | 1.70E-27 | 1.50E-26 | 1 |
| Human coxsackievirus B3 | 11.2 | 1.40E-26 | 1.10E-25 | 3 |
| Bathycoccus sp RCC1105 virus BpV2 | 7.1 | 2.90E-21 | 1.50E-20 | 7 |
| Human adenovirus A | 7 | 3.20E-21 | 1.60E-20 | 8 |
| ECHOvirus E18 | 6 | 1.60E-19 | 5.90E-19 | 15 |
| Lower in Women With T1D | ||||
| Coxsackievirus A10 | −11.5 | 1.70E-38 | 1.80E-36 | 2 |
| Brandmavirus UC1 | −9.3 | 2.90E-35 | 1.50E-33 | 4 |
| Phaeocystis globosa virus | −7.7 | 1.30E-31 | 3.50E-30 | 5 |
| Porcine picobirnavirus | −7.4 | 3.40E-22 | 1.90E-21 | 6 |
| Tomato mosaic virus | −7 | 2.30E-30 | 4.80E-29 | 9 |
| Ostreococcus lucimarinus virus OlV4 | −6.9 | 1.70E-29 | 2.20E-28 | 10 |
| Coxsackievirus A16 | −6.9 | 1.70E-29 | 2.20E-28 | 11 |
| Paramecium bursaria Chlorella virus | −6.8 | 7.70E-30 | 1.40E-28 | 12 |
| Coxsackievirus A5 | −6.4 | 4.30E-28 | 4.80E-27 | 13 |
| Coxsackievirus A14 | −6.4 | 4.60E-28 | 4.80E-27 | 14 |
Abbreviations: FD, fold difference; FDR, false discovery rate; T1D, type 1 diabetes.
DISCUSSION
We demonstrated that eukaryotic viruses are prevalent in the gut of women during pregnancy, and that women with T1D are more likely to harbor picobirnaviruses and tobamoviruses compared with women without T1D. Furthermore, we found significant differences in viral abundance between women with and without T1D, including 8 EV B types that were all present at a higher abundance in women with T1D. These results demonstrate a distinct profile of viruses in women with T1D in pregnancy.
The pathogenicity of picobirnaviruses in humans remains to be definitively established. A weak association with gastroenteritis in animals has been found, whereas in humans they are only considered as possible opportunistic pathogens [31]. Most recently, picobirnaviruses were detected at high levels in patients with human immunodeficiency virus [32] and graft-versus-host disease [33], leading to the proposal that they may serve as a biomarker of immunosuppression. Thus, it is plausible that a higher prevalence of picobirnaviruses in women with T1D could be reflective of impaired antiviral defence. Tobamoviruses are not known to be pathogenic to humans and are commonly thought to be introduced to the gut through diet [30, 34]. Therefore, their higher prevalence in women with T1D during pregnancy may reflect differences in diet or consumption of contaminated drinking water [35]. Alternatively, there may be other factors involved such as gut permeability and intestinal inflammation, which are both increased in individuals with T1D [36] and may prevent effective clearance of dietary viruses. Our frequent detection of tobamoviruses in feces is consistent with other virome studies, including a recent study of 5 mother-infant pairs [30, 37].
The predominance of EV-B types found in pregnant women with T1D is consistent with higher rates of EV-B observed in individuals with T1D versus healthy controls [38–40]. Furthermore, the greater abundance of CVB4 in women with T1D complements the body of molecular and epidemiological evidence implicating CVB4 in T1D pathogenesis [39, 41–44]. In contrast, EV-A types were more prevalent and present at a higher abundance in pregnant women without T1D (Figure 3). This contrasts results obtained from children in the Finnish Diabetes Prediction and Prevention (DIPP) study that found a higher rate of EV-A infections in cases who developed T1D compared with matched controls, during a time window more than 12 months before the first detection of islet autoantibody [45]. The result also contrasts our own findings in the Australian Viruses In the Genetically at Risk (VIGR) study [46], where we also found a predominance of EV-A viruses in feces of children before or at the time of the first islet autoantibody detection when compared with matched controls (Kim et al [47]).
