Abstract
Analysis of protein complexes provides insights into how the ensemble of expressed proteome is organized into functional units. While there have been advances in techniques for proteome-wide profiling of cytoplasmic protein complexes, information about human nuclear protein complexes are very limited. To close this gap, we combined native size exclusion chromatography (SEC) with label-free quantitative MS profiling to characterize hundreds of nuclear protein complexes isolated from human glioblastoma multiforme T98G cells. We identified 1794 proteins that overlapped between two biological replicates of which 1244 proteins were characterized as existing within stably associated putative complexes. co-IP experiments confirmed the interaction of PARP1 with Ku70/Ku80 proteins and HDAC1 (histone deacetylase complex 1) and CHD4. HDAC1/2 also co-migrated with various SIN3A and nucleosome remodeling and deacetylase components in SEC fractionation including SIN3A, SAP30, RBBP4, RBBP7, and NCOR1. Co-elution of HDAC1/2/3 with both the KDM1A and RCOR1 further confirmed that these proteins are integral components of human deacetylase complexes. Our approach also demonstrated the ability to identify potential moonlighting complexes and novel complexes containing uncharacterized proteins. Overall, the results demonstrated the utility of SEC fractionation and LC–MS analysis for system-wide profiling of proteins to predict the existence of distinct forms of nuclear protein complexes.
Keywords: glioblastoma multiforme, label-free quantitation, MS, protein complex, size exclusion chromatography
1. Introduction
Protein complexes are fundamental building blocks of cells and are required for the precise control and regulation of cellular pathways.[1] Only 16% of the predicted open reading frames in the human genome have been experimentally determined to encode subunits of protein complexes,[2] which likely encompass only a fraction. There are ≈20 000 individual protein coding genes in the human genome,[3] and there are no unifying rules for predicting which of these proteins will form complexes based on amino acid sequences.[4] To further complicate matters, the assembly of protein complexes is also altered by post-translational modifications.[5] Protein complexes are diverse in terms of composition, structure, stability, regulation, and function,[6] and complex size can range from a simple homomer, composed of self-interacting copies of a single type of subunit, to heteromers, composed of two or more distinct polypeptides.[7] Therefore, elucidating how ≈20 000 plus proteins encoded by the human genome are assembled into complexes and partitioned into different subcellular compartments in different cell types is a daunting task. Such data is important for defining the roles of individual proteins in association with their interacting partners and for understanding how protein complexes modulate phenotypic variations, cellular organization, and disease pathways.[8] A first step towards that goal is to generate high-quality protein complex maps using high-throughput technologies that place individual proteins within a specific functional module and cellular network.
Proteins are assembled into complexes through the interactions between protein subunits, which cannot be predicted from sequencing alone and must be determined empirically.[4] The yeast-two-hybrid (Y2H) method[9,10] and affinity purification-MS[11,12] are the two most commonly used methods for defining protein complexes. Chemical cross-linking or hydrogen– deuterium exchanges coupled with MS analysis have also been applied to obtain structural information of the protein complexes.[13,14] The Y2H method involves transcriptional activation and reporter gene expression to determine direct binary protein–protein interactions (PPIs). Although successfully applied to different organisms,[9,14,15] there are technical limitations of the Y2H method.[16] This method can only identify direct interactions between proteins, and the rates of false-positives and negatives are high.[17] In the affinity purification-MS method, the protein of interest is used as a “bait” to pull down its interacting proteins, and the co-purified proteins are identified by LC–MS/MS.[18] While this method has been used widely with increasingly high-throughput analysis and has provided great insight into protein–protein interactions, this method requires a tagged-bait protein. The process to generate tagged-bait proteins is often laborious, and the tags may impede complex formation or protein localization.[19] Further, introducing such a bait protein into the cells can alter the stoichiometry of proteins potentially skewing the composition of the endogenous complex.[20] Other studies have used bimolecular fluorescence complementation to determine if proteins interact. While this technique has numerous advantages including being used in live cells, there are a few drawbacks. This method can be time extensive to label the proteins of interest with the fluorescent protein fragments and optimize protein expression levels. Additionally, this technique requires that the tags have ample room to collide for the fluorescence emission,[21] which can miss countless interactions if the PPIs confine the movement of the fluorescent tag. Further, it is possible that labeling the proteins in particular positions can prevent complex formation. Hence, despite many successful studies of protein complexes using these techniques, characterization of protein complexes is still challenging.
Recently, we[4,22] and others[23–26] have developed alternative methods utilizing biochemical fractionation coupled with quantitative LC–MS profiling to analyze endogenous protein complexes in human cells or model organisms. These alternative methods utilize protein chromatography or native gel electrophoresis and MS-based proteomics profiling, and require no exogenous protein expression or tagging. Although recent application of size exclusion chromatography (SEC) combined with MS has been demonstrated for the analysis of native protein complexes in plants or mammalian cells, in-depth proteome-wide analysis of protein complexes in the human nucleus is still not well documented. Nuclear proteins constitute ≈14% of the human proteome,[24,27] and understanding how these nuclear proteins assemble into complexes can inform how cells mediate function and transcriptional regulation.
Many, if not most, nuclear proteins are multifunctional and can serve in multiple complexes. For example, the Brg-associated factors (BAF) chromatin remodeling complex is involved in transcriptional activation, DNA damage repair, DNA replication, and mitosis, and many subunits are shared with the closely related Polybromo-BAF (PBAF) complex, which has different cellular functions.[28] In addition, it is common to observe exchanges between subunits or paralogs in different cell types, or under different cellular conditions, with function-altering consequences.[29,30] While we know that exact protein complex composition can have important implications for biological activity, characterization of these complexes is a significant challenge. Having a tool to be able to study and identify nuclear complexes can shed light into the roles these multifunctional proteins have in the nucleus.
Epigenetic changes and misregulation of DNA damage repair processes are common in countless cancers and diseases. In particular, gliomas have been known to have alterations in DNA damage repair and chromatin regulation.[31] Here, we combined SEC fractionation of native proteins with high-throughput LC–MS profiling to characterize nuclear protein complexes isolated from the glioblastoma multiforme (GBM, grade IV glioma) cell line T98G to gain insight into these alterations. We demonstrate the utility and reproducibility of the approach for the proteome-wide analysis of native proteins without tagging. We also validate the identification of different protein complexes by co-immunoprecipitation (co-IP).
2. Experimental Section
Cell culture:
GBM T98G cells (ATCC) were cultured until confluent in Eagle’s minimum essential medium (Corning), containing 10% fetal bovine serum (Omega Scientific, Inc.), 1% penicillin/streptomycin (Corning), 1% non-essential amino acids (Corning), and 1% glutagro (Corning) at 37 °C, and 5% CO2.
Isolation of nuclear proteins:
Confluent 150 mm plates of T98G cells were washed once with PBS. Cells were lifted from the plate with 3 mL of buffer A (25 mm HEPES pH 7.8, 5 mm KCl, 25 mm MgCl2, 0.05 mm EDTA, 10% glycerol, 0.1% NP-40, protease inhibitor cocktail), and incubated on ice for 15 min to lyse the cell membrane. Intact nuclei were pelleted at 1000 × g, soluble cytoplasmic proteins were removed, and the nuclei pellet was re-suspended in MS compatible buffer (25 mm Tris-HCl pH 7.6, 300 mm NaCl, 1 mm EDTA, and protease inhibitor cocktail) and incubated on ice for 45 min. Chromatin was pelleted at 21 000 × g for 5 min, and the soluble nuclear proteins were transferred to a new microcentrifuge tube. Protein concentration was determined using BCA assay.
Size Exclusion Chromatography:
Nuclear cell lysates were fractionated using a Superdex 200 10/300 GL column (GE Health-care Life Sciences) using an ÄKTA fast protein LC system (Amer-sham Biosciences). The SEC column was equilibrated with two column volumes of the buffer (20 mm Tris-HCl, pH 7.5, 0.5 mM DTT, 1 mm EDTA, 100 mm NaCl, 5% glycerol). A total of 500 μL nuclear lysate (≈1 mg nuclear protein) was loaded onto the column, and the 20 SEC fractions (F15–34) that were within column resolution (and void volume) of 500 μL each were collected. Elution of proteins from the SEC column was performed using the same equilibration buffer at a flow rate of 0.2 mL min–1 and absorbance was monitored at 280 nm. Two biological replicates were processed identically. The column was calibrated using protein standards (MWGF1000, Sigma–Aldrich) covering a mass range from 29 kDa to 669 kDa. The void volume was measured with blue dextran. SEC separation was performed at 6 °C. Half of the sample volume was used for SDS–PAGE/immunoblotting and the remaining half was used for LC–MS/MS analysis (see below). SEC fractions were stored at −80 °C until further use.
Sample preparation for LC–MS/MS analysis:
Sample preparation was carried out as described previously.[4,22] Briefly, proteins were precipitated by adding five volumes of cold (–20 °C) acetone and incubated overnight at –20 °C. After centrifugation at 21,000 × g for 15 min at 4 °C, protein pellets were dissolved in 40 μL of 8 M urea, and protein concentration was determined using the BCA assay (Thermo Fisher Scientific). Samples were separately reduced and alkylated with DTT and iodoacetamide, respectively, prior to digestion with mass spec grade trypsin/LysC mix (Promega) at a 1:25 (w/w) enzyme-to-substrate ratio. After desalting with Pierce C18 spin columns (Pierce Biotechnology), peptides were suspended in 3% (v/v) ACN and 0.1% (v/v) formic acid (FA). Samples were loaded to the LC column by equal volume, not by equal amount. Peptides in each fraction were suspended in 80 μL of the buffer, which was determined based on the total peptide in the most concentrated SEC fraction. The final peptide concentration of that SEC fraction was 0.2 μg μL–1, and 5 μL was loaded to the column. For the rest of the fractions, 5 μL was also loaded, but the peptide concentration varied.
