Abstract
Among the age-dependent protein aggregation disorders, nine neurodegenerative diseases are caused by expansions of CAG repeats encoding polyglutamine (polyQ) tracts. We review the clinical, pathological, and biological features of these inherited disorders. We discuss insights into pathogenesis gleaned from studies of model systems and patients, highlighting work that informs efforts to develop effective therapies. An important conclusion from these analyses is that expanded CAG/polyQ domains are the primary drivers of neurodegeneration, with the biology of carrier proteins influencing disease-specific manifestations. Additionally, it has become apparent that CAG/polyQ repeat expansions produce neurodegeneration via multiple downstream mechanisms, involving both gain- and loss-of-function effects. This conclusion indicates that the likelihood of developing effective therapies targeting single nodes is reduced. The evaluation of treatments for premanifest disease will likely require new investigational approaches. We highlight the opportunities and challenges underlying ongoing work and provide recommendations related to the development of symptomatic and disease-modifying therapies and biomarkers that could inform future research.
Keywords: polyglutamine, neurodegeneration, trinucleotide repeat disorders, Huntington’s disease, spinal and bulbar muscular atrophy, spinocerebellar ataxia
INTRODUCTION
Repeat Expansion Diseases
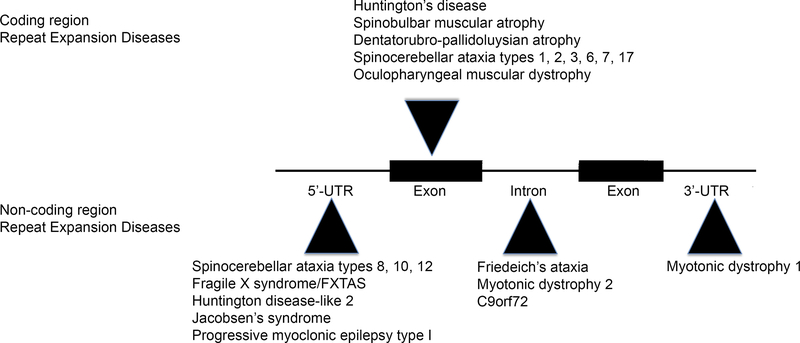
Microsatellites are repetitive DNA tracts that occur thousands of times throughout the human genome. The reiterated sequences making up these microsatellites vary in length from single nucleotides up to dodecamers or longer. These elements vary in length within the population, and expansions of microsatellites are known to cause a number of human diseases (Figure 1). The disease-causing repeat expansions may fall within noncoding sequences of genes, such as introns or untranslated regulatory regions flanking coding sequences (1). In such instances, the expanded microsatellite can become quite long, encoding thousands of repeated units. These elements induce cytotoxicity through diverse mechanisms. These include loss-of-function (LOF) effects via gene silencing and gain of function (GOF) via diverse mechanisms. The latter include the production of toxic RNAs from either sense or antisense strands that sequester critical factors involved in mRNA processing and the generation of potentially toxic aberrant polypeptides containing stretches of repetitive amino acids.
Figure 1.
Microsatellite repeat expansion diseases.
Microsatellite expansions of reiterated sequences of varying length occur in coding and non-coding regions of genes to cause human disease. Non-coding repeat expansions occur in 5’ or 3’ UTRs or introns and may encode thousands of repeated units. Coding region microsatellite expansions are restricted to trinucleotide repeats and tend to be more modest in length compared with non-coding region expansions. (Adapted from ref 150a).
Microsatellite expansions that fall within coding sequences are limited to trinucleotide repeats and tend to be modest in length compared with those occurring within noncoding regions. Almost all disease-causing coding region expansions consist of CAG repeats encoding glutamine. The exception is the GCG/polyalanine repeat expansion in the gene encoding polyA-binding protein-2, which is the cause of oculopharyngeal muscular dystrophy. We focus on the nine neurodegenerative disorders caused by expansions of polyglutamine (polyQ) tracts, including spinal and bulbar muscular atrophy (SBMA), Huntington’s disease (HD), dentatorubropallidoluysian atrophy (DRPLA), and six autosomal dominant forms of spinocerebellar ataxia (SCA1, 2, 3, 6, 7, and 17). As a mechanism of disease, polyQ repeat expansion appears to be unique to humans.
