Abstract
Background
Development of emergency department (ED) triage systems that accurately differentiate and prioritize critically ill from stable patients remains challenging. We used machine learning models to predict clinical outcomes, and then compared their performance with that of a conventional approach—the Emergency Severity Index (ESI).
Methods
Using National Hospital and Ambulatory Medical Care Survey (NHAMCS) ED data, from 2007 through 2015, we identified all adult patients (aged ≥ 18 years). In the randomly sampled training set (70%), using routinely available triage data as predictors (e.g., demographics, triage vital signs, chief complaints, comorbidities), we developed four machine learning models: Lasso regression, random forest, gradient boosted decision tree, and deep neural network. As the reference model, we constructed a logistic regression model using the five-level ESI data. The clinical outcomes were critical care (admission to intensive care unit or in-hospital death) and hospitalization (direct hospital admission or transfer). In the test set (the remaining 30%), we measured the predictive performance, including area under the receiver-operating-characteristics curve (AUC) and net benefit (decision curves) for each model.
Results
Of 135,470 eligible ED visits, 2.1% had critical care outcome and 16.2% had hospitalization outcome. In the critical care outcome prediction, all four machine learning models outperformed the reference model (e.g., AUC, 0.86 [95%CI 0.85–0.87] in the deep neural network vs 0.74 [95%CI 0.72–0.75] in the reference model), with less under-triaged patients in ESI triage levels 3 to 5 (urgent to non-urgent). Likewise, in the hospitalization outcome prediction, all machine learning models outperformed the reference model (e.g., AUC, 0.82 [95%CI 0.82–0.83] in the deep neural network vs 0.69 [95%CI 0.68–0.69] in the reference model) with less over-triages in ESI triage levels 1 to 3 (immediate to urgent). In the decision curve analysis, all machine learning models consistently achieved a greater net benefit—a larger number of appropriate triages considering a trade-off with over-triages—across the range of clinical thresholds.
Conclusions
Compared to the conventional approach, the machine learning models demonstrated a superior performance to predict critical care and hospitalization outcomes. The application of modern machine learning models may enhance clinicians’ triage decision making, thereby achieving better clinical care and optimal resource utilization.
Electronic supplementary material
The online version of this article (10.1186/s13054-019-2351-7) contains supplementary material, which is available to authorized users.
Keywords: Triage, Emergency department, Prediction, Machine learning, Mortality, Critical care, Hospitalization, Hospital transfer, Decision curve analysis
Background
Over the past two decades, the number of emergency department (ED) visits has increased by approximately 50% in the USA, with 138 million visits in 2014 [1]. This increase has contributed to ED crowding and delays in care [2–4]. The literature has demonstrated that delay in care results in greater morbidity and mortality for many disease conditions [3–7]. ED triage presents the first opportunity to promptly identify high-risk patients and efficiently allocate finite ED resources. Among various triage algorithms, the Emergent Severity Index (ESI) is the most commonly used algorithm in US EDs [8–10]. Despite its wide adoption, it heavily relies on clinical judgment, leading to high inter-rater variability and suboptimal predictive ability [9–13].
The advent of machine learning models has shown promise to improve predictive ability in various conditions (e.g., sepsis, unplanned transfers to intensive care unit) [14–16]. These approaches offer advantages in that they account for high-order, non-linear interactions between predictors and gain more stable prediction [17]. Recent studies have reported that the application of machine learning models may provide high predictive ability at ED triage in selected patient populations and settings—e.g., children [18], patients with asthma and COPD exacerbation [19], and in few urban EDs [11, 20, 21]. Despite this clinical and research promise, no study has yet examined the utility of modern machine learning models for predicting clinical outcomes in a large population of adult patients in the ED.
To address this knowledge gap, we used large ED visit data to develop machine learning models—by using routinely available triage data—to accurately predict clinical outcomes after ED triage. We also examined the predictive performance of these models in comparison to the model using the conventional five-level ESI algorithm.
Methods
Study design and setting
We used combined data from the ED component of the 2007–2015 National Hospital and Ambulatory Medical Care Survey (NHAMCS) [22]. NHAMCS collects a nationally representative sample of visits to non-institutional general and short-stay hospitals, excluding federal, military, and Veterans Administration hospitals, in the 50 states and the District of Columbia. The survey has been conducted annually since 1992 by the National Center for Health Statistics (NCHS). For example, a total of 21,061 ED visits were surveyed in 2015 and submitted electronically from 267 EDs, equivalent to a weighted national sample of 137 million ED visits. The details of the NHAMCS methods and procedures may be found in the NHAMCS data file [22]. We followed the reporting guideline from the TRIPOD (Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis) statement [23]. The institutional review board of Massachusetts General Hospital waived the review of this study.
Study samples
We identified all adult ED visits (aged ≥ 18 years) recorded in the 2007–2015 data. The study period was chosen because the information on respiratory rates and oxygen saturation levels were not available before 2007. We excluded patients with death on ED arrival, those who left before being seen or against medical advice, and those with missing information or data inconsistencies (i.e., systolic blood pressure > 300 mmHg, diastolic blood pressure > 200 mmHg, pulse rate > 300/min, respiratory rate > 80/min, oxygen saturation > 100%).
Predictors
As the predictors for the machine learning models, we included routinely available information at ED triage settings—i.e., patient age, sex, mode of arrival (walk-in vs. ambulance), triage vital signs (temperature, pulse rate, systolic and diastolic blood pressure, respiratory rate, and oxygen saturation), chief complaints, and patient comorbidities. Chief complaints were reclassified according to the Reason for Visit Classification for Ambulatory Care provided [22]. As the comorbidity classification, we adopted 30 Elixhauser comorbidity measures using the data of the International Classification of Diseases, Ninth Version, Clinical Modification (ICD-9-CM) codes [24, 25].
Outcomes
The primary outcome was critical care outcome, defined as either direct admission to an intensive care unit (ICU) or in-hospital death, as done in previous studies [11, 12, 19, 26]. The prompt and accurate prediction of the critical care outcome at ED triage enables clinicians not only to efficiently allocate ED resources but also to urgently intervene on high-risk patients. The secondary outcome was hospitalization, defined as either an admission to an inpatient care site or direct transfer to an acute care hospital [11, 12, 19].
Statistical analysis
In the training set (70% randomly selected samples), we developed a reference model and four machine learning models for each outcome. As the reference model, we fitted a logistic regression model using the conventional ESI as the predictor. NHAMCS uses the five-level ESI algorithm: immediate (level 1), emergent (level 2), urgent (level 3), semi-urgent (level 4), and non-urgent (level 5) [8]. While 7% of the EDs participating in the NHAMCS did not use this classification, NCHS systematically recoded all data into these five levels [22]. We also fitted logistic regression models using demographic and physiologic variables in the NHAMCS data (i.e., age, mean blood pressure, heart rate, and respiratory rate) and APACHE II scoring system [27] as physiologic score-based models.
