Abstract
β-Catenin is an evolutionarily conserved molecule in the canonical Wnt signaling pathway, which controls decisive steps in embryogenesis and functions as a crucial effector in the development of hair follicles. However, the molecular mechanisms underlying wool production have not been fully elucidated. In this study, we investigated the effects of ovine β-catenin on wool follicles of transgenic sheep produced by pronuclear microinjection with a skin-specific promoter of human keratin14 (k14). Both polymerase chain reaction and Southern blot analysis showed that the sheep carried the ovine β-catenin gene and that the β-catenin gene could be stably inherited. To study the molecular responses to high expression of β-catenin, high-throughput RNA-seq technology was employed using three transgenic sheep and their wild-type siblings. These findings suggest that β-catenin normally plays an important role in wool follicle development by activating the downstream genes of the Wnt pathway and enhancing the expression of keratin protein genes and keratin-associated protein genes.
Keywords: sheep, β-catenin, wool follicle, transgene, transcriptome
1. Introduction
The skin provides a barrier against environmental assault from foreign substances and organisms. The outer skin of mammals is covered by multilayered epithelial cells, including hair follicles and glandular structures [1]. Although the upper part of the hair follicle and the dermal papilla are formed during embryonic development, these structures are fixed in postnatal life [2]. The papilla is a large structure at the base of the hair follicle [3], and a series of cell signaling processes transfer signals from one cell to another between the epithelium and dermis during embryonic development and hair follicle morphogenesis [4]. In postnatal animals, the lower portion of the hair follicle is responsible for hair follicle cycles of anagen, catagen, and telogen, and molecular signatures in the dermal papilla are thought to induce the proliferation of stem-like cells in the epithelium of the hair follicle bulge and the migration of daughter cells to the lower portion of the hair follicle [2,5]. Previous research has shown that several key signals, such as fibroblast growth factor (FGF), bone morphogenic protein (BMP), and transforming growth factor β2 (TGF-β2) have been implicated in hair follicle induction [6]. Recent studies have shown that WNT signaling pathways, particularly the essential molecule β-catenin, play important roles in hair follicle formation and periodic growth [7].
As a multifunctional molecule, β-catenin plays various roles in WNT signaling and interacts with a number of proteins, including the actin-binding protein facin and presenilins. Moreover, β-catenin also has a role in Ca2+-dependent cell adhesion [8]. In the canonical WNT pathway, β-catenin contributes to the transduction of the signal to the nucleus and associates with transcription factors from the T-cell factor/lymphoid enhancer factor family to drive the transcription of WNT/β-catenin target genes. The mRNA expression level of β-catenin is increased during hair follicle formation in embryonic development. In contrast, blocking the expression of β-catenin disrupts the formation of normal hair follicles [9]. In vitro studies have indicated that two particular phenotypes are acquired by knockout of β-catenin: (1) placode formation is not observed during embryo development; and (2) hair loss appears in the first hair cycle [7]. Compared with wild-type mice, overexpression of a truncated β-catenin in the skin of mice was found to cause a rapid increase in the number and size of hair follicles, and the hair follicles in telogen could enter into the anagen in advance [10]. In the “bulge activation hypothesis”, when signals from dermal papilla transiently activate the quiescent hair follicle bulge stem cells to proliferate, the hair follicle turns into a new anagen phase; β-catenin plays an essential role in this process [3].
Aohan fine-wool sheep are a wool–meat breed generated from crossbreeding with Inner Mongolian local sheep and the former Soviet Union merino sheep [11]. These sheep are known for their slaughter rate, crude feed tolerance, and wool fineness. In recent years, the demand for wool has increased rapidly with the development of the wool textile industry. However, wool production cannot meet this increasing demand, and novel approaches for improving wool production are needed. As the origin of wool fibers, hair follicles have direct effects on the production and quality of ovine cashmere.
In the present study, we examined the role of the K14 gene promoter in directing the overexpression of β-catenin in the wool follicle in transgenic sheep and the effects of β-catenin overexpression on wool production in transgenic sheep.
2. Results
2.1. Transgenic Sheep
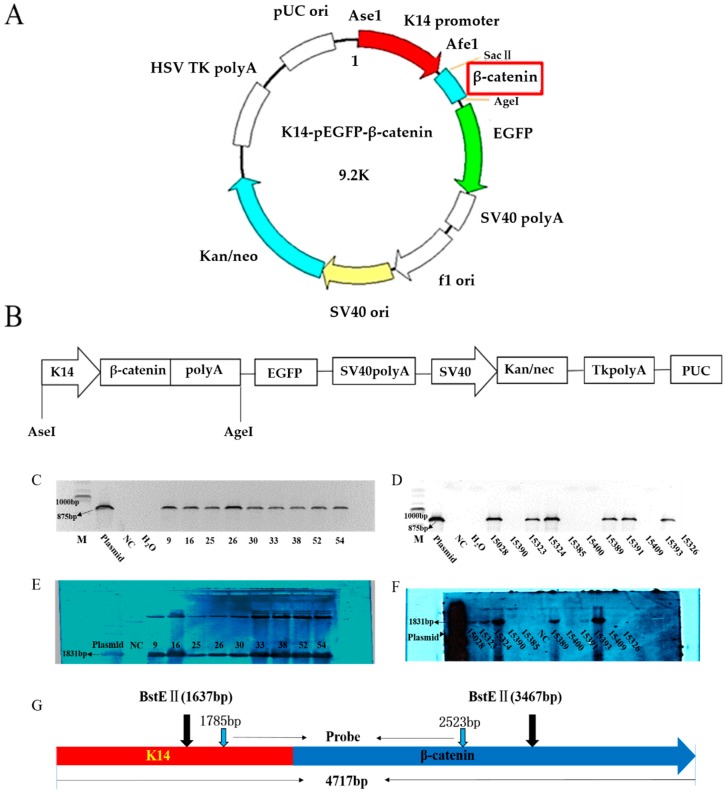
To investigate the functions of β-catenin in wool follicles, the pK14-β-catenin-enhanced green fluorescent protein (EGFP)-N1 plasmid was successfully constructed (Figure 1A). As shown in Figure 1B, the transgene fragments were generated by Ase1 and Age1 double enzyme digestion and then microinjected into Aohan fine-wool sheep zygotic pronuclei. In total, 155 fertilized oocytes were microinjected with a linearized transgene construct and transferred to 68 recipients; 62 lambs were produced, of which nine were transgenic as determined by PCR (Figure 1C) and Southern blot analysis (Figure 1E). One ram (B025) was used for artificial propagation by superovulation artificial insemination and embryo transfer. Twenty-six lambs were produced, six of which were transgenic as identified by PCR (Figure 1D) Southern blot analysis of DNA from the transgenic sheep further indicated the existence of foreign DNA in the transgenic sheep (Figure 1F). The transgenic sheep were healthy and showed no defects.
Figure 1.
Generation of ovine β-catenin transgenic sheep: (A) the recombinant plasmid structure of K14-β-catenin-N1-EGFP; (B) the stick diagram of K14-β-catenin-N1-EGFP; (C,D) the PCR identification of transgenic sheep; (E) Southern blot of DNA from F0 transgenic sheep skin (NC: sex- and age-matched wild-type control; plasmid: positive control); (F) Southern blot of DNA from F1 transgenic sheep skin; and (G) schematic diagram of restriction endonuclease digestion by Southern blotting.
