Abstract
Regulator of G protein signaling (RGS) proteins play a pivotal role in regulation of G protein-coupled receptor (GPCR) signaling and are there-fore becoming an increasingly important therapeutic target. Recently discovered thiadiazolidinone (TDZD) compounds that target cysteine residues have shown different levels of specificities and potencies for the RGS4 protein, thereby suggesting intrinsic differences in dynamics of this protein upon binding of these compounds. In this work, we investigated using atomistic molecular dynamics (MD) simulations the effect of binding of several small-molecule inhibitors on perturbations and dynamical motions in RGS4. Specifically, we studied two conformational models of RGS4 in which a buried cysteine residue is solvent-exposed due to side-chain motions or due to flexibility in neighboring helices. We found that TDZD compounds with aromatic functional groups perturb the RGS4 structure more than compounds with aliphatic functional groups. Moreover, small-molecules with aromatic functional groups but lacking sulfur atoms only transiently reside within the protein and spontaneously dissociate to the solvent. We further measured inhibitory effects of TDZD compounds using a protein-protein interaction assay on a single-cysteine RGS4 protein showing trends in potencies of compounds consistent with our simulation studies. ThermodynamicanalysesofRGS4 conformations in the apo-state and on binding to TDZD compounds revealed links between both conformational models of RGS4. The exposure of cysteine side-chains appears to facilitate initial binding of TDZD compounds followed by migration of the compound into a bundle of four helices, thereby causing allosteric perturbations of the RGS/Gα protein-protein interface.
Keywords: Regulators of G-protein signaling, Thiadiazolidinone inhibitors, Allosteric inhibitors, Molecular dynamics, Drug discovery, Protein-protein interactions
INTRODUCTION
G-protein coupled receptors (GPCRs) are membrane proteins of profound clinical relevance,1,2,3,4,5 as they mediate key roles in many cellular reactions to neurotransmitters and extracellular ligands.6 Signaling by GPCRs is negatively modulated by a family of proteins known as regulators of G protein signaling (RGS) proteins that serve as a critical node in controlling various cellular responses.7,8,9,10 The mechanism of action of RGS proteins is to bind to activated (GTP-bound) G protein α-subunits (Gα) and accelerate the rate of GTP-hydrolysis, resulting in conversion of GTP to GDP and deactivation of Gα-subunits. By this mechanism, RGS proteins rapidly dampen signaling by GPCRs, and therefore inhibitor candidates targeting RGS/Gα protein-protein interaction are potentially useful to enhance signaling by GPCRs11. Inhibiting protein-protein interactions, such as the one between an RGS protein and a Gα subunit, is particularly challenging,12,13,14,15 because of the lack of suitable binding pockets for small-molecules at the RGS/Gα protein-protein interface. Although progress has been made recently in the development of small molecules as covalent allosteric protein modifiers,16,17,18,19 application of small-molecule ligands for the inhibition of RGS proteins and protein-protein interactions is an active area of research.20,7,21,22,23,24,12,25,26,27,28
Specifically, high-throughput screens have revealed several classes of small molecule inhibitors acting through covalent modification of cysteine thiols on the RGS4 protein27,26,29,30, which is a well-characterized member of the R4 subfamily8. Among them, the thiadiazolidinone (TDZD) inhibitor CCG-50014 and its congeners (Fig. 1a) have shown high potency and specificity28. CCG-203769 is one of the congeners of CCG-50014 with two aliphatic side-chains, but with substantially improved solubility28. RGS4 is highly expressed in the striatum31,32, regulating synaptic plasticity in response to dopamine signaling33,34, and has been associated with Parkinson’s disease35. The effectiveness of CCG-203769 on RGS4 has been demonstrated through in-vivo studies, by reducing bradykinesia in a raclopride model of certain Parkinson’s-like motor deficits in mice36. Furthermore, RGS4 has also been implicated in cancer,37,38,39 heart rate control in the sinoatrial node40, suppression of the anticonvulsant action of adenosine41, and opioid receptor signaling42.
FIGURE 1.
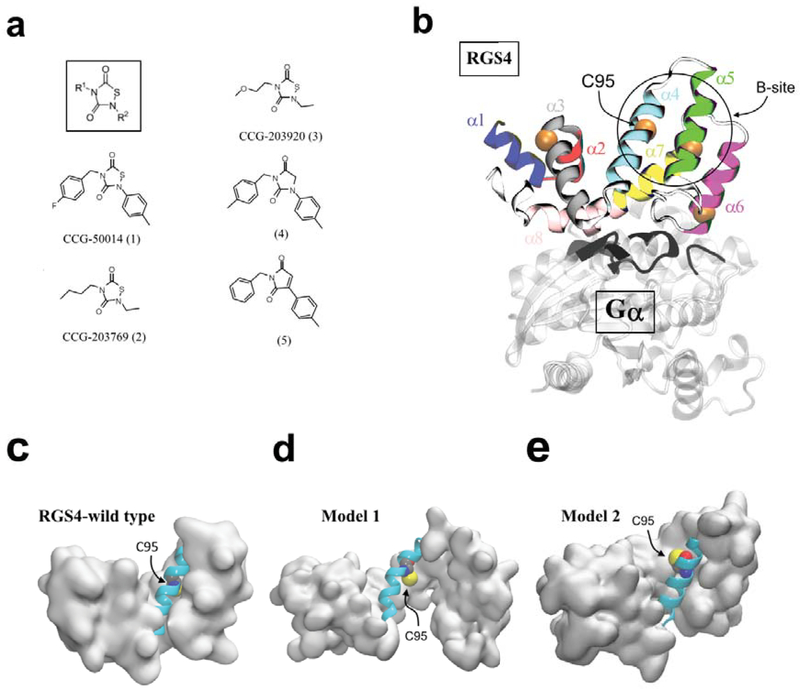
(a) Small-molecule structure with R1 and R2 functional groups (inset in box) along with the chemical structures of all small-molecules studied.28 (b) A cartoon representation of the RGS4-Gαi1 complex (PDB code 1AGR) is shown. Each of the 9 α-helices of RGS4 is colored and labeled, and the location of four cysteine residues are shown by their Cα-atoms as orange spheres with the C95 residue labeled. The Gαi1-subunit is shown in transparent white ribbons, and the loops of Gαi1 in the proximity of RGS4 are highlighted in black ribbons. (c, d, e) Cartoon representations of RGS4 conformations are shown for the wild-type apo-RGS4, and in its conformationally changed models (Model 145 and Model 248), respectively. Highlighted in cartoon representations are α4-helices as cyan cartoons along with the C95 residues as space-filling. The residue C95 is buried in the wild-type RGS4 structure but it is accessible in Models 1 and 2. For each model, the structure of RGS4 (except the α4-helix) is rendered in a white surface representation.
Structurally, RGS proteins are defined by a conserved ~120 residue-long box-domain with nine α-helices43. The crystal structure of RGS4 bound to Gαi1 (PDB code 1AGR) is shown in Fig. 1b44, highlighting the RGS domain with each helix distinctly colored. The crystal structure of RGS4 reveals the location of four cysteine residues (highlighted by orange spheres in Fig. 1b), three of which are solvent-exposed and one cysteine residue (labeled C95 in Fig. 1b) is buried. The C95-residue is also a conserved residue among many RGS proteins, thereby making it an attractive target for covalent modification. Although other cysteines in RGS4 play an important role in potent inhibition of RGS4 by covalent inhibitors,26 these residues are not conserved among all inhibited RGS proteins. Therefore, an understanding of the actions of TDZDs at C95 will be broadly applicable to other RGS proteins. Importantly, TDZD compounds can allosterically inhibit the RGS/Gα protein-protein interaction via the conserved and buried cysteine residue C95.26,27,45 Moreover, the activity of RGS4 is regulated allosterically by endogenous lipid modulators at a site far from the Gα interaction interface but near the C95 residue, also known as the “B-site” (the region marked by a circle in Fig. 1b)7. This allosteric site is also a promising drug target as many physiological regulators of RGS4 directly interact with the B-site46,47.