A recent study examining the intestinal virome changes that precede the development of autoimmunity in T1D-susceptible children detected circoviruses at a greater abundance and prevalence in controls, suggesting that infection with this virus may offer protection from the development of T1D [48]. Consistent with this hypothesis, circoviruses were exclusively detected in women without T1D in our investigation (Figure 1). However, our sample size was too small to detect a statistically significant difference in frequency, and the case participants examined in our study were all women who had a long-standing T1D.
We examined potential confounding factors, in addition to T1D, that may influence the risk of virus infection during pregnancy. In univariate analysis, younger maternal age was associated with picobirnaviruses, older maternal age and no tertiary education was associated with EVs, and low SES was associated with gyroviruses. In multivariable GEE models, all of the aforementioned relationships remained significant except between maternal age and picobirnaviruses. Glycemic control may also influence susceptibility to infection; however, the majority of women with T1D in our study achieved glycemic targets for pregnancy.
To the best of our knowledge, this is the first study to examine the longitudinal gut virome across all 3 trimesters of pregnancy, providing novel baseline data for future gut virome investigations. Another major strength of this study is the application of virome capture sequencing [11, 49], which is the most sensitive and comprehensive sequence-based virome characterization tool currently available for vertebrate-infecting viruses. This method specifically targets all known viruses capable of infecting humans and other vertebrates, significantly reducing sequences produced from host and bacterial background, allowing up to a 10 000-fold increase in the number of viral reads recovered compared with conventional virome sequencing methods. Furthermore, our method enabled the examination of both RNA and DNA viruses simultaneously [11]. Despite these strengths, the interpretation of our virome data is limited by the fact that sequencing cannot differentiate between the presence of viral genomes in the gut versus actively replicating viruses. In addition, the absence of a nonpregnant control group precluded the analysis of the effect of pregnancy on virus infection in this study.
Given the results of our recent systematic review and meta-analysis of 2992 women and children that demonstrated a significant association between maternal virus infection in pregnancy and T1D in the offspring [8], future studies could be aimed at examining the impact of maternal virus infections on the development of islet autoimmunity and T1D in the offspring. For this purpose, the offspring of women examined in this study are being followed longitudinal for these 2 outcomes as part of the ENDIA study, a prospective cohort study following at risk children. The characterization of the gut virome in these mother-infant pairs will allow identification of potential vertical transmission of viruses (currently underway). The impact of diet on the gut virome will also be examined. The virome of other potential sources of vertical transmission should also be investigated such as the oral, skin, breastmilk, and the vaginal virome, which has been recently shown to be of clinical importance for its potential contribution to preterm birth [50].
CONCLUSIONS
In conclusion, our findings provide novel insight into the diversity and dynamics of the gut virome during pregnancy and identify T1D and maternal age as key factors influencing virus infection in pregnancy. We show a novel potential association between T1D and picobirnaviruses and demonstrate a distinct profile of viruses during pregnancy in women with T1D, providing novel targets for prevention studies.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We thank Nishit P. Bhuva (Columbia University, NY) for performing the virome capture hybridization and submitting samples for sequencing; Cheng Guo (Columbia University, NY) for assisting in the processing of metagenomic data and generation of Basic Local Alignment Search Tool (BLAST) result summaries.
Financial support. This work was funded by the National Health and Medical Research Council (NHMRC) Practitioner fellowships (APP1045777 [to M. E. C.] and APP1044694 [to W. D. R.]), Juvenile Diabetes Research Foundation Australia (JDRF)/NHMRC Centre of Research Excellence for the Protection of Pancreatic Beta Cells (APP1078106), and the National Institutes of Health (Center for Research in Diagnostics and Discovery) (U19 AI109761 [to T. B. and W. I. L.]). The ENDIA Study is supported by JDRF Australia, the recipient of the Australian Research Council Special Research Initiative in Type 1 Juvenile Diabetes, The Leona M. and Harry B. Helmsley Charitable Trust, JDRF International, and the NHMRC.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
Presented in part: Immunology of Diabetes 2018 meeting, October 2018, London, UK.