2.2. Characterization of Transgene Expression in Transgenic Sheep and Their Wild-Type Siblings
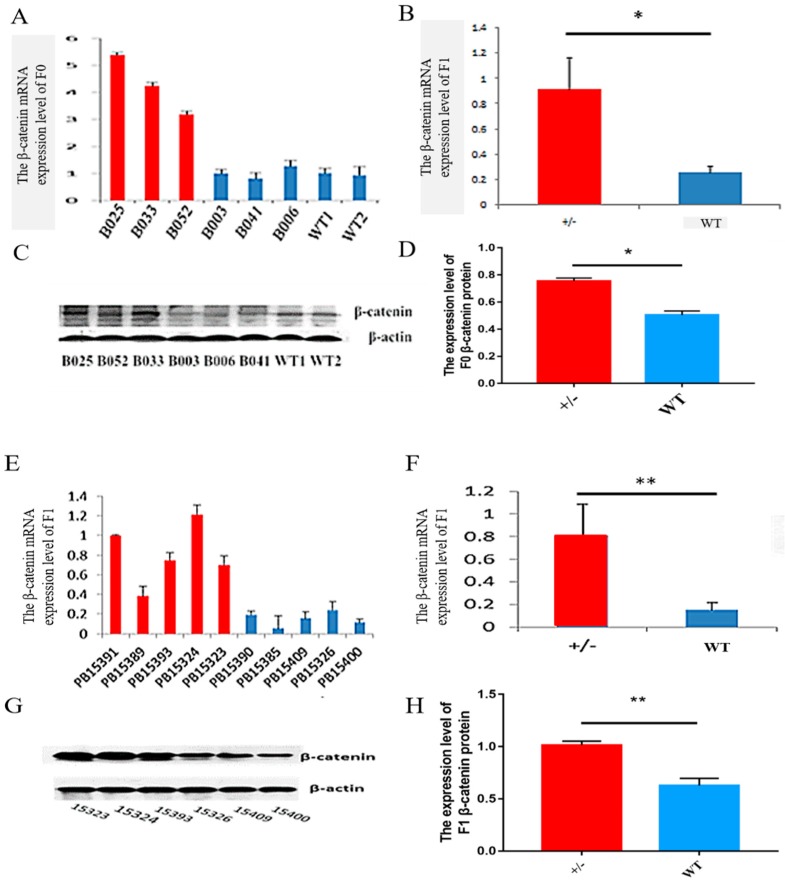
To compare transgene expression level, relative transgene mRNA levels were measured by quantitative reverse transcription polymerase chain reaction (qRT-PCR). The results indicate that relative transgene expression levels in the skin of F0 transgenic sheep were significantly higher than those in their wild-type siblings (P < 0.01; Figure 2A,B). We investigated the ovine β-catenin expression in F1 transgenic sheep skin tissues from line B025 for further study. The results show that transgene expression in the skin was significantly higher than that in their full siblings (P < 0.01; Figure 2E,F). Additionally, Western blotting results show that β-catenin protein in F0 (Figure 2C) and F1 (Figure 2G) was expressed at higher levels in transgenic sheep skin than in their wild-type siblings. ImageJ was used to analyze the protein expression level of F0 (Figure 2D) and F1 (Figure 2H).
Figure 2.
The expression of β-catenin in the skin of F0 and F1 sheep. The relative quantities of β-catenin mRNA in F0 (A,B) and F1 (E,F) were detected via qRT-PCR using the 2−ΔΔCT method, with GAPDH as the internal control. Each experimental group contained at least three replicates, and qRT-PCR was performed in triplicate for each sample. Bars with common lowercase letters are not significantly different at the level of 5%. WT1 and WT2 are wild-type control sheep. The analysis of β-catenin protein expression in F0 (C,D) and F1 (G,H) was performed using Western blot analysis at 12 months with β-actin as the internal control. The bands for β-catenin and β-actin proteins were quantified with ImageJ (http://rsb.info.nih.gov/ij). F0 transgenic sheep: B025, B033, and B052; F0 wild-type siblings: B003, B006, and B041; wild-type sheep: WT1 and WT2; F1 transgenic sheep: PB15391, PB15389, PB15393, PB15324, and PB15323; and F1 wild-type siblings: PB15390, PB15385, PB5409, PB15326, and PB15400. The significance of differences in β-catenin mRNA and protein expression levels between transgenic sheep and wild-type siblings was analyzed by paired Student’s t tests. * P < 0.05; ** P < 0.01 for comparisons between the two groups.
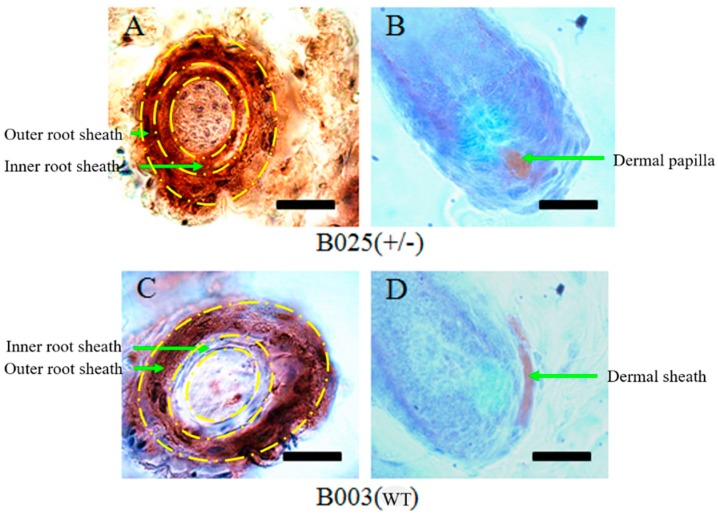
Immunohistochemical (IHC) assays were performed to determine the localization of β-catenin in the wool follicles (Figure 3). The IHC analysis showed that β-catenin protein in transgenic sheep skin was found in the inner root sheath and outer root sheath (Figure 3A). However, β-catenin was only expressed in the outer root sheath of wild-type siblings (Figure 3C). In addition, β-catenin protein was expressed in Dermal papilla of transgenic sheep (Figure 3B) compare with the Dermal sheath of wild-type sibling (Figure 3D).
Figure 3.
Localization of β-catenin protein in the wool follicle: (A,B) the expression of β-catenin in the inner and outer root sheath and hair dermal papilla in the transgenic sheep; and (C,D) the expression of β-catenin in the outer root sheath of the wool follicle in the wild-type sibling control. The brown granules show the localization of β-catenin in the wool follicle. Scale bars: 10 μm.
2.3. Advantage of Overexpressing Ovine β-Catenin in Wool Production
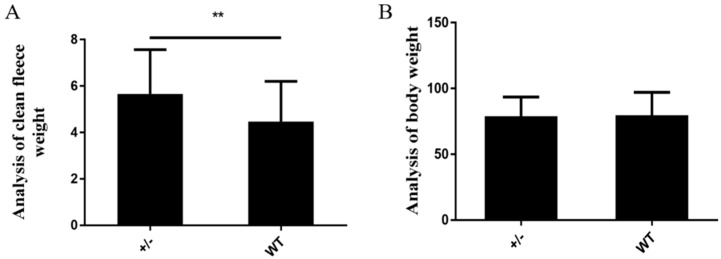
Clean fleece weight was obtained at approximately 14 months of age, and clean wool production on average was 26.49% greater (P = 0.0051) in the transgenic sheep than in their nontransgenic full siblings (Figure 4A) by Valency T tests. However, body weight did not differ significantly (P = 0.3354) between the transgenic sheep and their nontransgenic full siblings (Figure 4B). Table 1 gives the details of each sample.
Figure 4.
Comparison of clean fleece weights (A) and body weights (B) between the transgenic sheep and their wild-type siblings. The significance of differences in body weights and clean wool weights between transgenic sheep and wild-type siblings was analyzed by paired t-tests. ** P < 0.01 for comparisons between the two groups.
Table 1.