We have previously reported45 an open-state model of RGS4 (shown as Model 1 in Fig. 1d) which is conformationally different from apo-RGS4 (shown as Model 1 in Fig. 1c) and in which the flexibility in the α5-α6 helical pair facilitates access to the buried C95 residue for covalent-docking of the inhibitor CCG-50014 (compound 1 in Fig. 1a). On inhibitor binding, we found that the the α5-α6 helical pair remains perturbed and only partially relaxes toward the closed conformation of these helices in the apo-RGS4 structure. However, in the absence of inhibitor we observed that the protein largely reverts to a conformation similar to apo-RGS4. While we conducted only short time-scale (~40 ns) molecular dynamics (MD) simulations in that work, parallel NMR HSQC results identified several significant perturbations in residues surrounding the inhibitor binding site. Given the short time-scale of our earlier simulations, it is unclear to what extent the α5-α6 helical pair will remain perturbed by bound ligand in longer time-scale simulations. Moreover, differences in the degree of helical perturbations by CCG-50014 and its congeners with smaller side-chains28 (Fig. 1a) remain unknown as those small-molecules were not studied previously in the context of our open-state model (Model 1).
Furthermore, using μs time-scale MD simulations combined with hydrogen-deuterium exchange (HDX) studies of three RGS proteins (RGS4 and its homologues RGS8 and RGS19),48,49 we not only observed signatures of flexibility and partial unfolding in helices, but also found that the side-chains of buried cysteine residues (C95 in RGS4) are transiently exposed to solvent while apo proteins largely maintain a closed conformation. In our previous work,48 we did not carry out simulation studies of RGS proteins with inhibitors, but we hypothesized that inhibitors could potentially access the otherwise buried C95 residue in the cysteine-exposed closed conformation of RGS4 (Model 2 in Fig. 1e).
To determine whether ligand access to the buried cysteine residue (C95) was through large conformational changes (Model 1) or through small local fluctuations in the side-chain of C95 (Model 2), we here performed enhanced MD investigations of the mechanisms of interactions of 5 small-molecules (3 TDZD compounds with aromatic/aliphatic functional groups and 2 non-TDZD analogues lacking sulfur atoms; Fig. 1a) with the C95 residue of RGS4. We further performed a flow cytometry-based assay14 to measure concentrations of three TDZD compounds needed to inhibit the RGS4/Gα protein-protein interaction (specifically, the interaction between Gα and an RGS4 construct containing only a single-cysteine residue, RGS4 C95). Taken together, these studies highlight the role of cysteine exposure and global protein dynamics in recognition of TDZD compounds by RGS4, and suggest new venues for designing non-covalent small-molecules targeting RGS proteins.
MATERIALS AND METHODS
Molecular Dynamics (MD) Simulations
We performed two sets of classical all-atom and explicit-solvent MD simulations for RGS4/small-molecule complexes (Table S2) using the NAMD software50 and the CHARMM force-field with the CMAP correction51,52. We used VMD for system creation and post-simulation analysis.53 All small-molecules used in this work are reported in Table S1, and their force-fields were parameterized using the Multipurpose Atom-Typer for CHARMM (MATCH) tool.54 The first set of simulations (Set1 in Table S2) was for studying small-molecule analogues in complex with an open-state conformation of RGS4 (Model 1; Fig. 1d),45 and the second set of simulations (Set2 in Table S2) was for small-molecule analogues in complex with a closed-state conformation of RGS4 (Model 2; Fig. 1e).48
For simulations of Model 1 (Set1 in Table S2), the open-state conformation of RGS4 that was reported in our earlier work45 was used here as an initial state for docking of small-molecules. For specifically understanding the effect of binding of thiadiazolidinone (TDZD) and non-TDZD compounds in a binding pocket in the proximity of the cysteine residue C95 (Fig. 1d), a single-cysteine mutant of RGS4 was created, where all cysteine residues (three in total) except C95 were mutated to Ala. Following our previous protocol45, the open-state conformation of mutated RGS4 was then used to create 5 docked complexes with 5 compounds described in Table S1, where each TDZD compound is covalently bound to C95 and non-TDZD compounds are non-covalently docked in the same pocket where TDZD compounds are covalently docked (Fig. S1a, b, c, and Fig. S2a, b). For simulations of Model 2 (Set2 in Table S2), a protocol similar to Model 1 was followed where the initial state of RGS4 was a closed-conformation in which the key cysteine residue (C95) is surface-exposed (Fig. 1e) per our earlier work.48 The docking of compounds in Model 2 was further facilitated by the Internal Coordinate Mechanics (ICM) software55 to obtain their energetically favorable conformations.
We solvated all systems for Model 1 and Model 2 using explicit TIP3P water molecules,56 ionized using NaCl, and added all hydrogen atoms (see Table S2 for total number of atoms in each system). Each solvated and ionized system was then energy minimized for ~500–1000 cycles via conjugate-gradient optimization, and equilibrated via MD simulations, conducted with a time-step (Δt) of 2-fs, for 1 μs in the NPT ensemble where the Langevin thermostat with a damping coefficient of 5 ps−1 was used for temperature control and the Nosé-Hoover barostat was used for pressure control. Periodic boundary conditions were used throughout, non-bonded interactions were accounted with a cut-off of 10 Å where smooth switching was initiated at 8 Å, and long-range electrostatic interactions were handled using the Particle Mesh Ewald (PME) method. For each system, two (for TDZD compounds) or three (for non-TDZD compounds) independent MD simulations were carried out (Table S2). In addition to classical MD simulations, we used metadynamics as an enhanced sampling method (vide infra) to compare thermodynamics of conformational changes between open and closed-states in apo-RGS4 and RGS4/small-molecule complexes.
Metadynamics Simulations
Metadynamics is an enhanced sampling method for faster exploration of conformational space in a specified set of collective variables (CVs) by augmenting the forcefield with a history-dependent biasing potential (Vmeta) of the following form57,58:
| (1) |
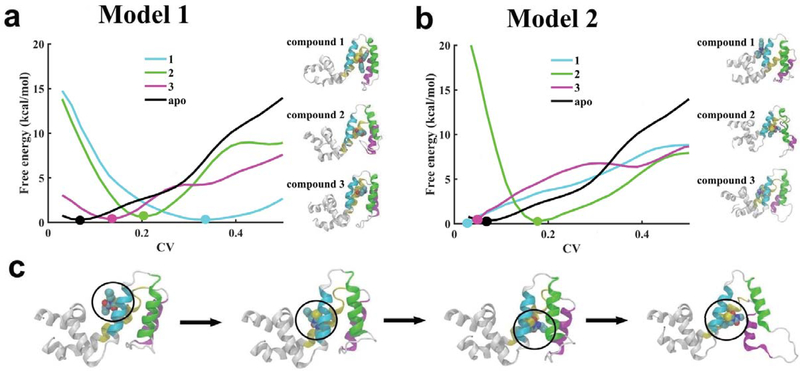
where ξi is the current value of the CV and ξi(t′) is the value of the CV at time t′. Vmeta is constructed as a sum of Ncv-dimensional repulsive Gaussian functions with a chosen height (W) and width (δ). The Gaussian functions can be added at a desired frequency τG. The three main parameters in metadynamics that control the efficiency and accuracy of the free energy reconstruction from converged metadynamics potential (Vmeta)59 are W, δ, and τG. The metadynamics method has been successfully applied to study many biophysical problems including computational drug design.60,61,62,63,64,65,66 In this work, we used an eigenvector as a CV67 that was computed based upon the atomic coordinates of the backbone C atoms of all residues in α4 through α7 helices. The eigenvector choice of a CV is a projection of the coordinates of a group of atoms (or more precisely, their deviations from the reference coordinates) onto a linear transformation between two end-states: open and closed conformations of RGS4. The vector is normalized, therefore the CV is 0 when the backbone C atoms are at the coordinates of the closed conformation of RGS4 and the CV is 1 when they are at the coordinates of the open conformation of RGS4. All metadynamics simulations were carried out with a time-step of 2-fs, and with W, δ, and τG values of 0.05 kcal/mol, 0.025 Å, and 2 ps, respectively. Overall, we carried out 7 metadynamics simulations: one simulation for apo-RGS4 (~0.25 μs long), and 3 simulations (∼0.15−0.45 μs long) each for Models 1 and 2 when bound to compounds 1, 2, and 3 (Fig. 1a and Table S1), respectively. The resulting free-energy profiles are shown in Fig. 6.
