Abstract
The human gut microbiota has adapted to the presence of antimicrobial peptides (AMPs) that are ancient components of immune defence. Despite its medical importance, it has remained unclear whether AMP resistance genes in the gut microbiome are available for genetic exchange between bacterial species. Here we show that AMP- and antibiotic-resistance genes differ in their mobilization patterns and functional compatibilities with new bacterial hosts. First, whereas AMP resistance genes are widespread in the gut microbiome, their rate of horizontal transfer is lower than that of antibiotic resistance genes. Second, gut microbiota culturing and functional metagenomics revealed that AMP resistance genes originating from phylogenetically distant bacteria have only a limited potential to confer resistance in Escherichia coli, an intrinsically susceptible species. Taken together, functional compatibility with the new bacterial host emerges as a key factor limiting the genetic exchange of AMP resistance genes. Finally, our results suggest that AMPs induce highly specific changes in the composition of the human microbiota with implications for disease risks.
Introduction
The maintenance of homeostasis between the gut microbiota and the human host tissues entails a complex co-evolutionary relationship1,2. Mucosal barriers covering the intestinal epithelium restrict microbes to the lumen, control the composition of commensal inhabitants, and ensure removal of pathogens3,4. Cationic host antimicrobial peptides (AMPs) have crucial roles in this process5. They are among the most ancient and efficient components of the innate immune defence in multicellular organisms and have retained their efficacy for millions of years5,6. As AMPs have a broad spectrum of activity, much effort has been put into finding potential antibacterial drugs among AMPs7,8.
However, therapeutic use of AMPs may drive bacterial evolution of resistance to our own immunity peptides9,10. Therefore, it is of central importance to establish whether genes that influence AMP resistance (i.e. AMP resistance genes) in the gut microbiome are available for genetic exchange with other bacterial species. Several lines of observation support the plausibility of this scenario. The gut bacterial community is a rich source of mobile antibiotic resistance genes11, and certain abundant gut bacterial species exhibit high levels of intrinsic resistance to AMPs12. Moreover, 2 even single genes can confer high AMP resistance in Bacteroidetes12. However, beyond the recent discovery of a horizontally spreading resistance gene family13,14, the mobility of AMP resistance-encoding genes across bacterial species has remained a terra incognita.
Here, we applied an integrated approach to systematically characterize the mobilization potential of the AMP resistance gene reservoir in the human gut microbiome. First, we examined the patterns of horizontal gene transfer events involving AMP resistance genes by analyzing bacterial genome sequences from the human gut and naturally occurring plasmid sequences from the human microbiome. Next, we experimentally compared the functional compatibility of AMP- versus antibiotic-resistance genes from the gut microbiome with a susceptible host, Escherichia coli (E. coli), by performing functional metagenomic selections and by culturing the gut microbiome in the presence of diverse AMPs and small-molecule antibiotics. Together, these analyses revealed that AMP resistance genes are less frequently mobilized owing to lack of functional compatibility with new bacterial hosts.
Results
Infrequent horizontal transfer of AMP resistance genes in the gut microbiota
We begin by asking whether the genetic determinants of resistance to AMPs and antibiotics, respectively, differ in their rate of horizontal transfer in the human gut microbiota. To systematically address this issue, we first collected previously characterized AMP- and antibiotic-resistance genes from literature and databases, yielding a comprehensive catalogue of 114 and 199 AMP-resistance and antibiotic-resistance gene families, respectively (see Methods and Supplementary Table 1). By definition, AMP resistance genes influence bacterial susceptibility to at least one AMP when mutated (see Methods). Next, we compared the frequencies of these previously identified AMP- and antibiotic-resistance genes in a catalogue of 37,853 horizontally transferred genes from 567 genome sequences of phylogenetically diverse bacterial species in the human gut microbiota15. This mobile gene catalogue relies on the identification of nearly identical genes that are shared by distantly related bacterial genomes and thereby provides a snapshot on the gene set subjected to recent horizontal gene transfer events in a representative sample of the human gut microbiome15. We identified homologs of the literature-curated resistance genes for which at least one transfer event was reported (i.e., those present in the mobile gene pool; see Methods and Supplementary Table 2).
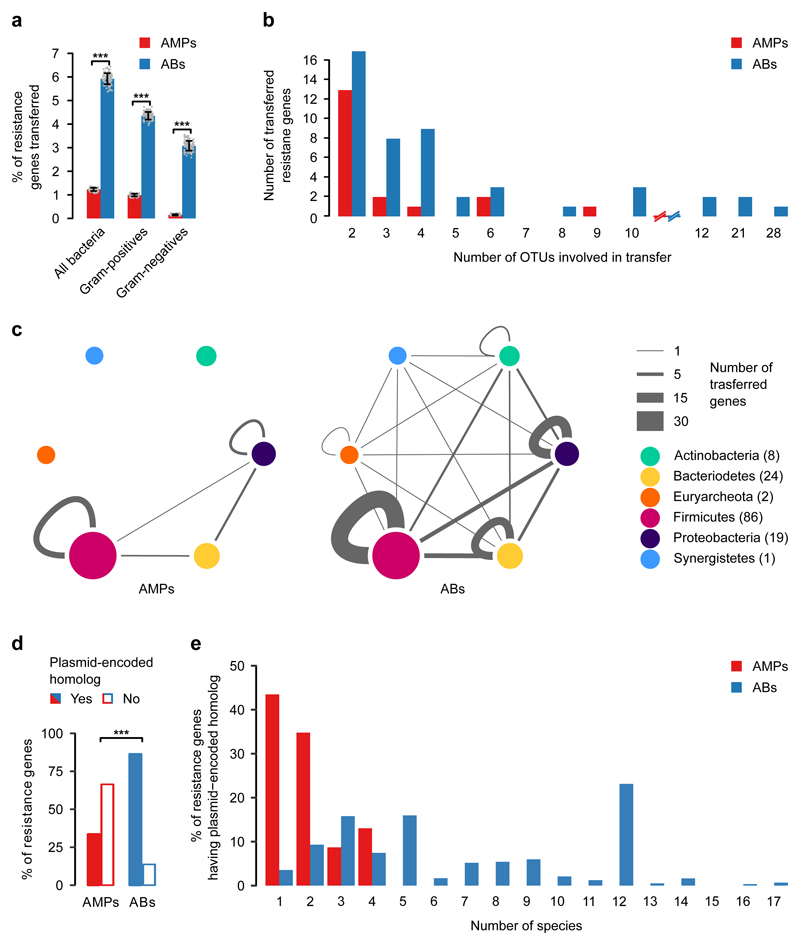
We found that the relative frequency of AMP resistance genes within the pool of mobile genes was 4.8-fold lower than that of antibiotic resistance genes, in spite of their similar frequencies in the genomes of the gut microbiota (Fig. 1a, Supplementary Fig. 1a, Supplementary Table 2). Notably, the relative underrepresentation of AMP resistance genes in the mobile gene pool cannot be simply attributed to the large physiological differences between Gram-negative and Gram-positive bacteria. When these two bacterial groups were considered individually, AMP resistance genes remained underrepresented in the mobile gene pool compared to antibiotic resistance genes (Fig. 1a). Moreover, the unique transferred AMP resistance genes were shared between fewer bacterial species, indicating fewer transfer events per gene (Fig. 1b). Notably, 65% of these transfer events mobilized AMP-transporting efflux pumps between bacteria within the Firmicutes phylum (Fig. 1c, Supplementary Fig. 1b, Supplementary Table 3).
Fig. 1. AMP resistance genes (AMPs) are less frequently transferred in the human gut microbiome than antibiotic resistance genes (ABs).
a) Percentage of AMP- (red bars) and antibiotic-resistance genes (blue bars) detected as horizontally transferred (i.e. present in the mobile gene pool). Resistance genes were identified using BLAST sequence similarity searches in the genome sequences of the gut microbiota (see Methods). *** indicates significant difference (p=2*10-16, 4*10-16, 2*10-13 from two-sided binomial test, in each case n=100 both for AMPs and ABs). Centre and error bars represent mean and SD calculated by randomly sampling 100 times from each of the 225 operational taxonomic units (OTUs), respectively (see Methods and Supplementary Table 2). b) Unique mobile AMP resistance genes were involved in half as many between-species transfer events as antibiotic resistance genes (p=0.01, two-sided negative binomial regression, n=19 and 50 for AMP- and antibiotic-resistance genes, respectively). The result remains when a non-parametric test is used, which is less sensitive to outliers (p=0.02, two-sided Mann-Whitney U-test). OTUs were generated as for Fig. 1a (See Supplementary Table 3). On the x-axes, the continuity of the scale breaks between 10 and 12. Above 12, only values with at least one transferred resistance gene are shown. c) Network representation of the mobile gene pool in the case of AMP- and antibiotic-resistance genes. Straight and curved lines represent genes that were shared between OTUs of different phyla and the same phylum, respectively. Line thickness represents the number of resistance genes shared between OTUs of six major phyla. Node size and numbers in parentheses indicate the number of OTUs in each phylum that shared at least one transferred AMP- or antibiotic-resistance gene. OTUs were generated as for Fig. 1a. d) Significantly less AMP resistance genes (46 out of 137) have a close homolog in naturally occurring plasmid sequences in the human microbiome as compared to antibiotic resistance genes (1867 out of 2163) (p=2.2*10-6, two-sided Fisher's exact test). n=137 and 2163 for AMPs and ABs, respectively (see Methods and Supplementary Table 4). e) Plasmid-encoded homologs of individual AMP resistance genes were found in significantly fewer species in the human microbiome as compared to antibiotic resistance genes (p=5.6*10-15, two-sided negative binomial regression, n=23 for AMPs and n=1772 for antibiotics, see Methods).
To further support the low mobility of AMP resistance genes, we next explored whether AMP resistance genes from our literature-curated list are associated with naturally occurring plasmid sequences or integrative conjugative elements (ICEs) in the human gut microbiome (see Methods). Strikingly, while 86% of the antibiotic resistance genes had close homologues on plasmids, only 33% of the AMP resistance genes did so (Fig. 1d, Supplementary Table 4). Moreover, the plasmids carrying individual antibiotic resistance genes were more wide-spread across bacterial species than those carrying AMP resistance genes (Fig. 1e). Notably, many of these plasmid-encoded AMP resistance genes were proteases and efflux pumps carried by virulence or multi-drug resistance plasmids in the human microbiome (Supplementary Table 4). Finally, as with plasmid sequences, we found that disproportionally fewer AMP resistance genes had homologs in ICEs than antibiotic resistance genes (Supplementary Fig. 2, Supplementary Table 4).