LC–MS/MS data collection and analysis:
Samples were analyzed by RP HPLC-ESI-MS/MS using the Dionex UltiMate 3000 RSLC Nano System (Thermo Fisher Scientific), which was directly connected to a Q Exactive HF Hybrid Quadrupole-Orbitrap MS (Thermo Fisher Scientific) and a Nanospray Flex Ion Source (Thermo Fisher Scientific). Purified peptides were loaded to the trap column (300 μm id × 5 mm) packed with 5 μm 100Å PepMap C18 medium and then separated on 15 cm analytical column (75 μm id) packed with 2 μm 100Å PepMap C18 medium (Thermo Fisher Scientific). For each sample, LC–MS/MS data were collected using a 120-min LC gradient. Mobile phase solvent A was 0.1% FA in water and solvent B was 0.1% FA in 80% ACN. Peptides were loaded to the trap column in loading buffer (2% ACN, 0.1% FA) for 5 min, and eluted with a linear 80 min gradient of 5–30% of buffer B, then changing to 45% of B at 91 min, 100% of B at 93 min at which point the gradient was held for 7 min before reverting back to 95% of A at 100 min. The columns were equilibrated at 95% of A for 20 min. The samples were loaded at a flow rate of 5 μL min–1 for 5 min, and eluted from the analytical column at a flow rate of 300 nL min–1. After each 120-min sample run, columns were washed 2 × with 30 min linear gradient of 5–45% of B to keep them clean and reduce sample carry over before running the next sample. Column temperature was maintained at 35 °C. The mass spectrometer was operated using standard data-dependent mode. MS data were acquired with a Top20 data-dependent MS/MS scan method. The full scan MS spectra were collected in the 400–1600 m/z range with a maximum injection time of 100 ms, a resolution of 120 000 at 200 m/z. Fragmentation of precursor ions was performed by high-energy C-trap dissociation with a normalized collision energy of 27 eV. MS/MS scans were acquired at a resolution of 15 000 at 200 m/z with an ion-target value of 1 × 105 and a maximum injection of 20 ms. The dynamic exclusion was set at 15 s to avoid repeated scanning of identical peptides. Instrument optimization and recalibration was carried out at the start of each batch run using calibration mix solution (Thermo Fisher Scientific). The performance of the instrument was also evaluated using Escherichia coli digest (Waters) at the start of each batch.
Data analysis:
All LC–MS/MS data were analyzed using MaxQuant software (v. 1.5.3.28)[32] with its built-in Andromeda search engine. The MS/MS spectra were searched against the human UniProt protein database (June 20, 2015) for protein identification and relative abundance profiling across SEC fractions. The minimal length of six amino acids was required in the database search. The database search was performed with the precursor mass tolerance set to 10 ppm and MS/MS fragment ions tolerance was set to 20 ppm. The database search was performed with enzyme specificity for trypsin and LysC, allowing up to two missed cleavages. Oxidation of methionine was defined as a variable modification, and carbamidomethylation of cysteine was defined as a fixed modification for database searches. The “unique plus razor peptides” were used for peptide quantitation. The false discovery rate of peptide and protein identification was set at 1%. We used the “match between runs” function with retention time window of 1 min to increase the number of peptides that can be used for protein quantification. This function allows the transfer of peptide identification between adjacent fractions in the absence of peptide sequencing by MS/MS spectra, utilizing their accurate mass and aligned retention time. Proteins labeled either as contaminants or reverse hits were removed from the analysis. Similarly, proteins identified without any quantifiable peak (zero intensity) and those identified by a single MS/MS count were also removed from the analysis. Proteins were clustered based on their elution profiles using hierarchical clustering in Data Analysis and Extension Tool (DAnTE)[33] and displayed as a heat map. DAnTE was also used to calculate Pearson correlation coefficients (PCCs) of protein elution between the two SEC fractions.
Gene Ontology (GO) analysis:
GO terms for the cellular component and molecular function were mapped using the Panther Gene Ontology Consortium Slim Cellular Component and Biological Processes analysis.[34] A total of 1515 IDs were mapped out of the 1794 to the Homo sapiens reference list. Bonferroni analysis was used and only significantly enriched genes were included (p < 0.05).
co-IP:
T98G cells were harvested as described for nuclear lysate isolation. Native cell lysate (200 μg) was diluted to 500 μL with dilution buffer (25 mM Tris pH 7.6, 150 mm NaCl, and 1 mm EDTA), incubated overnight at 4 °C with Protein G magnetic beads (Pierce) and 2 μg antibody. Following incubation, beads were washed three times with the dilution buffer and re-suspended in lithium dodecyl sulfate loading dye for immunoblot analysis.
SDS–PAGE/Immunoblotting:
Dried protein pellets in each SEC fraction were re-suspended in lithium dodecyl sulfate loading dye with beta-mercaptonal, boiled and ran on a 4–12% gradient gel (Invitrogen). Gels were transferred to PVDF membranes, blocked with 5% BSA (VWR) in PBS-t (0.1% Tween-20). After blocking, blots were incubated in primary antibody overnight at 4 °C and then incubated in the dark with LiCor goat anti-mouse and goat anti-rabbit fluorescence secondary antibodies (1:10 000) for an hour. Blots were imaged on LiCor Odyssey. Primary antibodies used: rabbit CBX8 (1:1000; Bethyl, A300–882A), mouse PARP1 (1:400; Santa Cruz, sc-8007), mouse PCNA (1:400; Santa Cruz, sc-56), mouse Ku70 (1:400; Santa Cruz, sc-17789) and mouse Ku86 (1:400; Santa Cruz, sc-5280), mouse HDAC1 (1:400; Santa Cruz, sc-81598), and mouse Mi2 (1:200; Santa Cruz, sc-55606).
PPI analysis by STRING:
The STRING 10.5 software was used for mapping protein interactors to the experimentally identified protein complexes.[35] Interacting proteins were identified by typing proteins names in the software along with the selection of the species under investigation (H. sapiens) in order to exclude false-positive PPI and functional annotations derived from investigations on other species. STRING determines and makes graphs of unbiased networks in which gene products are represented as nodes, and the biological relationship between two nodes is represented as an edge (line). Active interactions are supported by one or more evidences (text mining, experiments, gene fusion, database, co-occurrence, or co-expression) that are stored in the software internal database. The minimum required interaction score was set to 0.9 (high confidence) to exclude as many false positive interactions as possible, and the maximum interaction score was set to either five or ten depending on the target proteins.
Data availability:
All the raw LC–MS/MS data associated with these experiments have been deposited in Mass Spectrometry Interactive Virtual Environment (http://massive.ucsd.edu) with ID MSV000081520.
3. Results
3.1. Purity of Nuclear Protein Isolation
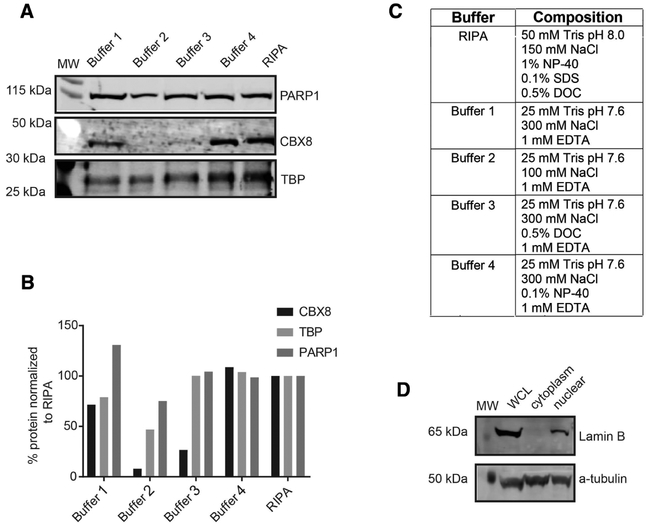
Standard nuclear protein isolation typically involves incubation in hypotonic buffer to disrupt the outer cell membrane and remove soluble cytosolic proteins, followed by a second lysis step in detergent to disrupt the nuclear membrane and release chromatin-bound proteins. Since detergent such as NP-40 in the lysis buffer can be problematic for LC–MS analysis, we first assessed the efficiency of alternate buffers for the nuclear lysis step by monitoring the elution of chromobox protein homolog 8 (CBX8), a chromatin-bound low abundant nuclear protein, into nuclear extracts using immunoblot analysis (Figure 1A). As observed in Figure 1A, a low salt buffer (100 mm NaCl, buffer 2) is capable of lysing the nuclei, however, it cannot elute chromatin-bound proteins. We determined that a high salt buffer (300 mm NaCl, buffer 1), hereinafter referred to as MS compatible buffer, with longer incubation times (45 min instead of 15 min) on ice provided the best nuclear lysis and elution of chromatin-bound proteins without detergent compared to the standard radio-immunoprecipitation assay buffer or other buffers with detergents (Figure 1A–C).