FIGURE 6.
(a and b) Free energy profiles are plotted against the collective variable (CV) for structural transitions (between open and closed states) in RGS4 when three TDZD compounds are docked in distinct pockets created in Models 1 and 2. Error bars corresponding to all free-energy profiles are shown in. For each model, thermodynamically favorable conformations of RGS4 bound to TDZD compounds are also shown as cartoons in panels a and b. (c) For compound 2 in Model 2, highlighted as cartoons are conformations of RGS4 showing spontaneous diffusion of compound 2 (CCG-203769; Fig. 1a) from its initially-docked position on the protein surface to within the α4-α7 helical bundle. The circle in panel c denotes the combined location of covalently-linked residue C95 and compound 2. For all panels, the protein backbone in snapshots is depicted in white ribbons except helices α4 through α7 that are uniquely colored as in Fig. 1b.
Protein Expression, Purification, and Flow Cytometry Assay
The Gα-subunitwasexpressedaspreviouslydescribed.68 ForRGS4expression, BL21(DE3) competent cells were transformed with a pET23d vector containing a 6x histidine-tagged, Δ51 N-terminally truncated rat RGS4 with all cysteines except C95 mutated to alanines. Protein expression was induced with 200 μM IPTG. After overnight expression at 25° C, the culture was centrifuged, resuspended in 50 mM HEPES and 100 mM NaCl with protease inhibitors and 1 mM DTT, and lysed by sonication. Lysate was separated by centrifugation at 45000 RCF for 1 hour. The supernatant was applied to a nickel affinity column, then eluted with 300 mM imidazole. Elutions were dialyzed into 50 mM HEPES and 100 mM NaCl, pH 7.4.
Flow Cytometry Protein Interaction Assay (FCPIA)
The Flow Cytometry Protein Interaction Assay (FCPIA) was performed as previously described.69 Briefly, biotinylated single-Cys RGS4 was biotinylated and immobilized on LumAvidin polystyrene microspheres (Luminex). Compounds were incubated with RGS-coated beads for 30 minutes. AlexaFluor 532-labeled Gα was incubated with beads in the presence of 50 μM GDP and AMF (150 μM AlCl3, 5 mM MgCl2, and 50 mM NaF) for 30 minutes. Bead fluorescence was read using Luminex 200 flow cytometer.
RESULTS AND DISCUSSION
To understand the effect of binding of small-molecules in each conformational model of RGS4 (Model 1 and Model 2; Fig. 1d,e), we docked TDZD as well as non-TDZD compounds (Fig. S1 and Fig. S2) in binding pockets created near the C95 residue and conducted several independent μs time-scale MD simulations (see methods and Table S2). The initial conformational state of RGS4 for docking was chosen to either represent flexibility in helices (open-state; Model 1)45 or cysteine-exposure (closedstate; Model 2)48. For each model, we performed two sets of independent simulations (each 1 μs long) for TDZD congeners having aromatic and aliphatic side-chains (compounds 1, 2, and 3 in Fig. 1a) covalently-docked at the residue C95 on the α4 helix (highlighted in cyan in Fig. 1d,e). We first describe results on various conformational metrics used to characterize the subsequent structural perturbations by three covalently-linking TDZD compounds in each model of RGS4.
Simulations of covalently-bound TDZD inhibitors: CCG-50014 (compound 1), CCG-203769 (compound 2), CCG-203920 (compound 3)
Root-mean-squared-fluctuation/deviation (RMSF/RMSD) analyses:
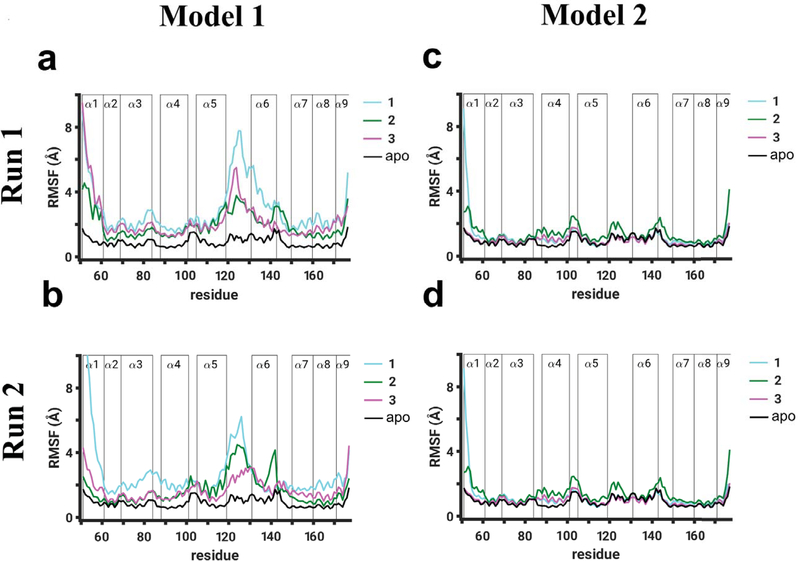
To resolve residue-level perturbations on binding of TDZD compounds (1, 2, and 3) in each model of RGS4, we calculated RMSF per residue from two independent sets of simulations for each model (cyan, green, and magenta traces in Fig. 2) and compared these values with RMSF per residue values of apo-RGS4 from our previous work (black traces in Fig. 2).48 In addition to higher fluctuations expected in free terminal helices (α1 and α9), we observed in both simulations of Model 1 (panels a and b in Fig. 2) that all compounds induced significant perturbations (∼4–8 Å higher than in apo-RGS4) in helices α5 and α6, and in the α5-α6 interhelical loop which directly contacts the Gα-subunit in the RGS4/Gα complex (Fig. 1b). Moreover, the RMSF values show that compound 1 (with aromatic functional groups) perturbs the α5-α6 interhelical loop more than compounds 2 and 3 (with aliphatic functional groups) (cyan vs. green and magenta traces in Fig. 2a,b). The perturbations in this interhelical loop also appear to propagate to structural motifs flanking this loop, namely the C-terminus of the α5 helix and/or the N-terminus of the α6 helix. For compound 1 in comparison to compounds 2 and 3, we also observed marginally higher perturbations in the α3-α4 interhelical loop, another structural region of RGS4 that is known to directly contact the Gα-subunit in the RGS4/Gα complex (Fig. 1b). However, in Model 2 (panels c and d in Fig. 2), where all three compounds are covalently-docked on the protein surface (Fig. S1), we observed no significant perturbations in any structural motif of RGS4 as the RMSF values are comparable to apo-RGS4.
FIGURE 2.
Root-mean-squared-fluctuation (RMSF) per residue are shown for Model 1 (panels a and b) and Model 2 (panels c and d) of RGS4. The RMSF values are reported from two independent 1 μs long simulation runs (Run 1 and Run 2) for each model, where simulations were conducted with TDZD analogues (compounds 1, 2, and 3 in Fig. 1a) covalently-bound to the C95 residue of RGS4. As a baseline reference, the RMSF values of the RGS4 structure without any compound (apo-form; black traces) are also shown from our previous work.48 The vertical bars labeled α1 through α9 demarcate the locations of residues in 9 α-helices of RGS4.