Overall, these results suggest that AMP resistance genes are less frequently transferred across bacterial species in the human microbiota.
Short genomic fragments from the gut microbiota rarely confer AMP resistance
One possible reason for the low mobilization of AMP resistance genes could be that AMP resistance is an intrinsic property of certain bacteria shaped by multi-gene networks10. Genes involved in AMP resistance may display strong epistatic interactions, and therefore they may have little or no impact on resistance individually. If it was so, horizontal gene transfer of single genes or transcriptional units encoded by short genomic fragments would not provide resistance in the recipient bacterial species. Indeed, AMPs interact with the cell membrane, a highly interconnected cellular structure, and membership in complex cellular subsystems has been shown to limit horizontal gene transfer16,17.
To investigate this scenario, we experimentally compared the ability of short genomic fragments to transfer resistance phenotypes towards AMPs versus antibiotics. To this end, we applied an established functional metagenomic protocol11,18 to identify random 1.5-5 kb long DNA fragments in the gut microbiome that confer resistance in an intrinsically susceptible E. coli strain. Importantly, the length distributions of the known AMP-reistance and antibiotic-resistance genes are well within this fragment size range (Supplementary Fig. 3), indicating that our protocol is suitable to capture single resistance genes for both AMPs and antibiotics. Metagenomic DNA from human gut fecal samples was isolated from two unrelated, healthy individuals who have not taken any antibiotics for at least one year. The resulting DNA samples were cut and fragments between 1.5-5 kb were shotgun cloned into a plasmid to express the genetic information in Escherichia coli K-12. About 2 million members from each library, corresponding to a total coverage of 8 Gb (the size of ~2000 bacterial genomes), were then selected on solid culture medium in the presence of one of 12 diverse AMPs and 11 antibiotics (Supplementary Table 5) at concentrations where the wild-type host strain is susceptible. Finally, using a third-generation long-read sequencing pipeline19, the number of unique DNA fragments conferring resistance (i.e. resistance contigs) was determined.
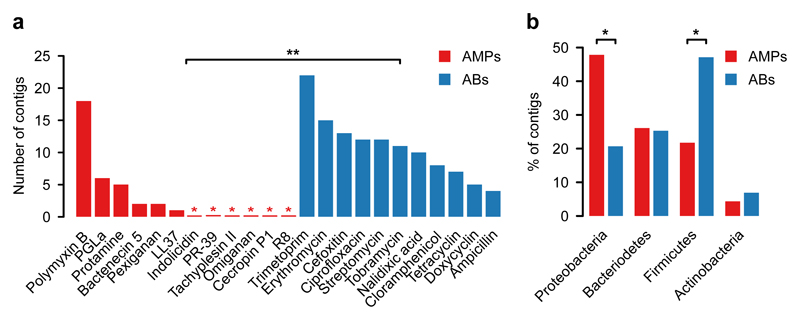
In agreement with prior studies11,20, multiple resistant clones emerged against all tested antibiotics (Fig. 2a, Supplementary Table 6). In sharp contrast, no resistance was conferred against half of the AMPs tested, and, in general, the number of unique AMP resistance contigs (N=34) was substantially lower than the number of unique antibiotic resistance contigs (N=119) (Fig. 2a, Supplementary Table 6). Polymyxin B – an antimicrobial peptide used as a last-resort drug in the treatment of multidrug-resistant Gram-negative bacterial infections21 – is a notable exception to this trend, with a relatively high number of unique resistance contigs (Fig. 2a). Indeed, a resistance gene (mcr-1) against Polymyxin B is rapidly spreading horizontally worldwide, representing an alarming global healthcare issue22. In contrast to Polymyxin B, we detected only one unique contig conferring resistance to LL37, a human AMP abundantly secreted in the intestinal epithelium23 (Supplementary Table 6). These specific AMP resistance genes are involved in cell surface modification, peptide proteolysis and regulation of outer membrane stress response (Table 1, Supplementary Table 6 and Supplementary Fig. 4).
Fig. 2. In E. coli, short genomic fragments from the human gut microbiota confer AMP resistance (AMPs) less frequently than antibiotic resistance (ABs).
a) Functional selection of metagenomic libraries with 12 AMPs (red bars) resulted in fewer distinct resistance-conferring DNA contigs than with 11 conventional small-molecule antibiotics (ABs, blue bars). p=0.002 from two-sided negative binomial regression, n=34 (AMP resistance contigs), n=119 (AB resistance contigs). Red asterisks indicate zero values and ** indicates a significant difference between AMPs and antibiotics. b) Phylum-level distribution (%) of the AMP- (red bars) and antibiotic-resistance contigs (blue bars). In the case of AMPs, significantly more resistance contigs are originating from the Proteobacteria phylum (p=0.015, two-tailed Fisher's exact test, n=110), while contigs originating from the Firmicutes phylum are underrepresented (p=0.033, two-tailed Fisher's exact test, n=110). *Significant difference between AMPs and antibiotics for a given phylum.
Table 1.
List of putative AMP resistance genes identified from our functional metagenomic screens. These resistance genes could be functionally annotated based on a literature-curated catalogue of resistance genes (Supplementary Table 1). NA stands for Not available.
| Resistance gene | Resistance gene function | Classification of resistance function | Estimated source organism | AMP |
|---|---|---|---|---|
| IpxFa | Lipid A 4'- phosphatase | Target alteration | Parabacteroides merdae | Polymyxin B, Pexiganan |
| IpxFb | Lipid A 4'- phosphatase | Parabacteroides sp. | Polymyxin B, Pexiganan | |
| IpxFc | Lipid A 4'- phosphatase | NA | Polymyxin B | |
| arnTEF | 4-amino-4-deoxy-L-arabinose modification of Lipid-A | Escherichia coli MS 107-1 | Polymyxin B | |
| pmrAB (qseBC) | Response regulator | Regulation | Sutterella wadsworthensis 2_1_59BFAA | Polymyxin B |
| pmrD | Signal transduction protein | Escherichia coli MS 107-1 | Polymyxin B | |
| ompT | Protease 7 precursor | Agent inactivation | Escherichia coli MS 196-1 | Bactenecin 5, LL37, Pexiganan |
characterized in this work (Fig. 4b, Fig. 4c and Fig.Supplementary Fig. 11); annotated as un-decaprenyl pyrophosphate phosphatase in Supplementary Table 6
annotated as bcrC in Supplementary Table 6
annotated as ybjG in Supplementary Table 10
If lack of functional compatibility with the host cell prevents AMP resistance genes from exerting their phenotypic effects, then DNA fragments identified in our screen should more often come from phylogenetically closely related bacteria. 53% of the contigs showed over 95% sequence identity to bacterial genome sequences from the HMP database24 (see Methods, Supplementary Fig. 5), allowing us to infer the source taxa with high accuracy (Supplementary Table 6). Indeed, while antibiotic resistance contigs were overrepresented from Firmicutes, the most abundant phylum in the gut, AMP resistance contigs originated excessively from Proteobacteria, which are phylogenetically close relatives of the host E. coli (Fig. 2b, Supplementary Fig. 6). Notably, this trend was not driven by Polymyxin B only but was valid for the rest of the AMPs as well (Supplementary Fig. 6).
Whereas these patterns are consistent with the hypothesis that the genetic determinants of AMP resistance are difficult to transfer via short genomic fragments owing to a lack of functional compatibility with the new host, another explanation is also plausible. In particular, AMP resistant bacteria might be relatively rare in the human gut microbiota, therefore, AMP resistance genes from these bacteria might simply remain undetected. However, as explained below, we can rule out this alternative hypothesis.
AMP-resistant gut bacteria are abundant and phylogenetically diverse
To assess the diversity and the taxonomic composition of gut bacteria displaying resistance to AMPs and antibiotics, we carried out anaerobic cultivations and selections of the gut microbiota using a state-of-the-art protocol25. To this end, fecal samples were collected from 7 healthy individuals (i.e. Fecal 7 mix, see Methods). As expected25, the cultivation protocol allowed representative sampling of the gut microbiota: we could cultivate 65-74% of the gut microbial community at the family level in the absence of any drug treatment (Supplementary Fig. 7, Supplementary Table 7). Next, the same fecal samples were cultivated in the presence of one of 5-5 representative AMPs and antibiotics(Supplementary Table 8). We applied drug dosages that retained 0.01 to 0.1% of the total cell populations from untreated cultivations (Supplementary Table 8) and assessed the taxonomic composition of these cultures by 16S rRNA sequencing (see Methods).