PolyQ disorders share several important clinical and pathological characteristics despite the fact that their only common genetic feature is the pathogenic repeat expansion; the affected genes are otherwise unrelated (Table 1) (2). All are neurodegenerative disorders, with disease onset typical in midlife and slowly progressive phenotypes. All polyQ diseases exhibit threshold phenomena with very high or complete penetrance occurring once repeat numbers exceed disease-specific limits. While the mutant genes are widely expressed by many cells and tissues, these diseases exhibit preferential degeneration of distinct cell types. In each case, the expanded repeat is unstable and may increase in length as it is passed from one generation to the next, particularly with male transmission. Intergenerational repeat expansion underlies genetic anticipation, wherein disease manifestations occur at earlier ages in subsequent generations. The age of onset is inversely correlated with CAG tract length, but this relationship is complex and is more pronounced in some noncoding repeat disorders, notably myotonic dystrophy type 1. Less common cases of childhood-onset disease occur in children with very long repeats. Expanded polyQ mutations are thought to lead to cellular toxicity as a result of aberrantly conformed mutant proteins. Based on the dominant pattern of inheritance and other data, proteotoxicity unrelated to the normal function of the mutant protein (toxic GOF) is thought to be common to all of these disorders and a significant contributor to cellular dysfunction and death. Eight of the polyQ diseases are inherited in an autosomal dominant manner, with the penetrance of SBMA, caused by a CAG repeat expansion in the X-linked androgen receptor (AR) gene, depending upon male levels of circulating androgens. This aspect of SBMA pathogenesis leads to the disease occurring only in men.
Table 1.
Polyglutamine (polyQ) repeat expansion diseases
| Disease | Locus; gene | Protein | Normal Q tract length | Pathogenic polyQ tract | Normal function | Additional notes |
|---|---|---|---|---|---|---|
| SBMA | Xq11-q12; AR | Androgen receptor | 6–36 | 38–≤70 | Transcription factor | Hormone-dependent phenotype |
| HD | 4p16.3; HTT | Huntingtin | 6–35 | 36–121 | Likely scaffold protein | Reduced penetrance with ≤38 Qs |
| DRPLA | 12p13; ATN1 | Atrophin-1 | 3–38 | 49–88 | Transcriptional corepressor | |
| SCA1 | 6p23; ATXN1 | Ataxin-1 | 6–34 | 39–88 | Likely gene expression | Histidine interruptions in Q tracts ≥20 |
| SCA2 | 12q24; ATXN2 | Ataxin-2 | 14–31 | 32–77 | RNA metabolism | Most common normal allele contains interrupted CAG/CAA |
| SCA3 | 14q24-q31; ATXN3 | Ataxin-3 | 12–40 | 55–86 | Deubiquitinase | |
| SCA6 | 19p13; CACNA1A | Cav2.1 | 4–18 | 21–33 | Calcium channel | Bicistronic RNA generates transcription factor α 1ACT |
| SCA7 | 3p21-p12; ATXN7 | Ataxin-7 | 7–18 | 38–200 | SAGA complex | |
| SCA17 | 6p27; TBP | TATA-binding protein | 25–43 | 45–63 | Transcription factor |
Abbreviations: DRPLA, dentatorubropallidoluysian atrophy; HD, Huntington’s disease; SBMA, spinal and bulbar muscular atrophy; SCA, spinocerebellar ataxia.
Several salient clinical and pathological aspects of polyQ disorders are summarized in Table 2. Despite the commonalities noted above, each disorder is distinguished by the preferential vulnerability of specific cell populations, a feature that underlies distinctive clinical manifestations. In the central nervous system (CNS), neurons are key target cells, and cytotoxicity is manifested by synaptic loss, atrophy of dendritic arborizations, abnormal axonal swellings, and irregularities of nuclear contours. Intranuclear and cytoplasmic protein aggregates are often present and contain all or part of the disease-causing proteins, including the polyQ tract. Additional proteins may localize to these aggregates, including ubiquitin and ubiquitin-binding proteins, molecular chaperones, proteasome components, and, in the case of intranuclear inclusions, transcriptional coregulators. Frank neuron loss is associated with evidence of neuroinflammation, including the activation of microglia and astrocytes.
Table 2.