Analysis of wool production and body weight in F1.
| Ear Tag | Donor | Donor | Sex | Body | Yearling | Clean | Clean | GMO (+/−) |
|---|---|---|---|---|---|---|---|---|
| rams | ewes | Weight (kg) | Shearing (kg) | Fleece Yield (%) | Fleece Weight (kg) | Non-GMO (WT) | ||
| 15323 | B025 | ZA94822 | ♂ | 85.35 | 10.85 | 50.14 | 5.44019 | +/− |
| 15409 | ♂ | 88.5 | 8.4 | 58.63 | 4.92492 | WT | ||
| 15324 | B025 | ZA94822 | ♀ | 54.6 | 4.45 | 63.48 | 2.609035 | +/− |
| 15326 | ♀ | 50 | 2.7 | 53.92 | 1.58301 | WT | ||
| 15389 | B025 | ZA94863 | ♂ | 82.6 | 12.3 | 58.64 | 7.21149 | +/− |
| 15385 | ♂ | 84.85 | 8.85 | 60.78 | 5.188755 | WT | ||
| 15391 | B025 | ZA96588 | ♂ | 93.2 | 12.55 | 42.29 | 7.358065 | +/− |
| 15390 | ♂ | 95.15 | 10.55 | 45.98 | 6.185465 | WT |
2.4. Analysis of Wool Follicle Density of Transgenic Sheep
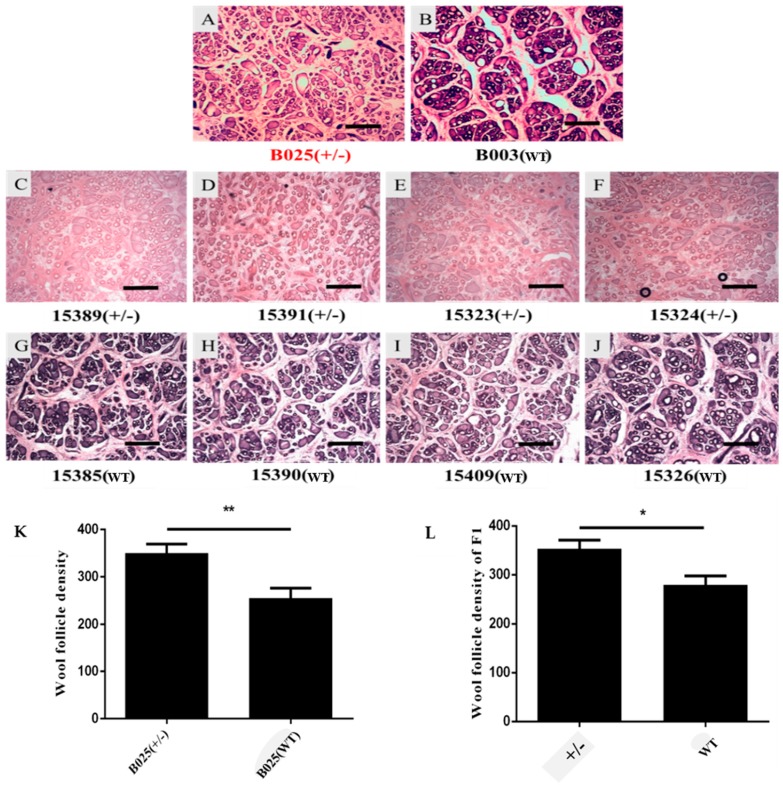
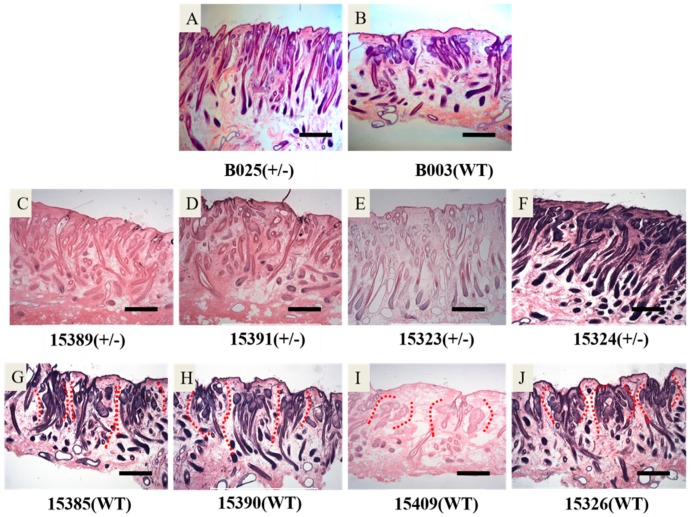
The frozen sections of skin showed different wool follicle growth patterns and wool follicle densities between transgenic sheep and their nontransgenic full siblings. The wool follicles of transgenic sheep showed a more uniform distribution than that in nontransgenic sheep (Figure 5A–J). This difference was also observed on longitudinal sections of skin (Figure 6A–J). This wool follicle phenotype characteristic led to the higher wool follicle density in transgenic sheep than in nontransgenic full siblings by wool follicle density analysis (Figure 5K,L). There were no significant difference between transgenic sheep and nontransgenic full siblings in the wool length (Figure S1A) and diameter (Figure S1B).
Figure 5.
Analysis of wool follicle density in F0 and F1: (A,B) Hematoxylin–eosin staining showing the hair follicle morphological characteristics of a transgenic sheep (B025) and its wild-type sibling (B003); (C–F) transgenic sheep of F1; (G–J) wild-type siblings longitudinally corresponding to the transgenic sheep of F1; (K) wool follicle density comparison of the F0 pair; and (L) wool follicle density comparison of the F1 pairs. * P < 0.05, ** P < 0.01. Scale bars: 100 μm.
Figure 6.
Developmental characteristics of hair follicles between transgenic sheep and their nontransgenic full siblings: (A) K14-β-catenin transgenic ram (B025); (B) the nontransgenic full sibling of B025, B003; (C–J) slices from the offspring of the ram B025 generated by synchronous estrus, superovulation, and artificial insemination; (C–F) slices from the F1 transgenic sheep; and (G–J) slices from the wild-type siblings longitudinally corresponding to the transgenic sheep of F1.
2.5. Identification of Expressed Transcripts in the Sheep Skin Transcriptome
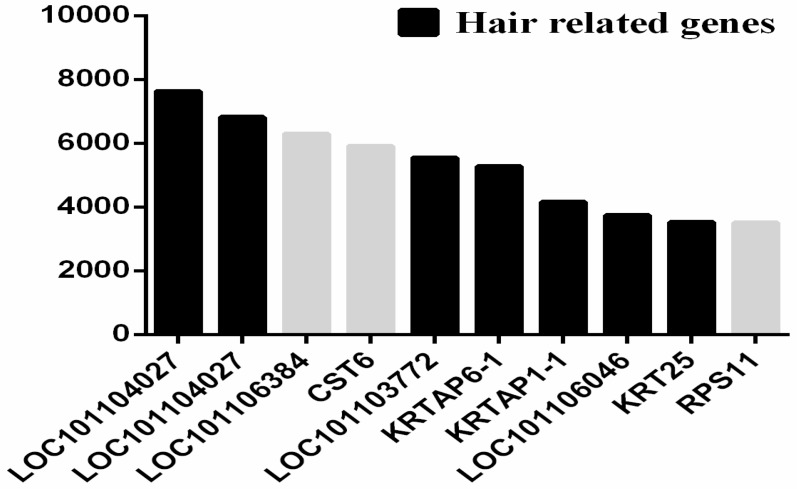
In this study, 58,356,618–63,795,138 raw reads were generated for each sample (Table S1). We obtained 20,532 transcripts from the groups after quality control (Table S2). About 85% of the total reads were mapped to the sheep reference genome, and about 75% of the total reads were mapped to the reference genome only at one site (Table S3). Information for read density on chromosomes is provided in the Supplementary Materials (Table S3). The top 10 annotated transcripts, ranging from 3509 to 7628 fragments per kilobase of exons per million fragments mapped (FPKM) reads (Figure 7), were ranked by abundance and included the keratin protein genes or keratin-associated protein genes LOC101112404 (keratin-associated protein 3-2), LOC101104027 (keratin-associated protein 7-1-like), LOC101103772 (keratin-associated protein 11-1), KRTAP6-1, KRTAP1-1, KRT25, and LOC101106046 (keratin-associated protein 13-1-like).
Figure 7.
Expression of the top 10 most highly expressed genes in sheep skin. The x-axis shows gene ID, and the y-axis shows gene expression level (FPKM).