We further report that the Cα-RMSD traces for helices α4 through α7 (Figs. S3 and S4) and their average RMSD from both simulations (Fig. S5), measured relative to the crystallographic conformation of apo-RGS4 (PDB: 1AGR), highlight that all compounds induce greater perturbations when covalently-docked in Model 1 in comparison to Model 2. Also, compounds with aromatic functional groups (compound 1) induce larger perturbations than compounds with aliphatic functional groups (compound 2 and 3). The fluctuations of residues located at the interaction site with the Gα subunit was shown in our previous work to result in significant weakening of the RGS/Gα protein-protein interaction in the presence of compound 1 (CCG-50014).45 As also observed here, this was largely attributed to structural rearrangements of the α5-α6 helical pair and the loop connecting them. Specifically, we observed significant allosteric perturbations in two residues (T124 and E126) of the α5-α6 interhelical loop in which we previously reported perturbations on binding of compound 1 based upon NMR HSQC data.45 We also observed ring-ring interactions (Fig. S6) between compound 1 and the side-chains of neighboring aromatic residues some of which (e.g. F91) were reported as highly perturbed on binding of compound 1 in our previous work.45
Buried Surface Area (BSA) analysis:
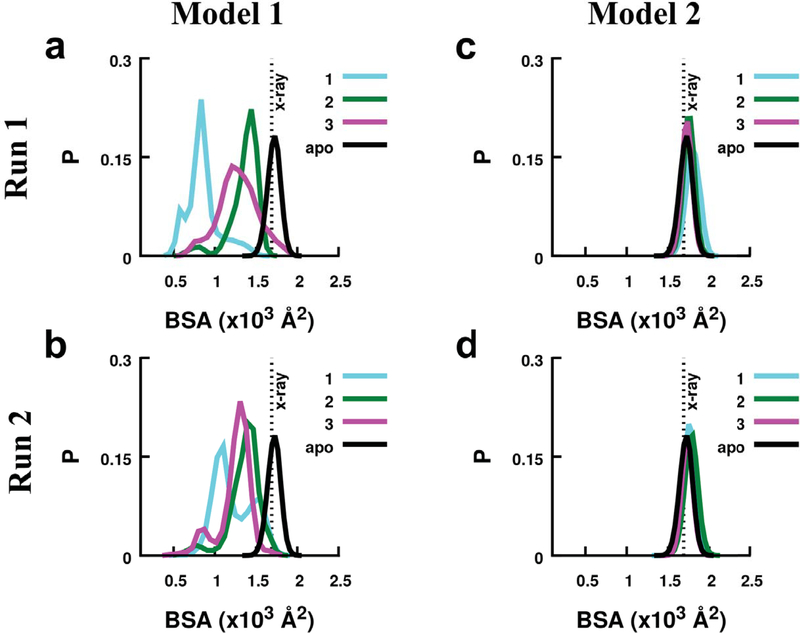
To further investigate conformational changes on binding of compounds in each model of RGS4, we measured the BSA between the α5-α6 helical pair and the rest of the RGS4 structure from two independent simulations (Fig. 3 and Fig. S7). In simulations of Model 1 (Fig. 3a, b), we observed that compound 1 produces a greater shift in the peaks of the BSA-histograms than do compounds 2 and 3 compared to the BSA value in the crystal structure (vertical dotted lines labeled x-ray in Fig. 3) or in the apo-RGS4 simulation (black traces in Fig. 3a, b). The BSA traces for all compounds in Model 1 (Fig. S7a,b) show deviations from the BSA values in the crystallographic or apo-RGS4 simulation indicating that the α5 and α6 helices only partially relax toward their closed conformation in the crystal structure. In both simulations of compound 2 with aliphatic functional groups, the BSA traces (green traces in Fig. S7a,b) show a gradual increase in the BSA toward crystallographic values indicating a nearly complete closure of the α5-α6 helical pair. In simulations of Model 2, the peaks of the BSA-histograms (Fig. 3c, d) and the BSA traces show no significant deviation from the crystal structure values (Fig. S7c,d), thereby indicating a closed conformation of RGS4 when bound to TDZD compounds. Consistent with RMSD/RMSF trends, these results show that the binding of compounds in Model 1 perturbs the RGS4 structure significantly more than their binding in Model 2. Furthermore, in Model 1, the α5-α6 helical pair, which is critical for RGS/Gα binding, only partially relaxes toward the crystallographic conformation in simulations of compound 1, but significantly recovers in simulations of compound 2. This suggests that the compounds with aliphatic functional groups (compounds 2 and 3), when covalently-docked within the α4-α7 helical bundle, are more easily accommodated than those with aromatic functional groups (compound 1).
FIGURE 3.
The histograms of the buried surface area (BSA) between the α5-α6 helical pair and the rest of RGS4 are shown for Model 1 (panels a and b) and Model 2 (panels c and d). Data are shown for simulations of each model conducted with TDZD congeners (compounds 1, 2, and 3). The vertical dotted lines in panels indicate the values of BSA in the RGS4 crystal structure (PDB: 1AGR). The BSA traces for apo-RGS4 computed from a simulation reported in our previous work48 are also shown (black traces).
Salt-bridging interactions:
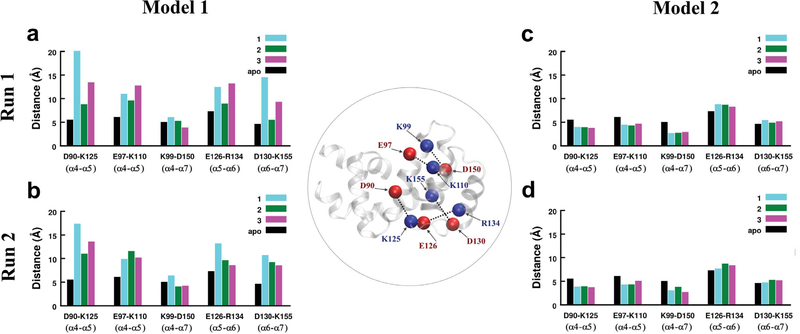
Theα4-α7 helicalbundleinRGS4hasseveralcharged amino-acids (K, R, D, and E) that likely form stable or intermittent salt-bridges due to electrostatic interactions. To understand the ability of TDZD compounds to perturb interactions between charged residues and thereby between helices, we investigated potential perturbations in several salt-bridge forming residue pairs: D90-K125 (α4-α5), E97-K110 (α4-α5), K99-D150 (α4-α7), E126-R134 (α5-α6), and D130-K155 (α6-α7). In Fig. 4, we show histograms of average distances between the center-of-mass of these residue pairs from simulations with compounds (cyan, green, magenta bars) and without compounds (black bars; apo-RGS4). On comparing these data for Model 1, we observed that all compounds perturb salt-bridges between the helical pairs α4-α5 (D90-K125/E97-K110), α5-α6 (E126-R134), and α6-α7 (D130-K155), but only compound 1 perturbs the α4-α7 salt-bridge (K99-D150) marginally higher than compounds 2 and 3; perturbations of compounds 2 and 3 are comparable to apo-RGS4. For salt-bridges between the helical pairs α4-α5 (D90-K125) and α6-α7 (D130-K155), compound 1 perturbs salt-bridges more than compounds 2 and 3 in both simulations of Model 1 (Fig. 4a,b). The α5-α6 salt-bridge (E126-R134) is also perturbed more by compound 1 in comparison to compound 2 in both simulations (Fig. 4a,b), and in comparison to compound 3 in the second simulation (Fig. 4b). However, compound 2 or compound 3 could perturb one of the α4-α5 salt-bridges (E97-K110) marginally more than compound 1 (Fig. 4a,b).
FIGURE 4.
The histograms of average distances between the centers of mass of residues involved in five salt-bridge-forming residue pairs are shown from two independent simulations of Model 1 and Model 2 for three TDZD compounds. The data for an apo-RGS4 simulation from our previous work48 are also shown (black histograms). The Cα-atoms of all residues involved in salt-bridges are shown and labeled as red/blue spheres on the RGS4 structure (inset in circle).