Next, using machine learning approaches, we developed four additional models: (1) logistic regression with Lasso regularization (Lasso regression), (2) random forest, (3) gradient boosted decision tree, and (4) deep neural network. First, Lasso regularization is one of the models that shrinks regression coefficients toward zero, thereby effectively selecting important predictors and improving the interpretability of the model. Coefficients of Lasso regression are the values that minimize the residual sum of square plus shrinkage penalty [17, 28, 29]. We used a 10-fold cross-validation to yield the optimal of regularization parameter (lambda) minimizing the sum of least square plus shrinkage penalty by using R glmnet package [28, 30]. Second, random forest is an ensemble of decision trees from bootstrapped training samples, and random samples of a certain number of predictors are selected to tree induction. We used R ranger and caret packages to construct random forest models [31, 32]. Third, gradient boosted decision tree is also an ensemble method which constructs new tree models predicting the errors and residuals of previous models. When adding the new models, this model uses a gradient descent algorithm to minimize a loss function [33]. We used R xgboost package to construct gradient boosted decision tree models [34]. Lastly, deep neural network model is composed of multiple processing layers. Outcomes are modeled by intermediate hidden units, and each hidden unit consists of the linear combination of predictors which are transformed into non-linear functions [17]. We used six-layer feedforward model with adaptive moment estimation optimizer and tuned hyperparameters (e.g., the number of hidden units, batch size, learning rate, learning rate decay, and dropout rate) using R Keras package [35, 36]. In these machine learning models, we used several methods to minimize potential overfitting—e.g., (1) Lasso regularization, (2) out-of-bag estimation, (3) cross-validation, and (4) dropout, Ridge regularization, and batch normalization in each model. To examine the importance of each predictor in the random forest models, we used permutation-based variable importance that is determined by the normalized average value of difference between prediction accuracy of the out-of-bag estimation and that of the same measure after permutating each predictor. In the gradient boosting decision tree models, we also computed the importance that is summed over iterations [32].
In the test set (the remaining 30% sample), we computed the prediction performance of each model that was derived above. As the prediction performance, we computed (1) the area under the receiver-operating-characteristics curve (AUC), (2) net reclassification improvement, (3) confusion matrix results (i.e., sensitivity, specificity, positive predictive value, and negative predictive value), and (4) net benefit through decision curve analysis. To compare the receiver-operating-characteristics curve (ROC) between models, Delong’s test was used [37]. The net reclassification improvement was used to quantify whether a new model provides clinically relevant improvements in prediction [38]. The decision curve analysis incorporates the information about the benefit of correctly triaging patients (true positives) and the relative harm of the over-triages (false positives)—i.e., the net benefit—over a range of threshold probability of the outcome (or clinical preference) [39–42]. We graphically demonstrated the net benefit of each model through a range of threshold probabilities of the outcome as a decision curve. All analyses were performed with R version 3.5.1.
Results
During 2007–2015, the NHAMCS recorded 209,800 adult ED visits. Of these, we excluded 97 ED visits with death on arrival, 6350 visits who left before being seen, or against medical advice, 67,674 visits with missing information, and 209 visits with data inconsistencies, leaving the analytic cohort of 135,470 ED visits. The patient characteristics between the analytic and non-analytic cohorts were generally similar (Additional file 1). In the analytic cohort, the median age was 46 years (IQR 29–60 years) and 43.1% were women (Table 1). Overall, 2782 ED visits (2.1%) had critical care outcome; pneumonia, chest pain, acute cerebrovascular disease, and congestive heart failure are the most common diagnoses (Table 2). Additionally, 22,010 ED visits (16.2%) had hospitalization outcome; nonspecific chest pain, abdominal pain, pneumonia, and other lower respiratory diseases are the most common diagnoses.
Table 1.
Predictor variables and outcomes in 135,470 adult emergency department visits
| Variable | n = 135,470 | |
|---|---|---|
| Age (year), median (IQR) | 46 | (29–60) |
| Female sex | 58,450 | (43.1) |
| Mode of arrival | ||
| Ambulance | 26,820 | (19.8) |
| Emergency Severity Index | ||
| 1 (immediate) | 2628 | (1.9) |
| 2 (emergent) | 16,908 | (12.5) |
| 3 (urgent) | 65,917 | (48.7) |
| 4 (semi-urgent) | 41,007 | (30.3) |
| 5 (non-urgent) | 9010 | (6.7) |
| Vital signs | ||
| Temperature (F), median (IQR) | 98.1 | (97.6–98.5) |
| Pulse rate (bpm), median (IQR) | 85 | (74–97) |
| Systolic blood pressure (mmHg), standard deviation (SD) | 136 | (23.2) |
| Diastolic blood pressure (mmHg), standard deviation (SD) | 79 | (14.5) |
| Respiratory rate (per min), median (IQR) | 18 | (16–20) |
| Oxygen saturation (%), median (IQR) | 98 | (97–99) |
| Common chief complaints | ||
| Musculoskeletal-related complaints | 21,499 | (15.9) |
| Gastrointestinal-related complaints | 20,947 | (15.5) |
| General complaints (e.g., fever) | 20,581 | (15.2) |
| Injuries | 16,731 | (12.4) |
| Respiratory-related complaints | 13,539 | (10.0) |
| Neurological-related complaints | 9828 | (7.3) |
| Urological-related complaints | 6869 | (5.1) |
| Psychiatry-related complaints | 4379 | (3.2) |
| Treatment-related complaints (e.g., side effects) | 3368 | (2.5) |
| Eye and ear-related complaints | 2952 | (2.2) |
| Skin-related complaints | 2902 | (2.1) |
| Intoxication | 1980 | (1.5) |
| Elixhauser comorbidity measures (≥ 1) | 18,249 | (13.5) |
| Clinical outcomes | ||
| Critical care outcome* | 2782 | (2.1) |
| Hospitalization outcome† | 22,010 | (16.2) |
Data are presented as number (percentage) of visits unless otherwise indicated
Abbreviations: ED emergency department, IQR interquartile range, SD standard deviation
*Direct admission to intensive care unit (ICU) or in-hospital death
† Admission to an inpatient care site or direct transfer to an acute care hospital
Table 2.