Figure 1.
Nuclear sample preparation. A) Immunoblot analysis of different nuclei lysis conditions using the nuclear protein PARP1 and the chromatin-bound proteins CBX8 and TATA-binding protein (TPB) to assess lysis. B) Quantitation of the immunoblot normalized to radio-immumoprecipitation assay, a high detergent lysis buffer. C) Different buffer compositions tested. D) Immunoblot analysis of whole cell lysate (WCL), cytoplasmic, and nuclear fractions to demonstrate enrichment of nuclear proteins against lamin B1 (nuclear) and α-tubulin cytoplasmic antibodies.
We then sought to determine the purity of the nuclear fraction using our buffer conditions. To do so, we isolated whole cell lysate (WCL), the cytosolic fraction and the nuclear fraction and assessed expression of lamin B1, a nuclear protein, and α-tubulin, a cytosolic protein via immunoblot. As expected, lamin B1 and α-tubulin were present in the WCL. The nuclear fraction indeed enriched for nuclear proteins because lamin B1 was detected in this fraction and not the cytosolic fraction. The α-tubulin was detected both in the cytoplasm and the nuclear fraction with the highest signal in the cytoplasm. This suggested that our nuclear lysate was not completely devoid of cytosolic proteins, most likely due to their high abundance. We chose to accept the reduced purity of our sample and used reliable knowledge-based data sets and bioinformatics tools to identify cytosolic contaminants. Although, we have focused on the nuclear fraction, this approach is equally suitable for other organelles.
3.2. Workflow Overview
The goal of our work was to apply an LC–MS/MS-based proteomics workflow to identify mammalian nuclear protein complexes. Aryal et al. previously described a workflow that combined SEC and label-free quantitative LC–MS profiling to globally monitor Arabidopsis cytoplasmic proteins,[4,22] which we have adapted for mammalian nuclear proteins in this study (see Figure 2 for basic workflow). Briefly, nuclear lysate was isolated from harvested T98G cells as outlined in the Methods. The nuclear lysate was fractionated by size into 20 SEC fractions on a Superdex 200 10/300 GL column in an ÄKTA fast protein LC system. Proteins in each fraction were prepared for LC– MS/MS analysis on a Thermo QExactive Orbitrap MS. Peptides were identified using MaxQuant and quantified by intensity-based label-free quantitation. Overlapping peptides and proteins in the two biological replicates were used for further analysis and to predict protein complexes. Protein intensity (elution) profiles were used to calculate the apparent molecular weight (Mapp) of the proteins based on our standard curve. The apparent ratio (Rapp), defined as the ratio of the Mapp to the Mmono (monomer mass),[4,22] was used to predict proteins eluting as putative complexes through the SEC column. Proteins with an Rapp score of 2 or larger were considered to be in a complex.
Figure 2.
Workflow of nuclear complex profiling. Harvested T98G cells were lysed to break the cell membrane and centrifuged to collect nuclear fraction. Following the lysis of nuclear membrane, nuclear extracts were separated by size exclusion chromatography. Peptides were identified in each fraction with MaxQuant search. Relative protein abundance in each SEC fraction was determined using intensity-based label-free quantitation. Mapp were calculated for proteins identified in both biological replicates based on their elution profiles. The Mapp was divided by Mmono to determine Rapp. Proteins with an Rapp ≥ 2 in both the replicate were considered to be in a complex
3.3. Reproducibility of SEC Fractionation and LC–MS/MS Analysis
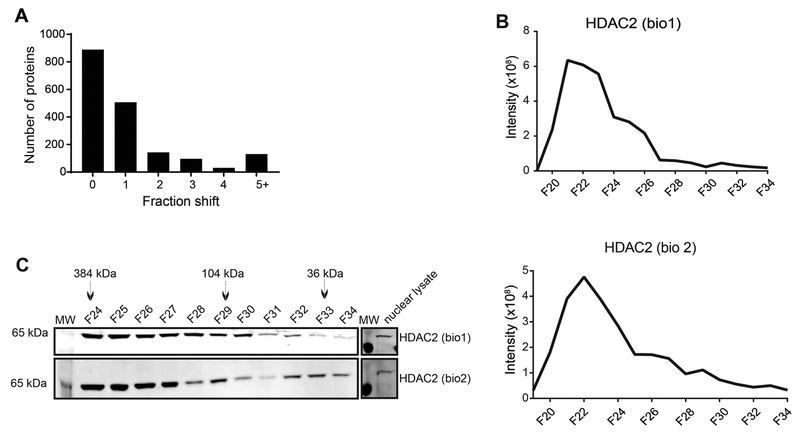
Reproducibility of SEC fractionation and LC–MS analysis is important for successfully predicting protein complexes using this approach. Reproducibility of SEC fractionation was tested by comparing peak shifts of the identified proteins between the two independent SEC separations and detected >90% of the proteins within zero to one fraction shift (Figure 3A) indicating SEC reproducibility. To test the variability of LC–MS runs, we calculated the coefficient of variation of MS1 intensity and retention time of peptides by analyzing three independent technical replicates of the SEC fraction 24 from biological replicate 2. Combined together, we identified 3575 peptides mapping to 590 proteins/protein families of which 2978 peptides (83.3%) and 529 proteins (89.7%) were identified in all the three technical replicates (Figure S1A and B, Supporting Information). This demonstrates reproducibility of peptide and protein identification. The average CV of peptide retention time was 0.8% and of peptide MS1 intensity was 15.8% (Figure S1C and D, Supporting Information). This confirms reproducibility of our LC– MS platform for peptide quantitation. Accuracy of MS1 intensity was also evaluated by comparing the results with the spectral counts (data not shown). MS1 intensity provided smaller CV than the spectral counts. To further assess the accuracy of our SEC profiling strategy for predicting protein complexes, we compared the MS1-based abundance profiles (Figure 3B) for histone deacetylase complex 2 (HDAC2) using an orthogonal protein immunoblot analysis (Figure 3C). There was agreement between the two methods in assigning the peak signal for the protein. After removing the void (F15–21), both LC–MS and immunoblot results revealed that the elution of HDAC2 was predominately in fractions 23–26 in both the replicates (Figure 3B and C). Fractions 23–26 correspond approximately to 500–228 kDa, which is significantly larger than the monomeric mass of HDAC2 (≈65 kDa). This suggests that our isolation and fractionation scheme maintains complex integrity, fractionates proteins by size, and is reproducible.
Figure 3.
Reproducibility of SEC fractionation. A) Histogram showing the shift in peak elution fraction of proteins between the two replicates. The peak elution of [2248]90% of the proteins was determined within zero to one fraction shift. B) MS1 intensity profile of HDAC2 in the two biological replicates. C) Immunoblot analysis of SEC fractions for verification. HDAC2 elutes in higher molecular weight fractions F24–26. Nuclear lysate control (15 μg) to demonstrate expression level. MS1 intensity correlates to immunoblot analysis for HDAC2 in both biological replicates (Note: fraction F21 and lower are void, unresolved complexes)
3.4. Global Analysis of Nuclear Protein Complexes
Following the validation of the MS1-based relative abundance profiling for estimating functional masses of the identified proteins via immunoblot, we prepared fractions 19 to 34 for LC–MS analysis from two independent nuclear extractions as outlined in the Methods. To have an accurate profile of nuclear proteins, we identified the SEC fraction with the highest concentration of peptide and re-suspended the sample to be at a concentration of 0.2 μg μL–1. The subsequent fractions were re-suspended in the same volume (80 μL) and ran through the Q Exactive Orbitrap HF MS. We identified 13 400 peptides mapping to 1794 proteins that overlapped between the two biological replicates (Tables S1 and S2, Supporting Information). The 1794 proteins identified in both the replicates were used for further analysis. Detailed information of the peptides and proteins is provided in Tables 1–3, Supporting Information.
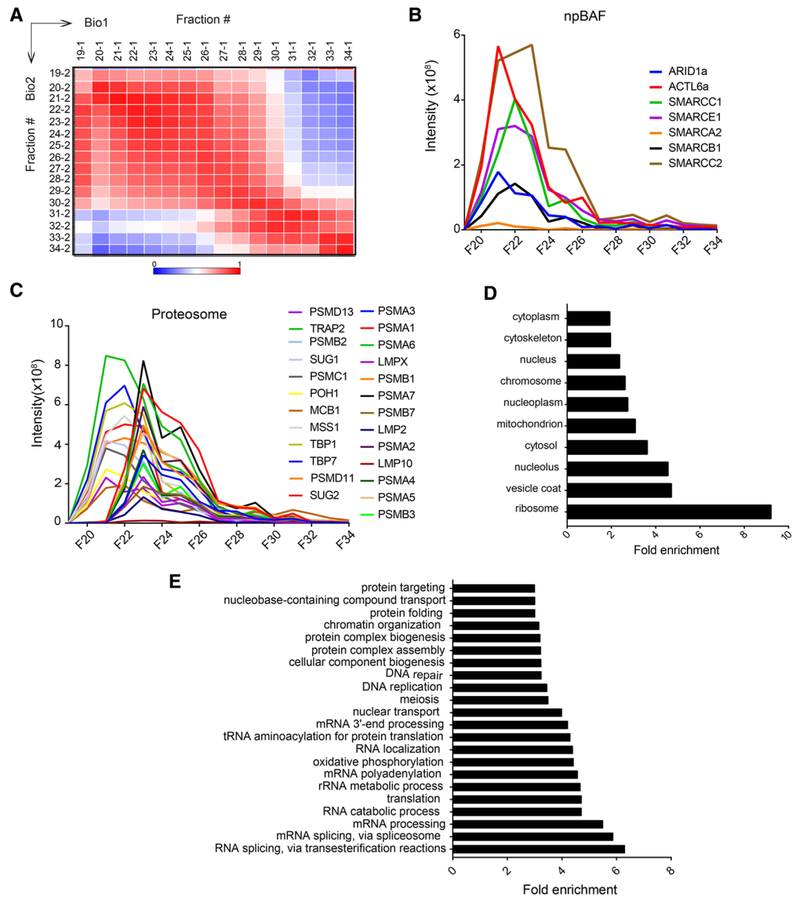
To compare elution profiles of proteins in Bio1 and Bio2, we calculated PCCs of the protein intensities across all SEC fractions. As expected, PCC showed highest correlation coefficients along the diagonal (Figure 4A), suggesting that protein elution profiles were most strongly correlated in identical and/or adjacent SEC fractions. However, many proteins eluting in high mass SEC fractions had higher correlation signals expanded to multiple adjacent fractions. This is due to the large size of the complexes eluting in those fractions that could not be resolved properly by the column.