The data from two simulations of Model 2 (Fig. 4c,d) reveal no significant perturbations in these salt-bridging interactions although salt-bridges between the helical pairs α4-α5 (D90-K125/E97-K110) and α4-α7 (K99-D150) are marginally stabilized in comparison to apo-RGS4. These data suggest that in Model 1 compounds with aromatic functional groups destabilize interhelical salt-bridging interactions more than the compounds with aliphatic functional groups. Moreover, the perturbations are allosteric since two salt-bridges significantly perturbed by compound 1 (D90-K125 and D130-K155) have residues K125 and D130 that are located in the α5-α6 interhelical loop, away from the docking site residue (C95). As highlighted above, this loop directly participates in the protein-protein interaction between RGS4 and the Gα-subunit (Fig. 1b). Overall, the mode of binding of TDZD compounds in Model 2 resulted in largely insignificant perturbations in the RGS4 structure in comparison to binding of these compounds in Model 1.
Simulations of non-covalent (non-TDZD) ligands 4 and 5
We further studied compounds 4 and 5 that are analogues of compound 1 with aromatic functional groups but lacking sulfur atoms (Fig. 1a). While compound 1 is a known inhibitor of the wild-type RGS4/Gα protein-protein interaction, compounds 4 and 5 do not inhibit this interaction.28 The mechanistic basis of this observation remains unknown. To test the ability of compounds 4 and 5 to perturb the RSG4 structure, we conducted MD simulations after docking both compounds in each model of RGS4 (Fig. S1 and S2). For Model 1, we conducted three independent 1 μs long MD simulations of each compound (Table S2) and observed that in two out of three simulations, these compounds reside within the α4-α7 helical bundle throughout 1 μs (see snapshots corresponding to Run1 and Run2 in Fig. S8), but in one simulation, each compound diffuses out of the helical bundle into the solvent after transiently residing within the helices (see snapshots corresponding to Run3 in Fig. S8). For Model 2 as well, we conducted three independent simulations of each non-TDZD compound (Table S2). From these simulations, we observed non-specific interactions of compounds (with residues on the protein surface in the vicinity of the docking site) that lead to their rapid dissociation into the solvent (Fig. S9). We discontinued these trajectories after the dissociation of each compound. We therefore present analyses from three trajectories of each non-TDZD compound for Model 1 where compounds can transiently reside within the protein.
The data from RMSF, BSA, and salt-bridge measurements from the first two simulations of each compound in Model 1 (where compounds remain bound) are presented in Fig. 5 and from the third simulations (where compounds dissociate) are shown in Fig. S10. Additionally, the BSA traces from all simulations are shown in Fig. S11, and the RMSD data are shown in Fig. S12 and Fig. S13. The RMSF data (Fig. 5a,d and Fig. S10a) show that both compounds in their transiently bound states perturb the α5 and α6 helices as well as the α5-α6 interhelical loop, and the perturbations by compound 5 are marginally higher than compound 4. The peaks of the BSA histograms (Fig. 5b,e and Fig. S10b) for each compound are shifted away from the values in the crystal structure or in the apo-RGS4 simulation, thereby indicating an open conformation of these helices. Accordingly, 4 of 5 interhelical salt-bridges (D90-K125, E97-K110, E126-R134, and D130-K155) are significantly perturbed, while the α4α7 (K99-D150) is marginally stabilized (Fig. 5c,f and Fig. S10c). This is consistent with the observation that in two simulations of Model 1, both non-TDZD compounds reside within the α4-α7 helical bundle. On dissociation of compound 4 in the third simulation, we observed decreased fluctuations in the α5-α6 interhelical loop and a shift of the BSA-histogram peak toward the crystallographic or apo-RGS4 values (yellow traces in Fig. S10d,e vs. Fig. S10a,b), after compound 5 moved out of helices even though it continued to interact with the protein surface, thereby perturbing the α5-α6 interhelical loop and neighboring helices (magenta traces in Fig. S10d,e). The compounds continue to perturb salt-bridging interactions (Fig. S10c,f) so long as they reside within the helices or near the protein surface on dissociation from the pockets.
FIGURE 5.
The data from RMSF (panel a/d), BSA (panel b/e), and salt-bridging interactions (panel c/f) are shown from two simulations of each non-TDZD compound in Model 1 (compound 4, yellow trace; compound 5, magenta trace). Other details in panels a/d, b/e, and c/f are similar to Figs. 2, 3, and 4, respectively.
In each model, simulations of the non-TDZD compounds (4 and 5) show spontaneous dissociation, consistent with the hypothesis that these two compounds do not reside inside RGS4 for longer time-scales, as they cannot covalently bind to the key cysteine residue (C95). However, despite the transient residence of non-TDZD compounds within the protein domain and their subsequent spontaneous dissociation to the solvent, their ability to perturb the RGS4 structure while bound suggests the potential of non-covalent compounds (possibly with higher binding affinities) as promising candidates for developing the next generation of RGS inhibitors.
Thermodynamic analyses of apo-RGS4 and RGS4/TDZD complexes
To understand thermodynamics of conformational changes in RGS4 in the apo-form as well as when bound to TDZD small-molecules (compounds 1, 2, and 3 in Fig. 1a and Table S1), we carried out seven independent enhanced sampling MD simulations using the metadynamics method57 (see methods). Specifically, we resolved free-energy profiles for structural transitions between the open and closed-states in apoRGS4 and in Models 1 and 2 of RGS4 when bound to the three TDZD compounds (Fig. 6a and 6b). The free-energy profiles were resolved in a multidimensional projection of atomic Cartesian coordinates on an eigenvector reaction coordinate (also referred to as a collective variable, CV) such that the CV spans a range between 0 and 1 for closed and open conformations, respectively. The reference conformational states defining the CV were chosen to represent the apo-RGS4 crystal structure (closed conformation of RGS4) and the conformation reported in our previous work (open conformation of RGS4).45
We observed that the free-energy profile for a structural transition from a closed to open-state in apo-RGS4 (black trace in Fig. 6a,b) has a global minimum (marked by a black filled circle in Fig. 6a,b) very close to the initial value of CV = 0, which indicates that RGS4 in its apo-state thermodynamically favors a closed-state, as also seen in the crystal structure. On comparing free-energy profiles of protein bound to compound 1 (cyan traces in Fig. 6a,b), we observed that the global minimum in Model 1 (marked by a cyan filled circle in Fig. 6a) is located at a CV value of ~0.35, while in Model 2 the global minimum (marked by a cyan filled circle in Fig. 6b) is located very close to zero. This suggests that open-like RGS4 states are thermodynamically favored when compound 1 is docked in the binding pocket in Model 1 in comparison to Model 2, where thermodynamically favored conformations are similar to the closed-state, as in apo-RGS4.
However, from the free-energy profiles for compound 2 in both models (green traces in Fig. 6a,b), we observed that the global minimum (marked by a green filled circle in Fig. 6a,b) is located in the vicinity of a CV value of ~0.20. This suggests that open-like RGS4 states likely exist for compound 2 not only in Model 1 but also in Model 2. The ability of compound 2 to stabilize open-like conformations in Model 2 is explained by the observation that the compound 2 spontaneously diffuses during the metadynamics simulation from its initially-docked position on the protein surface to its final position within the α4-α7 helical bundle (snapshots in Fig. 6c), thereby acquiring conformations similar to Model 1. Importantly, the diffusion of compound 2 from the protein surface to within this helical bundle is driven both by the flexibilities in helices and burial of the side-chain of key cysteine residue C95.
For compound 3, the location of the global minimum in each free-energy profile (marked by a magenta filled circle in Fig. 6a,b) indicates a conformational stability behavior similar to compound 1 in that the open-like RGS4 states are thermodynamically favored in Model 1 and a closed state is favored in Model 2. However, the global minimum for compound 3 in Model 1 is located at a CV value of ~0.14 smaller than the CV value of ~0.35 for the global minimum for compound 1 (magenta vs. cyan filled circles in Fig. 6a). This suggests that perturbations to the RGS4 structure (relative to the closed state) are smaller for compound 3 (with aliphatic functional groups) than for compound 1 (with aromatic functional groups). Overall, the shift in the free-energy minimum from the higher to lower CV values on binding of TDZD compounds in Model 1 (CV = 0.35, 0.20 and 0.14 for compounds 1, 2, and 3, respectively) is consistent with the perturbation trends observed in classical MD simulations (Fig. 2, 3, 4).