References
- 1. Guleria I, Sayegh MH. Maternal acceptance of the fetus: true human tolerance. J Immunol 2007; 178:3345–51. [DOI] [PubMed] [Google Scholar]
- 2. Racicot K, Kwon JY, Aldo P, et al. Understanding the complexity of the immune system during pregnancy. Am J Reprod Immunol 2014; 72:107–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jamieson DJ, Theiler RN, Rasmussen SA. Emerging infections and pregnancy. Emerg Infect Dis 2006; 12:1638–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sykes L, MacIntyre DA, Yap XJ, et al. The Th1:th2 dichotomy of pregnancy and preterm labour. Mediators Inflamm 2012; 2012:967629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Scott GM, Chow SS, Craig ME, et al. Cytomegalovirus infection during pregnancy with maternofetal transmission induces a proinflammatory cytokine bias in placenta and amniotic fluid. J Infect Dis 2012; 205:1305–10. [DOI] [PubMed] [Google Scholar]
- 6. Lin SF, Kuo CF, Chiou MJ, Chang SH. Maternal and fetal outcomes of pregnant women with type 1 diabetes, a national population study. Oncotarget 2017; 8:80679–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sappenfield E, Jamieson DJ, Kourtis AP. Pregnancy and susceptibility to infectious diseases. Infect Dis Obstet Gynecol 2013; 2013:752852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Allen DW, Kim KW, Rawlinson WD, Craig ME. Maternal virus infections in pregnancy and type 1 diabetes in their offspring: systematic review and meta-analysis of observational studies. Rev Med Virol 2018; 28:e1974. [DOI] [PubMed] [Google Scholar]
- 9. Penno MA, Couper JJ, Craig ME, et al. Environmental determinants of islet autoimmunity (ENDIA): a pregnancy to early life cohort study in children at-risk of type 1 diabetes. BMC Pediatr 2013; 13:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Conceição-Neto N, Zeller M, Lefrère H, et al. Modular approach to customise sample preparation procedures for viral metagenomics: a reproducible protocol for virome analysis. Sci Rep 2015; 5:16532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Briese T, Kapoor A, Mishra N, et al. Virome capture sequencing enables sensitive viral diagnosis and comprehensive virome analysis. MBio 2015; 6:e01491–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Luo R, Liu B, Xie Y, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 2012; 1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li D, Liu CM, Luo R, et al. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015; 31:1674–6. [DOI] [PubMed] [Google Scholar]
- 15. Mundry M, Bornberg-Bauer E, Sammeth M, Feulner PG. Evaluating characteristics of de novo assembly software on 454 transcriptome data: a simulation approach. PLoS One 2012; 7:e31410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol 2011; 29:24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kearse M, Moir R, Wilson A, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012; 28:1647–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. von Elm E, Altman DG, Egger M, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) Statement: guidelines for reporting observational studies. Int J Surg 2014; 12:1495–9. [DOI] [PubMed] [Google Scholar]
- 19. Pink B. Socio-Economic Indexes for Areas (SEIFA) - Technical Paper. In: Australian Bureau of Statistics, ed. Canberra: Commonwealth of Australia; 2011. [Google Scholar]
- 20. Department of Education and Training. Low socio-economic status (SES) - postcode measure Available at: http://heimshelp.education.gov.au/sites/heimshelp/resources/glossary/pages/glossaryterm?title=Low-socio-economic-status-(SES)-postcode-measure. Accessed 1 December 2017.