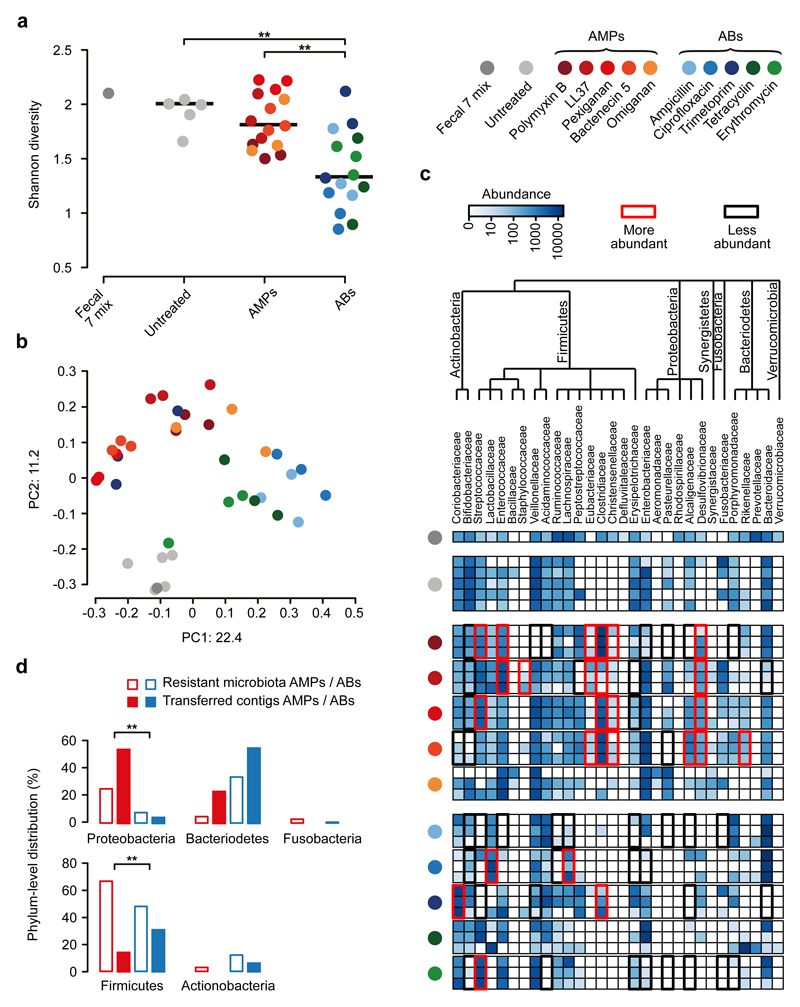
Remarkably, the diversities of the AMP-treated and the untreated bacterial cultures did not differ significantly from each other (Fig. 3a), despite marked differences in their taxonomic compositions (Fig. 3b). AMP-treated samples contained several bacterial families from the Firmicutes and Actinobacteria phyla, which are phylogenetically distant from E. coli (Fig. 3c). Notably, exposure to AMP stress provided a competitive growth advantage to bacterial families that remained undetected in the untreated samples (Fig. 3c). The examples include Desulfovibrionaceae, Clostridiaceae and Eubacteriaceae. Desulfovibrionaceae is a clinically relevant bacterial family that is linked to ulcerative colitis26 – an inflammatory condition with elevated AMP levels27, while Clostridiaceae and Eubacteriaceae have key roles in maintaining gut homeostasis28 (Fig. 3c). In sharp contrast, the diversity of antibiotic-treated cultures dropped significantly compared to both the untreated and the AMP-treated cultures (Fig. 3a and Supplementary Fig. 8). Several bacterial families had a significantly lower abundance in the antibiotic-treated cultures than in the untreated ones (Fig. 3c). These results indicate that the human gut is inhabited by a large number of bacterial families across all major phyla in the gut that exhibit intrinsic resistance to AMPs.
Fig. 3. Culturing reveals that short genomic DNA fragments from the gut microbiota have a limited potential to transfer AMP resistance to E. coli.
a) Diversities of the cultured microbiota and the original fecal sample (Fecal 7 mix). Data represents Shannon alpha diversity indexes at family level based on 16S rRNA profiling of V4 region. AMP/antibiotic (AB) treatments are colour-coded. Untreated samples were grown in the absence of any AMP or AB. ** indicates a significant difference from two-sided Mann-Whitney U test, p=0.005 for Untreated vs ABs, p=3*10-4 for AMPs vs ABs. Sample sizes were 5, 15 and 15 for Untreated, AMPs and ABs, respectively. Central horizontal bars represent median values. b) A PCoA (principal coordinate analysis) plot based on unweighted UniFrac distances34 separates the AMP- and antibiotic-resistant, and the untreated microbial communities (p=0.001, PERMANOVA test, n=36). c) Differential abundance analyses between the untreated and the AMP- and antibiotic-resistant microbiota at the family level (see Methods). Brackets depict a significant increase (red) or decrease (black) in abundance of a given family as a consequence of AMP or antibiotic (AB) treatment based on pairwise two-sided negative binomial tests (see Methods). Sample sizes were 5 and 3-3 for Untreated and AMPs and ABs, respectively. The phylogenetic tree is based on the assignment of the NCBI Taxonomy database and created by using ete3 toolkit37. d) Phylum-level distributions of resistant gut bacteria and resistance DNA contigs originating from them. Compared to their relative frequencies in the drug-treated cultured microbiota (based on colony numbers), the phylogenetically close Proteobacteria contributed disproportionally more AMP- (red bars) than antibiotic-resistance contigs (AB, blue bars), whereas the opposite pattern was seen for the distantly related Firmicutes. Asterisks indicate significant interaction terms in logistic regression models, p=0.018 (*) and p=0.003 (**) for Proteobacteria and Firmicutes, respectively (for more details see Methods and Supplementary Table 11). Sample sizes were 22651 and 24336 for the total colony number of AMP-resistant and AB-resistant microbiota, respectively,and 33 and 54 for the number of transferred AMP-resistant and AB-resistant contigs, respectively.
Human microbiota harbours a large reservoir of AMP resistance genes
Next, we assessed if the high taxonomic diversity in the AMP-resistant microbiota corresponds to a diverse reservoir of AMP resistance genes. To this end, we annotated previously identified AMP- and antibiotic-resistance genes in a set of gut bacterial genomes15 representing bacterial families that were detected in our culturing experiments upon AMP- and antibiotic-selection, respectively (Fig. 3c, for details, see Methods). Remarkably, 65% of our literature- curated AMP resistance gene families (Supplementary Table 1) were represented in at least one of these genomes (Supplementary Table 2), which is similar to that of antibiotic resistance gene families (58%). Finally, AMP resistance gene families, on average, were 39% more widespread in these species than the same figure for antibiotic resistance genes (Supplementary Fig. 9). Thus, the human gut harbours diverse AMP-resistant bacteria and a large reservoir of AMP resistance genes.
Phylogenetic constraints on the functional compatibility of AMP resistance genes
We next directly tested whether the shortage of AMP resistance DNA fragments from distantly related bacteria can be explained by the low potential of genomic fragments to transfer AMP resistance phenotypes to E. coli. To this end, we constructed metagenomic libraries from the AMP- and antibiotic-resistant microbiota cultures. From each AMP and antibiotic treatments, two biological replicates were generated (see Methods), resulting in 10-10 libraries, covering 25.6 Gb and 14 Gb DNA, respectively (Supplementary Table 9). These metagenomic libraries were next screened on the corresponding AMP- or antibiotic-containing solid medium. Finally, the phylogenetic sources of the resulting AMP- and antibiotic-resistance contigs were inferred with high confidence (Supplementary Fig. 10, Supplementary Table 10). Compared to their relative frequencies in the drug-treated cultured microbiota, the phylogenetically close Proteobacteria contributed disproportionally more AMP- than antibiotic-resistance DNA fragments, whereas the opposite pattern was seen for the distantly related Firmicutes (Fig. 3d, Supplementary Table 11). Taken together, phylogenetically diverse gut bacterial species show AMP resistance, but there is a shortage of transferable AMP resistance DNA fragments from phylogenetically distant relatives of E. coli.
Pervasive genetic background dependence of AMP resistance genes
Finally, we present evidence that DNA fragments that confer resistance to AMPs and were isolated from our screens show stronger genetic background dependence than those conferring resistance to antibiotics.
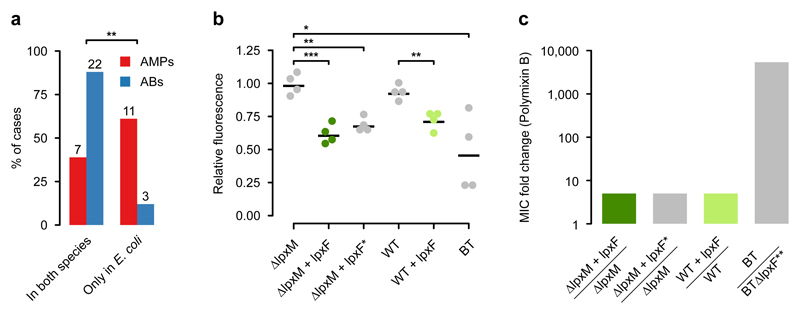
To systematically test the genetic background-dependency of AMP resistance genes, we examined how DNA fragments that provide AMP- or antibiotic-resistance in E. coli influence drug susceptibility in a related Enterobacter species, Salmonella enterica. We analyzed a representative set of 41 resistance-conferring DNA fragments derived from our screens (Supplementary Table 12) by measuring the levels of resistance provided by them in both E. coli and S. enterica. Strikingly, while 88% of the antibiotic resistance DNA fragments provided resistance in both host species, only 38.9% of AMP resistance DNA fragments did so (Fig. 4a, Supplementary Table 12). Thus, the phenotypic effect of AMP resistance genes frequently depends on the genetic background, even when closely related hosts are compared.
Fig. 4. AMP resistance DNA fragments provide host-dependent phenotypic effects.
a) A significantly lower proportion of AMP resistance DNA fragments (AMPs, red bars) conferred resistance in both E. coli and S. enterica compared to antibiotic resistance DNA fragments (ABs, blue bars), suggesting weaker between-species conservation of the AMP resistance phenotypes. Asterisks indicate significant difference, p=0.0011, two-sided Fisher's exact test, n=16 for AMPs, n=25 for ABs. b) The IpxF ortholog from Parabacteroides merdae, isolated in our screen (marked as lpxFa here and in Table 1; represented with green colour) and a previously characterized fully functional IpxF from Francisella novicida35 (marked as lpxF*) decrease the net negative surface charge of ΔlpxM E. coli to a similar extent, and close to the level of wild-type Bacteroides thetaiotaomicron (BT) expressing its native lpxF. LpxFa has a similar effect both in ΔlpxM and wild-type E. coli (dark and light green dots). The fluorescence signal is proportional to the binding of the FITC-labeled poly-L-lysine polycationic molecules. Less poly-L-lysine binding reflects a less negative net cell surface charge36. *, **, *** indicate significant differences (p=0.03, 0.001 and 0.0004, respectively, Welch two-sample t-test, n=4 biological replicates, central horizontal bars represent mean values). Corresponding microscopic pictures are shown in Supplementary Fig. 11. c) LpxFa increases Polymyxin B resistance of both ΔlpxM and wild-type E. coli only five-fold (dark and light green bars) (n=3), to the same extent as LpxF from Francisella novicida (marked as lpxF*) (n=3). In contrast, lpxF in its original host, B. thetaiotaomicron (marked as lpxF**) provides a 5000-fold increment in Polymyxin B resistance12.