The 20 most common emergency department diagnoses for critical care and hospitalization outcome
| Critical care outcome | Hospitalization outcome | ||||
|---|---|---|---|---|---|
| CCS* | Diagnostic category | n | CCS* | Diagnostic category | n |
| 122 | Pneumonia | 161 | 102 | Nonspecific chest pain | 1836 |
| 102 | Nonspecific chest pain | 161 | 251 | Abdominal pain | 900 |
| 109 | Acute cerebrovascular disease | 138 | 122 | Pneumonia | 892 |
| 108 | Congestive heart failure (non-hypertensive) | 133 | 133 | Other lower respiratory diseases | 732 |
| 133 | Other lower respiratory diseases | 125 | 108 | Congestive heart failure (non-hypertensive) | 626 |
| 153 | Gastrointestinal hemorrhage | 101 | 127 | Chronic obstructive pulmonary disease and bronchiectasis | 570 |
| 106 | Cardiac dysrhythmias | 95 | 245 | Syncope | 556 |
| 131 | Respiratory failure, insufficiency, and arrest | 90 | 259 | Residual codes (unclassified) | 554 |
| 2 | Septicemia | 90 | 657 | Mood disorders | 535 |
| 259 | Residual codes (unclassified) | 86 | 197 | Skin and subcutaneous tissue infections | 531 |
| 55 | Fluid and electrolyte disorders | 82 | 106 | Cardiac dysrhythmias | 530 |
| 127 | Chronic obstructive pulmonary disease and bronchiectasis | 70 | 109 | Acute cerebrovascular disease | 483 |
| 100 | Acute myocardial infarction | 64 | 153 | Gastrointestinal hemorrhage | 466 |
| 101 | Coronary atherosclerosis and other heart disease | 62 | 159 | Urinary tract infections | 463 |
| 50 | Diabetes mellitus with complications | 57 | 55 | Fluid and electrolyte disorders | 459 |
| 233 | Intracranial injury | 53 | 659 | Schizophrenia and other psychotic disorders | 391 |
| 242 | Poisoning by other medications and drugs | 45 | 101 | Coronary atherosclerosis and other heart disease | 319 |
| 251 | Abdominal pain | 43 | 660 | Alcohol-related disorders | 285 |
| 245 | Syncope | 40 | 252 | Malaise and fatigue | 270 |
| 159 | Urinary tract infections | 40 | 246 | Fever of unknown origin | 258 |
Abbreviation: CCS Clinical Classification Software
*The principal diagnoses (> 14,000 ICD-9-CM diagnosis codes) were consolidated into 285 mutually exclusive diagnostic categories using the Agency for Healthcare Research and Quality Clinical Classifications Software (CCS) [50], as done previously [51]
Predicting critical care outcome
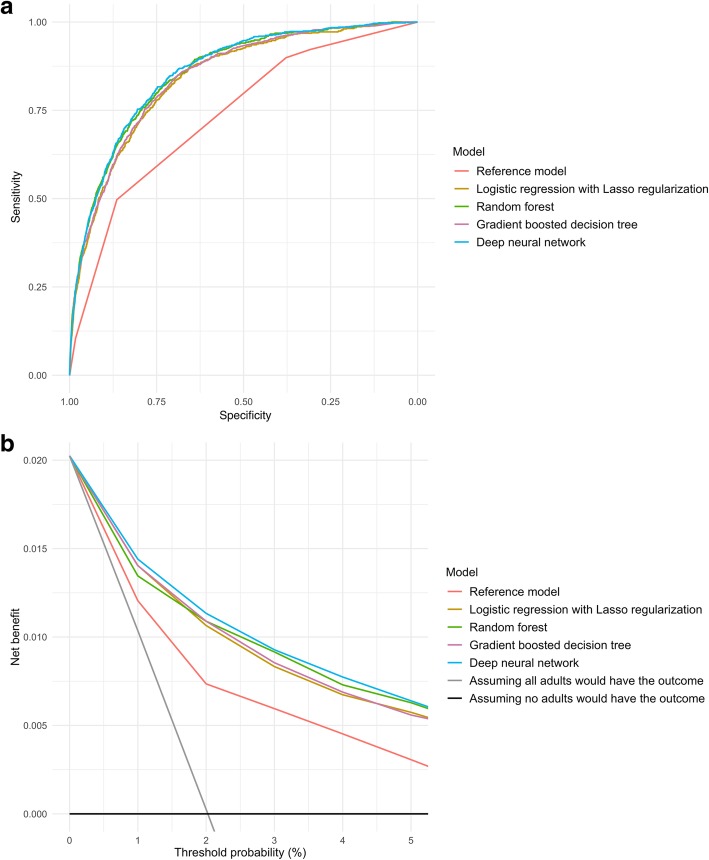
In the prediction of critical care outcome, the discriminatory abilities of all models are shown in Fig. 1a and Table 3. Compared with the reference model, all four machine learning models demonstrated a significantly higher AUC (all P < 0.001). For example, compared to the reference model (AUC 0.74 [95%CI 0.72–0.75]), the AUC was higher in the gradient boosted decision tree (0.85 [95%CI 0.83–0.86]) and deep neural network (0.86 [95%CI 0.85–0.87]) models. Likewise, compared with the reference model, all machine learning models also achieved significant net reclassification improvement (e.g., P < 0.001 in the deep neural network model).
Fig. 1.
Prediction ability of the reference model and machine learning models for intensive care use and in-hospital mortality in the test set. a Receiver-operating-characteristics (ROC) curves. The corresponding values of the area under the receiver-operating-characteristics curve (AUC) for each model are presented in Table 2. b Decision curve analysis. X-axis indicates the threshold probability for critical care outcome and Y-axis indicates the net benefit. Compared to the reference model, the net benefit for all machine learning models was larger over the range of clinical threshold
Table 3.
Prediction performance of the reference and machine learning models in the test set
| Outcome and model | AUC | P value* | NRI† | P value† | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|---|---|
| Critical care outcome | ||||||||
| Reference model | 0.74 (0.72–0.75) | Reference | Reference | Reference | 0.50 (0.47–0.53) | 0.86 (0.82–0.87) | 0.07 (0.05–0.08) | 0.988 (0.988–0.988) |
| Lasso regression | 0.84 (0.83–0.85) | < 0.001 | 0.39 (0.32–0.46) | < 0.001 | 0.75 (0.72–0.78) | 0.77 (0.75–0.80) | 0.06 (0.06–0.07) | 0.993 (0.993–0.994) |
| Random forest | 0.85 (0.84–0.87) | < 0.001 | 0.07 (0.003–0.14) | 0.04 | 0.86 (0.83–0.88) | 0.68 (0.68–0.71) | 0.05 (0.05–0.06) | 0.996 (0.996–0.996) |
| Gradient boosted decision tree | 0.85 (0.83–0.86) | < 0.001 | 0.32 (0.25–0.38) | < 0.001 | 0.75 (0.73–0.79) | 0.77 (0.75–0.80) | 0.06 (0.06–0.07) | 0.993 (0.993–0.994) |
| Deep neural network | 0.86 (0.85–0.87) | < 0.001 | 0.73 (0.67–0.79) | < 0.001 | 0.80 (0.77–0.83) | 0.76 (0.73–0.78) | 0.06 (0.06–0.07) | 0.995 (0.994–0.995) |
| Hospitalization outcome | ||||||||
| Reference model | 0.69 (0.68–0.69) | Reference | Reference | Reference | 0.87 (0.86–0.87) | 0.42 (0.39–0.43) | 0.23 (0.22–0.23) | 0.94 (0.94–0.94) |
| Lasso regression | 0.81 (0.80–0.81) | < 0.001 | 0.53 (0.50–0.55) | < 0.001 | 0.71 (0.70–0.72) | 0.76 (0.75–0.77) | 0.36 (0.35–0.37) | 0.93 (0.93–0.93) |
| Random forest | 0.81 (0.81–0.82) | < 0.001 | 0.66 (0.63–0.68) | < 0.001 | 0.77 (0.76–0.78) | 0.71 (0.70–0.72) | 0.34 (0.33–0.35) | 0.94 (0.94–0.94) |
| Gradient boosted decision tree | 0.82 (0.82–0.83) | < 0.001 | 0.63 (0.61–0.66) | < 0.001 | 0.75 (0.73–0.76) | 0.75 (0.74–0.76) | 0.37 (0.36–0.38) | 0.94 (0.94–0.94) |
| Deep neural network | 0.82 (0.82–0.83) | < 0.001 | 0.68 (0.65–0.70) | < 0.001 | 0.79 (0.78–0.80) | 0.71 (0.69–0.72) | 0.35 (0.34–0.36) | 0.95 (0.94–0.95) |
Abbreviations: AUC area under the curve, NRI net reclassification improvement, PPV positive predictive value, NPV negative predictive value
*P value was calculated to compare the area under the receiver-operating-characteristics curve (AUC) of the reference model with that of each machine learning model
†We used continuous NRI and its P value
Additionally, compared with the reference model, all machine learning models demonstrated a higher sensitivity—e.g., 0.50 [95%CI 0.47–0.53] in the reference model vs. 0.86 [95%CI 0.83–0.88] in the random forest model; Table 3. As a trade-off, the specificity of the reference model appeared higher than that of machine learning models—e.g., 0.82 [95%CI 0.82–0.86] in the reference model vs. 0.68 [95%CI 0.68–0.71] in the random forest model. Given the low prevalence of the critical care outcome, all models had high negative predictive values—e.g., 0.988 [95%CI 0.988–0.988] in the reference model vs. 0.996 [95%CI 0.996–0.996] in the random forest model. The AUC of the physiologic score-based model was 0.75 [95%CI 0.74–0.77]. Other predictive performance measures included sensitivity of 0.68 [95%CI 0.65–0.71] and specificity of 0.72 [95%CI 0.71–0.72].