2.6. Functional Analysis of Differentially Expressed Genes (DEGs)
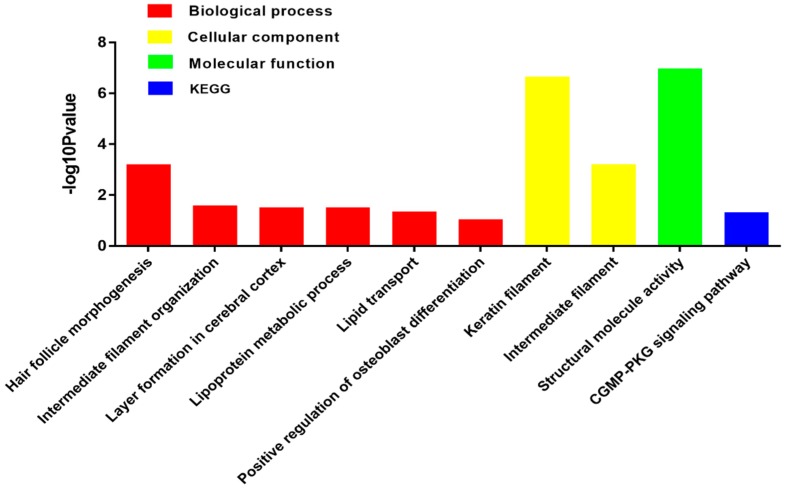
To elucidate the biological processes in transgenic sheep, gene ontology (GO) assignments were performed. Based on sequence homology, 76 identified upregulated DEGs and 37 downregulated genes were categorized into major functional groups, including biological processes (BPs), cellular components (CCs), and molecular functions (MFs; Figure 8). Among these terms, the first functional categories of BPs, CCs, and MFs were related to hair follicle development (Figure 7). Of these upregulated genes, FN1 and matrix metalloproteinase-7 (MMP-7) were found to be direct target genes of β-catenin [3].
Figure 8.
Functional categorization of DEGs based on known genes in the Uniprot database. The x-axis shows GO and KEGG terms; the y-axis shows the negative log10Pvalue.
2.7. Identification of DEGs between the F0 Transgenic Sheep and Wild-Type Siblings
Approximately 113 DEGs between the two groups were identified by paired t-tests. By applying the criteria of ≥2-fold change and P < 0.05 as cut off values, 76 upregulated and 37 downregulated DEGs were screened out. Further annotation showed that 11 keratin protein genes or keratin-associated protein genes were upregulated in the transgenic sheep, whereas none of these keratin and keratin-associated protein genes were found in downregulated genes in transgenic sheep. In addition, β-catenin and its target genes (MMP-7 and FN1) were significantly upregulated in transgenic sheep compared with that in their wild-type siblings (Table 2).
Table 2.
Hair-related important DEGs between transgenic sheep and wild-type siblings.
| Gene Name | Description | Reference |
|---|---|---|
| β-catenin | β-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin | [7] |
| MMP-7 | MMP-7 or matrilysin (β-catenin target gene) is expressed in hair placode keratinocytes | [11] |
| FN1 | β-catenin target gene | [12] |
| KRT79 | Keratin 79 identifies a novel population of migratory epithelial cells that initiate hair canal morphogenesis and regeneration | [13] |
| KRT2.11 | KRT2.11 transcripts are present in wool follicle RNA and are expressed in follicle cortical keratinocytes located above the dermal papilla | [14] |
| KRT8 | KRT8 is an epithelial marker | [15] |
| KRT71 | Mutations in KRT71 are observed in mice, rats, and dogs and are linked to a wavy coat phenotype | [16] |
| KRT5 | KRT5 is a marker of basal and undifferentiated keratinocytes and is increased in the epidermis of alopecic mice | [17] |
| KRTAP1-1 | The ovine KRTAP1-4 gene is clustered with the KRTAP1-1 and KRTAP1-3 genes on chromosome 11 and appears to be associated with wool staple | [18] |
| KRT25 | A homozygous missense variant in type I keratin KRT25 causes autosomal recessive woolly hair | [19] |
2.8. Validation of DEGs by Real-Time PCR
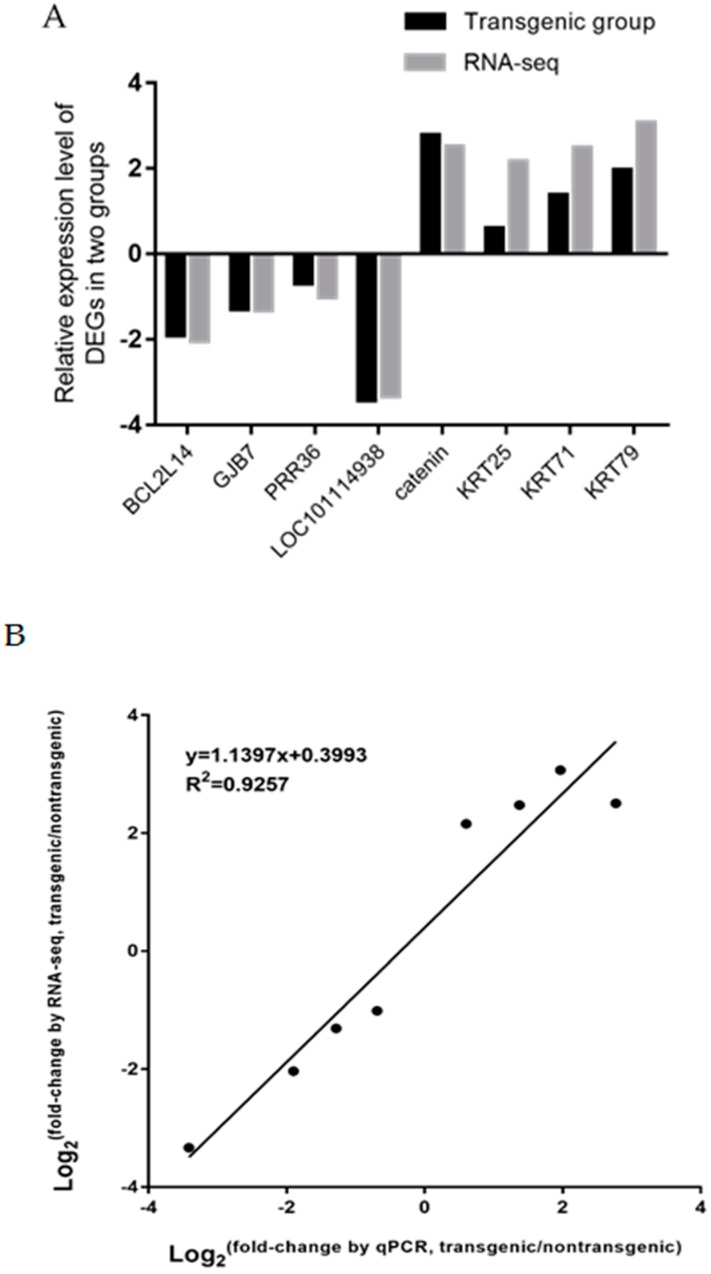
Eight DEGs were selected for validating the DEGs identified from RNA-seq data, including four upregulated genes (β-catenin, KRT25, KRT71, and KRT79) and four downregulated genes (MPC1, KRTDAP, ASAP2, and ASB7). The expression patterns of DEGs between these two groups were confirmed by the results of RT-PCR (Figure 9A). The correlation of fold change between RNA-seq and qRT-PCR was analyzed, and the correlation coefficient was 0.9257 (Figure 9B).
Figure 9.
The expression levels of DEGS validated by qPCR (A); and comparison between RNA-seq and qPCR by correlational analyses (B).