As an example, we finally focused on a putative ortholog of a previously characterized AMP resistance gene, lpxF12. LpxF is a key determinant of AMP resistance in Bacteroidetes, a member of the human gut microbiota. By decreasing the net negative surface charge of the bacterial cell, it provides a 5000-fold increment in Polymyxin B resistance in these species12,29. To test the impact of one of the lpxF orthologs identified in our screen (denoted as lpxFa in Table 1) on AMP resistance in a new bacterial species, we expressed it in wild-type and a mutant E. coli strain (ΔlpxM) as well. Similarly to Bacteroidetes, the ΔlpxM strain uniquely synthesizes penta-acylated lipid A molecules, the natural substrate of lpxF, and is therefore especially suitable for testing the impact of the isolated lpxF ortholog30. Reassuringly, surface charge measurements confirmed that the lpxFa is fully functional in E. coli (Fig. 4b, Supplementary Fig. 11). However, it provided a mere five-fold increase in Polymyxin B resistance in E. coli, even in the ΔlpxM background (Fig. 4c). Notably, unlike the previously characterized lpxF from Francisella novicida30, the lpxF ortholog isolated from our screen was active both on the penta- and hexa-acylated lipid A molecules (Fig. 4b and 4c). Therefore, substrate specificity alone is unlikely to limit transfer of lpxFa into E. coli or other Enterobacteriaceae species. The compromised resistance phenotype conferred by lpxFa in the new host shows that the function of other genes is also required to achieve the high AMP resistance level seen in the donor bacterium.
Discussion
This work systematically investigated the mobility of AMP- versus antibiotic-resistance genes in the gut microbiome. We report that AMP resistance genes are less frequently transferred between members of the gut microbiota than antibiotic resistance genes (Fig. 1). In principle, this pattern could be explained by at least two independent factors: shortage of relevant selection regimes during the recent evolutionary history of the gut microbiota and lack of functional compatibility of AMP resistance genes upon transfer to a new host. We focused on testing the second possibility due to its experimental tractability and relevance to forecast the mobility of resistance genes upon AMP treatment. In a series of experiments, we showed that phylogenetically diverse gut bacteria display high levels of AMP resistance (Fig. 3), yet the underlying resistance genes individually often fail to confer resistance upon transfer to E. coli (Fig. 2 and Fig. 3). Furthermore, we demonstrated that the AMP resistance conferred by 1.5-5 kb long genomic fragments often depends on the genetic background of the recipient bacterium (Fig. 4). Together, these results support the notion that horizontal acquisition of AMP resistance is constrained by phylogenetic barriers owing to functional incompatibility with the new host cell31.
An important issue is whether the simultaneous transfer of multiple AMP resistance genes carried by longer DNA segments is feasible and can provide resistance to recipient bacteria. While this is certainly a realistic possibility, bioinformatic analyses support the conclusions derived from the metagenomic screens: AMP resistance genes are relatively rare in the mobile gene pool and on plasmids in nature (Fig. 1).
We speculate that the large differences in functional compatibility between antibiotic- and AMP-resistance genes might be caused by the latter being more often parts of highly interconnected cellular subsystems, such as cell envelope biosynthesis pathways. We note that the compromised benefit may not be the only manifestation of functional incompatibility and the exclusive reason for the limited presence of AMP resistance genes in the mobile gene pool and on plasmids (Fig. 1). It is also plausible that some AMP resistance genes have severe deleterious side effects in the new host in addition to conferring a compromised resistance. For example, the introduction of lpxF into bacterial pathogens reduces virulence in mice, probably because it perturbs the stability of the bacterial outer membrane in enterobacterial species32. Future works should elucidate whether AMP resistance genes are especially prone to induce deleterious side effects compared to antibiotic resistance ones. Clearly, deciphering the biochemical underpinnings of functional incompatibility of AMP resistance genes remains an area for future research.
Our results also provide mechanistic insights into the functional capacity of AMPs to control the composition and stability of the gut microbiome over evolutionary timescales. Specifically, as phylogenetic barriers limit the horizontal transfer of AMP resistance mechanisms, the exact dosages and combinations of AMPs could prove to be critical for the long-term advantage of gut bacterial species involved in human health. Indeed, our work indicates that specific AMP stresses can lead to an increase in the amount of bacteria linked to ulcerative colitis (Desulfovibrionaceae).
Another important and unresolved issue is why natural AMPs that are part of the human innate immune system have remained effective for millions of years without detectable resistance in several bacterial species. One possibility, supported by our work, is that the acquisition of resistance through horizontal gene transfer from human gut bacteria is limited, most likely due to compromised functional compatibility in the recipient bacteria. We do not wish to claim, however, that AMPs in clinical use would generally be resistance-free. In agreement with the prevalence of Polymyxin B resistance DNA fragments (Fig. 2a), a plasmid conferring colistin resistance is spreading globally33. Rather, our work highlights major differences in the frequencies and the mechanisms of resistance across AMPs, with the ultimate aim to identify antimicrobial agents less prone to resistance.
Materials and Methods
Establishing a comprehensive AMP resistance gene dataset
Even though several databases have been created for antibiotic resistance genes, a comprehensive list of AMP resistance genes has not been compiled so far. Therefore, we carried out a systematic literature mining in PubMed NCBI and Google Scholar with the keywords “antimicrobial peptide” + “resistance”. From the identified publications genes with experimentally confirmed influence on AMP susceptibility were included in our manually curated AMP resistance gene dataset (Supplementary Table 1). Altogether 138 AMP resistance genes were identified. As a next step, the compiled AMP resistance genes were classified into resistance gene families (orthologous gene groups or orthogroups) by applying the EggNOG-mapper software (version 0.12.7) on the bacterial EggNOG 4.5.1 database37. Then, AMP resistance genes were classified into broad functional categories analogous to the classification of antibiotic resistance genes in The Comprehensive Antibiotic Resistance Database (CARD)38. To obtain a comparable dataset for known antibiotic resistance genes we downloaded the CARD database38. Genes associated with AMP resistance in CARD were filtered out and the remaining antibiotic resistance genes from CARD were grouped into resistance gene families in the same way as AMP resistance genes using the EggNOG database (Supplementary Table 1).
Analysis of the mobile gene pool of the gut microbiota
A previously published mobile gene catalogue of the human gut microbiota15 was analyzed to compare the patterns of horizontal gene transfer events involving AMP- and antibiotic-resistance genes across a wide range of bacteria. This mobile gene catalogue relies on the identification of nearly identical genes in distantly related bacterial genomes and thereby provides a snapshot on the gene set subjected to recent horizontal gene transfer events in a representative sample of the gut microbiota. The goal in our analysis was to determine the presence/absence pattern of the AMP- and antibiotic-resistance genes not only in the mobile gene pool but also in the 567 genomes from which the mobile gene pool was derived. In this way, not only the horizontally transferred resistance genes were identified but also those that have not been detected in such transfer events, but were present in the gut microbiome. To this end, the genomes and proteomes used by Brito et al.15 were downloaded from the Human Microbiome Project (HMP) database (https://www.hmpdacc.org/HMRGD/) and from Fijicomp website (http://www.fijicomp.org). DNA sequences derived from the latter database were used for ORF prediction with Prodigal software (version 2.6.3,39). Then, a sequence similarity search was applied to the compiled list of proteins encoded in the analyzed genomes to identify those that were present in the mobile gene pool as well. The sequence similarity search between the mobile gene pool and the proteins from the genomes was carried out with the blastx option of the Diamond software (version 0.9.10,40) with 50% sequence coverage and 100% sequence identity (parameters were chosen to reproduce the original publication of the mobile gene pool15). Out of the 37,870 unique mobile genes in the mobile gene pool, we identified 37,184 in the genomes (98.28%).
Next, both the antibiotic- and AMP-resistance genes were identified among the mobile genes and among those that have not been detected in the mobile gene pool but were present in the genomes. For this functional annotation, a BLAST search was carried out against the antibiotic resistance genes from the CARD database with the blastp option of the Diamond software with strict parameters (e-value < 10-5, > 40% identity at the protein level and 80% query sequence coverage) (Supplementary Table 2). In a similar vein, AMP resistance genes were identified by performing blastp sequence similarity search against the manually curated list of AMP resistance genes (Supplementary Table 1). Antibiotic- and AMP-resistance genes in our databases were classified into resistance gene families by the EggNOG-mapper software on the bacterial EggNOG database (Supplementary Table 2). For the annotated resistance gene list in the mobile gene pool, see Supplementary Table 3.
To compare the relative frequency of the AMP- and antibiotic-resistance genes in the mobile gene pool (Fig. 1a), we restricted the analysis to one genome per species. This was necessary to avoid sampling bias since different species were represented by unequal numbers of genomes in the dataset. To this aim, 16S rRNA sequences were determined for each genome (for HMP genomes they were downloaded from the Silva database41, while for Fijicomp genomes they were identified directly in the genomes using the RNAmmer software (version 1.2, 42). Then, genomes with lower than 2% 16S rRNA gene dissimilarities were collapsed into genome groups ('species' or OTUs) using average linkage clustering as it is described in the publication of the mobile gene pool15. Each such genome group was represented by one randomly chosen genome for the statistical analysis presented in Fig. 1a (note that the random sampling was repeated 100 times, yielding an estimate of standard error). Resistance genes in the mobile gene pool that resulted in a BLAST hit both from the AMP- and the antibiotic-resistance databases were excluded from the analysis. The remaining resistance genes annotated in the mobile gene pool were counted and plotted as the percentage of the total number of annotated resistance genes in the genome sequences (“All bacteria” from Fig. 1a). Additionally, the analysis was separately performed for the Gram-negative and Gram-positive bacterial genomes as well (“Gram-positives” and “Gram-negatives” in Fig. 1a). For the network representation of the mobile gene pool, we used Cytoscape43.
For each unique mobile AMP- or antibiotic-resistance gene we also estimated the minimum number of independent transfer events (Fig. 1b) by counting the number of genome groups (i.e. OTUs) in which the gene is present in the mobile gene pool15 (Supplementary Table 3).
Identification of homologs of the literature-curated AMP- and antibiotic-resistance genes in naturally occurring plasmid sequences and in integrative conjugative elements (ICEs)
For the identification of plasmid-encoded AMP resistance genes, we used two independent databases: a plasmid-encoded protein database from NCBI Reference Sequence Database (Refseq)44 (ftp://ftp.ncbi.nih.gov/refseq/release/plasmid/) and a curated plasmid database containing 2097 entire plasmid sequences from the Enterobacteriaceae bacteria45 (https://figshare.com/s/18de8bdcbba47dbaba41). From the Refseq database, protein sequences were downloaded and both the antibiotic- and AMP-resistance proteins were identified with the blastp algorithm using our literature-curated lists of resistance genes. Hits were accepted only if they showed >40% sequence similarity over 80% of the length of the subject protein, with an e-value less than 10-5. Since this Refseq dataset contains plasmid-encoded proteins from various sources in addition to the human microbiota, we filtered our hits using a previously compiled list of species from the human microbiota46. Thus, only those resistance gene hits were retained that are present on plasmids from human-associated bacteria. From the Enterobacteriaceae-specific plasmid database, we downloaded the translated nucleotide sequences for all 6 reading frames and carried out the similarity search as above using blastp with 80% query coverage, and >40% sequence identity. For this dataset, we manually checked the presence of the plasmid-encoded resistance genes in the human microbiome using the NCBI database (Supplementary Table 4). Finally, we took the union of these two datasets to calculate the percentage of resistance genes residing on plasmids (Fig. 1d).