Figure 4.
Protein identification and relative abundance profiling. A) Heatmap of Pearson correlation values of relative protein abundances (intensities) across SEC fractions between both biological replicates. The correlations coefficients were calculated using DAnTE.[33] Proteins identified in both replicates (fractions 19–34) were used for the analysis. B) Elution profiles of the npBAF complex demonstrating subunits co-elute in a single fraction. C) Elution profiles of the 19S and 20S proteasome subunits. Distinct elution peaks of 19S and 20S in fraction 21 and 23, respectively, demonstrates the elution shift of the different size complexes. Elution profiles of only Bio1 sample are shown. Elution profiles of npBAF and proteasome complex subunits were similar for both the replicates. D) Cellular component analysis of the proteins identified in both biological replicates. Components shown are significantly enriched in our analysis. E) Biological processes analysis of the proteins identified in both replicates. Processes shown are significantly enriched in our analysis
3.5. Protein Complex Elution Profiles
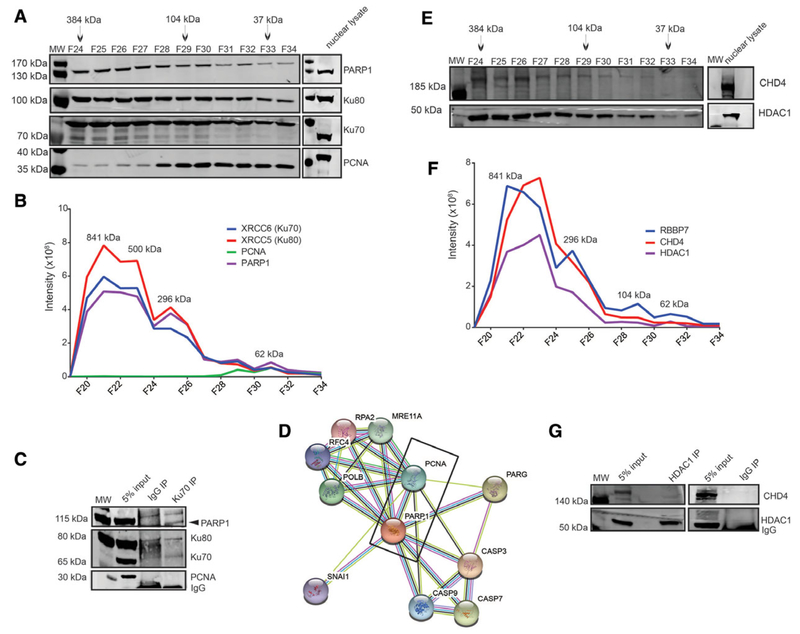
Using knowledge of known nuclear protein complex subunits, we validated our SEC co-fractionation by comparing the elution profiles (Table S2, Supporting Information). As expected, subunits of stable protein complexes displayed similar elution profiles. For example, we identified components of the neural progenitor BAF (npBAF) complex including ARID1A, ACTL6, SMARCC1, SMARCA2, SMARCB1, SMARCE1, and SMARCC2, which migrated together through the SEC column and coeluted as a single peak in fraction 22 (Figure 4B). The npBAF complex is an ATP-dependent chromatin-remodeling complex necessary for the development of the mammalian nervous system.[36] Components of the nucleosome remodeling deacetylase (NuRD) and SIN3A chromatin remodeling complexes also co-eluted in fraction 21 or 22 (Figure S3A and B, Supporting Information). HDAC1 and HDAC2, which are components of both complexes,[37] eluted with the other SIN3A components including SAP30, SIN3A, RBBP4, RBBP7, and NCOR1 at a high mass SEC fraction (Figure S3A and Table S2, Supporting Information) suggesting their physical interaction to form a large complex assembly. Moreover, HDAC1/2 also eluted together with the NuRD components GATAD2B, CHD4, MTA2, MTA3, and MBD3 (Figure S3B and Table S2, Supporting Information). Further, these SEC co-elution profiles correspond to the majority of the HDAC1 and HDAC2 interacting proteins in SIN3A and NuRD complexes as proposed by the STRING database (Figure S3C and D, Supporting Information). The elution of HDAC1/2 also matched with histone lysine demethylase (KDM1A) and corepressor RCOR1 (Figure S4A and Table S2, Supporting Information). Both KDM1A and RCOR1 are the known interactors of HDAC1/2.[11] From our analysis, we were able to identify the relative expression of HDAC1, HDAC2, and HDAC3 that co-eluted at a fraction with an apparent mass (Mapp) of 649 kDa. Among the three histone deacetylases, HDAC1 was the most abundant and HDAC3 was the least abundant in their expression levels.
Many other nuclear proteins such as Lamin A/C, Lamin B1, and components of nuclear pore complex (NUP155, NUP93, NUP62, NUP188, and NUP50) were also eluting in high mass SEC fractions (Table S2, Supporting Information). We also identified several interactors of Nibrin (NBN) (Figure S4B, Supporting Information), a component of the MRE11/RAD50/NBN (MRN) complex. MRN participates in the early steps of DNA damage sensing and double stand DNA breaks repair.[38] The MRE11A,RAD50, ATM, SMC1A, and TERF2 proteins identified in this study were co-eluting together with NBN and peak at fraction 22 with an estimated Mapp of ≈ 649 kDa. The co-elution pro files also matched with the MRN interactome map gleaned from the STRING analysis (Figure S4C, Supporting Information). The ability of NBN to interact with MRE11, RAD50, ATM, H2A.X, CHEK2, BRCA1, SMC1, CtIP, SP100, and 53BP1 has been confirmed previously by immunoblot analysis.[39]
Further, the elution profiles of proteasome subunits are often used to validate the proteomic profiling results. The 26S proteasome is responsible for the degradation of the majority of proteins in eukaryotic cells.[40] It is located both in the nucleus and the cytoplasm[41] and is made up of two sub-complexes: a catalytic core particle (CP) known as the 20S proteasome and the regulatory particle (RP) known as 19S proteasome. The 19S proteasome binds to one or both ends of the 20S protea-some to form an enzymatically active proteasome. The highly abundant 20S CP is a heteromeric assembly of 28 α- and β-subunits, all of roughly 25 to 30 kDa size, and has a predicted mass of 700 kDa.[40] We identified 14 of the CP subunits co-eluting as a single peak in fraction 23 (Figure 4C), with an estimated mass of ≈ 500 kDa. The 26S proteasome holoenzyme consists of two RPs capping each end of the barrel-shaped 20S CP.[42] We identified 11 of the 19 RP subunits, which had a much higher relative abundance compared with the CP subunits (Figure 4C). All of the identified RP subunits eluted two fractions earlier than the CP subunit peak (Figure 4C), consistent with the larger size of the 26S holoenzyme. Consistent elution peaks of these large complex subunits again suggested the accuracy of our strategy for predicting protein complexes. Overall results suggest that our data (Table S2, Supporting Information) may contain many large novel complexes not reported previously.
3.6. Global Characterization of Identified Proteins
We performed GO analysis of our protein list for both biological processes and cellular components. The cellular components analysis demonstrated enrichment for ribosomal, nuclear, and cytoplasmic proteins (Figure 4D). This aligns with our lysate fractionation immunoblot (Figure 1D). Functional classification showed enriched biological processes including histone modification, DNA repair, mRNA processing, splicing, cell cycle, chromatin remodeling, DNA replication, transcription, and nuclear transport, among other nuclear functions (Figure 4E). Detection of many abundant nuclear proteins suggests that the proteins identified in both biological replicates are enriched for nuclear proteins.