3. Discussion
Hair follicle morphogenesis is a fine example of the exchange of information between epithelium and mesenchyme, which is the basis of organogenesis [4]. The molecular mechanisms of these interactions are still largely unclear, but are likely to involve several signaling molecules, including WNTs [20], FGFs [21], ectodysplasin A receptors [22,23], BMPs [24], Sonic hedgehog proteins [25], TGF-βs [26], and insulin-like growth factors [27]. Among these mechanisms, the Wnt pathway plays an essential role in many aspects of development and tumorigenesis [28]. Nuclear localization of β-catenin is downstream of the Wnt pathway, which is localized in the cytoplasm and membrane [29]. A conditional mutation in β-catenin in the epidermis and hair follicles demonstrated that the formation of placodes, which generate hair follicles, was blocked [7], suggesting that activation of the canonical Wnt signaling pathway was necessary for hair follicle formation.
To better understand the effects of β-catenin on wool development, stable and heritable skin-specific β-catenin transgenic sheep were obtained for the first time. In our study, the wool follicle density was significantly greater in transgenic sheep than in their wild-type siblings of the same sex. Consistent with our study, an overall increase of 300% ± 30% in the hair germ density was found in β-catenin transgenic mice skin relative to that in control skin [10].
Furthermore, we found that the wool follicles in transgenic sheep skin were distributed more homogeneously than their wild-type siblings by skin observation on cross-sections; this was an abnormal wool follicle growth characteristic because the wool follicles were accustomed to growth in clumps, called wool follicle groups [30]. The overexpression of β-catenin was thought to induce new wool follicle outgrowth and promote the development of wool follicles such that the classical development characteristics (wool follicle groups) of wool follicles would be disrupted. The same conclusion was reached by analysis of longitudinal sections of skin between transgenic sheep and their wild-type siblings, which was similar to a study in which activation of the β-catenin signal in the adult mouse epidermis was sufficient to induce new hair follicles [31] and ectopic hair outgrowth [32].
To analyze differences in the activity of Wnt signaling between transgenic sheep and negative control sheep, the localization of β-catenin was observed by IHC staining. The results show that β-catenin mRNA and protein were significantly upregulated in transgenic sheep skin compared with those in wild-type siblings of the same sex. Interestingly, our work suggested more active expression of β-catenin protein in the dermal papilla of transgenic wool follicles compared with that in nontransgenic full siblings according to the results of IHC staining. Furthermore, IHC staining results show that β-catenin protein was not only expressed in the outer root sheath but also in the inner root sheath (IRS) of transgenic sheep wool follicles. In contrast, expression was observed only in the outer root sheath of nontransgenic full siblings. At present, we have not found any evidence to explain the possible reasons this differential expression pattern occurred; however, KRT5, KRT25, and KRT71 were found to be expressed in the Henle layer, Huxley layer, and IRS cuticle [33]. In addition, these three keratin protein genes were mainly expressed in transgenic sheep compared with their nontransgenic full siblings. These findings suggested that there may be a connection between β-catenin and keratin protein genes and a link between hair shaft shape as natural traits and keratin protein genes [34]. In addition, previous studies have shown that the dermal papilla is of prime importance in hair follicle morphogenesis. Papillae can induce hair growth after implantation into follicles [35,36] and can interact with the skin epidermis to form new hair follicles [35]. Accordingly, we suspect that β-catenin can promote the development of dermal papilla and IRS in transgenic sheep wool follicles, allowing the growth state to be more active than that in wool follicles of wild-type sibling sheep.
There is a complex interactive process between molecular signaling transduction and hair morphogenesis. The WNT pathway plays an essential role in the formation of hair placodes during embryogenesis [37], and β-catenin is required for the differentiation of skin stem cells in the adult, making it the key molecule in the Wnt pathway. In the absence of β-catenin, these stem cells are unable to adopt the fate of hair keratinocytes and instead differentiate into epidermal keratinocytes [7]. Additionally, inactivation of the β-catenin gene in the dermal papilla of anagen hair follicles gives rise to obviously reduced proliferation of progenitors (dermal sheath), which produce the hair shaft [38]. However, few studies have examined sheep wool follicle development and changes in the molecular network in transgenic sheep overexpressing β-catenin. In our study, high-throughput sequencing was used to obtain DEGs from three pairs of full sibling sheep of the same sex. Transcripts encoding 113 DEGs were identified, with 72 significantly upregulated genes in transgenic sheep relative to those in nontransgenic sheep, including β-catenin and its direct or indirect target genes (MMP-7, FN1, KRT79, KRT2.11, KRT8, KRT71, KRT5, KRT25, and KRTAP1-1). This result show that the WNT pathway was more active in transgenic sheep than in nontransgenic sheep and that DEGs, associated with hair follicle development were mainly upregulated in transgenic sheep relative to those in nontransgenic sheep. In addition, GO analysis was used to identify functional categories of those DEGs obtained from the global analysis for in-depth study. For the upregulated genes in transgenic sheep, analysis of the top ranking GO terms revealed an enrichment in genes related to “hair follicle morphogenesis” and “keratin filament”, which was further evidence that the β-catenin skin-specific transgenic sheep model was successfully constructed.
4. Materials and Methods
4.1. Animals and Treatments
Three pairs of full siblings (one pair of males and two pairs of females), at 12 months of age, were selected and divided into transgenic (+/−) and nontransgenic (WT) groups. Skin tissue was obtained from the scapular region of each sheep and frozen in liquid nitrogen directly. All of the procedures and experiments performed in this study were approved by the Animal Care and Use Committee of China Agricultural University (Approval no. XK257).
4.2. K14-β-Catenin-EGFP Plasmid Construction
The pEGFP-N1 vector was digested with restriction enzymes AseI and AfeI to remove the CMV promoter and then inserted into the human K14 promoter (~2.0 kb) between the AseI and AfeI sites to generate the skin-specific expression plasmid K14-EGFP. Subsequently, the K14-EGFP plasmid was digested with restriction enzymes SacII and BamHI, and the complete coding sequence of the ovine β-catenin gene was inserted between the SacII and BamHI sites. The resulting K14-β-catenin-EGFP plasmid was then verified by sequencing with the primers shown in Table 1 (SinoGenoMax Co. Ltd., Beijing, China).
4.3. Generation of Transgenic Sheep
All of the procedures and experiments performed in this study were approved by the Animal Care and Use Committee of China Agricultural University (Approval no. XK257). Linear DNA fragments were obtained by double digestion of the K14-β-catenin-IRES-EGFP plasmid with AseI/Age1 and re-suspended in sterile ddH2O at a concentration of 20 ng/μL. The linearized construct was injected into the pronuclei of the fertilized eggs flushed from donor sheep. The K14-β-catenin-IRES-EGFP transgene-positive founder ram was mated with the wild-type to obtain F1 generation sheep using artificial insemination, superovulation, and embryo transfer.
4.4. Positive Identification of F0 and F1 Transgenic Sheep
Genomic DNA extracted from skin tissues of transgenic sheep using the TIANamp Genomic DNA Kit (TIANGEN, Beijing, China) was used for PCR and Southern blotting analysis. PCR analysis was performed using specific primers (forward primer: 5′-CTGCCTGGATTTCTCTTTGA-3′; reverse primer: 5′-CCATTGTCCACGCAAGATTT-3′). The 875-bp PCR product contained the partial human K14 promoter sequence and partial ovine β-catenin sequence. PCR amplification was carried out as follows: denaturation for 5 min at 95 °C; then 35 cycles of 30 s for 95 °C, 30 s for 58 °C, and 72 °C for 1 min; 72 °C for 10 min; and hold at 12 °C. The positive sheep identified by PCR were subjected to Southern blot analysis.
For Southern blot analysis, 25 μg genomic DNA obtained from skin tissues was digested with the restriction enzyme BstEII at 60 °C for 20 h. A 739-bp probe, amplified by F6 (CAAGAAAGCCCAAAACAC) and R6 (TAGCGTCTCAGGGAACAT) from the promoter and β-catenin sequences, was labeled with a PCR digoxigenin probe synthesis kit (Roche, Basel, Switzerland). Hybridization and washing were performed using a DIG-High Prime DNA Labeling and Detection Starter Kit II (Roche) according to the manufacturer’s instructions.