With regard to the number of actual and predicted outcomes stratified by ESI level (Table 4), the reference model correctly predicted critical care outcomes in the triage levels 1 and 2 (immediate and emergent: 49.6% of all critical care outcomes). However, it also over-triaged a large number of patients in these high-acuity categories and failed to predict all critical care outcomes in the levels 3 to 5—i.e., under-triaging 50.4% of critically ill patients. In contrast, the machine learning models successfully predicted 71.3–81.6% of the actual outcomes in the triage levels 3 to 5. Likewise, the decision curve analysis (Fig. 1b) also demonstrated that the net benefit of all machine learning models surpassed that of the reference model throughout the threshold ranges, indicating machine learning-based prediction would more accurately identify patients at high risk with taking the trade-off with over-triages into consideration.
Table 4.
The number of actual and predicted outcomes of prediction models in the test set
| Conventional 5 triage levels (ESI) | Actual number of critical care outcome, n (%) | Reference model | Lasso regression | Random forest | Gradient boosted tree | Deep neural network | |||||
| Number of correctly identified outcome | Number of predicted outcome | Number of correctly identified outcome | Number of predicted outcome | Number of correctly identified outcome | Number of predicted outcome | Number of correctly identified outcome | Number of predicted outcome | Number of correctly identified outcome | Number of predicted outcome | ||
| 1: Immediate (n = 768) | 86 (11.1) | 86 | 768 | 72 | 366 | 79 | 460 | 74 | 373 | 76 | 393 |
| 2: Emergent (n = 5046) | 323 (6.4) | 323 | 5046 | 241 | 2175 | 290 | 2970 | 249 | 2165 | 264 | 2387 |
| 3: Urgent (n = 19,700) | 331 (1.7) | 0 | 0 | 244 | 5395 | 269 | 7482 | 239 | 5278 | 255 | 5698 |
| 4: Semi-urgent (n = 12,344) | 64 (0.5) | 0 | 0 | 45 | 1498 | 52 | 2283 | 44 | 1464 | 46 | 1505 |
| 5: Non-urgent (n = 2783) | 19 (0.7) | 0 | 0 | 15 | 308 | 17 | 457 | 14 | 300 | 16 | 315 |
| Overall (n = 40,641) | 823 (2.0) | 409 | 5814 | 617 | 9742 | 707 | 13,652 | 620 | 9580 | 657 | 10,298 |
| Conventional 5 triage levels (ESI) | Actual number of hospitalization outcome, n (%) | Reference model | Lasso regression | Random forest | Gradient boosted tree | Deep neural network | |||||
| Number of correctly identified outcome | Number of predicted outcome | Number of correctly identified outcome | Number of predicted outcome | Number of correctly identified outcome | Number of predicted outcome | Number of correctly identified outcome | Number of predicted outcome | Number of correctly identified outcome | Number of predicted outcome | ||
| 1: Immediate (n = 768) | 319 (41.5) | 319 | 768 | 213 | 393 | 259 | 460 | 241 | 434 | 252 | 471 |
| 2: Emergent (n = 5046) | 1810 (35.9) | 1810 | 5046 | 1398 | 2702 | 1500 | 3039 | 1482 | 2875 | 1563 | 3188 |
| 3: Urgent (n = 19,700) | 3628 (18.4) | 3628 | 19,700 | 2528 | 7320 | 2716 | 8307 | 2626 | 7512 | 2791 | 8484 |
| 4: Semi-urgent (n = 12,344) | 717 (5.8) | 0 | 0 | 466 | 2103 | 509 | 2584 | 488 | 2154 | 524 | 2515 |
| 5: Non-urgent (n = 2783) | 173 (6.2) | 0 | 0 | 105 | 434 | 121 | 577 | 117 | 441 | 120 | 547 |
| Overall (n = 40,641) | 6647 (16.4) | 5757 | 25,514 | 4710 | 12,952 | 5105 | 14,967 | 4954 | 13,416 | 5250 | 15,205 |
Abbreviations: ESI Emergency Severity Index, ICU intensive care unit
Predicting hospitalization outcome
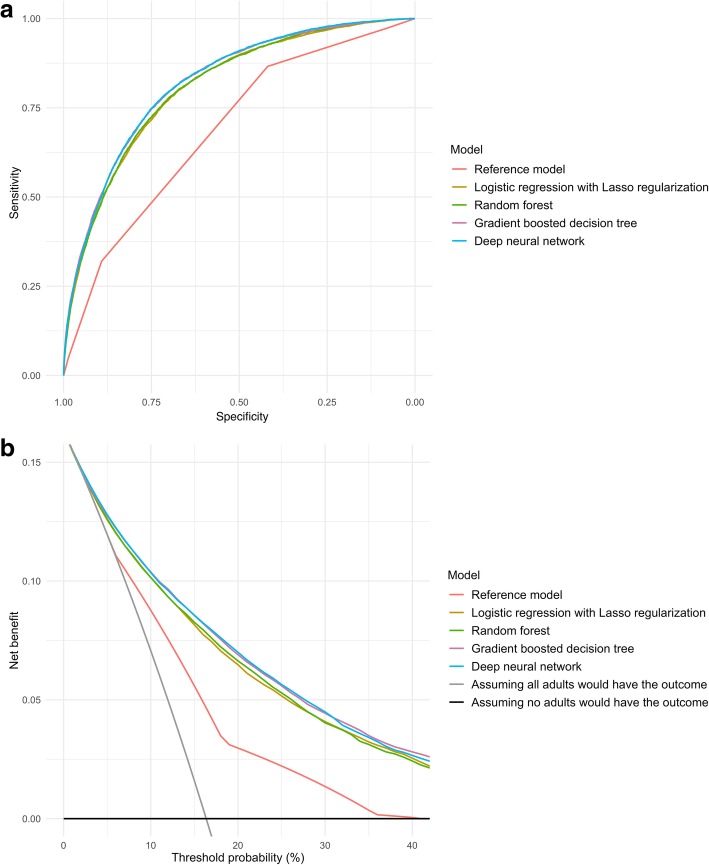
In the prediction of hospitalization outcome, the discriminatory abilities of models are shown in Fig. 2a and Table 3. Compared with the reference model, all four machine learning models demonstrated a significantly higher AUC (P < 0.001). For example, compared to the reference model (AUC 0.69 [95%CI 0.68–0.69]; Table 3), the AUC was higher in the gradient boosted decision tree (0.82 [95%CI 0.82–0.83]) and deep neural network (0.82 [95%CI 0.82–0.83]) models. Likewise, compared with the reference model, all machine learning models achieved significant net reclassification improvement (e.g., P < 0.001 in the random forest model).