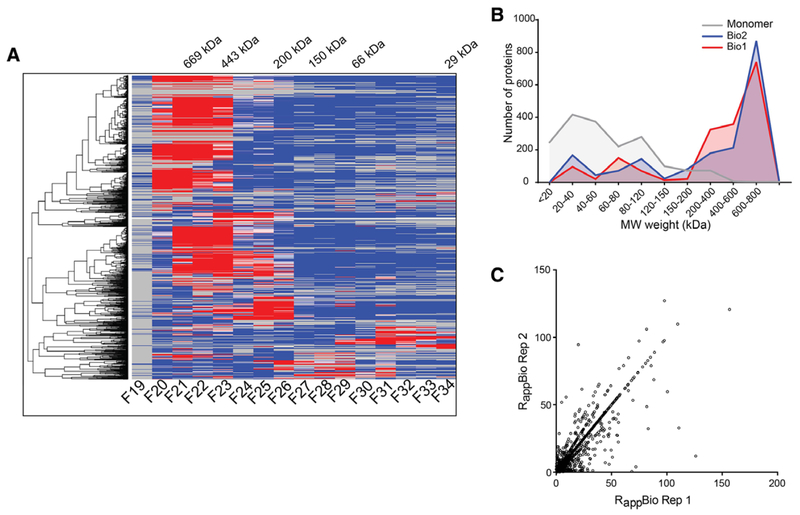
To identify novel protein complex subunits, we performed distance-based clustering analyses of the intensity profiles. The clustering results were plotted as a heat map (Figure 5A). A high-resolution image of the Figure 5A that can be searched by locus IDs is available as Figure S5, Supporting Information (Figure S5A, Supporting Information for the replicate 1 and S5B for the replicate 2).
Figure 5.
Cluster analysis and protein complex identification. A) Hierarchical clustering of protein elution profiles. A total of 1794 nuclear proteins identified in both the biological replicates were clustered based on their SEC elution profiles and displayed as a heatmap. Only biological replicate 1 is shown. Numbers on the top show the molecular masses of protein standards used to calibrate the column. A searchable high resolution heatmap of proteins is shown in Figure S5A, Supporting Information for Bio1 and S5B for Bio2. B) Distribution of monomer and Mapp (kDa) of all the 1794 proteins in both biological replicates. C) Scatter plot showing the distribution of calculated Rapp for proteins identified in both biological replicates
Most proteins showed a single major SEC peak allowing effective clustering based on intensity profiles. However, not surprisingly, more than half of the proteins had their elution peaks in the first three high mass SEC fractions indicating that the majority of the nuclear complexes exist as large oligomeric complexes. A small subset of the proteins also exhibited multiple elution peaks or broad elution profiles. For example, as shown later in Figure 6B, components of the non-homologous end joining (NHEJ) complex PARP1, Ku80, and Ku70, a known heterooligomeric complex, had three distinct peaks. The dominant first peak had a Mapp of ≈499 kDa and a second and third peak at ≈ 296 kDa and ≈ 104 kDa, respectively. The peaks at the later fractions (fraction 29 and 31) also co-elute with PCNA. This may indicate the presence of both the monomeric and/or variety of oligomeric sub-complexes and suggest their involvement in multiple functions. Such proteins performing additional activities are referred to as “moonlighting” proteins or multitasking,[43] and our experimental system can identify such proteins. This is biologically relevant; however, high-throughput analysis of proteins existing in multiple oligomerization states requires deconvolution of the chromatogram into its constituent peaks. This was not possible in this study due to insufficient resolution of the SEC column. As a result, we used global maximum of the MS1 intensity of each identified protein as the peak for Mapp calculation and protein complex prediction. We compared our Mapp values with some published apparent masses in the literatures,[2,37,42,44–46] and found good agreement between our results and published masses (Table 1).
Figure 6.
Validation of NHEJ and NuRD complex. A) SEC immunoblot profile from biological replicate 1 demonstrating a co-elution of PARP1, Ku80, and Ku70. Nuclear lysate control (15 μg) to demonstrate protein expression levels. B) Elution profiles of PARP1, PCNA, Ku80, and Ku70 components across SEC fractions. The immunoblot profile matches the intensity profiles for each protein across SEC fractions. C) Immunoblot of Ku70 IP staining with antibodies against PARP1, Ku70, PCNA, and Ku80. D) STRING interactome of PARP1 indicating an interaction with PCNA. E) SEC immunoblot profile from Bio1 showing a co-elution of CHD4 and HDAC1 components of the NuRD complex. Nuclear lysate control (15 μg) to demonstrate protein expression. F) SEC elution profiles of CHD4, HDAC1, and RBBP7 components of the NuRD complex. G) Immunoblot of HDAC1 IP staining with antibodies against HDAC1 and Mi2 (CHD3/4). Similar results were observed for Bio2
Table 1.
Predicted sizes compared to experimentally determined complexes. Numbers in parenthesis after complex names indicate the reference/s of the known complex size. Mapp; Apparent mass determined based on SEC elution profile of the proteins.
| Subunit (Complex name) | Known size [kDa] (Literature) | Mapp [kDa] (This study) |
|---|---|---|
| ENL (Super Elongation Complex) (44) | 337 | 649 |
| EZH2 (PRC2) (46) | 444 | 500 |
| HDAC3 (NCOR) (45) | 501 | 649 |
| CHD4 (NuRD) (37) | 505 | 649 |
| SIN3a (SIN3a/HDAC) (37) | 398 | 649 |
| PSA1 (20S Proteasome) (42) | 700 | 500 |
| TRAP1 (19S proteasome) (42) | 900 | 649 |
| TRAP1 (19S proteasome) (42) | 301 | 228 |
| SSRP1 (FACT) (2) | 195 | 228 |
| MSH6 (MSH2/6-PCNA) (2) | 285 | 296 |
Figure 5B depicts the distribution of the predicted monomer (Mmono) and the Mapp of all the identified proteins in Bio1 and Bio2. We observed that the distribution of the Mmono was con-centrated more in the lower mass ranges than the distribution of the Mapp. When we quantitated the number of proteins in different molecular weight ranges for the monomeric weight and apparent weight, we found that while over 50% of the proteins have the Mmono less than 40 kDa, over 50% of the proteins have Mapp ranges greater than 500 kDa. We calculated the apparent ratio (Rapp) of proteins as the ratio of the Mapp to the monomeric mass (Mmono), and used the Rapp score to predict whether a protein is eluting as a complex or a monomer.[4,26,47] Previously, we have shown that unless a protein is a rod-shape, the Rapp for a monomer would never be greater than approximately 1.5.[4,26,47] Therefore, proteins with an Rapp of 2 or higher in both the biological replicates with elution patterns within two fractions between the two independent SEC fractionations were considered members of a protein complex. More than 70% of the proteins were identified as putative complex members (Figure 5C and Table S2, Supporting Information). We acknowledge that the Rapp has limitations, and in some cases, we are likely to misidentify large proteins as monomeric in cases where they interact with proteins that are relatively small. However, our Rapp predictions agree very well with the oligomerization state of known protein complexes (Table 1). For example, we observe that proteins such as ENL have an Mapp around 649 kDa, which is consistent with it being a member of the super elongation complex (ENL/ELL/EAF/PTEFb/AFF[44]).
3.7. Protein Complex Validation
To complement LC–MS profiles, we performed immunoblot analysis of several protein complex subunits identified in this study (Figure 6). We identified the DNA damage proteins PARP1, PCNA, Ku80, and Ku70. PARP1, Ku80, and Ku70 co-eluted together from the column at a molecular weight above 400 kDa predominately, however, PCNA did not share a common elution profile in the high molecular weight fraction (Figure 6A and B). The elution profile observed by immunoblot closely correlated with the LC–MS profile based on protein abundance (Figure 6A and B). Signals at the low molecular weight range (fraction 31 or higher) likely represent monomeric forms as complexes would be predicted to elute in a higher molecular weight fraction. We also wanted to determine if co-eluting proteins in our profiling experiment could be identified independently by co-IP. co-IP with Ku70 antibody demonstrated interactions with Ku80 and PARP1 (Figure 6C). Interestingly, we could not detect PCNA in our Ku70 co-IP. Based on these observations, it is tempting to think that PARP1, Ku70, and Ku80 might belong to a protein complex without PCNA, which is also confirmed by the results of Ku70 IP. However, evidence of a PARP1/PCNA interaction has also been shown previously[48] (Figure 6D). Therefore, this might also be a result of a low abundant interaction in comparison to the amount of Ku70 expressed in the cells. In addition, we also identified the members of the NuRD complex in our analysis with an Mapp of approximately 500 kDa (Figure 6E and F). This corresponds to the approximate mass of a variation of the NuRD complex composed of HDAC1, CHD4, RBBP7, MTA1, P66, and MDB3.[30] The immunoblot analysis of the SEC co-fractionation for CHD3/4 and HDAC1 (Figure 6D) were consistent with the LC–MS profiles (Figure 6E). To test this further, we next performed co-IP experiments to examine whether CHD4 and HDAC1 can be detected as components of NuRD complex. The IP product of HDAC1 pull-down was blotted against HDAC1 and CHD3/4 antibodies (Figure 6G) and validated that these proteins are indeed in a complex. The co-IPs of both the complexes together with the immunoblots of SEC fractions and LC–MS intensity profiles confirm that the method outlined here can be used to identify nuclear complexes. Collectively, these results validate the use of SEC co-fractionation of native proteins with the quantitative LC– MS for proteome-wide profiling of proteins and thus to predict the existence of distinct forms of protein complexes.