4.5. Analysis of Transgene Expression of F0 and F1 Sheep
Total RNA was isolated from the skin of transgenic sheep using an RNAprepPure Tissue Kit (TIANGEN), according to the manufacturer’s instructions. The first-strand cDNA was synthesized from 1–2 μg of purified total RNA using an ImProm-II Reverse Transcription System (Promega, Beijing, China). PCR was performed using the primers shown in Table 1. Tissue-specific expression analysis of β-catenin was performed using qRT-PCR with an iQ5 real-time PCR detection system (Bio-Rad, Hercules, CA, USA) relative to the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which was used as a housekeeping gene (internal reference). The relative β-catenin expression levels were assessed in triplicate and calculated using the 2−ΔΔCt method.
4.6. Protein Analysis of F0 and F1 Sheep
For Western blot analysis, protein from F0 and F1 sheep skin tissues was extracted using RIPA buffer (BeYoTime, Suzhou, China) according to the manufacturer’s instructions. A BCA Protein Assay Kit from BeYoTime was used to determine the protein concentrations, and bovine serum albumin was used as a standard. Equal amounts of total cellular protein (20 μg/lane) were resolved on sodium dodecyl sulfate-polyacrylamide gels and transferred onto polyvinyl-difluoride membranes. The membranes were hybridized with 1:1000 dilutions of anti-β-catenin antibodies (cat no. E247; Abcam, Cambridge, UK) or antiβ-actin antibodies (cat. no. AA128; BeYoTime) and incubated at 37 °C for 1 h after blocking with blocking buffer for 1 h at room temperature. After washing five times with Tris-buffered saline with Tween20 (TBST) for 5 min, membranes were incubated with horseradish peroxidase-conjugated secondary antibody (1:10,000) for 1 h at 37 °C, washed six times with TBST for 5 min each, and visualized using an enhanced chemiluminescence system (cat. no. P0018S; BeYoTime).
4.7. Immunohistochemistry
Frozen sections of skin were washed with distilled water three times, incubated in 3% hydrogen peroxide solution for 20 min at room temperature, washed with distilled water again, and blocked with blocking buffer for 30 min at room temperature. The sections were incubated with primary mouse anti-human β-catenin antibodies (Abmart, Shanghai, China; 1:200) overnight at 4 °C. After rinsing with phosphate-buffered saline (PBS), the sections were treated with goat anti-mouse immunoglobulin G conjugated to horseradish peroxidase (Zhongshan GoldenBridge Biotechnology, Beijing, China) for 1 h at room temperature in the dark. After washing with PBS three times, the sections were incubated with 3, 3′-diaminobenzidine (DAB) for 10 min at room temperature in the dark. After DAB staining, slides were rinsed three times in deionized water for 30 s and then stained with hematoxylin. After treatment with hydrochloric acid, ethyl alcohol, and dimethylbenzene, the sections were embedded in Permount Mounting Medium and observed under a microscope.
4.8. Clean Fleece Weight
At yearling shearing, body weight was recorded, the fleece was weighed, and a sample was taken for scouring. Clean dry fleece weight and yield were then assessed. To measure clean dry fleece weight, greasy wool samples were kept in a conditioned environment (20 °C, 65% relative humidity (RH)) for 24 h, washed in a sample wool scourer with nonionic detergent at 65 °C, dried at 60 °C for 20 min, maintained in a conditioned environment at 20 °C and 65% RH for 24 h, and then weighed. Yield was calculated as clean weight divided by greasy weight and expressed as a percentage.
4.9. RNA Extraction and Transcriptome Sequencing
RNA was extracted from skin using TRIzol Reagent (Invitrogen, San Diego, CA, USA), following the manufacturer’s protocol. Next, 1% agarose gels were used to determine the quality of the RNA. RNA concentration was measured using a Qubit RNA Assay Kit in a Qubit 2.0 Fluorometer (Life Technologies, CA, USA). A NanoPhotometer (IMPLEN, Munchen, Germany) and RNA Nano 6000 Assay kit for a Bioanalyzer 2100 system (Agilent Technologies, Foster city, CA, USA) were used to check the RNA purity, and RNA samples passing the quality tests were used for further analyses.
Three micrograms of RNA were used for library construction. We use an IlluminaTruseq RNA sample preparation Kit (Illumina, San Diego, CA, USA) to produce sequencing libraries. Poly-T oligo-attached magnetic beads were used to purify mRNA from total RNA. The mRNAs were broken into short fragments, and random oligonucleotides and Superscript II were used to generate the first-strand cDNA. Second-strand cDNA was synthesized using DNA polymerase I and RNase H. In addition, sequencing adaptors were used to purify and modify double-stranded cDNAs, and a cDNA library was created using suitable fragments, which were selected by gel purification and enriched by PCR amplification. Subsequently, the library was used for sequencing on an IlluminaHiseq 2000 platform (Illumina, San Diego, CA, USA).
4.10. Sequence Reads Mapping, Assembly, and Annotation
Raw reads in the fastq format were first processed through in-house Perl scripts. During this step, clean reads were obtained by removing reads containing adapters, reads containing poly-N sequences, and low-quality reads from raw data. At the same time, Q30, GC-content, and sequence duplication levels of the clean data were calculated. All high-quality clean data were used for subsequent analyses.
The sheep reference genome and gene model annotation files were downloaded from the genome website (http://www.sheephapmap.org/news/OARv2p0.php). Bowtie v0.12.8 was used to build an index of the reference genome [39], and TopHat v1.4.0 was used when paired-end clean reads were aligned to the reference genome [40]. Clean reads were aligned to the reference genome using SOAP2 [41]. To eliminate the PCR interference and ambiguous mapping, duplicated reads and multimapped reads were filtered from the alignment results.
4.11. Quantification and Analysis of DEGs
The gene expression levels were estimated by HTSeqv0.5.3 (http://www-huber.embl.de/users/anders/HTSeq) for each sample. The FPKM was obtained according to the mapped transcript fragments, transcript length, and sequencing depth. Differences in gene expression between control and case groups were analyzed based on the DESeq R package (1.10.1) [42]. To control the false discovery rate, the Benjamini and Hochberg’s approach were used to adjust the P values [43]. Genes with an adjusted P value of less than 0.05 found by DESeq were regarded as differentially expressed.
4.12. GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analysis of DEGs
GO enrichment analysis of differentially expressed transcripts was performed using the GOseq R package [44], such that the gene length bias could be adjusted. To discover and cluster the biological functions of DEGs, the KEGG pathways were analyzed by Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 (https://david.ncifcrf.gov/). DAVID provides a comprehensive set of functional annotation tools for investigators to understand biological meaning behind large lists of genes. Functional groups with at least two DEGs in the background terms were selected, and those with a P value of less than 0.05 were considered significantly overexpressed. The top 10 functional groups, such as MF, CC, and BP, were selected to show in the picture. KOBAS [44] software was used to test the statistical enrichment of DEGs in KEGG pathways.
4.13. Real-Time PCR Validation of DEGs
DEGs selected from RNA-seq data were validated using qPCR, which was performed with SYBR Premix ExTaq (TaKaRa, Dalian, China) on an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). cDNA synthesis was performed using Quantscript RT Kit Quant cDNA (Tiangen Biotech, Beijing, China) with approximately 400 ng of total RNA as the template. SYBR Premix ExTaq (Takara, Dalian, China) was used for real-time PCR analysis on an ABI7500 Real-Time PCR System (Applied Biosystems) with cycling conditions of 95 °C held for 30 s; 42 cycles at 95 °C for 5 s and 59 °C for 20 s; followed by the programmed dissociation analysis from 95 °C to 60 °C to verify the amplification authenticity. The β-actin gene was used as a reference control. Each plate was repeated three times in independent runs for all references. Eight genes were chosen (β-catenin, KRT25, KRT71, KRT79, MPC1, KRTDAP, ASAP2, and ASB7) for the validation. PCR was performed using the primers shown in Table 3, all of which were self-designed except for KRT71 [45]. Gene expression was evaluated by the 2−ΔΔCt method [46].