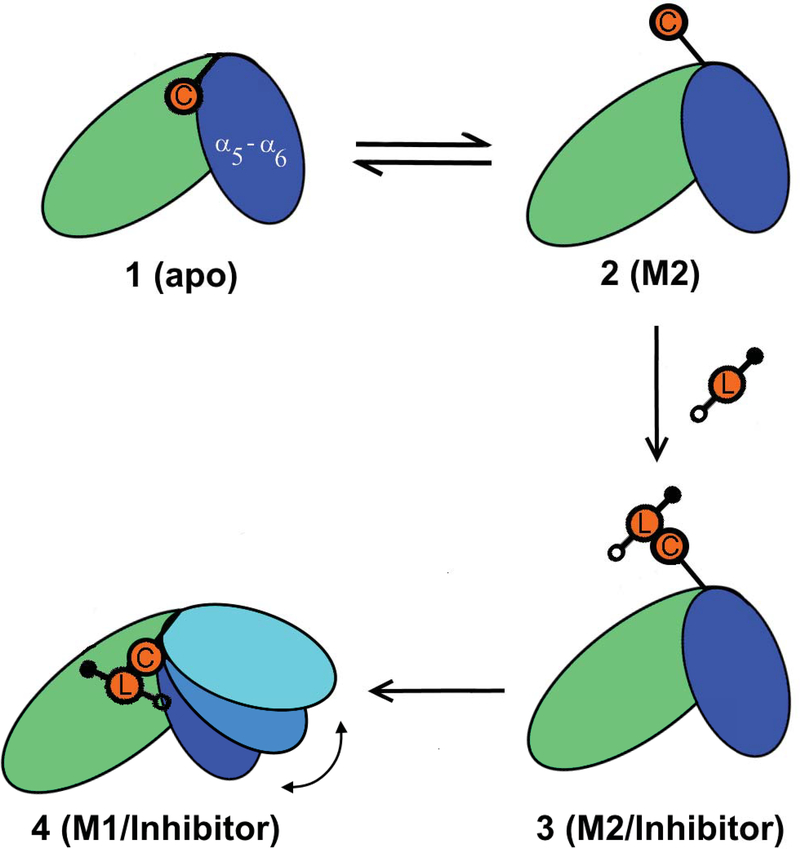
The possibility of binding of TDZD analogues (1, 2, and 3) to C95 in a closed-state conformation of RGS4 due to a transient exposure of C95 (Model 2)48 shows that multiple binding mechanisms may exist by which these small-molecules can access the otherwise buried cysteine residue C95. In Model 2, all compounds are docked on the protein surface (Fig. S1 and S2) located near the “B-site” of RGS4,7 entirely outside of the α4-α7 helical bundle. Therefore, the initial binding of TDZD analogues to C95 is not dependent on a significant opening of the α5-α6 helical pair. This mechanism of small-molecule recognition by C95 is distinct from our previously suggested mechanism45 that highlighted the flexibility in the α5-α6 helical pair as a potential route for compound binding. However, our enhanced sampling metadynamics simulations showed that compounds covalently-bound to the exposed C95 residue in Model 2 (e.g. compound 2) could translocate from the protein surface to the core of the α4-α7 helical bundle (Fig. 6c) and stabilize open-like states similar to Model 1 while remaining covalently-bound, thereby suggesting that Model 2 can evolve toward Model 1 (Fig. 7). Therefore, we propose that these mechanisms of binding of compounds may not be mutually exclusive. This substantiates the importance of coupling between local conformational flexibilities in protein side-chains (e.g. exposure of C95) with global protein dynamics (e.g. flexibilities in RGS4 helices) to facilitate small-molecule recognition and allow allosteric inhibition of the protein-protein interface.
FIGURE 7.
A schematic highlighting the proposed mechanism of binding of TDZD small-molecules to RGS4 is shown. In this scheme, the exposure of C95 (orange circle labeled C) in the apo-RGS4 conformation (panels 1 and 2) allows initial covalent recognition (panel 3) of small-molecules (orange circle labeled L flanked by filled/empty circles indicating R1 and R2 functional groups) and a subsequent migration of compounds to the core of the α4-α7 helical bundle causing allosteric structural perturbations in helices (panel 4), especially in residues in the RGS/Gα protein-protein interface.
FunctionalanalysisoftheinhibitionoftheRGS4-C95/Gαprotein-protein interaction by TDZD compounds
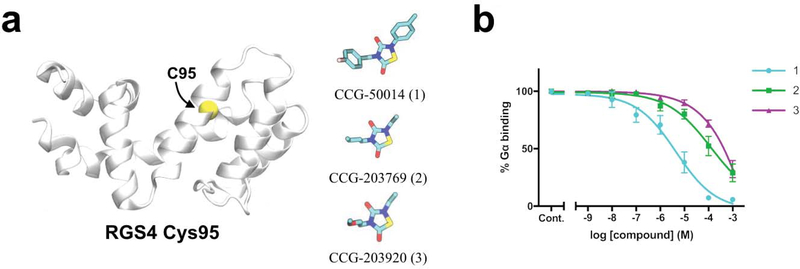
To further investigate the effect of binding of TDZD compounds 1, 2 and 3 on the interaction of single-cysteine RGS4 (RGS4 C95) and Gα in vitro, we utilized a flow cytometry protein interaction assay (FCPIA) (see methods).14A single-cysteine (RGS4 C95) mutant was used to limit compound binding to C95, such that the results reflect compound action at the same cysteine to which compound was covalently docked in simulations. The concentration-response curves (Fig. 8) show that the single-cysteine protein is inhibited by compound 1 with an IC50 value of ∼4.5 μM, and barely inhibited by compounds 2 and 3 (Fig. 8b). The magnitude of structural perturbations and deviations from the native RGS4 conformation (Fig. 2, 3, 4, and 6) induced by the compounds in modeling and simulation studies correlate well with the results of inhibition experiments. The use of single-cysteine mutants resulted in lower potencies of inhibition by each compound compared to wild-type proteins.28 Notably, the difference in potencies between aromatic compound (1) and aliphatic compounds (2 and 3) is more pronounced in single-cysteine RGS4 C95 than in WT RGS4. This may be because in WT proteins, differences in dynamics between compounds bound to C95 are masked by the action of compounds at other cysteines. Compound 1 causes the greatest perturbations in the RGS4 structure and also showed the greatest potency to inhibit RGS4/Gα binding. Compounds 2 and 3 caused smaller perturbations in the RGS4 structure and accordingly showed a reduced ability to inhibit RGS4/Gα binding.
FIGURE 8.
(a) The structures of the single-cysteine RGS4 construct (RGS4 C95) and three compounds used in the flow cytometry protein-protein interaction assay are shown. (b) Inhibition of the RGS4 C95/Gα protein-protein interaction by compounds 1, 2 and 3 over a range of concentrations is shown.
CONCLUSIONS
In this work, we have presented modeling and simulation studies predicting structural perturbations in the RGS4 protein on binding to various small-molecules. We found that compounds with the aromatic functional groups significantly perturb the protein structure in comparison to those with the aliphatic functional groups. Non-covalent compounds only transiently perturb the protein as these compounds spontaneously dissociate to the solvent. We further report thermodynamic analyses of RGS4/small-molecule complexes and suggest that two distinct modes of binding can lead to a twostep mechanism in which compounds are initially recognized by the exposed sidechains of conserved and buried cysteine residues (C95 in RGS4) followed by their migration to the helical core of RGS4 that leads to significant allosteric perturbations in those RGS4 residues that are located in the RGS4/Gα protein-protein interface. These findings will inform future drug development efforts focused on the discovery of non-covalent compounds capable of inducing similar allosteric perturbations.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NSF Grants 1508595 (HV) and 1507588 (RRN), and the NIH Training Grant T32 GM092715 (VSS, RRN). This work was also supported in part by an NIH COBRE Center of Integrated Biomedical and Bioengineering Research (CIBBR, P20GM113131) through an Institutional Development Award (IDeA; to HV) from the National Institute of General Medical Sciences. We are grateful for computational support in part from Trillian, a Cray XE6m-200 supercomputer supported by the NSF MRI program under Grant PHY-1229408, and the NSF-supported (ACI-1053575) Extreme Science and Engineering Discovery Environment (XSEDE)70 Comet resource at San Diego Supercomputer Center (SDSC) underGrantTG-MCB160183 (HV).