To estimate the species level distribution of the plasmid-encoded resistance proteins we used the Refseq dataset only as it gives information on the identity of species from which the plasmids were isolated. Specifically, for each AMP- and antibiotic-resistance gene that resulted in plasmid-encoded homologs, we counted the number of species that bore at least one plasmid-encoded homolog of the given AMP- and antibiotic-resistance gene (Fig. 1e).
For the identification of AMP resistance genes that are associated with ICEs, we used a database that contains 16,820 cargo genes associated with ICE sequences from human gut bacterial genome sequences (Intestinal Microbiome Mobile Element Database, (https://immedb.org/))47. Nucleotide sequences were downloaded and following translation of the sequences with tblastn algorithm both the antibiotic- and AMP-resistance proteins were identified using our literature-curated lists of resistance genes with the same BLAST parameters as before (>40% sequence similarity over at least 80% of the length of the subject protein). Since the literature-curated lists of resistance genes contain proteins from various sources in addition to the human gut microbiota, only those homologs were considered in the set of non-ICE-associated resistance genes that were detected in the genome sequences of the HMP database (Supplementary Table 4).
Construction of gut metagenomic libraries
To sample the gut resistome, we applied a previously established small-insert shotgun metagenomic protocol11 with small modifications. This method identifies small genomic fragments that decrease drug susceptibility when random genomic fragments are expressed from a multicopy plasmid with an inducible promoter. For the construction of the metagenomic libraries, human stool samples were obtained from two healthy unrelated individuals, who have not taken any antibiotics for at least one year prior to sample donation. Throughout the whole study, we have complied with all relevant ethical regulations. The protocol related to human fecal sample collection was approved by the Ethical Review Board of the Albert Szent-Györgyi Health Centre, University of Szeged (approval ID: 42/2017-SZTE). Written informed consent from each participant was obtained before fecal sample collection. Protocols related to human fecal sample handling were approved by the Ministry of Agriculture (Hungary) (approval ID: TMF/146-9/2017). Gut community DNA was isolated immediately after sample donation, using ZR Fecal DNA MiniPrep™ kit (Zymo Research) according to the manufacturer’s instructions (http://www.zymoresearch.com/downloads/dl/file/id/91/d6010i.pdf). Subsequently, 10 μg of metagenomic DNA from each sample was partially digested with 0.25 U MluCI restriction enzyme (New England BioLabs) in 10x CutSmart Buffer (New England BioLabs) at 37°C for 20 minutes, followed by heat inactivation at 85°C for 20 min. MluCI is a four-base cutter restriction enzyme that produces overhangs complementary to the ones that EcoRI produces. By varying incubation time or the enzyme concentration, the size range of the resulting DNA fragments can be set. The fragmented DNA was size selected by electrophoresis on a 1 % (m/V) agarose gel in 1X Tris-Acetate-EDTA (TAE) buffer. A gel slice corresponding to 1500-5000 bp was excised from the gel and DNA was isolated using a GeneJET Gel Extraction and DNA Cleanup Micro Kit (Thermo Scientific). 5 μg of pZErO-2 plasmid DNA (ThermoFisher) was digested with 25 U EcoRI restriction enzyme (Fermentas) in 10x EcoRI Buffer (Fermentas) for 2 hrs, followed by 20 minutes heat inactivation at 65°C. After purification with DNA Clean & Concentrator™-5 kit (Zymo Research), digested pZErO-2 plasmid was dephosphorylated with FastAP alkaline phosphatase (Thermo Scientific) as follows: 4 μg plasmid DNA was incubated with 4U enzyme in 10x FastDigest buffer at 37°C for 1 hr, followed by 5 minutes heat inactivation at 74°C and purification with DNA Clean & Concentrator™-5 kit (Zymo Research). DNA was ligated into pZErO-2 at the EcoRI site using the Rapid DNA ligation kit (Thermo Scientific). The ligation reaction was performed in 15 μl total volume using a 5:1 insertvector ratio: 4.5 μl (310 ng) digested and gel purified DNA insert, 0.65 μl (62 ng) EcoRI-cut pZErO-2 vector, 3 μL 5X ligation buffer, 0.75 μL 10 mM ATP, 4.1 μL dH2O, 2 μL T4 DNA Ligase (5 U/μl). The ligation mixture was incubated at 16°C overnight, followed by heat inactivation at 65°C for 10 minutes.
Prior transformation, the ligation mixture was purified with DNA Clean & Concentrator™-5 kit (Zymo Research). 3.5 μL of the resulting ligation mixture was transformed by electroporation into 50 μL of electrocompetent E. coli DH10B™ cells (Invitrogen). Electroporation was carried out with a standard protocol for a 1 mm electroporation cuvette. Cells were recovered in 1 mL SOC medium, followed by 1-hour incubation at 37°C. 500 μL of the recovered cells was plated onto square Petri dishes containing Luria Bertani (LB) agar supplemented with 50 μg/mL kanamycin. In order to assess the library size (number of colony forming units (CFU)), 1 μL of the electroporated cells was saved for plating onto a separate Petri dish containing Luria Bertani (LB) agar supplemented with 50 μg/mL kanamycin. From each plate, 10 clones were randomly picked for colony PCR in order to confirm the presence and the size distribution of the inserts. PCR was performed using the Sp6-T7 primer-pair (Supplementary Table 13) flanking the EcoRI site of the multiple cloning site of the pZErO-2 vector. The sizes of the PCR products were determined by gel electrophoresis and the average insert size was calculated as 2-3 kb. The size of each library was determined by multiplying the average insert size by the number of total colony forming units (CFU). The size distributions of the libraries varied between 4.4 – 16 Gb coverage with this protocol, which is in line with a previously published state-of-the-art protocol11,18. The resulting colonies from the Petri dishes were collected and the plasmid library was isolated using InnuPREP Plasmid Mini Kit (Analytic Jena). 30-60 ng of isolated plasmid library was transformed by electroporation into 40 μL electrocompetent E. coli BW25113 (prepared as described in 47). This E. coli strain was used for the functional selections (see below). After electroporation, cells were recovered in 1 mL of LB medium for 1 hour at 37°C. Special care was taken to achieve high electroporation efficiency in order to cover 10-100 times the original library size. In this way, we ensured that most library members are electroporated from the plasmid library. The 1 mL recovered cell culture was added to 9 ml of LB medium supplemented with 50 μg/mL kanamycin, and grown at 37 °C for 2-3 hours until it reached the 7.5-10 x 108 cell density (OD600: 1.5-2). Cell aliquots were frozen in 20% glycerol and kept at -80°C for subsequent functional selection experiments.
Metagenomic libraries were generated from the uncultured microbiota (i.e. the total DNA extracted from the stool samples) and from the cultured microbiota (i.e. the genomic DNA extracted from the cultured pooled microbiota), too. For details see the section “Cultivation of the gut microbiota under anaerobic conditions and DNA extraction”.
Functional metagenomic selections for AMP- and antibiotic-resistance genes
Functional selections for resistance were carried out on solid plates containing one of the 12 antimicrobial peptides (AMPs) or 11 antibiotics (Supplementary Table 5). Instead of the plating assay that is commonly used in the field11, we applied a modified gradient plate assay49, where bacteria are exposed to a concentration gradient of the antimicrobial instead of a single concentration. We found that this strategy improves reproducibility of AMP selections, where changes in the resistance levels are relatively small compared to that in the case of antibiotics. The growth medium in these plates was a modified minimal salt medium (MS) with reduced salt concentration (1 g (NH4)2SO4, 3 g KH2PO4, 7 g K2HPO4, 100 μl MgSO4 (1 M), 540 μl FeCl3 (1 mg/ml), 20 μl thiamine (50 mg/ml), 20 ml casamino acids (BD) (10 % (m/V)), 5 ml glucose (40 % (m/V)) in a final volume of 1 L), since most AMPs are not effective in vitro in the presence of high salt concentrations. In the case of the AMP containing plates, the solidifying agent was changed to 1.5 % (m/V) of low melting point agarose (UltraPure™ LMP Agarose (Invitrogen)) from 1.5 % (m/V) agar, in order to prevent any heat-induced structural damage of the peptides during plate pouring. Antibiotics and AMPs were purchased from Sigma and ProteoGenix, respectively. Onto each of the gradient plates (Tray plates, SPL Life Sciences) 2x108 cells were plated out from the thawed stocks of E. coli BW25113 bearing the meta-genomic plasmid libraries. In this way, each metagenomic library member was represented about 10-100 times on each plate. We found this necessary for a good reproducibility for our experiments. Subsequently, plates were incubated at 30°C for 24 hours. For each functional selection, a control plate was prepared where the same number of E. coli BW25113 was plated out. These cells contained the pZErO-2 plasmid with a random metagenomic DNA insert that has no effect on AMP- and antibiotic-resistance. This control plate showed the minimum inhibitory concentration (MIC) of the antimicrobial without the effect of a resistance plasmid. The empty plasmid was not applicable as a control because in the absence of a DNA insert the CcdB toxic protein is expressed from the plasmid. In order to isolate the resistant clones from the library plates, sporadic colonies were identified above the MIC level (defined using the control plate) by visual inspection. These clones were collected by scraping them into 2 ml of LB broth and stored subsequently at -80°C.
Validation of the resistance-conferring metagenomic DNA fragments
Following selection of the metagenomic libraries, the putative resistance phenotypes conferred by the plasmid selections were confirmed for a representative fraction of the colonies. From each selection at least 20 colonies were picked and the MIC increase was determined by a standard broth microdilution method50, as it is described in the section “Quantification of the resistance gains that metagenomic contigs provide”. For these measurements, the same control plasmid was used as in the functional selections. MIC values were determined after 16-24 hours of incubation at 30°C with a continuous shaking at 240 rpm. Plasmids from validated resistant clones were retransformed into the BW25113 E. coli strain, followed by a second MIC determination in order to exclude the possibility that the increase in the MIC was induced by genomic mutations. Plasmids not showing MIC increase in the validation protocol were excluded from further analysis. Mostly the clones situated closer than 1 cm to the MIC level on the gradient plates did not confer resistance during validation. To avoid these false positive resistance plasmids, colonies at the borderline were not collected for further analysis without confirming their phenotype. The rest of the colonies were collected by scraping them into 2 ml of LB broth. Bacterial samples were stored at -80 °C in 20 % (m/V) glycerol. When only a few clones were on the plates, all were tested for resistance to make sure that we do not lose potential hits. We encountered such situations only in the case of AMPs. If the number of resistant clones on a plate was less than or equal to 20, plasmids were isolated individually from the MIC validated clones and sent for Sanger bidirectional sequencing with the Sp6-T7 primer pair (Supplementary Table 13). When the resistance-conferring insert was longer than what the initial sequencing covered, we applied a primer walking strategy to sequence the middle part of the insert.