Fig. 2.
Prediction ability of the reference model and machine learning models for hospitalization in the test set. a Receiver-operating-characteristics (ROC) curves. The corresponding values of the area under the receiver-operating-characteristics curve (AUC) for each model are presented in Table 2. b Decision curve analysis. X-axis indicates the threshold probability for hospitalization outcome and Y-axis indicates the net benefit. Compared to the reference model, the net benefit for all machine learning models was larger over the range of clinical threshold
While all the machine learning models demonstrated a lower sensitivity (e.g., 0.87 [95%CI 0.86–0.87] in the reference model vs. 0.71 [95%CI 0.70–0.72] in Lasso regression, Table 3), they yield a higher specificity (e.g., 0.42 [95%CI 0.39–0.43] in the reference model vs. 0.76 [95%CI 0.75–0.77] in Lasso regression model). The AUC of the physiologic score-based model was 0.71 [95%CI 0.71–0.72]. Other predictive performance measures included sensitivity of 0.63 [95%CI 0.62–0.65] and specificity of 0.69 [95%CI 0.68–0.69].
With regard to the number of actual and predicted outcomes stratified by ESI (Table 4), the reference model over-triaged a large number of patients in the triage levels 1 to 3 and failed to predict all hospitalization outcomes in the levels 4 and 5—i.e., under-triaging 13.4% of hospitalized patients. In contrast, the machine learning models successfully predicted 64.2–72.4% of the actual outcomes in the levels 4 and 5. Likewise, the decision curve analysis (Fig. 2b) also demonstrated that the net benefit of all machine learning models surpassed that of the reference model throughout the threshold ranges.
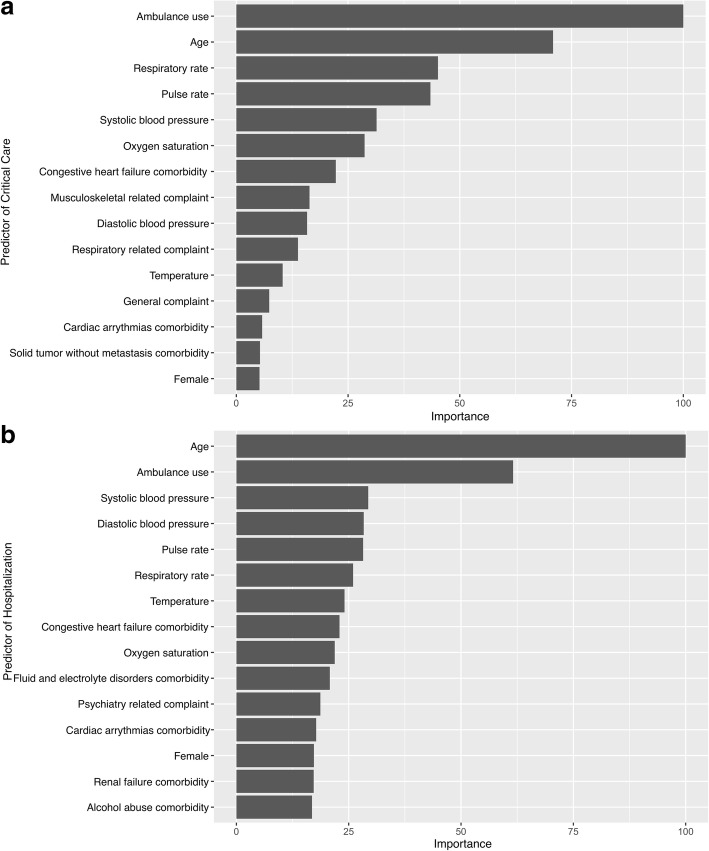
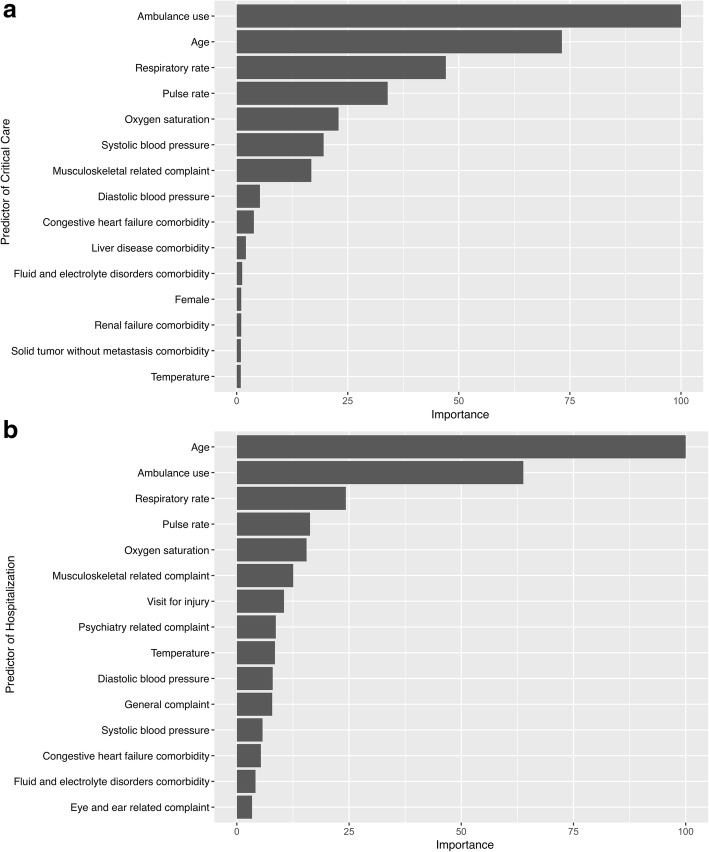
Variable importance
To gain insights into the relevance of each predictor, Figs. 3 and 4 summarize the 15 most important predictors of random forest and gradient boosted decision tree models for each outcome. In the random forest models, ambulance use, age, vital signs, and comorbidities (e.g., congestive heart failure) were most important predictors for the critical care (Fig. 3a) and hospitalization (Fig. 3b) outcomes. The variable importance was similar in the gradient boosted decision tree models (Fig. 4).