Funding information
NSF. Grant Numbers: 1508595,1507588 and PHY-1229408; NIH. Grant Number: GM092715; NSF-supported (ACI-1053575) Extreme Science and Engineering Discovery Environment (XSEDE). Grant Number: TG-MCB160183
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- 1.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov 2002;1(9):727–730. [DOI] [PubMed] [Google Scholar]
- 2.Carrieri A, Perez-Nueno VI, Lentini G, Ritchie DW. Recent trends and future prospects in computational GPCR drug discovery: from virtual screening to polypharmacology. Curr Top Med Chem 2013;13(9):1069–1097. [DOI] [PubMed] [Google Scholar]
- 3.Nickols HH, Conn PJ. Development of allosteric modulators of GPCRs for treatment of CNS disorders. Neurobiol Dis 2014;61:55–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Congreve M, Dias JM, Marshall FH. Structure-based drug design for G protein-coupled receptors. Prog Med Chem 2014;53:1–63. [DOI] [PubMed] [Google Scholar]
- 5.Christopoulos A Advances in G protein-coupled receptor allostery: from function to structure. Mol Pharmacol 2014;86(5):463–478. [DOI] [PubMed] [Google Scholar]
- 6.Venkatakrishnan A, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G-protein-coupled receptors. Nature 2013;494(7436):185–194. [DOI] [PubMed] [Google Scholar]
- 7.Zhong H, Neubig RR. Regulator of G protein signaling proteins: novel multifunctional drug targets. J Pharmacol Exp Ther 2001;297(3):837–845. [PubMed] [Google Scholar]
- 8.Neubig RR, Siderovski DP. Regulators of G-protein signalling as new central nervous system drug targets. Nat Rev Drug Discov 2002;1(3):187–197. [DOI] [PubMed] [Google Scholar]
- 9.Hollinger S, Hepler JR. Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol Rev 2002;54(3):527–559. [DOI] [PubMed] [Google Scholar]
- 10.Kimple AJ, Bosch DE, Giguère PM, Siderovski DP. Regulators of G-protein signaling and their Gα substrates: promises and challenges in their use as drug discovery targets. Pharmacol Rev 2011;63(3):728–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berman DM, Kozasa T, Gilman AG. TheGTPase-activatingproteinRGS4stabilizes the transition state for nucleotide hydrolysis. J Biol Chem 1996;271(44):27209–27212. [DOI] [PubMed] [Google Scholar]
- 12.Blazer LL, Neubig RR. Small molecule protein–protein interaction inhibitors as CNS therapeutic agents: current progress and future hurdles. Neuropsychopharmacology 2009;34(1):126–141. [DOI] [PubMed] [Google Scholar]
- 13.Arkin MR, Wells JA. Small-molecule inhibitors of protein–protein interactions: progressing towards the dream. Nat Rev Drug Discov 2004;3(4):301–317. [DOI] [PubMed] [Google Scholar]
- 14.Roman DL, Talbot JN, Roof RA, Sunahara RK, Traynor JR, Neubig RR. Identification of small-molecule inhibitors of RGS4 using a high-throughput flow cytometry protein interaction assay. Mol Pharmacol 2007;71(1):169–175. [DOI] [PubMed] [Google Scholar]
- 15.London N, Raveh B, Schueler-Furman O. Druggable protein-protein interactions–from hot spots to hot segments. Curr Opin Chem Biol 2013;17(6):952–959. [DOI] [PubMed] [Google Scholar]
- 16.Kalgutkar AS, Dalvie DK. Drug discovery for a new generation of covalent drugs. Expert Opin Drug Discov 2012;7(7):561–581. [DOI] [PubMed] [Google Scholar]
- 17.Mullard A Protein–protein interaction inhibitors get into the groove. Nat Rev Drug Discov 2012;11(3):173–175. [DOI] [PubMed] [Google Scholar]
- 18.Morelli X, Bourgeas R, Roche P. Chemical and structural lessons from recent successes in protein–protein interaction inhibition (2P2I). Curr Opin Chem Biol 2011;15(4):475–481. [DOI] [PubMed] [Google Scholar]
- 19.O Villoutreix B, M Labbe C, Lagorce D, Laconde G, Sperandio O. A leap into the chemical space of protein-protein interaction inhibitors. Curr Pharm Des 2012;18(30):4648–4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arkin MR, Tang Y, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem Biol 2014;21(9):1102–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sjögren B, Neubig RR. Thinking outside of the “RGS box”: new approaches to therapeutic targeting of regulators of G protein signaling. Mol Pharmacol 2010;78(4):550–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roman DL, Traynor JR. Regulators of G Protein Signaling (RGS) Proteins as Drug Targets: Modulating G-Protein-Coupled Receptor (GPCR) Signal Transduction: Miniperspective. J Med Chem 2011;54(21):7433–7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dripps IJ, Wang Q, Neubig RR, Rice KC, Traynor JR, Jutkiewicz EM. The role of regulator of G protein signaling 4 in delta-opioid receptor-mediated behaviors. Psychopharmacology 2017;234(1):29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sjögren B Regulator of G protein signaling proteins as drug targets: current state and future possibilities. Adv Pharmacol Sci 2011;62:315. [DOI] [PubMed] [Google Scholar]
- 25.Blazer LL, Roman DL, Chung A, Larsen MJ, Greedy BM, Husbands SM, et al. Reversible, allosteric small-molecule inhibitors of regulator of G protein signaling proteins. Mol Pharmacol 2010;78(3):524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roman DL, Blazer LL, Monroy CA, Neubig RR. Allosteric Inhibition of the Regulator of G Protein Signaling–Gα Protein–Protein Interaction by CCG-4986. Mol Pharmacol 2010;78(3):360–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blazer LL, Zhang H, Casey EM, Husbands SM, Neubig RR. A nanomolar-potency small molecule inhibitor of regulator of G-protein signaling proteins. Biochemistry 2011;50(15):3181–3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turner EM, Blazer LL, Neubig RR, Husbands SM. Small molecule inhibitors of regulators of G Protein signaling (RGS) proteins. ACS Med Chem Lett 2011;3(2):146–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storaska AJ, Mei JP, Wu M, Li M, Wade SM, Blazer LL, et al. Reversible inhibitors of regulators of G-protein signaling identified in a high-throughput cell-based calcium signaling assay. Cell Signal 2013;25(12):2848–2855. http://www.sciencedirect.com/science/article/pii/S0898656813002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roman DL, Ota S, Neubig RR. Polyplexed flow cytometry protein interaction assay: a novel high-throughput screening paradigm for RGS protein inhibitors. J Biomol Screen 2009;14(6):610–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gold SJ, Ni YG, Dohlman HG, Nestler EJ. Regulators of G-protein signaling (RGS) proteins: region-specific expression of nine subtypes in rat brain. J Neurosci 1997;17(20):8024–8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larminie C, Murdock P, Walhin JP, Duckworth M, Blumer KJ, Scheideler MA, et al. Selective expression of regulators of G-protein signaling (RGS) in the human central nervous system. Mol Brain Res 2004;122(1):24–34. [DOI] [PubMed] [Google Scholar]
- 33.Lerner TN, Kreitzer AC. RGS4 is required for dopaminergic control of striatal LTD and susceptibility to parkinsonian motor deficits. Neuron 2012;73(2):347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen W, Plotkin J, Francardo V, Ko W, Xie Z, Li Q, et al. M4 Muscarinic Receptor Signaling Ameliorates Striatal Plasticity Deficits in Models of L-DOPA-Induced Dyskinesia. Neuron 2015;88(4):762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ko WKD, Martin-Negrier ML, Bezard E, Crossman AR, Ravenscroft P. RGS4 is involved in the generation of abnormal involuntary movements in the unilateral 6OHDA-lesioned rat model of Parkinson’s disease. Neurobiol Dis 2014;70:138–148. [DOI] [PubMed] [Google Scholar]