- 21. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 2014; 10:e1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010; 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robinson MD, Smyth GK. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics 2007; 23:2881–7. [DOI] [PubMed] [Google Scholar]
- 25. Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 2008; 9:321–32. [DOI] [PubMed] [Google Scholar]
- 26. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc 1995; 57:289–300. [Google Scholar]
- 27. Schep AN, Kummerfeld SK. iheatmapr: interactive complex heatmaps in R. J Open Source Softw 2017; 2, 359. doi: 10.21105/joss.00359. [DOI] [Google Scholar]
- 28. Colwell RK, Elsensohn JE. EstimateS turns 20: statistical estimation of species richness and shared species from samples, with non‐parametric extrapolation. Ecography; 2014; 37:609–13. doi: 10.1111/ecog.00814. [DOI] [Google Scholar]
- 29. Colwell RK, Chao A, Gotelli NJ, et al. Models and estimators linking individual-based and sample-based rarefaction, extrapolation and comparison of assemblages. J Plant Ecol 2012; 5:3–21. [Google Scholar]
- 30. Zhang T, Breitbart M, Lee WH, et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol 2006; 4:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smits SL, van Leeuwen M, Schapendonk CM, et al. Picobirnaviruses in the human respiratory tract. Emerg Infect Dis 2012; 18:1539–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Giordano MO, Martinez LC, Rinaldi D, et al. Diarrhea and enteric emerging viruses in HIV-infected patients. AIDS Res Hum Retroviruses 1999; 15:1427–32. [DOI] [PubMed] [Google Scholar]
- 33. Legoff J, Resche-Rigon M, Bouquet J, et al. The eukaryotic gut virome in hematopoietic stem cell transplantation: new clues in enteric graft-versus-host disease. Nat Med 2017; 23:1080–5. [DOI] [PubMed] [Google Scholar]
- 34. Phan TG, Li L, O’Ryan MG, et al. A third gyrovirus species in human faeces. J Gen Virol 2012; 93:1356–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haramoto E, Kitajima M, Kishida N, et al. Occurrence of pepper mild mottle virus in drinking water sources in Japan. Appl Environ Microbiol 2013; 79:7413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vaarala O, Atkinson MA, Neu J. The “perfect storm” for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes 2008; 57:2555–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Asnicar F, Manara S, Zolfo M, et al. Studying vertical microbiome transmission from mothers to infants by strain-level metagenomic profiling. mSystems 2017; 2:e00164–16. doi: 10.1128/mSystems.00164-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Laitinen OH, Honkanen H, Pakkanen O, et al. Coxsackievirus B1 is associated with induction of β-cell autoimmunity that portends type 1 diabetes. Diabetes 2014; 63:446–55. [DOI] [PubMed] [Google Scholar]
- 39. Yin H, Berg AK, Tuvemo T, Frisk G. Enterovirus RNA is found in peripheral blood mononuclear cells in a majority of type 1 diabetic children at onset. Diabetes 2002; 51:1964–71. [DOI] [PubMed] [Google Scholar]
- 40. Oikarinen S, Tauriainen S, Hober D, et al. Virus antibody survey in different European populations indicates risk association between coxsackievirus B1 and type 1 diabetes. Diabetes 2014; 63:655–62. [DOI] [PubMed] [Google Scholar]
- 41. Schulte BM, Bakkers J, Lanke KH, et al. Detection of enterovirus RNA in peripheral blood mononuclear cells of type 1 diabetic patients beyond the stage of acute infection. Viral Immunol 2010; 23:99–104. [DOI] [PubMed] [Google Scholar]
- 42. Coleman TJ, Gamble DR, Taylor KW. Diabetes in mice after Coxsackie B 4 virus infection. Br Med J 1973; 3:25–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Adams SF. The seasonal variation in the onset of acute diabetes: the age and sex factors in 1000 diabetic patients. Arch Intern Med 1926; 37:861–4. [Google Scholar]
- 44. Dotta F, Censini S, van Halteren AG, et al. Coxsackie B4 virus infection of beta cells and natural killer cell insulitis in recent-onset type 1 diabetic patients. Proc Natl Acad Sci U S A 2007; 104:5115–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Honkanen H, Oikarinen S, Nurminen N, et al. Detection of enteroviruses in stools precedes islet autoimmunity by several months: possible evidence for slowly operating mechanisms in virus-induced autoimmunity. Diabetologia 2017:1–8. [DOI] [PubMed] [Google Scholar]
- 46. Yeung WC, Al-Shabeeb A, Pang CN, et al. Children with islet autoimmunity and enterovirus infection demonstrate a distinct cytokine profile. Diabetes 2012; 61:1500–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kim KW, Horton JL, Pang IC, et al. Higher abundance of enterovirus A species in the gut of children with islet autoimmunity. Sci Rep 2019. doi: 10.1038/s41598-018-38368-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhao G, Vatanen T, Droit L, et al. Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children. Proc Natl Acad Sci U S A 2017; 114:E6166–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wylie TN, Wylie KM, Herter BN, Storch GA. Enhanced virome sequencing using targeted sequence capture. Genome Res 2015; 25:1910–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wylie KM, Wylie TN, Cahill AG, et al. The vaginal eukaryotic DNA virome and preterm birth. Am J Obstet Gynecol 2018; 219:189.e1–189.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.