Fig. 3.
Variable importance of predictors in the random forest models. The variable importance is a scaled measure to have a maximum value of 100. The predictors with a variable importance of the top 15 are shown. a Critical care outcome. b Hospitalization outcome
Fig. 4.
Variable importance of predictors in the gradient boosted decision tree models. The variable importance is a scaled measure to have a maximum value of 100. The predictors with a variable importance of top 15 are shown. a Critical care outcome. b Hospitalization outcome
Discussion
Based on the data of 135,470 adult ED visits, we applied four modern machine learning approaches (i.e., Lasso regression, random forest, gradient boosted decision tree, and deep neural network) to the routinely available triage information. Compared to the conventional model, based on ESI algorithm [8], these machine learning models demonstrated a superior performance in predicting critical care and hospitalization outcomes, including improved AUCs and net reclassification. Additionally, the machine learning models had a higher sensitivity for the critical care outcome with a reduced number of under-triaged critically ill patients, and had a higher specificity for the hospitalization outcome with fewer over-triages. Moreover, the decision curve analysis revealed that all machine learning models yielded a larger net benefit—the trade-off between appropriate triages and over-triages—throughout the wide range of thresholds. To date, this is the first investigation that has comprehensively examined the utility of modern machine learning models for predicting clinical outcomes in a large population of adult patients in the ED.
The major goals of ED triage are to accurately differentiate high-risk patients from more-stable patients and to efficiently allocate finite ED resources. Prior studies have documented that current triage algorithms (e.g., ESI) have a suboptimal predictive ability to identify critically ill patients, low inter-rater agreement, and high variability within a same triage level [9–13]. While the use of a complete set of information—such as detailed data on past and present illnesses, physical examinations, and repeated measurements during the ED course—may improve prediction abilities, it is impractical at ED triage settings because of the limited information and time available. An alternative approach to enhance clinicians’ prediction abilities is to utilize advanced machine learning models. Recently, the machine learning models have been applied to outcome predictions in diverse medical fields—e.g., mortality in patients with sepsis [14], cardiac complications in patients with acute chest pain [43], rehospitalization in patients with congestive heart failure [44], critical care and hospitalization outcomes in children [18] and in adults with asthma and COPD exacerbation [19], and unplanned transfer to ICU [15]. The current study corroborates the promise suggested by these recent studies and extends them by demonstrating superior predictive abilities of modern machine learning models over the conventional model in a large population of adults in the ED.
ED triage systems seek for an optimal balance between under-triages and over-triages. The present study showed that, compared to the conventional ESI approach, our machine learning models demonstrated a higher sensitivity in predicting ICU admission and in-hospital mortality. Indeed, the models correctly identified critically ill patients who would be inappropriately under-triaged into lower-acuity ESI levels (levels 3 to 5), supporting the advantages of machine learning-based prediction at the ED triage where rapid identification of patients at high risk is paramount. By contrast, patients who are going to be admitted do not always need excessive recourse in the ED (e.g., patients with cellulitis who are admitted to an ED observation unit). Therefore, predictions of hospitalization outcome using a high-sensitivity (and low-specificity) model would lead to over-triages and excessive resource utilization. However, our machine learning models yielded a higher specificity in predicting hospitalization with a reduced number of over-triaged patients, particularly in the higher-acuity ESI levels (levels 1 to 3), who may not utilize excessive resource. Additionally, the utility of machine learning-based prediction is further buttressed by the greater net benefit observed in the decision curve analysis—which incorporates the trade-off between over- and under-triages [39, 45]—across the wide range of clinical thresholds.
The reasons for the improvement in predictive abilities observed in the machine learning models are likely multifactorial. First, the ESI algorithm heavily relies on subjective clinical assessment of anticipated ED resource use that leads to modest performance and large variabilities between providers [8–10]. Second, advanced machine leaning approaches are adept at handling high-order interactions between the predictors and non-linear relationships with the outcome [17, 28]. Third, while overfitting in conventional models is often problematic, our machine learning models adopted multiple rigorous approaches to mitigate overfitting, such as regularization, cross-validation, and dropout. Although our machine learning models achieved the superior predictive ability, the performance was not perfect. This is attributable, at least partly, to the limited set of predictors, subjectivity of data (e.g., visit reasons), various clinical factors after ED triage (e.g., quality and timeliness of ED management and patients’ clinical responses), differences in patients’ health behaviors, providers’ practice patterns, and availability of ED resources. Yet, in the era of health information technology, machine learning-based prediction has a scalable advantage—e.g., updating prediction models through an automated extraction of electronic health record data and integration with digital images, natural language processing, and continuous monitoring of physiological data [46–48]. This scalability had been unattainable in the conventional models where decisions were made based on fixed rules encoding knowledge. Taken together, our findings and recent developments suggest that machine learning approaches are indispensable next-generation assistive technology to further advance clinical decision-making abilities [49].
The current study has several potential limitations. First, we excluded samples with missing information. Yet, the analytic and non-analytic cohorts were generally comparable in the patient demographics, ED presentation, and outcomes. These similarities argue against substantial selection bias. Second, the quality of data is important in data-driven machine learning-based prediction. Although survey data may have some misclassification and ascertainment bias, NHAMCS has a coding error rate of < 1% in their 10% quality control sample [22]. Third, NHAMCS data do not collect some helpful clinical variables (e.g., chronic medications, socioeconomic status, health behaviors). However, the goal of the present investigation is not to develop prediction models using a broad set of predictors but to derive machine learning models using a limited set of predictors that are routinely available at current ED triage settings. Finally, the indication and clinical threshold of ICU admission, hospitalization, and hospital transfer depend on the local healthcare resource and may vary between different EDs and clinicians. However, the decision curve analysis demonstrated that the net benefit of all machine learning models was consistently greater than that of the reference model across the wide range of threshold probabilities (or clinical preferences). This finding supports the generalizability of prediction models.
Conclusions
Based on the analysis of 135,470 adult ED visit data, we developed the machine learning models using ED triage data. These models yielded a superior performance in predicting critical care and hospitalization outcomes over the conventional ESI-based model. Particularly, the machine learning models would reduce the number of critically ill who are under-triaged by the conventional approach. Furthermore, the models would also decrease over-triaging hospitalization outcomes that lead to excessive resource allocation to less-sick patients. Moreover, the machine learning models also yielded a greater net benefit across wide ranges of threshold probabilities. While external validations are necessary, the current study lends substantial support to the application of machine learning-based predication to ED triage as a decision support technology. Machine learning models—as assistive technologies—offer new avenues for enhancing the clinician’s ED triage decision making, which will, in turn, improve patient care and optimize resource utilization in already-stressed emergency care systems.