- 36.Blazer LL, Storaska AJ, Jutkiewicz EM, Turner EM, Calcagno M, Wade SM, et al. Selectivity and Anti-Parkinson’s Potential of Thiadiazolidinone RGS4 Inhibitors. ACS Chem Neurosci 2015;6(6):911–919. [DOI] [PubMed] [Google Scholar]
- 37.Tatenhorst L, Senner V, Püttmann S, Paulus W. Regulators of G-protein signaling 3 and 4 (RGS3, RGS4) are associated with glioma cell motility. J Neuropathol Exp Neurol 2004;63(3):210–222. [DOI] [PubMed] [Google Scholar]
- 38.Nikolova DN, Zembutsu H, Sechanov T, Vidinov K, Kee LS, Ivanova R, et al. Genome-wide gene expression profiles of thyroid carcinoma: identification of molecular targets for treatment of thyroid carcinoma. Oncol Rep 2008;20(1):105–121. [PubMed] [Google Scholar]
- 39.Hurst JH, Mendpara N, Hooks SB. Regulator of G-protein signalling expression and function in ovarian cancer cell lines. Cell Mol Biol Lett 2008;14(1):153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cifelli C, Rose RA, Zhang H, Voigtlaender-Bolz J, Bolz SS, Backx PH, et al. RGS4 regulates parasympathetic signaling and heart rate control in the sinoatrial node. Circ Res 2008;103(5):527–535. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y, Liu Y, Cottingham C, McMahon L, Jiao K, Greengard P, et al. Neurabin scaffolding of adenosine receptor and RGS4 regulates anti-seizure effect of endogenous adenosine. J Neurosci 2012;32(8):2683–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Q, Liu-Chen LY, Traynor JR. Differential modulation of μ-and δ-opioid receptor agonists by endogenous RGS4 protein in SH-SY5Y cells. J Biol Chem 2009;284(27):18357–18367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tesmer JJ. Structure and function of regulator of G protein signaling homology domains. Prog Mol Biol Transl Sci 2009;86:75–113. [DOI] [PubMed] [Google Scholar]
- 44.Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF 4–activated G iα1: stabilization of the transition state for GTP hydrolysis.Cell 1997;89(2):251–261. [DOI] [PubMed] [Google Scholar]
- 45.Vashisth H, Storaska AJ, Neubig RR, Brooks CL III. Conformational dynamics of a regulator of G-protein signaling protein reveals a mechanism of allosteric inhibition by a small molecule. ACS Chem Biol 2013;8(12):2778–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Popov SG, Krishna UM, Falck J, Wilkie TM. Ca2+/Calmodulin reverses phosphatidylinositol 3, 4, 5-trisphosphate-dependent inhibition of regulators of G protein-signaling GTPase-activating protein activity. J Biol Chem 2000;275(25):18962–18968. [DOI] [PubMed] [Google Scholar]
- 47.Ishii M, Fujita S, Yamada M, Hosaka Y, Kurachi Y. Phosphatidylinositol 3, 4, 5-trisphosphate and Ca2+/calmodulin competitively bind to the regulators of G-protein-signalling (RGS) domain of RGS4 and reciprocally regulate its action. Biochem J 2005;385(1):65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shaw VS, Mohammadiarani H, Vashisth H, Neubig RR. Differential Protein Dynamics of Regulators of G-Protein Signaling: Role in Specificity of Small-Molecule Inhibitors. J Am Chem Soc 2018;140(9):3454–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mohammadiarani H, Shaw VS, Neubig RR, Vashisth H. Interpreting Hydrogen–Deuterium Exchange Events in Proteins Using Atomistic Simulations: Case Studies on Regulators of G-Protein Signaling Proteins. J Phys Chem B 2018;122(40):9314–9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, et al. Scalable molecular dynamics with NAMD. J Comput Chem 2005;26(16):1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J Phys Chem B 1998;102(18):3586–3616. [DOI] [PubMed] [Google Scholar]
- 52.MacKerell AD, Feig M, Brooks CL. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem 2004;25(11):1400–1415. [DOI] [PubMed] [Google Scholar]
- 53.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph Model 1996;14(1):33–38. [DOI] [PubMed] [Google Scholar]
- 54.Yesselman JD, Price DJ, Knight JL, Brooks CL. MATCH: An atom-typing toolset for molecular mechanics force fields. J Comput Chem 2012;33(2):189–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abagyan R, Totrov M, Kuznetsov D. ICM—–a new method for protein modeling and design: applications to docking and structure prediction from the distorted native conformation. J Comp Chem 1994;15(5):488–506. [Google Scholar]
- 56.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. JChemPhys 1983;79(2):926–935. [Google Scholar]
- 57.Laio A, Parrinello M. Escaping free-energy minima. Proc Natl Acad Sci USA 2002;99(20):12562–12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laio A, Gervasio FL. Metadynamics: a method to simulate rare events and reconstruct the free energy in biophysics, chemistry and material science. Rep Prog Phys 2008;71(12):126601. [Google Scholar]
- 59.Crespo Y, Marinelli F, Pietrucci F, Laio A. Metadynamics convergence law in a multidimensional system. Phys Rev E 2010;81(5). [DOI] [PubMed] [Google Scholar]
- 60.Limongelli V, Marinelli L, Cosconati S, La Motta C, Sartini S, Mugnaini L, et al. Sampling protein motion and solvent effect during ligand binding. Proc Natl Acad Sci USA 2012;109(5):1467–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Granata D, Camilloni C, Vendruscolo M, Laio A. Characterization of the free-energy landscapes of proteins by NMR-guided metadynamics. Proc Natl Acad Sci USA 2013;110(17):6817–6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barducci A, Bonomi M, Prakash MK, Parrinello M. Free-energy landscape of protein oligomerization from atomistic simulations. Proc Natl Acad Sci USA 2013;110(49):E4708–E4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mohammadiarani H, Vashisth H. all-atom structural Models of the Transmembrane Domains of insulin and Type 1 insulin-like growth Factor receptors. Front Endocrinol 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mohammadiarani H, Vashisth H. Insulin mimetic peptide S371 folds into a helical structure. J Comput Chem 2017;38(15):1158–1166. [DOI] [PubMed] [Google Scholar]
- 65.Biarnés X, Bongarzone S, Vargiu AV, Carloni P, Ruggerone P. Molecular motions in drug design: the coming age of the metadynamics method. J Comput Aided Mol Des 2011;25(5):395–402. [DOI] [PubMed] [Google Scholar]
- 66.Incerti M, Russo S, Callegari D, Pala D, Giorgio C, ZanottiI, et al. Metadynamics for perspective drug design: computationally driven synthesis of new protein–protein interaction inhibitors targeting the EphA2 receptor. J Med Chem 2017;60(2):787–796. [DOI] [PubMed] [Google Scholar]
- 67.Fiorin G, Klein ML, Hénin J. Using collective variables to drive molecular dynamics simulations. Mol Phys 2013;111(22–23):3345–3362. [Google Scholar]
- 68.Lee E, Linder ME, Gilman AG. [12] Expression of G-protein a subunits in Es-cherichia coli In: Iyengar R, editor. Heterotrimeric G Proteins, vol. 237 of Methods Enzymol. Academic Press; 1994. p. 146–164. [DOI] [PubMed] [Google Scholar]
- 69.Blazer LL, Roman DL, Muxlow MR, Neubig RR. Use of Flow Cytometric Methods to Quantify Protein-Protein Interactions. Curr Protoc Cytom 2010;p. 13–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Towns J, Cockerill T, Dahan M, Foster I, Gaither K, Grimshaw A, et al. XSEDE: Accelerating Scientific Discovery. Comput Sci Eng 2014. Sept;16(5):62–74. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.