3.8. Post-Translational Modifications of Nuclear Protein Complexes
Post-translational modifications (PTMs) of proteins play a critical role in protein oligomerization and sub-cellular location. Phosphorylation and acetylation can modulate many structural properties or binding affinities between proteins by creating or removing binding sites.[25] In addition, evidence of a cross-talk between phosphorylation and lysine acetylation also has been reported.[49]
To gather some insights into whether PTMs can affect the way proteins interact to form complexes, we analyzed the LC–MS/MS dataset for mapping any phosphorylation and acetylation using MaxQuant. For high confidence, we restricted our PTM analysis to peptides and sites that have been identified in both the biological replicates. We identified 162 phosphorylated peptides matching to 108 proteins that overlapped between the two biological replicates (Table S3, Supporting Information). We also identified 93 acetylated peptides that mapped to 87 proteins that were also common between the replicates (Table S3, Supporting Information). Of the 93 acetylated peptides, 25 peptides and 18 proteins were also identified as phosphorylated. Identification of low number of modified peptides was expected as we did not perform any enrichment of the modified peptides. Intensity profiles of the modified peptides matched with the intensity profiles of the proteins, and modified peptides were mostly detected in high mass SEC fractions (Figure S6, Supporting Information). This may indicate the existence of modified proteins in a complex form. Data also indicated that if the protein abundance increases, the probability of identifying modified peptides also increases (Figure S6, Supporting Information). Many of these modified proteins were nuclear proteins and were functionally diverse including mRNA processing, translational elongation, translational termination, protein transport, RNA splicing, mitosis, chromatin modification, nucleosome assembly, glycolysis, and protein degradation (Table S3, Supporting Information). The neuroblast differentiation associated protein (AHNAK) was phosphorylated as well as acetylated in multiple sites. AHNAK is a large (700 kDa) structural scaffold protein and may play a role in neuronal differentiation and tumor metastasis. The eukaryotic translation initiation (EIF3C, EIF3F, EIF3CL, EIF5B) and elongation factor subunits (EEF1D) were also identified as phosphorylated and acetylated proteins. Interaction between AHNAK and EEF1D has been reported previously.[35,50]
4. Discussion
In this study, we applied an integrated SEC co-fractionation of native proteins with quantitative MS-based proteomics to obtain a system-wide analysis of the diverse nuclear protein complexes using GBM T98G model cells. GBM is one of the most devastating human cancers in adults. It comprises 15.1% of all primary brain tumors and 46.1% of primary malignant brain tumors in the United States.[51] The use of MS-based proteomics to characterize protein complexes in GBM cells provides the cellular context of protein interactions and the pathways that integrate them. This information is important to develop new strategies or improve the existing strategies for the treatment of GBM patients.
We identified 1794 proteins present in both biological replicates of GBM T98G nuclear lysate. GO analysis indicates that they are enriched for nuclear proteins and biological functions such as DNA damage repair and replication. We used our Mapp measurements of proteins in the nuclear fraction to predict whether identified proteins exist in stable protein complexes using Rapp.[4,22] More than 1200 proteins reported in this study had an Rapp ≥ 2 suggesting the majority of nuclear proteins are in a complex. Over 50% of these complexes have an Mapp larger than 500 kDa suggesting that most nuclear proteins exist as large multi-subunit complexes. Protein profiles revealed that a subset of the proteins had more than one distinct peak, which suggests that a protein can exist in multiple oligomeric states in the cell and may participate in multiple cellular functions. Hierarchical clustering analysis by the similarity of elution profiles identified proteins that co-elute with known components of protein complexes. These clusters may represent previously undetected interacting partners or entirely new clusters. Using these profiles, we have demonstrated examples of proteins that form multiple, separable complexes and confirmed these findings in detail for npBAF, SIN3A, and NuRD interaction networks. Using multiple examples, we have confirmed that the conclusions derived from LC–MS-based protein identification, quantitation, and complex predictions are supported by parallel antibody-based protein detection using immunoblot. Furthermore, we performed independent co-IP experiments to validate two complexes identified by their elution profiles, an NHEJ complex comprised PARP1, Ku70, and Ku80, and the NuRD complex comprised HDAC1 and CHD4.
4.1. The Importance of the Nucleus and its Challenges
The nucleus, the control center of the cell,[52] houses three meters of DNA and is responsible for DNA replication, transcription, DNA damage repair, and ribosomal assembly,[53] all critical processes for determining the cell’s function and fate. Within the nucleus, DNA is organized into a higher order structure known as chromatin that is composed of DNA wrapped around his-tone protein octamers. Over the past few decades, research has shown that this chromatin structure is highly regulated allowing for transcription, replication, and repair to occur. Proper regulation of chromatin structure is essential for the cell’s function and fate. In these processes, proteins are critical, and unlike signaling mechanisms in the cytoplasm, the proteins that regulate chromatin structure primarily work in the context of complexes.[54]
Fink et al.[27] estimate that nuclear proteins comprise approximately 14% of the proteome. The low abundance of nuclear proteins can make it challenging to study them. As observed by our immunoblot (Figure 1D), our nuclear preparation was enriched for nuclear proteins but also contaminated with cytosolic proteins. The GO analysis in particular suggests that we have an enrichment for ribosomal, mitochondrial, and many cytosolic proteins in addition to nuclear proteins. However, this is common, and likely due to the high abundance of cytosolic, mitochondrial, and ribosomal proteins in the cell. It might also be possible that many proteins partition between the cytosol and the nucleus. Torrente et al., utilized three different approaches to isolate nuclear proteins and identified only 30 to 45% nuclear proteins.[55] In any case, this level of enrichment allows for a much more thorough identification of nuclear complex profiling than using WCL.
Our experimental system is expected to detect proteins that are partitioned between the cytosol and the nucleus; indeed, there were a number of known cytosolic proteins as apparent subunits of nuclear complexes. It might be possible that these cytosolic proteins may either be regulated by nuclear localization, or they may have independent functions in the nucleus. As an example, we identified several 20S proteasome subunits, which are known to have a nuclear localization signal.[41] The positive and negative charge clusters can serve to regulate the translocation of these proteasomes from the cytoplasm to the nucleus, and tyro-sine phosphorylation, which adds the negative charge to the cluster, has been suggested to play an additional role.[56] The β-type 20S proteasome subunits encoded by PSMB9 (LMP2), PSMB10 (MECL-1), and PSMB8 (LMP7) genes are known as immunoproteasome (β1i, β2i, and β5i) subunits. We identified all three subunits with the other 20S proteasomes (Figure 3C and Table S2, Supporting Information). GFP-labeled LMP2 was shown to incorporate with the immunoproteasomes, which was transported slowly and unidirectionally to the nucleus.[41] Increased nuclear localization of proteasomes in HeLa and PtK2 cells during the progression of cell cycle has also been reported.[57] Therefore, it is possible that our experimental system may be able to identify cytosolic proteins that also function in the nucleus.
4.2. Validation of Disease-Relevant Complexes
HDACs are chromatin modifiers and play an important role in the epigenetic regulation of gene expression.[58] Evidence suggests a link between misregulated HDAC activity and neurode-generative diseases such as Parkinson’s disease.[59] Inhibition of misregulated HDAC activities is an effective strategy for therapeutic treatments, and numerous studies are focusing to reveal HDAC inhibitor specificities and molecular actions.[37,60] Therefore, mammalian HDAC family enzymes are emerging as important drug targets. However, very little is known about the specific molecular mechanism of HDAC inhibitors, and this molecular mechanism may involve the interaction of HDAC complexes and their PTMs. Our SEC co-fractionation, LC–MS profiling, and complementary co-IP experiments confirmed interaction of HDAC1/2 with the various components of SIN3A and NuRD complex components.
Additionally, histone methylation and demethylation processes are important for gene transcription and changes in histone methylation are also linked to many cancers and neurodegenerative diseases.[61] The lysine specific demethylase KDM1A is also implicated in brain function. RCOR1 has a specific role to repress transcription of neuron-specific genes in non-neuronal cells. SEC co-fraction of KDM1A and RCOR1 with HDAC1/2/3 confirm that both KDM1A and RCOR1 are integral components of the human deacetylase complex.[45,62]
4.3. Identification of Potential Novel Protein Complexes
The method we outlined here to profile nuclear complexes can be used not only to identify known complexes but also for the identification of novel complexes. Our protein search identified numerous unannotated proteins with an Rapp value greater than 2 in both replicates. One particular protein is DERP12. A monomer of DERP12 is 38 kDa, however, our analysis indicates that DERP12 is in a complex that is over 100 kDa. Interestingly, DERP12 has been reported to be differentially expressed in ovarian cancer following treatment with the chemotherapy cisplatin,[63] and DERP12 may serve as a biomarker for chemoresistance in ovarian cancers. Additionally, we identified the protein GPatch8 in our analysis to have an Rapp greater than 2. GPatch8, a 164 kDa RNA binding protein, has an Mapp over 500 kDa. According to the STRING database, GPatch8 is predicted to interact with Death Inducer-obliterator 1, while BioGRID indicates 40 interactions identified via high-throughput methods, however, very little has been validated. Interestingly, high-throughput genomic analyses in GBM suggest that high expression of GPatch8 correlates to a longer survival time.[64] Further characterization of these potential protein complexes is necessary, but this demonstrates how utilizing this approach can define novel complexes that may have biological significance.
Furthermore, our hierarchical clustering is another way to identify putative complexes. Proteins are clustered by their elution profile, therefore those that cluster together are likely to be in a complex together. Again, further validation studies to confirm such interactions are necessary.