Amplification of the resistance-conferring metagenomic DNA fragments
Plasmid pools from the scraped resistant clones were PCR amplified for subsequent PacBio sequencing51. To this aim, first, the plasmid pools were isolated from each metagenomic selection using InnuPREP Plasmid Mini Kit (Analytic Jena). Then, these plasmid pools served as templates for subsequent PCR amplification reactions to amplify the inserts from the pooled plasmids. These PCR reactions were performed with barcoded Sp6 and T7 specific primers as forward and reverse primers, respectively. The 16-base long PacBio barcode dual-end labelling scheme was used to label each plasmid pool for sample identification in the subsequent PacBio sequencing. Primer sequences are shown in Supplementary Table 13. PCR reactions consisted of 30 ng of template DNA, Q5 Hot Start High-Fidelity 2x Master Mix (New England BioLabs), 0.2 μM barcoded primers in a final reaction volume of 25 μl. Following an optimization process, the number of PCR cycles was reduced to 15 in order to minimize amplification bias. The following thermocycler conditions were used: 98°C for 5 minutes, 15 cycles of 98°C for 15 seconds + 69°C for 30 seconds + 72°C for 2 minutes, and 72°C for 7 minutes. The amplified metagenomic inserts were then cleaned using the DNA Clean & Concentrator™-5 kit (Zymo Research) and their concentration was measured with Qubit fluorometer (Invitrogen). In order to get rid of the short amplicons (e.g. primer dimers), which may interfere with the sequencing process, barcoded amplicons were mixed at an equimolar ratio and the sample was gel extracted following electrophoreses using 1% agarose gel. The sample was purified (Zymoclean™ Gel DNA Recovery Kit) prior to sequencing.
PacBio sequencing and data analysis
Sequences of the pooled PCR products were obtained from the Norwegian Sequencing Centre at the University of Oslo, Norway. The library was prepared using Pacific Biosciences Amplicon library preparation protocol. Samples were sequenced with Pacific Biosciences RS II instrument using P6-C4 chemistry and MagBead loading in one SMRT cell. 61,641 reads were obtained with a mean length of 20,961 bp. Reads were filtered without demultiplexing using RS_subreads.1 pipeline on SMRT Portal (version 2.3.0) using default settings (number of passes 1, minimum accuracy 0.9). Following barcode detection and demultiplexing, reads were collapsed to consensus sequences using the long amplicon analysis pipeline of the SMRT Portal with default settings. We validated our sequencing effort on a mock sample containing 9 previously sequenced DNA contigs that originate from our metagenomic selections. Reassuringly, out of the 9 sequences 8 were present in the PacBio sequencing result with at least 99% sequence identity. The single non-detected contig was the longest one (4500 bp) which may indicate a bias of the pipeline toward shorter insert sizes.
Functional annotation and resistance gene identification on the metagenomic contigs
To functionally analyze the ORFs on the assembled contigs from the metagenomic selections, ORFs were predicted and annotated using the Prokka suite (version 1.11, 52) with the bacterial prediction settings and an e-value threshold of 10-5. Within Prokka, Prodigal 9subscript was modified to run without -c parameter to identify highly probable ORFs, even if the ends were not closed. This was necessary because some contigs may have been shortened by a few residues during the cloning process or in the assembly due to low coverage, without losing their functionality. Other parameters were kept as default. Next, ORFs on the metagenomic contigs were functionally annotated with our resistance gene lists introduced in the section “Analysis of the mobile gene pool of the gut microbiota”. Specifically, an ORF was classified as an antibiotic resistance gene when sequence similarity search using blastp53 against the CARD8 database38, resulted in an annotation with an e-value<10-5, identity >30% and coverage >80%. Here, the minimum sequence identity was lower than in the case of the analysis of the mobile gene pool, since the experimentally observed resistance phenotype provided an extra confidence for the annotation. Similarly, AMP resistance genes were identified by performing blastp sequence similarity search against the manually curated list of AMP resistance genes (Supplementary Table 1). We note that 55.4% of the antibiotic resistance contigs carried a homolog of a known antibiotic resistance gene and 23.5% of the 34 unique AMP resistant DNA contigs encoded a gene that has been associated with AMP resistance in the literature. This difference suggests that more AMP resistance genes await discovery. To estimate the identity of the donor organisms from which the assembled DNA contig sequences originated from, a nucleotide sequence similarity search was carried out for the entire DNA contigs as query sequences against the genome sequences from the HMP database24 using blastn, with an e-value threshold of <10-10. Taxonomy was assigned with the ete3 toolkit37.
Cultivation of the gut microbiota under anaerobic conditions and DNA extraction
In order to compare the abundance and phylogenetic distribution of the AMP- and antibiotic-resistant gut bacteria, we applied a recently established anaerobic cultivation protocol of the human gut microbiota with small modifications25. For this purpose, human fecal samples were obtained from seven healthy unrelated volunteers, who had not taken any antibiotics for at least one year prior to sample donation. Ethical rules were observed throughout the whole study. Following defecation, stool samples were immediately placed into uncapped 50 ml plastic tubes (Sarstedt), deposited in anaerobic bags (Oxoid, Thermo Scientific) and samples were transferred into the anaerobic chamber within 1 hour after sample collection. All anaerobic experiments were performed in a Bactron IV anaerobic chamber (Shel Lab) filled with an atmosphere of 95% nitrogen and 5% hydrogen, with palladium catalysts. Two grams of the fecal samples were suspended in 20 ml of modified Gifu Anaerobic Medium (GAM) Broth (HyServe). After 10 minutes of incubation, letting the solid particles settle down, the supernatants were supplemented with 20% glycerol, aliquoted and stored at -80°C. Prior to the cultivation of the microbiota, thawed aliquots from the samples of the 7 individuals were combined in an equal ratio (we refer to this sample mix as “Fecal 7 mix” sample) in the anaerobic chamber. Then this Fecal 7 mix sample was plated out anaerobically in the presence and absence of one of the five AMPs or one of the five antibiotics that were active in the culturing medium (Supplementary Table 5). The culturing medium was modified Gifu Anaerobic Medium (GAM), considering that the best reconstruction of the composition and architecture of the human gut bacterial community could be obtained using this medium25. In the case of AMP containing plates, the solidifying agent was low melting agarose instead of agar for the same reason as before (see section “Functional metagenomic selections for AMP- and antibiotic-resistance genes”). Each AMP and antibiotic was applied in three different concentrations on separate plates in three replicates.
To mimic the thermal conditions encountered by the gut bacteria in the human intestinal tract, plates were incubated at 37°C in the anaerobic chamber for 4 days. After a 4-day time interval, we observed a plateau in the number of appearing colonies. Following colony counts the plates had to fulfil two requirements to be selected for subsequent analysis. First, colony numbers needed to be in the range of 0.01-0.1% as compared to the colony numbers in the absence of any AMP and antibiotic treatment (we refer to these samples as Untreated). Second, colony numbers needed to be high, but still in the countable range (1000-5000 colonies). To be in this range in the case of the untreated samples as well the Fecal 7 mix sample was plated out in a concentration that is 1000-fold more dilute as what was applied in the case of AMP and antibiotic selections. For Trimetoprim the selection pressure could not be increased that much to select for the most resistant 0.01-0.1% of the population since 0.49% of the colonies were able to grow even at the solubility limit of this antibiotic. From the selected plates resistant colonies were scraped and the pooled colonies from each plate were used to isolate genomic DNA with ZR Fecal DNA MiniPrep™ kit (Zymo Research) according to the manufacturer's instructions. The isolated DNA samples were subsequently used for both small-insert shotgun library constructions (referred to them as libraries from cultured microbiota, Supplementary Table 10) and for 16S rRNA based community profiling. To estimate the relative resistance level of the gut microbiota compared to E. coli BW25113, we used the colony counts from the anaerobic cultivation experiments at each AMP and antibiotic concentration, i.e. the resistance level of the gut microbiota against an AMP and antibiotic is defined as the AMP and antibiotic concentration at which only 0.1-0.01% of the untreated microbiota population could grow (see details above), analogously to a MIC value determination for a single species. The susceptibility of E. coli was estimated in the same way as for the gut microbiota by plating out anaerobically the same number of cells as in the case of the Fecal 7 mix sample for each AMP and antibiotic treatment. Then we determined the AMP and antibiotic concentrations at which only 0.1-0.01% of the E. coli cells could grow. The relative resistance level of the gut microbiota was defined as the resistance level of the gut microbiota divided by the resistance level of E. coli (Supplementary Table 8).
16S rRNA-based bacterial community profiling
To determine the taxonomic composition of the AMP- and antibiotic-resistant gut bacterial communities, we sequenced and analyzed the V4 region of the 16S rRNA genes from the Fecal 7 mix samples cultivated in the presence of individual AMPs and antibiotics. We also determined the phylogenetic composition of the uncultivated Fecal 7 mix sample. The V4 region of the 16S rRNA gene was PCR amplified with dual-indexed Illumina primer-pairs, using different combinations of barcoded forward and reverse primers (v4.SA501-505 and v4.SA701-706, respectively, Supplementary Table 13) as previously described54. The primers consist of the appropriate Illumina adapters, an 8-nt index sequence, a 10-nt pad sequence, a 2-nt linker and specific sites for the V4 region. The PCR reactions consisted of 1.5 μl (30 ng) of template DNA, 10 μl of Phusion HF buffer (Thermo Fisher Scientific), 4 μl of 2.5 mM deoxynucleotide triphosphates mix (dNTPs), 0.5 μl of Phusion DNA polymerase (2 U/μl) (Thermo Fisher Scientific), 1-1 μl of primers, 10 μM each, 3 μl DMSO (100%) and 29 μl of nuclease-free H2O in a final reaction volume of 50 μl. The following thermocycler conditions were used: 95 °C for 2 minutes, 25 cycles of 95 °C for 20 seconds + 56 °C for 15 seconds + 72 °C for 5 minutes, and 72 °C for 10 minutes. Following gel electrophoreses of the PCR products, the 400 bp long amplicons were extracted from the gel (Thermo Scientific GeneJET Gel Extraction Kit) and, following a second purification step (Zymo Research DNA Clean & Concentrator™-5 Kit), were sequenced using MiSeq Illumina platform. To prepare the samples for sequencing, the amplicons were quantified using a fluorometric method (Qubit dsDNA BR Assay Kit, Thermo Fischer Scientific) and libraries were mixed with Illumina PhiX in a ratio of 0.95 to 0.05. Sequencing on the Illumina MiSeq instrument was carried out with a v2 500 cycle sequencing kit (Illumina). 100 μM stock custom sequencing primers53 were mixed with standard read1, index read and read2 sequencing primers included in the MiSeq cartridge.