Additional file
Comparison of predictor variables and outcomes between the analytic and non-analytic cohort. (DOCX 16 kb)
Acknowledgements
Not applicable
Funding
None
Availability of data and materials
The datasets analysed during the current study are publicly available: https://www.cdc.gov/nchs/ahcd/datasets_documentation_related.htm
Abbreviations
- AUC
Area under the receiver-operating-characteristics curve
- CDC
Centers for Disease Control and Prevention
- ED
Emergency department
- ESI
Emergency Severity Index
- NHAMCS
National Hospital and Ambulatory Medical Care Survey
- NPV
Negative predictive value
- PPV
Positive predictive value
- TRIPOD
Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis
Authors’ contributions
YR contributed to the design of the study, data analysis, and interpretation of the results and writing the manuscript. TG contributed to the design of the study and interpretation of the results and critically reviewed the manuscript. MKF, DFMB, and CAC contributed to the acquisition of and maintaining the data and critically reviewed the manuscript. KH contributed to the analysis of the data, design of the study, and interpretation of the result and critically reviewed the manuscript. All authors finally approved the final version of manuscript.
Ethics approval and consent to participate
The institutional review board of Massachusetts General Hospital waived review of this study.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yoshihiko Raita, Email: Yoshihiko_Raita@hms.harvard.edu.
Tadahiro Goto, Email: tag695@mail.harvard.edu.
Mohammad Kamal Faridi, Email: MKFARIDI1@mgh.harvard.edu.
David F. M. Brown, Email: Brown.David@mgh.harvard.edu
Carlos A. Camargo, Jr., Email: ccamargo@partners.org
Kohei Hasegawa, Email: khasegawa@mgh.harvard.edu.
References
- 1.HCUPnet. https://hcupnet.ahrq.gov Accessed 28 Nov 2018.
- 2.Emergency department wait times, crowding and access. American College of Emergency Physicians News Room. http://newsroom.acep.org/2009-01-04-emergency-department-wait-times-crowding-and-access-fact-sheet Accessed 1 Dec 2018.
- 3.Sun BC, Hsia RY, Weiss RE, Zingmond D, Liang L-J, Han W, et al. Effect of emergency department crowding on outcomes of admitted patients. Ann Emerg Med. 2013;61(6):605–611.e6. doi: 10.1016/j.annemergmed.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaieski DF, Agarwal AK, Mikkelsen ME, Drumheller B, Cham Sante S, Shofer FS, et al. The impact of ED crowding on early interventions and mortality in patients with severe sepsis. Am J Emerg Med. 2017;35(7):953–960. doi: 10.1016/j.ajem.2017.01.061. [DOI] [PubMed] [Google Scholar]
- 5.Gruen RL, Jurkovich GJ, McIntyre LK, Foy HM, Maier RV. Patterns of errors contributing to trauma mortality. Ann Surg. 2006;244(3):371–380. doi: 10.1097/01.sla.0000234655.83517.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hasegawa K, Sullivan AF, Tsugawa Y, Turner SJ, Massaro S, Clark S, et al. Comparison of US emergency department acute asthma care quality: 1997-2001 and 2011-2012. J Allergy Clin Immunol. 2015;135(1):73–80. doi: 10.1016/j.jaci.2014.08.028. [DOI] [PubMed] [Google Scholar]
- 7.Rathore SS, Curtis JP, Chen J, Wang Y, Nallamothu BK, Epstein AJ, et al. Association of door-to-balloon time and mortality in patients admitted to hospital with ST elevation myocardial infarction: national cohort study. BMJ. 2009;338:b1807. doi: 10.1136/bmj.b1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emergency Severity Index (ESI): A Triage Tool for Emergency Department https://www.ahrq.gov/professionals/systems/hospital/esi/index.html. Accessed 1 Dec 2018.
- 9.Mistry B, Stewart De Ramirez S, Kelen G, PSK S, Balhara KS, Levin S, et al. Accuracy and reliability of emergency department triage using the Emergency Severity Index: An International Multicenter Assessment. Ann Emerg Med. 2018;71(5):581–587.e3. doi: 10.1016/j.annemergmed.2017.09.036. [DOI] [PubMed] [Google Scholar]
- 10.Arya R, Wei G, McCoy JV, Crane J, Ohman-Strickland P, Eisenstein RM. Decreasing length of stay in the emergency department with a split Emergency Severity Index 3 patient flow model. Acad Emerg Med. 2013;20(11):1171–1179. doi: 10.1111/acem.12249. [DOI] [PubMed] [Google Scholar]
- 11.Levin S, Toerper M, Hamrock E, Hinson JS, Barnes S, Gardner H, et al. Machine-learning-based electronic triage more accurately differentiates patients with respect to clinical outcomes compared with the Emergency Severity Index. Ann Emerg Med. 2018;71(5):565–574.e2. doi: 10.1016/j.annemergmed.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Dugas AF, Kirsch TD, Toerper M, Korley F, Yenokyan G, France D, et al. An electronic emergency triage system to improve patient distribution by critical outcomes. J Emerg Med. 2016;50(6):910–918. doi: 10.1016/j.jemermed.2016.02.026. [DOI] [PubMed] [Google Scholar]
- 13.McHugh M, Tanabe P, McClelland M, Khare RK. More patients are triaged using the Emergency Severity Index than any other triage acuity system in the United States. Acad Emerg Med Off J Soc Acad Emerg Med. 2012;19(1):106–109. doi: 10.1111/j.1553-2712.2011.01240.x. [DOI] [PubMed] [Google Scholar]
- 14.Taylor RA, Pare JR, Venkatesh AK, Mowafi H, Melnick ER, Fleischman W, et al. Prediction of in-hospital mortality in emergency department patients with sepsis: a local big data-driven, machine learning approach. Acad Emerg Med Off J Soc Acad Emerg Med. 2016;23(3):269–278. doi: 10.1111/acem.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wellner B, Grand J, Canzone E, Coarr M, Brady PW, Simmons J, et al. Predicting unplanned transfers to the intensive care unit: a machine learning approach leveraging diverse clinical elements. JMIR Med Inform. 2017;5(4):e45. doi: 10.2196/medinform.8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desautels T, Das R, Calvert J, Trivedi M, Summers C, Wales DJ, et al. Prediction of early unplanned intensive care unit readmission in a UK tertiary care hospital: a cross-sectional machine learning approach. BMJ Open. 2017;7(9):e017199. doi: 10.1136/bmjopen-2017-017199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhn M, Johnson K. Applied predictive modeling. New York: Springer-Verlag; 2013. [Google Scholar]
- 18.Goto T, Camargo C, Faridi M, Freishtat R, Hasegawa K. Machine learning-based prediction of clinical outcomes for children during emergency department triage. JAMA Netw Open. 2019;2(1):e186937. doi: 10.1001/jamanetworkopen.2018.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goto T, Camargo CAJ, Faridi MK, Yun BJ, Hasegawa K. Machine learning approaches for predicting disposition of asthma and COPD exacerbations in the ED. Am J Emerg Med. 2018;36(9):1650–1654. doi: 10.1016/j.ajem.2018.06.062. [DOI] [PubMed] [Google Scholar]
- 20.Hong WS, Haimovich AD, Taylor RA. Predicting hospital admission at emergency department triage using machine learning. PLoS One. 2018;13(7):e0201016. doi: 10.1371/journal.pone.0201016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Kim J, Patzer RE, Pitts SR, Patzer A, Schrager JD. Prediction of emergency department hospital admission based on natural language processing and neural networks. Methods Inf Med. 2017;56(5):377–389. doi: 10.3414/ME17-01-0024. [DOI] [PubMed] [Google Scholar]
- 22.NAMCS/NHAMCS Ambulatory Health Care Data 2015 https://www.cdc.gov/nchs/ahcd/index.htm. Accessed 22 Nov 2018.