4.4. Limitations and Future Applications
While the Q Exactive HF Orbitrap has greater sensitivity than the older generations of mass spectrometers, we indeed experienced some limitations of peptide detection. This is likely due to the low expression levels of certain nuclear proteins. For example, the Polycomb proteins that were identified by immunoblot analysis and co-IP MS were not detected in this study. This is likely due to the high abundance of other nuclear proteins as well as contamination from the cytosol and the mitochondria. To overcome such limitation, it will be necessary to continue improving nuclear enrichment protocols to remove background and release more chromatin-bound proteins. Furthermore, there are additional challenges of predicting nuclear and other sub-cellular protein complexes from the profiling data. As we observed in this study, a significantly large number of proteins peaked in the column void due to their large complex sizes and the limited mass range of the SEC column used. While we were able to profile smaller and/or sub-complexes of these larger complexes, as it is common for nuclear proteins to be a part of several different complexes,[37] further experiments with a larger SEC column such as Superose 6 or orthogonal separation of SEC fractions using other chromatographic separation techniques, such as ion exchange chromatography and/or hydrophobic interaction chromatography, are necessary to resolve the larger complexes. Finally, when predicting the Mapp of proteins, it is important to consider that an Rapp ≥ 2 can be an artifact of large PTMs, such as poly-ubiquitin. Such modifications can alter the molecular weight of the protein and may suggest it is in a larger complex. Other analyses such as IP or MS-based PTM identification can aid in separating out highly modified proteins from those in complexes.
It has been found that chromatin regulators are critical in proper development, cell lineage specification, DNA damage repair, and much more. In some instances, these regulating complexes have subunits with numerous paralogs creating compositional and functional diversity. The methodology outlined in this article allows us to profile and identify cell-type specific nuclear complexes. Being able to identify which composition is in a particular cell type is the starting point to understanding the biological function of that specific complex and how its function may vary from different compositions of the same complex. Furthermore, in various cancers, it has been demonstrated that nuclear proteins can have different interacting partners altering transcriptional function and driving oncogenesis. This technique will allow us to define complexes of interest in healthy cells and then observe complex compositional changes in the disease state. This can enable us to identify oncogenic complexes.
In conclusion, we expect that this study provides useful data for future studies that are more focused or targeted to specific protein complexes in humans. This study also provides an initial framework to investigate how phosphorylation and acetylation affect protein complex formation and function. Clustering proteins into different groups based on the similar elution profiles allows the identification of known protein complexes as well as many currently unknown protein complexes. This SEC approach can be used and expanded to characterize the localization and dynamics of nuclear protein complexes in cells and tissues as a function of different physiological or disease states, and to obtain insights into potential oncogenic mechanisms.
Supplementary Material
Significance Statement.
In this study, we applied a protein complex analysis technique for human cells that couples cell fractionation and chromatographic separation of endogenous protein complexes with label-free LC–MS/MS protein abundance profiling across chromatography fractions. Applying this technique to crude nuclear fractions isolated from human Glioblastoma Multiforme (grade IV glioma) cell line T98G, we identified hundreds of proteins with apparent masses indicative of the existence of stable multi-subunit protein complexes. Many of these protein complexes appear to be novel. Several nuclear protein complexes identified by this method were confirmed by independent Western Blot and co-immunoprecipitation (co-IP) experiments. Interactions of PARP1 with Ku70/Ku80, and HDAC1 with CHD4, were validated using co-IP and Western Blot analyses. This study shows the utility of using label-free MS1 profiles to simultaneously analyze the oligomerization states of endogenous nuclear proteins to form complexes, and sets the stage for the addition of other techniques that should allow the determination of protein complex composition and dynamics under different conditions. The whole pipeline is simple to implement and can be expanded to any organisms and cell types to predict the existence of distinct forms of nuclear protein complexes.
Acknowledgements
K.E.C., E.C.D., and U.K.A. designed research; K.E.C. and U.K.A. performed fractionation experiments; K.E.C. performed sample preparation, co-IP, and immunoblotting experiments; V.H. and U.K.A. collected all LC–MS data; V.H., K.E.C., T.S.P., and U.K.A. performed LC–MS data analysis and interpretation; K.E.C., E.C.D., and U.K.A. wrote the Manuscript. E.C.D. and U.K.A. mentored and supervised the project. All authors read the manuscript, made their comments, and approved the final version. This research was supported by #UL1 TR001108 from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award to ECD and UKA. KEC is supported by the Indiana CTSI Predoctoral Fellowship (grant #UL1TR001108). Size Exclusion Chromatography Fractionation and LC–MS experiments were performed at the Purdue Proteomics Facility, Bindley Bioscience Center.
Abbreviations
- AP-MS
affinity purification MS
- BAF
Brg-associated factors
- CBX8
chromobox homolog 8
- co-IP
co-immunoprecipitation
- CP
catalytic core particle
- DAnTE
Data Analysis and Extension Tool
- FA
formic acid
- FPLC
fast protein LC
- GBM
glioblastoma multiforme
- GO
gene ontology
- HDAC
histone deacetylase complex
- KDM1A
lysine demethylase 1a
- Mapp
apparent mass
- Mmono
monomeric mass
- MRN
MRE11/RAD50/NBN
- NBN
nibrin
- NHEJ
non-homologous end joining
- npBAF
neural progenitor BAF
- NuRD
nucleosome remodeling deacetylase
- PBAF
Polybromo Brg-associated factors
- PCC
Pearson correlation coefficients
- PTM
post-translational modification
- Rapp
apparent ratio
- RP
regulatory particle
- SEC
size exclusion chromatography
- WCL
whole cell lysate
- Y2H
yeast two-hybrid
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Katelyn E. Connelly, Department of Medicinal Chemistry and Molecular Pharmacology, Purdue University, 201 S. University Street, 47907, West Lafayette, IN, USA
Emily C. Dykhuizen, Department of Medicinal Chemistry and Molecular Pharmacology, Purdue University, 201 S. University Street, 47907, West Lafayette, IN, USA
Uma K. Aryal, Purdue Proteomics Facility, Bindley Biosciences Center, Discovery Park, Purdue University, 1203 W. State Street, 47907, West Lafayette, IN, USA
References
- [1].a) Hartwell LH, Hopfield JJ, Leibler S, Murray AW, Nature 1999, 402, C47; [DOI] [PubMed] [Google Scholar]; b) Good MC, Zalatan JG, Lim WA, Science 2011, 332, 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ruepp A, Waegele B, Lechner M, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Mewes HW, Nucleic Acids Res. 2010, 38, D497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ, Nat. Genet 2008, 40, 1413. [DOI] [PubMed] [Google Scholar]
- [4].Aryal UK, Xiong Y, McBride Z, Kihara D, Xie J, Hall MC, Szymanski DB, Plant Cell 2014, 26, 3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Strumillo M, Beltrao P, Bioorg. Med. Chem 2015, 23, 2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nooren IM, Thornton JM, EMBO J. 2003, 22, 3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Marsh JA, Hernandez H, Hall Z, Ahnert SE, Perica T, Robinson CV, Teichmann SA, Cell 2013, 153, 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Menche J, Sharma A, Kitsak M, Ghiassian SD, Vidal M, Loscalzo J, Barabasi AL, Science 2015, 347, 1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jansen R, Yu H, Greenbaum D, Kluger Y, Krogan NJ, Chung S, Emili A, Snyder M, Greenblatt JF, Gerstein M, Science 2003, 302, 449. [DOI] [PubMed] [Google Scholar]
- [10].Rolland T, Tasan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, Kamburov A, Ghiassian SD, Yang X, Ghamsari L, Balcha D, Begg BE, Braun P, Brehme M, Broly MP, Carvunis AR, Convery-Zupan D, Corominas R, Coulombe-Huntington J, Dann E, Dreze M, Dricot A, Fan C, Franzosa E, Gebreab F, Gutierrez BJ, Hardy MF, Jin M, Kang S, Kiros R, Lin GN, Luck K, MacWilliams A, Menche J, Murray RR, Palagi A, Poulin MM, Rambout X, Rasla J, Reichert P, Romero V, Ruyssinck E, Sahalie JM, Scholz A, Shah AA, Sharma A, Shen Y, Spirohn K, Tam S, Tejeda AO, Trigg SA, Twizere JC, Vega K, Walsh J, Cusick ME, Xia Y, Barabasi AL, Iakoucheva LM, Aloy P, De Las Rivas J, Tavernier J, Calderwood MA, Hill DE, Hao T, Roth FP, Vidal M, Cell 2014, 159, 1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Altelaar AF, Munoz J, Heck AJ, Nat. Rev. Genet 2013, 14, 35. [DOI] [PubMed] [Google Scholar]