Table 3.
Primers for real-time polymerase chain reaction.
| Gene | Primer Sequence (5′→3′) | Tm (°C) | |
|---|---|---|---|
| β-catenin | F1 | AGCGTCGTACATCTATGGG | 58 |
| R1 | ATAATCCTGTGGCTTGACC | ||
| KRT25 | F7 | AACAATATGAGAGCCGAGTA | 57 |
| R7 | AACAATATGAGAGCCGAGTA | ||
| KRT71 | F8 | TCATCGACAAGGTGAGGTTCC | 59 |
| R8 | CTGTCCGCCTGTTGATTTCTT | ||
| KRT79 | F9 | GCAGACATACTCCACCAA | 56 |
| R9 | GTTGACCGAGATGCTCTT | ||
| MPC1 | F10 | GCCATCAATGACATGAAGAA | 52 |
| R10 | CACCTTGTAGGCGAATCT | ||
| KRTDAP | F11 | GAGGAAGAGACCACCATTG | 52 |
| R11 | CGTGCCAGTTCAGGAATT | ||
| ASAP2 | F12 | CGTTCCTCAAGTTCTCAGT | 55 |
| R12 | GCTCCTTCTCCATCTCCT | ||
| ASB7 | F13 | ACTTAATCGGAGGCTTCAC | 55 |
| R13 | GGAGGAACATTCGGCAAT |
5. Conclusions
In our study, heritable, stable expression of ovine β-catenin in the wool skin of transgenic sheep was obtained. New wool follicles were induced by high expression of β-catenin in sheep. The wool follicle density and fleece weight were increased. These sheep were expected to be an excellent resource for breeding of sheep producing high-quality wool. The results of high-throughput RNA-seq showed that β-catenin and some of its direct or indirect target genes were upregulated in transgenic sheep, demonstrating the high activity of the Wnt pathway in these transgenic sheep. Thus, our findings established a new animal model for exploring the molecular mechanism of wool follicle development and for additional analysis of the Wnt pathway. Further studies are still needed to elucidate the mechanisms of β-catenin gene expression in the inner root and outer root of the sheath of transgenic sheep; such studies may reveal new approaches for regulating hair follicle development by the Wnt pathway.
Acknowledgments
We thank Rui Li from Inner Mongolia Caoyuan Jinfeng Company Ltd., for providing samples. We appreciate Zhengxing Lian, Guoshi Liu, and their teams from China Agricultural University and Weiheng Rong from Inner Mongolia Academy of Agricultural and Animal Husbandry Sciences for support with the field experiments.
Abbreviations
| bp | Base pairs |
| DEGs | Differentially expressed genes |
| FPKM | Fragments per kilobase of exon per million fragments mapped |
| GO | Gene ontology |
| BP | Biological process |
| CC | Cellular component |
| MF | Molecular function |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| qRT-PCR | Quantitative reverse transcription polymerase chain reaction |
| RNA-Seq | RNA sequencing |
| IHC | Immunohistochemical |
Supplementary Materials
Supplementary Materials can be found at http://www.mdpi.com/1422-0067/20/3/620/s1. Table S1: Data quality analysis in investigated samples. Table S2: Total transcript data analyzed in the investigated samples. Table S3: Summary of Illumina sequencing and mapping. Figure S1: Analysis of wool length (A) and diameter (B) of F0 and F1 sheep.
Author Contributions
J.W., K.C. and Z.Y. performed the experiments. J.W. drafted this manuscript. T.L. and G.H. analyzed the data. D.H., Y.Y., J.C., X.D. (Xiaotian Deng) and X.Y. collected samples. X.D. (Xuemei Deng) conceived and designed the experiments and modified the manuscript. All authors read and approved the final version of the manuscript.
Funding
This research was funded by programs of the Major Project for Cultivation Technology of New Varieties of Genetically Modified Organisms of the Ministry of Agriculture (grant Nos. 2016ZX08008-001 and 2013ZX08008-001).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Watt F.M. Stem cell fate and patterning in mammalian epidermis. Curr. Opin. Genet. Dev. 2001;11:410–417. doi: 10.1016/S0959-437X(00)00211-2. [DOI] [PubMed] [Google Scholar]
- 2.Lavker R.M., Sun T.T., Oshima H., Barrandon Y., Akiyama M., Ferraris C., Chevalier G., Favier B., Jahoda C.A., Dhouailly D., et al. Hair follicle stem cells. J. Investig. Dermatol. Symp. Proc. 2003;8:28–38. doi: 10.1046/j.1523-1747.2003.12169.x. [DOI] [PubMed] [Google Scholar]
- 3.Jahoda C.A., Oliver R.F., Reynolds A.J., Forrester J.C., Gillespie J.W., Cserhalmi-Friedman P.B., Christiano A.M., Horne K.A. Trans-species hair growth induction by human hair follicle dermal papillae. Exp. Dermatol. 2001;10:229–237. doi: 10.1034/j.1600-0625.2001.100402.x. [DOI] [PubMed] [Google Scholar]
- 4.Hardy M.H. The secret life of the hair follicle. Trends Genet. 1992;8:55–61. doi: 10.1016/0168-9525(92)90350-D. [DOI] [PubMed] [Google Scholar]
- 5.Oshima H., Rochat A., Kedzia C., Kobayashi K., Barrandon Y. Morphogenesis and renewal of hair follicles from adult multipotent stem cells. Cell. 2001;104:233–245. doi: 10.1016/S0092-8674(01)00208-2. [DOI] [PubMed] [Google Scholar]
- 6.Millar S.E. Molecular mechanisms regulating hair follicle development. J. Investig. Dermatol. 2002;118:216–225. doi: 10.1046/j.0022-202x.2001.01670.x. [DOI] [PubMed] [Google Scholar]
- 7.Huelsken J., Vogel R., Erdmann B., Cotsarelis G., Birchmeier W. Beta-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 2001;105:533–545. doi: 10.1016/S0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 8.Ozawa M., Baribault H., Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J. 1989;8:1711–1717. doi: 10.1002/j.1460-2075.1989.tb03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchs E., Merrill B.J., Jamora C., DasGupta R. At the roots of a never-ending cycle. Dev. Cell. 2001;1:13–25. doi: 10.1016/S1534-5807(01)00022-3. [DOI] [PubMed] [Google Scholar]
- 10.Gat U., DasGupta R., Degenstein L., Fuchs E. De Novo hair follicle morphogenesis and hair tumors in mice expressing a truncated beta-catenin in skin. Cell. 1998;95:605–614. doi: 10.1016/S0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- 11.Rasali D.R., Shrestha J.N.B., Crow G.H. Development of composite sheep breeds in the world: A review. Can. J. Anim. Sci. 2006;86:1–24. [Google Scholar]
- 12.Konigshoff M., Balsara N., Pfaff E.M., Kramer M., Chrobak I., Seeger W., Eickelberg O. Functional Wnt Signaling Is Increased in Idiopathic Pulmonary Fibrosis. PLoS ONE. 2008;3:e2142. doi: 10.1371/journal.pone.0002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veniaminova N.A., Vagnozzi A.N., Kopinke D., Do T.T., Murtaugh L.C., Maillard I., Dlugosz A.A., Reiter J.F., Wong S.Y. Keratin 79 identifies a novel population of migratory epithelial cells that initiates hair canal morphogenesis and regeneration. Development. 2013;140:4870–4880. doi: 10.1242/dev.101725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powell B.C., Beltrame J.S. Characterization of a hair (wool) keratin intermediate filament gene domain. J. Investig. Dermatol. 1994;102:171–177. doi: 10.1111/1523-1747.ep12371758. [DOI] [PubMed] [Google Scholar]
- 15.Veraitch O., Kobayashi T., Imaizumi Y., Akamatsu W., Sasaki T., Yamanaka S., Amagai M., Okano H., Ohyama M. Human induced pluripotent stem cell-derived ectodermal precursor cells contribute to hair follicle morphogenesis in vivo. J. Investig. Dermatol. 2013;133:1479–1488. doi: 10.1038/jid.2013.7. [DOI] [PubMed] [Google Scholar]