- 23.Moons KGM, Altman DG, Reitsma JB, Ioannidis JPA, Macaskill P, Steyerberg EW, et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162(1):W1–73. doi: 10.7326/M14-0698. [DOI] [PubMed] [Google Scholar]
- 24.Quan H, Sundararajan V, Halfon P, Fong A, Burnand B, Luthi J-C, et al. Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. Med Care. 2005;43(11):1130–1139. doi: 10.1097/01.mlr.0000182534.19832.83. [DOI] [PubMed] [Google Scholar]
- 25.icd: Comorbidity Calculations and Tools for ICD-9 and ICD-10 Codes. https://CRAN.R-project.org/package=icd Accessed 1 Dec 2018.
- 26.Mirhaghi A, Kooshiar H, Esmaeili H, Ebrahimi M. Outcomes for emergency severity index triage implementation in the emergency department. J Clin Diagn Res. 2015;9(4):OC04–OC07. doi: 10.7860/JCDR/2015/11791.5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knaus WA, Draper EA, Wagner DP, Zimmerman JE. APACHE II: a severity of disease classification system. Crit Care Med. 1985;13(10):818–829. doi: 10.1097/00003246-198510000-00009. [DOI] [PubMed] [Google Scholar]
- 28.James G, Witten D, Hastie T, Tibshirani R. An introduction to statistical learning: with applications in R. New York: Springer-Verlag; 2013. [Google Scholar]
- 29.Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc Ser B Methodol. 1996;58(1):267–288. [Google Scholar]
- 30.glmnet: Lasso and Elastic-Net regularized generalized linear models. https://CRAN.R-project.org/package=glmnet. Accessed 1 Dec 2018.
- 31.ranger: A Fast Implementation of Random Forests. https://CRAN.R-project.org/package=ranger. Accessed 29 Nov 2018.
- 32.caret Package. http://topepo.github.io/caret/index.html. Accessed 1 Dec 2018.
- 33.Natekin A, Knoll A. Gradient boosting machines, a tutorial. Front Neurorobot. 2013. 10.3389/fnbot.2013.00021. [DOI] [PMC free article] [PubMed]
- 34.xgboost: Extreme gradient boosting. https://CRAN.R-project.org/package=xgboost. Accessed 1 Dec 2018.
- 35.R Interface to “Keras”. https://keras.rstudio.com/. Accessed 1 Dec 2018.
- 36.Kingma DP, Ba J. Adam: A method for stochastic optimization. ArXiv14126980 Cs http://arxiv.org/abs/1412.6980. Accessed 1 Dec 2018.
- 37.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 38.Pencina MJ, D’Agostino RB, D’Agostino RB, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2008;27(2):157–172. doi: 10.1002/sim.2929. [DOI] [PubMed] [Google Scholar]
- 39.Van Calster B, Wynants L, Verbeek JFM, Verbakel JY, Christodoulou E, Vickers AJ, et al. Reporting and interpreting decision curve analysis: a guide for investigators. Eur Urol. 2018;74(6):796–804. doi: 10.1016/j.eururo.2018.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fitzgerald M, Saville BR, Lewis RJ. Decision curve analysis. JAMA. 2015;313(4):409–410. doi: 10.1001/jama.2015.37. [DOI] [PubMed] [Google Scholar]
- 41.Steyerberg EW, Vickers AJ. Decision curve analysis: a discussion. Med Decis Mak. 2008;28(1):146–149. doi: 10.1177/0272989X07312725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Mak. 2006;26(6):565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu N, Koh ZX, Chua EC-P, Tan LM-L, Lin Z, Mirza B, et al. Risk scoring for prediction of acute cardiac complications from imbalanced clinical data. IEEE J Biomed Health Inform. 2014;18(6):1894–1902. doi: 10.1109/JBHI.2014.2303481. [DOI] [PubMed] [Google Scholar]
- 44.Mortazavi BJ, Downing NS, Bucholz EM, Dharmarajan K, Manhapra A, Li S-X, et al. Analysis of machine learning techniques for heart failure readmissions. Circ Cardiovasc Qual Outcomes. 2016;9(6):629–640. doi: 10.1161/CIRCOUTCOMES.116.003039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rousson V, Zumbrunn T. Decision curve analysis revisited: overall net benefit, relationships to ROC curve analysis, and application to case-control studies. BMC Med Inform Decis Mak. 2011;11:45. doi: 10.1186/1472-6947-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ting DSW, Cheung CY-L, Lim G, Tan GSW, Quang ND, Gan A, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017;318(22):2211–2223. doi: 10.1001/jama.2017.18152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kolachalama VB, Singh P, Lin CQ, Mun D, Belghasem ME, Henderson JM, et al. Association of pathological fibrosis with renal survival using deep neural networks. Kidney Int Rep. 2018;3(2):464–475. doi: 10.1016/j.ekir.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Priesol AJ, Cao M, Brodley CE, Lewis RF. Clinical vestibular testing assessed with machine-learning algorithms. JAMA Otolaryngol-Head Neck Surg. 2015;141(4):364–372. doi: 10.1001/jamaoto.2014.3519. [DOI] [PubMed] [Google Scholar]
- 49.Obermeyer Z, Emanuel EJ. Predicting the future - big data, machine learning, and clinical medicine. N Engl J Med. 2016;375(13):1216–1219. doi: 10.1056/NEJMp1606181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clinical Classifications Software (CCS) for ICD-9-CM. https://www.hcup-us.ahrq.gov/toolssoftware/ccs/ccs.jsp Accessed 28 Jan 2019.
- 51.Hasegawa K, Gibo K, Tsugawa Y, Shimada YJ, Camargo CA. Age-related differences in the rate, timing, and diagnosis of 30-day readmissions in hospitalized adults with asthma exacerbation. Chest. 2016;149(4):1021–1029. doi: 10.1016/j.chest.2015.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of predictor variables and outcomes between the analytic and non-analytic cohort. (DOCX 16 kb)
Data Availability Statement
The datasets analysed during the current study are publicly available: https://www.cdc.gov/nchs/ahcd/datasets_documentation_related.htm