After sequencing, 16S rRNA reads were demultiplexed and processed with the Mothur software (version 1.36.1, 55). The average counts per sample were 21,979. To filter out the low-read-counts we followed the protocol of Rettedal et al. 25 The number of sequences per sample was equalized to 20,000 read counts using random re-sampling with a custom R script. Reassuringly, 20,000 read counts are well above the threshold where phylogenetic diversities show saturation in the samples (see the rarefaction curves of samples in Supplementary Fig. 12, that were calculated using the vegan (version 2.4-3, 56) R package). Sequences were merged at the level of 97% sequence identity and taxonomically assigned using the Silva ribosomal RNA database41. OTUs were classified at the family level since the V4 region allows accurate identification only down to this level57 (Supplementary Table 7). After removal of reads that could not be classified, 362 OTUs remained. To evaluate the reproducibility of the cultivation and sequencing, we generated seven replicates from the untreated samples. Samples referred to as “Untreated 1-5” originate from independent cultivation experiments started from different aliquots of the same five frozen samples (Supplementary Fig. 7, Supplementary Fig. 12). In the case of “Untreated 5, 5i and 5ii” samples cultivations were started from the same sample and cultures were grown in parallel (Supplementary Fig. 7, Supplementary Fig. 12).
To quantify within-sample diversity from 16S rRNA data, we used the vegan R package to calculate the most commonly used alpha diversity indices (Shannon index: see Fig. 3a; Fisher’s and Inverse Simpson indices: see Supplementary Fig. 8). Unweighted Unifrac distances (Fig. 3b) were computed by the Phyloseq (version 1.22.3 R package58).
To identify differentially abundant bacterial families in the resistance microbiota, we applied the edgeR (version 3.16.5 R package59) on the family-level 16S rRNA abundance data of the cultured microbiota samples as suggested earlier60. To this end, abundances were normalized using the TMM (Trimmed Mean of M-values) method61 and then untreated and AMP and antibiotic treated samples were compared using negative binomial tests in a pairwise manner. We used the Benjamini-Hochberg FDR correction method to correct the p-values for multiple testing62.
Comparing the prevalence of AMP- and antibiotic-resistance genes in gut microbial genomes
In order to assess if the large taxonomic diversity in the AMP resistant microbiota corresponds to a diverse reservoir of AMP resistance genes, we compared the prevalence of previously described AMP- and antibiotic-resistance gene families in a representative set of gut microbial genomes as follows. Genome sequences used for the analysis of the mobile gene pool (Supplementary Table 2) were filtered to retain only those sequences that belong to one of the AMP- and antibiotic-resistant bacterial families (Supplementary Table 7). In this analysis, we used only the genome sequences from the Human Microbiome Project since in the Fijicomp cohort the family-level taxonomic assignments of the genome sequences were sporadic. In the remaining dataset 24 AMP- and 26 antibiotic-resistant bacterial families were represented with 222 and 219 genome sequences, respectively. To avoid sampling bias due to unequal representation of species among the genome sequences, one genome sequence was randomly picked from each genome group (genomes with lower than 2% 16S rRNA gene dissimilarities, for details, see section “Analysis of the mobile gene pool of the gut microbiota”). Then, for each known AMP- and antibiotic-resistance gene family, the number of genomes with at least one positive annotation was counted and plotted (Supplementary Fig. 9).
Comparing phylum-level distributions of the resistant microbiota and the transferring resistance contigs
To compare the phylum-level distributions of resistant gut bacteria and the resistance contigs originating from them (Fig. 3d), we carried out logistic regression analyses on count data (Supplementary Table 11) as follows. For each phylum, we fitted a logistic regression model to predict if a resistant gut microbiota colony or resistance-conferring contig belongs to that particular phylum or to the rest of phyla. Thus, each entry in the dataset represented either a colony from the drug-treated cultivation experiment or a resistance contig detected in the functional metagenomics screen. The predictor variables of the models were i) whether the entry was a resistance contig or a cultivated colony, ii) the type of treatment (AMP or antibiotic) and iii) the interaction of these two variables. As we were interested in whether a given phylum contributed disproportionally more (less) AMP than antibiotic resistance contigs compared to its relative frequency in the drug-treated cultured microbiota, we tested if the interaction term of the logistic regression model was significant (using the glm function of R).
Comparison of resistance levels in E. coli and S. enterica
In order to investigate whether the level of resistance provided by AMP resistance genes depends more on the genetic background of the recipient bacterium than in the case of antibiotic resistance genes, we measured how DNA fragments that provide AMP- or antibiotic-resistance to E. coli influence susceptibility in a related Enterobacter species, S. enterica Serovar Typhimurium LT2. For this purpose, we used a representative set of plasmids carrying resistance DNA fragments that were isolated in our screens from AMP and antibiotic treatments (Supplementary Table 12). Special care was taken to avoid the inclusion of multiple plasmids carrying resistance genes with the same function or bias toward certain AMPs and antibiotics. To this end, from each AMP and antibiotic selection we chose 1-5 plasmids carrying resistance genes with different functions. This resulted in 16 plasmids and 25 AMPs and antibiotics. The provided resistance levels (MIC fold changes) were measured for both species with standard micro-dilution assay in the same way as described in the section below.
Quantification of the resistance gains that metagenomic contigs provide
To investigate the resistance gains that contigs provide for the recipient bacteria against AMP and antibiotic treatments, we carried out minimum inhibitory concentration (MIC) measurements with selected contigs from the uncultured and cultured microbiota. The resistance gains were quantified by measuring minimum inhibitory concentrations (MIC) with a standard broth microdilution method48. Briefly, 5000 E. coli and S. enterica cells grown overnight in MS medium with 50 μg/ml kanamycin were used to inoculate wells of a 96-well plate. Three rows on the plate were inoculated with the strain in question and three rows with control cells. As a control, E. coli BW25113 or S. enterica Serovar Typhimurium LT2 strains carrying the control plasmid was used as in the functional metagenomic selection experiments. Prior to inoculation, each well on the plate was pre-filled with 100 μL modified MS medium supplemented with the proper concentration of AMP or antibiotic. On the plate, each AMP and antibiotic was represented in 11 different concentrations (3 wells /concentration /clone or control). Three wells per genotype contained only medium to check the growth in the absence of any antimicrobial. MIC values were determined by measuring OD600 after 16-24 hours of incubation at 30°C with a continuous shaking at 240 rpm. To minimize potential evaporation of the medium and consequent plate edge effects, 96-well plates were incubated at 30°C following standard laboratory procedures of MIC measurements.
Functional analysis of a putative LpxF from the metagenomic selections
The aim of the functional characterization was twofold. First, to support the functional prediction for the identified LpxF orthologs with a biochemical assay. Second, to estimate quantitatively the extent to which these LpxF orthologs reduce the net negative surface charge of the bacterial cell as compared to a previously characterized LpxF from Francisella novicida that removes >90 % of the 4’-phosphate groups from the pentaacylated lipid A molecules and hence alters the charge of the outer membrane35. To estimate the surface charge of bacterial cells, we used a fluorescein isothiocyanate (FITC)-labeled poly-L-lysine (PLL) (FITC-PLL) (Sigma-Aldrich®) based assay. Poly-L-lysine is a polycationic molecule, widely applied to study the interaction between charged bilayer membranes and cationic peptides36. The following strains were used in this measurement: E. coli BW25113 (WT), E. coli BW25113 carrying the pZErO-2 plasmid with the LpxF ortholog from Parabacteroides merdae (denoted as lpxFa in Table 1), identified in our functional selection experiments (WT + lpxF), E. coli BW25113 ΔlpxM (ΔlpxM), E. coli BW25113 ΔlpxM carrying the pZErO-2 plasmid with the LpxF ortholog from Parabacteroides merdae (ΔlpxM + lpxF), E. coli BW25113 ΔlpxM carrying the pWSK29 plasmid with LpxF from Francisella novicida35 (ΔlpxM + lpxF*) and Bacteroides thetaiotaomicron. We measured the phenotypic effect of both LpxF carrying plasmids on ΔlpxM genetic background, since LpxF from F. novicida cannot carry out its biological function in wild-type E. coli, only when the lipid A molecules are tetra- or pentaacylated as it is in the case of ΔlpxM E. coli35. B. thetaiotaomicron intrinsically expresses the BT1854 lpxF ortholog (lpxF**) which is responsible for the high level of resistance of this strain against Polymyxin B12. Prior to the surface charge measurements, cells were grown overnight in TYG (Tryptone Yeast Extract Glucose) medium63 in anaerobic conditions. We grew all the strains in TYG medium to allow the comparability with B. thetaiotaomicron. Cells were washed twice with phosphate-buffered saline (PBS) then resuspended to a cell density of OD600 = 1. Cells were incubated with 2 μl of 5 mg/ml FITC-PLL and 100 μl of 1 μg/ml 4,6-diamidino-2-phenylindole (DAPI) for 10 minutes, followed by centrifugation (4500 rpm, 5’). DAPI was used in order to identify the live cells. Cells were washed twice with PBS then diluted 100-fold in 100 μl of PBS and transferred into a black clear-bottom 96-well microplate (Greiner Bio-One). Prior to fluorescent microscopy analysis, cells were collected to the bottom of the plate by centrifugation (4500 rpm, 10'). Pictures were taken with a PerkinElmer Operetta microscope using a 60x high-NA objective to visualize the cells. Images of two channels (DAPI and FITC-PLL) were collected from ten sites of each well. Mean fluorescent intensity for each well was calculated using the Harmony® High Content Imaging and Analysis Software. Experiments were carried out in 4 biological replicates.