- [12].a) Dunham WH, Mullin M, Gingras AC, Proteomics 2012, 12, 1576; [DOI] [PubMed] [Google Scholar]; b) Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B, Nat. Biotechnol 1999, 17, 1030. [DOI] [PubMed] [Google Scholar]
- [13].a) Politis A, Stengel F, Hall Z, Hernandez H, Leitner A, Walzthoeni T, Robinson CV, Aebersold R, Nat. Methods 2014, 11, 403; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Walzthoeni T, Leitner A, Stengel F, Aebersold R, Curr. Opin. Struct. Biol 2013, 23, 252. [DOI] [PubMed] [Google Scholar]
- [14].a) Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St Onge P, Ghanny S, Lam MH, Butland G, Altaf-Ul AM, Kanaya S, Shilatifard A, O’Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF, Nature 2006, 440, 637; [DOI] [PubMed] [Google Scholar]; b) Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM, Nature 2000, 403, 623. [DOI] [PubMed] [Google Scholar]
- [15].Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, Hyman AA, Mann M, Cell 2015, 163, 712. [DOI] [PubMed] [Google Scholar]
- [16].a) Jansen R, Gerstein M, Curr. Opin. Microbiol 2004, 7, 535; [DOI] [PubMed] [Google Scholar]; b) Wodak SJ, Pu S, Vlasblom J, Seraphin B, Mol. Cell Proteomics 2009, 8, 3. [DOI] [PubMed] [Google Scholar]
- [17].Deane CM, Salwinski L, Xenarios I, Eisenberg D, Mol. Cell Proteomics 2002, 1, 349. [DOI] [PubMed] [Google Scholar]
- [18].Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G, Nature 2006, 440, 631. [DOI] [PubMed] [Google Scholar]
- [19].Werner JN, Chen EY, Guberman JM, Zippilli AR, Irgon JJ, Gitai Z, Proc. Natl. Acad. Sci. USA 2009, 106, 7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].a) Gardiner K, Behav. Genet 2006, 36, 439; [DOI] [PubMed] [Google Scholar]; b) Huang CJ, Das U, Xie W, Ducasse M, Tucker HO, Aging (Albany NY) 2016, 8, 3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kerppola TK, Annu. Rev. Biophys 2008, 37, 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Aryal UK, McBride Z, Chen D, Xie J, Szymanski DB, J. Proteomics 2017, 166, 8. [DOI] [PubMed] [Google Scholar]
- [23].a) Dong M, Yang LL, Williams K, Fisher SJ, Hall SC, Biggin MD, Jin J, Witkowska HE, J. Proteome. Res 2008, 7, 1836; [DOI] [PubMed] [Google Scholar]; b) Kristensen AR, Gsponer J, Foster LJ, Nat. Methods 2012, 9, 907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, Wang PI, Boutz DR, Fong V, Phanse S, Babu M, Craig SA, Hu P, Wan C, Vlasblom J, Dar VU, Bezginov A, Clark GW, Wu GC, Wodak SJ, Tillier ER, Paccanaro A, Marcotte EM, Emili A, Cell 2012, 150, 1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kirkwood KJ, Ahmad Y, Larance M, Lamond AI, Mol. Cell Proteomics 2013, 12, 3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Liu X, Yang WC, Gao Q, Regnier F, Chromatogr J. A 2008, 1178, 24. [DOI] [PubMed] [Google Scholar]
- [27].Fink JL, Karunaratne S, Mittal A, Gardiner DM, Hamilton N, Mahony D, Kai C, Suzuki H, Hayashizaki Y, Teasdale RD, Genome Biol. 2008, 9, R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hodges C, Kirkland JG, Crabtree GR, Cold Spring Harb. Perspect Med. 2016, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Connelly KE, Dykhuizen EC, Biochim. Biophys. Acta 2017, 1860, 233. [DOI] [PubMed] [Google Scholar]
- [30].Ho L, Crabtree GR, Nature 2010, 463, 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].a) Broustas CG, Lieberman HB, Radiat Res 2014, 181, 111; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mack SC, Hubert CG, Miller TE, Taylor MD, Rich JN, Nat. Neurosci 2016, 19, 10; [DOI] [PMC free article] [PubMed] [Google Scholar]; Solis-Paredes M, Eguia-Aguilar P, Chico-Ponce de Leon F, Sadowinski-Pine S, Perezpena-Diazconti M, Arenas-Huertero F, Childs. Nerv. Syst 2014, 30, 123. [DOI] [PubMed] [Google Scholar]
- [32].a) Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M, Mol. Cell Proteomics 2014, 13, 2513; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cox J, Mann M, Nat. Biotechnol 2008, 26, 1367; [DOI] [PubMed] [Google Scholar]; c) Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M, J. Proteome Res. 2011, 10, 1794. [DOI] [PubMed] [Google Scholar]
- [33].Polpitiya AD, Qian WJ, Jaitly N, Petyuk VA, Adkins JN, Camp DG 2nd, Anderson GA, Smith RD, Bioinformatics 2008, 24, 1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD, Nucleic Acids Res. 2017, 45, D183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ,von Mering C, Nucleic Acids Res. 2017, 45, D362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ronan JL, Wu W, Crabtree GR, Nat. Rev. Genet 2013, 14, 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Delcuve GP, Khan DH, Davie JR, Clin. Epigenetics 2012, 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].a) D’Amours D, Jackson SP, Nat. Rev. Mol. Cell Biol 2002, 3, 317; [DOI] [PubMed] [Google Scholar]; b) Wen J, Cerosaletti K, Schultz KJ, Wright JA, Concannon P, Oncogene 2013, 32, 4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Cilli D, Mirasole C, Pennisi R, Pallotta V, D’Alessandro A, Antoccia A, Zolla L, Ascenzi P, di Masi A, PLoS One 2014, 9, e114651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Finley D, Annu. Rev. Biochem 2009, 78, 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wojcik C, DeMartino GN, Int. J. Biochem. Cell Biol 2003, 35, 579. [DOI] [PubMed] [Google Scholar]
- [42].Tanahashi N, Murakami Y, Minami Y, Shimbara N, Hendil KB, Tanaka K, J. Biol. Chem 2000, 275, 14336. [DOI] [PubMed] [Google Scholar]
- [43].Butler GS, Overall CM, Nat. Rev. Drug Discov 2009, 8, 935. [DOI] [PubMed] [Google Scholar]
- [44].Luo Z, Lin C, Shilatifard A, Nat. Rev. Mol. Cell Biol 2012, 13, 543. [DOI] [PubMed] [Google Scholar]
- [45].You A, Tong JK, Grozinger CM, Schreiber SL, Proc. Natl. Acad. Sci. USA 2001, 98, 1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Aranda S, Mas G, Di Croce L, Sci. Adv 2015, 1, e1500737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gao Q, Madian AG, Liu X, Adamec J, Regnier FE, J Chromatogr. A 2010, 1217, 7661. [DOI] [PubMed] [Google Scholar]
- [48].Sekimoto T, Oda T, Pozo FM, Murakumo Y, Masutani C, Hanaoka F, Yamashita T, Mol. Cell 2010, 37, 79. [DOI] [PubMed] [Google Scholar]
- [49].van Noort V, Seebacher J, Bader S, Mohammed S, Vonkova I, Betts MJ, Kuhner S, Kumar R, Maier T, O’Flaherty M, Rybin V, Schmeisky A, Yus E, Stulke J, Serrano L, Russell RB, Heck AJ, Bork P, Gavin AC, Mol. Syst. Biol 2012, 8, 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wan C, Borgeson B, Phanse S, Tu F, Drew K, Clark G, Xiong X, Kagan O, Kwan J, Bezginov A, Chessman K, Pal S, Cromar G, Papoulas O, Ni Z, Boutz DR, Stoilova S, Havugimana PC, Guo X,Malty RH, Sarov M, Greenblatt J, Babu M, Derry WB, Tillier ER,Wallingford JB, Parkinson J, Marcotte EM, Emili A, Nature 2015, 525, 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C,Wolinsky Y, Kruchko C, Barnholtz-Sloan JS, Neuro Oncol. 2015, 17 Suppl 4, iv1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Spector DL, Annu. Rev. Cell Biol 1993, 9, 265. [DOI] [PubMed] [Google Scholar]
- [53].Lamond AI, Earnshaw WC, Science 1998, 280, 547. [DOI] [PubMed] [Google Scholar]
- [54].Kadoch C, Crabtree GR, Sci. Adv 2015, 1, e1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Torrente MP, Zee BM, Young NL, Baliban RC, LeRoy G, Floudas CA, Hake SB, Garcia BA, PLoS One 2011, 6, e24747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Tanaka K, Yoshimura T, Tamura T, Fujiwara T, Kumatori A, Ichihara A, FEBS Lett. 1990, 271, 41. [DOI] [PubMed] [Google Scholar]
- [57].Palmer A, Mason GG, Paramio JM, Knecht E, Rivett AJ, Eur. J. Cell Biol. 1994, 64, 163. [PubMed] [Google Scholar]
- [58].Bolden JE, Peart MJ, Johnstone RW, Nat. Rev. Drug Discov. 2006, 5, 769. [DOI] [PubMed] [Google Scholar]
- [59].Garber K, Nat. Biotechnol 2007, 25, 17. [DOI] [PubMed] [Google Scholar]
- [60].Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M, Science 2009, 325, 834. [DOI] [PubMed] [Google Scholar]
- [61].Maes T, Mascaro C, Ortega A, Lunardi S, Ciceri F, Somervaille TC, Buesa C, Epigenomics 2015, 7, 609. [DOI] [PubMed] [Google Scholar]
- [62].Joshi P, Greco TM, Guise AJ, Luo Y, Yu F, Nesvizhskii AI, Cristea IM, Mol. Syst. Biol 2013, 9, 672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Song J, Shih Ie M, Chan DW, Zhang Z, Neoplasia 2009, 11, 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Xiang Y, Zhang CQ, Huang K, BMC Bioinformatics 2012, 13, S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the raw LC–MS/MS data associated with these experiments have been deposited in Mass Spectrometry Interactive Virtual Environment (http://massive.ucsd.edu) with ID MSV000081520.