- 16.Harel S., Christiano A.M. Keratin 71 mutations: From water dogs to woolly hair. J. Investig. Dermatol. 2012;132:2315–2317. doi: 10.1038/jid.2012.291. [DOI] [PubMed] [Google Scholar]
- 17.Nanashima N., Ito K., Ishikawa T., Nakano M., Nakamura T. Damage of hair follicle stem cells and alteration of keratin expression in external radiation-induced acute alopecia. Int. J. Mol. Med. 2012;30:579–584. doi: 10.3892/ijmm.2012.1018. [DOI] [PubMed] [Google Scholar]
- 18.Gong H., Zhou H., Hickford J.G. Polymorphism of the ovine keratin-associated protein 1–4 gene (KRTAP1-4) Mol. Biol. Rep. 2010;37:3377–3780. doi: 10.1007/s11033-009-9925-4. [DOI] [PubMed] [Google Scholar]
- 19.Ansar M., Raza S.I., Lee K., Irfanullah, Shahi S., Acharya A., Dai H., Smith J.D., Shendure J., Bamshad M.J., Nickerson D.A., et al. A homozygous missense variant in type I keratin KRT25 causes autosomal recessive woolly hair. J. Med. Genet. 2015;52:676–680. doi: 10.1136/jmedgenet-2015-103255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andl T., Reddy S.T., Gaddapara T., Millar S.E. WNT signals are required for the initiation of hair follicle development. Dev. Cell. 2002;2:643–653. doi: 10.1016/S1534-5807(02)00167-3. [DOI] [PubMed] [Google Scholar]
- 21.Rosenquist T.A., Martin G.R. Fibroblast growth factor signalling in the hair growth cycle: Expression of the fibroblast growth factor receptor and ligand genes in the murine hair follicle. Dev. Dyn. 1996;205:379–386. doi: 10.1002/(SICI)1097-0177(199604)205:4<379::AID-AJA2>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 22.Fujimoto A., Kimura R., Ohashi J., Omi K., Yuliwulandari R., Batubara L., Mustofa M.S., Samakkarn U., Settheetham-Ishida W., Ishida T., et al. A scan for genetic determinants of human hair morphology: EDAR is associated with Asian hair thickness. Hum. Mol. Genet. 2008;17:835–843. doi: 10.1093/hmg/ddm355. [DOI] [PubMed] [Google Scholar]
- 23.Botchkarev V.A., Fessing M.Y. Edar signaling in the control of hair follicle development. J. Investig. Dermatol. Symp. Proc. 2005;10:247–251. doi: 10.1111/j.1087-0024.2005.10129.x. [DOI] [PubMed] [Google Scholar]
- 24.Kulessa H., Turk G., Hogan B.L. Inhibition of Bmp signaling affects growth and differentiation in the anagen hair follicle. EMBO J. 2000;19:6664–6674. doi: 10.1093/emboj/19.24.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiang C., Swan R.Z., Grachtchouk M., Bolinger M., Litingtung Y., Robertson E.K., Cooper M.K., Gaffield W., Westphal H., Beachy P.A., et al. Essential role for Sonic hedgehog during hair follicle morphogenesis. Dev. Biol. 1999;205:1–9. doi: 10.1006/dbio.1998.9103. [DOI] [PubMed] [Google Scholar]
- 26.Oshimori N., Fuchs E. Paracrine TGF-beta signaling counterbalances BMP-mediated repression in hair follicle stem cell activation. Cell Stem Cell. 2012;10:63–75. doi: 10.1016/j.stem.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weger N., Schlake T. Igf-I signalling controls the hair growth cycle and the differentiation of hair shafts. J. Investig. Dermatol. 2005;125:873–882. doi: 10.1111/j.0022-202X.2005.23946.x. [DOI] [PubMed] [Google Scholar]
- 28.Wodarz A., Nusse R. Mechanisms of Wnt signaling in development. Annu. Rev. Cell Dev. Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- 29.Playford M.P., Bicknell D., Bodmer W.F., Macaulay V.M. Insulin-like growth factor 1 regulates the location, stability, and transcriptional activity of beta-catenin. Proc. Natl. Acad. Sci. USA. 2000;97:12103–12108. doi: 10.1073/pnas.210394297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badreldin A., Shafei M.M., Marai I.F.M., Kamar G.A.R. Histological Structure of Wool Follicle Group in Fat-Tailed Sheep. Acta Anat. 1961;44:363–373. doi: 10.1159/000141734. [DOI] [PubMed] [Google Scholar]
- 31.Lo Celso C., Prowse D.M., Watt F.M. Transient activation of beta-catenin signalling in adult mouse epidermis is sufficient to induce new hair follicles but continuous activation is required to maintain hair follicle tumours. Development. 2004;131:1787–1799. doi: 10.1242/dev.01052. [DOI] [PubMed] [Google Scholar]
- 32.Tao Y., Yang Q., Wang L., Zhang J., Zhu X., Sun Q., Han Y., Luo Q., Wang Y., Guo X., et al. Beta-catenin activation in hair follicle dermal stem cells induces ectopic hair outgrowth and skin fibrosis. J. Mol. Cell Biol. 2018 doi: 10.1093/jmcb/mjy032. [DOI] [PubMed] [Google Scholar]
- 33.Langbein L., Rogers M.A., Praetzel-Wunder S., Helmke B., Schirmacher P., Schweizer J. K25 (K25irs1), K26 (K25irs2), K27 (K25irs3), and K28 (K25irs4) represent the type I inner root sheath keratins of the human hair follicle. J. Investig. Dermatol. 2006;126:2377–2386. doi: 10.1038/sj.jid.5700494. [DOI] [PubMed] [Google Scholar]
- 34.Yoshida H., Taguchi H., Hachiya A., Kitahara T., Boissy R.E., Visscher M.O. The natural trait of the curvature of human hair is correlated with bending of the hair follicle and hair bulb by a structural disparity in the root sheath. J. Dermatol. Sci. 2014;75:195–199. doi: 10.1016/j.jdermsci.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Oliver R.F. Ectopic regeneration of whiskers in the hooded rat from implanted lengths of vibrissa follicle wall. J. Embryol. Exp. Morphol. 1967;17:27–34. [PubMed] [Google Scholar]
- 36.Ibrahim L., Wright E.A. Inductive capacity of irradiated dermal papillae. Nature. 1977;265:733–744. doi: 10.1038/265733a0. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y., Andl T., Yang S.H., Teta M., Liu F., Seykora J.T., Tobias J.W., Piccolo S., Schmidt-Ullrich R., Nagy A., et al. Activation of beta-catenin signaling programs embryonic epidermis to hair follicle fate. Development. 2008;135:2161–2172. doi: 10.1242/dev.017459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Enshell-Seijffers D., Lindon C., Kashiwagi M., Morgan B.A. Beta-catenin Activity in the Dermal Papilla Regulates Morphogenesis and Regeneration of Hair. Dev. Cell. 2010;18:633–642. doi: 10.1016/j.devcel.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trapnell C., Pachter L., Salzberg S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li R., Yu C., Li Y., Lam T.W., Yiu S.M., Kristiansen K., Wang J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 42.Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benjamini Y., Hochberg Y. Controlling the False Discovery Rate—A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B Met. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 44.Mao X.Z., Cai T., Olyarchuk J.G., Wei L.P. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- 45.Kang X., Liu Y., Zhang J., Xu Q., Liu C., Fang M. Characteristics and Expression Profile of KRT71 Screened by Suppression Subtractive Hybridization cDNA Library in Curly Fleece Chinese Tan Sheep. DNA Cell Biol. 2017;36:552–564. doi: 10.1089/dna.2017.3718. [DOI] [PubMed] [Google Scholar]
- 46.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T) (-Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.