Supplementary Material
Acknowledgements
We thank Dezső Módos, Dávid Fazekas, Károly Kovács, Ave Tooming-Klunderud, József Sóki and Edit Urbán for their technical support. Funding: This work was supported by the ‘Lendület’ programme of the Hungarian Academy of Sciences (B.P. and C.P.), the Wellcome Trust (B.P.), The European Research Council H2020-ERC-2014-CoG 648364- Resistance Evolution (C.P.), GINOP-2.3.2-15-2016-00014 (EVOMER, C.P. and B.P.), GINOP-2.3.2-15-2016-00020 (MolMedEx TU-MORDNS, C.P.) and GINOP-2.3.2-15-2016-00026 (iChamber, B.P.), National Research, Development and Innovation Office, Hungary NKFIH grant K120220 (B.K.), NKFIH grant FK124254 (O.M.) and NKFIH grant KH125616 (B.P.), and a PhD fellowship from the Boehringer Ingelheim Fonds (A.N.). BK holds a Bolyai Janos Scholarship. The Pacific Biosciences sequencing service was provided by the Norwegian Sequencing Centre, a national technology platform hosted by the University of Oslo and supported by the "Functional Genomics" and "Infrastructure" programs of the Research Council of Norway and the Southeastern Regional Health Authorities.
Footnotes
Data availability
GenBank accession numbers for the PacBio sequencing data are MH883365-MH883616. 16S rRNA sequencing reads are available in the Sequence Read Archive (SRA) (BioProject PRJNA494380). All data generated or analysed during this study are included in this published article and its supplementary information files. For each Fig., the availability of the analysed data is provided in the Fig. legend. All accession numbers with information on the associated samples are provided as Supplementary Data.
Author contributions
B.K. and C.P. conceived the project, B.K., O.M., E.A., A.N., B.P. and C.P. planned experiments and data analyses. O.M., M.S., P.K.J. and R.T. performed the experiments. I.N. performed Illumina sequencing, A.B. and T.M. were responsible for fecal sample collection. B.K., O.M., E.A., A.G., F.P., G.F., I.L., B.B., B.M.V and M.B. analyzed the experimental data and carried out bioinformatic analyses. B.K., O.M., B.P., and C.P. wrote the manuscript.
Competing interests
B.M.V., B.B. and I.N. had consulting positions at SeqOmics Biotechnology Ltd. at the time the study was conceived. SeqOmics Biotechnology Ltd. was not directly involved in the design and execution of the experiments or in the writing of the manuscript. This does not alter the author's adherence to all the Nature policies on sharing data and materials. The rest of the authors declare no competing interests.
References
- 1.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicholson JK, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 3.Littman DR, Pamer EG. Role of the Commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011;10:311–323. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:600–600. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 6.Peschel A, Sahl HG. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat Rev Microbiol. 2006;4:529–536. doi: 10.1038/nrmicro1441. [DOI] [PubMed] [Google Scholar]
- 7.Hancock R, Patrzykat A. Clinical development of cationic antimicrobial peptides: from natural to novel antibiotics. Curr Drug Target - Infectious Disord. 2002;2:79–83. doi: 10.2174/1568005024605855. [DOI] [PubMed] [Google Scholar]
- 8.Hancock REW, Sahl H-G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24 doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 9.Kubicek-Sutherland JZ, et al. Antimicrobial peptide exposure selects for Staphylococcus aureus resistance to human defence peptides. J Antimicrob Chemother. 2017;72:115–127. doi: 10.1093/jac/dkw381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersson DI, Hughes D, Kubicek-Sutherland JZ. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist Updat. 2016;26:43–57. doi: 10.1016/j.drup.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 11.Sommer MOA, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–1131. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cullen TW, et al. Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science. 2015;347:170–5. doi: 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y-Y, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, et al. Newly identified colistin resistance genes, mcr-4 and mcr-5, from upper and lower alimentary tract of pigs and poultry in China. PLoS One. 2018;13 doi: 10.1371/journal.pone.0193957. e0193957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brito IL, et al. Mobile genes in the human microbiome are structured from global to individual scales. Nature. 2016;535:435–439. doi: 10.1038/nature18927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jain R, Rivera MC, Lake JA. Horizontal gene transfer among genomes: the complexity hypothesis. Proc Natl Acad Sci U S A. 1999;96:3801–6. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen O, Gophna U, Pupko T. The complexity hypothesis revisited: connectivity rather than function constitutes a barrier to horizontal gene transfer. Mol Biol Evol. 2011;28:1481–1489. doi: 10.1093/molbev/msq333. [DOI] [PubMed] [Google Scholar]
- 18.Forsberg KJ, et al. Bacterial phylogeny structures soil resistomes across habitats. Nature. 2014;509 doi: 10.1038/nature13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Helm E, et al. Rapid resistome mapping using nanopore sequencing. Nucleic Acids Res. 2017;45:e61. doi: 10.1093/nar/gkw1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pehrsson PC, et al. Interconnected microbiomes and resistomes in low-income human habitats. Nature. 2016;533(7602):212–216. doi: 10.1038/nature17672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaye KS, Pogue JM, Tran TB, Nation RL, Li J. Agents of last resort. Infect Dis Clin North Am. 2016;30:391–414. doi: 10.1016/j.idc.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Zhi C, Lv L, Yu L-F, Doi Y, Liu J-H. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16:292–293. doi: 10.1016/S1473-3099(16)00063-3. [DOI] [PubMed] [Google Scholar]
- 23.Dürr UHN, Sudheendra US, Ramamoorthy A. LL-37, the only human member of the cathelicidin family of antimicrobial peptides. 2006;1758(9):1408–25. doi: 10.1016/j.bbamem.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 24.Methé BA, et al. A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rettedal EA, Gumpert H, Sommer MOA. Cultivation-based multiplex phenotyping of human gut microbiota allows targeted recovery of previously uncultured bacteria. Nat Commun. 2014;5 doi: 10.1038/ncomms5714. 4714. [DOI] [PubMed] [Google Scholar]
- 26.Rowan F, Docherty N, Murphy M. Desulfovibrio bacterial species are increased in ulcerative colitis. Dis Colon. 2010;53(11):1530–6. doi: 10.1007/DCR.0b013e3181f1e620. [DOI] [PubMed] [Google Scholar]
- 27.Wehkamp J, et al. Inducible and constitutive β-defensins are differentially expressed in Crohn’s disease and ulcerative colitis. Inflamm Bowel Dis. 2003;9:215–223. doi: 10.1097/00054725-200307000-00001. [DOI] [PubMed] [Google Scholar]
- 28.Lopetuso LR, Scaldaferri F, Petito V, Gasbarrini A. Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathogens. 2013;5(23) doi: 10.1186/1757-4749-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coats SR, To TT, Jain S, Braham PH, Darveau RP. Porphyromonas gingivalis resistance to Polymyxin B is determined by the lipid A 4′-phosphatase, PGN_0524. Int J Oral Sci. 2009;1:126–135. doi: 10.4248/IJOS.09062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ingram BO, Masoudi A, Raetz CRH. Escherichia coli mutants that synthesize dephosphorylated lipid A molecules. Biochemistry. 2010;49:8325–8337. doi: 10.1021/bi101253s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porse A, Schou TS, Munck C, Ellabaan MMH, Sommer MOA. Biochemical mechanisms determine the functional compatibility of heterologous genes. Nat Commun. 2018;9 doi: 10.1038/s41467-018-02944-3. 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kong Q, et al. Phosphate groups of lipid A are essential for Salmonella enterica serovar Typhimurium virulence and affect innate and adaptive immunity. Infect Immun. 2012;80:3215–24. doi: 10.1128/IAI.00123-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang R, et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat Commun. 2018;9 doi: 10.1038/s41467-018-03205-z. 1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, McGrath SC, Cotter RJ, Raetz CRH. Expression cloning and periplasmic orientation of the Francisella novicida lipid A 4’-phosphatase LpxF. J Biol Chem. 2006;281:9321–30. doi: 10.1074/jbc.M600435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossetti FF, et al. Interaction of poly(L-lysine)-g-poly(ethylene glycol) with supported phospholipid bilayers. Biophys J. 2004;87:1711–21. doi: 10.1529/biophysj.104.041780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huerta-Cepas J, Serra F, Bork P. ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol. 2016;33:1635–8. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McArthur AG, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57:3348–57. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hyatt D, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 41.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lagesen K, et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–8. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Leary NA, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orlek A, et al. A curated dataset of complete Enterobacteriaceae plasmids compiled from the NCBI nucleotide database. Data Br. 2017;12:423–426. doi: 10.1016/j.dib.2017.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smillie CS, et al. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480:241–244. doi: 10.1038/nature10571. [DOI] [PubMed] [Google Scholar]
- 47.Jiang X, et al. Comprehensive analysis of mobile genetic elements in the gut microbiome reveals a phylum-level niche-adaptive gene pool. bioRxiv. 2017 doi: 10.1371/journal.pone.0223680. 214213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook J, Russell DW. Molecular Cloning : A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 49.Szybalski W. Genetic studies on microbial cross resistance to toxic agents. IV. cross resistance of Bacillus megaterium to forty-four antimicrobial drugs. Appl Microbiol. 1954;2:57–63. doi: 10.1128/am.2.2.57-63.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiegand I, Hilpert K, Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 51.Rhoads A, Au KF. PacBio Sequencing and its applications. Genomics Proteomics Bioinformatics. 2015;13:278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. [Google Scholar]
- 53.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oksanen J, et al. vegan: community ecology package. 2017 [Google Scholar]
- 57.Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods. 2007;69:330–339. doi: 10.1016/j.mimet.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061217. e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jonsson V, Österlund T, Nerman O, Kristiansson E. Statistical evaluation of methods for identification of differentially abundant genes in comparative metagenomics. BMC Genomics. 2016;17:78. doi: 10.1186/s12864-016-2386-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Source J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 63.Bacic MK, Smith CJ. Laboratory maintenance and cultivation of bacteroides species. Curr Protoc Microbiol. 2008 doi: 10.1002/9780471729259.mc13c01s9. Chapter 13, Unit 13C.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fisher RA, Corbet AS, Williams CB. The relation between the number of species and the number of individuals in a random sample of an animal population. J Anim Ecol. 1943;12:42. [Google Scholar]
- 65.Simpson EH. Measurement of diversity. Nature. 1949;163:688–688. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.