Clinical and pathological features of polyglutamine (polyQ) diseases
| Disease | Neuropathology | Clinical features |
|---|---|---|
| SBMA | Preferential site of vulnerability: lower motor neurons, skeletal muscle Loss of motor neurons of spinal cord and brainstem cranial nerve motor nuclei, except CNs III, IV, and VI Less severe involvement of dorsal root ganglia Skeletal muscle shows neurogenic and myopathic features Nuclear inclusions stain for androgen receptor and ubiquitin |
Males affected; females clinically asymptomatic Weakness, atrophy, and fasciculations of bulbar, facial, and limb muscles, with prominent facial fasciculations Progressive dsyphagia Deep tendon reflexes diminished; no pathological reflexes Diminished vibration sensation in legs Gynecomastia and testicular atrophy may be present |
| HD | Preferential site of vulnerability: striatum Disease progresses to involve entire brain, with significant gross atrophy (Vonsattel Grade 4) Striatal medium spiny neurons most affected; gliosis Neuronal nuclear and cytoplasmic aggregates that stain for huntingtin, polyQ, and ubiquitin |
Personality changes and mood disorders Progressive cognitive impairment Chorea Speech difficulties and dysphagia Impaired gait, balance, and coordination Epilepsy (in juvenile HD) |
| DRPLA | Preferential site of vulnerability: dentatorubral and pallidoluysian systems Neuron loss; gliosis in globus pallidus (especially lateral), dentate nucleus, and subthalamic nucleus Neuronal intranuclear inclusions that stain for atrophin-1, polyQ, and ubiquitin; diffuse staining for polyQ; cytoplasmic polyQ aggregates |
Clinical heterogeneity Onset age <20 years: epilepsy, myoclonus, progressive intellectual deterioration Onset age >20 years: cerebellar ataxia, choreoathetosis and dementia; clinically resembles HD or SCAs |
| SCAs | SCAs 1, 2, and 7: atrophy of basis pontis (pontine gray matter), middle cerebellar peduncles, cerebellar white matter and folia, and inferior olives SCA3: pontine atrophy, degeneration of dentate nucleus and substantia nigra; sparing of cerebellar cortex and olive SCAs 6 and 17: cerebellar cortical atrophy, especially anterior–superior vermis SCA17 pathological analysis more limited, with cortical, striatal, and thalamic involvement reported Spinal cord posterior columns and spinocerebellar tract show varying degrees of degeneration (especially in SCAs 1, 2, 7, and 17) Neuronal nuclear or cytoplasmic inclusions stain for disease-causing protein, polyQ, and ubiquitin |
All show ataxia of the trunk and extremities, gaze nystagmus, cerebellar dysarthria, ataxic speech specific features of specific disease types.**okay] SCA1: pyramidal tract signs, dysphagia, amyotrophy SCA2: slow eye movements, hyporeflexia, postural tremor, chorea SCA3: clinical heterogeneity, with weakness and spasticity, sensory loss, amyotrophy, ataxia, parkinsonism SCA6: pure cerebellar signs SCA7: retinal degeneration, oculomotor signs, extrapyramidal signs SCA17: dementia, parkinsonism, involuntary movements, hyperreflexia |
Abbreviations: CN, cranial nerve; DRPLA, dentatorubropallidoluysian atrophy; HD, Huntington’s disease; SBMA, spinal and bulbar muscular atrophy; SCA, spinocerebellar ataxia.
In addition to GOF proteotoxicity, polyQ tract expansions likely alter the normal function of the mutant proteins, possibly contributing to disease-specific manifestations. One striking example is the effect of CAG repeat length on AR function as a ligand-activated transcription factor (3). Variations in repeat length, even within the nonpathogenic range, influence the activity of the AR, with shorter repeats associated with more transcriptionally active receptors. This phenomenon is associated with a variety of phenotypic outcomes in men, including modifying risk for the development or progression of prostate cancer. As the CAG repeat increases in length, AR activity diminishes, with pathological expansions in people with SBMA associated with signs of partial androgen insensitivity, such as gynecomastia, testicular atrophy, and diminished fertility.
Some polyQ domains may have pathogenic effects via interactions with other genes, even when repeat numbers are below pathological thresholds. Elden and colleagues (4) discovered that yeast and Drosophila orthologs of ataxin-2 (ATXN2; the protein causing SCA2) modified the toxicity of TAR DNA binding protein 43 (TDP-43), an RNA binding protein mutated in familial amyotrophic lateral sclerosis that accumulates in both sporadic and familial disease. An association between intermediate-length ATXN2 alleles (≥27 Q) and amyotrophic lateral sclerosis was demonstrated in several North American and European cohorts (4–6). The mechanism underlying this effect remains unclear, although both proteins are implicated in RNA processing.
Disease Epidemiology and Costs
PolyQ diseases are rare disorders whose impact is disproportionate to their prevalence. Among polyQ diseases, the epidemiology of HD is the best studied. In the best-ascertained populations, in Western Europe and North America, prevalence estimates approximate 5–6/100,000 population, with considerable variation among studies (7, 8). Likely reflecting the availability of definitive genetic testing, a greater awareness of HD, and possibly longer survival of people with HD, recent prevalence estimates have tended to be higher. In limited studies, prevalence estimates in non-European populations are significantly lower. In some studies of Asian populations, prevalence estimates are an order of magnitude lower than estimates in populations of European descent. Fragmentary data suggest that the same is true for African populations. Relatively careful studies in the United Kingdom and Canada suggest prevalence rates of 12–15/100,000 among individuals of European descent. In a study based on the United Kingdom’s General Practice Research Database, prevalence approximates 12/100,000 (9). In British Columbia, the overall prevalence is estimated at approximately 13/100,000, with an estimated prevalence among those of European descent of approximately 17/100,000 and much lower estimated prevalence rates among individuals of Asian or First Nations descent (10).
Prevalence estimates for other polyQ diseases are sketchy. In a systematic review of prevalence studies, Ruano et al. (11) estimated that the worldwide average prevalence of dominantly inherited cerebellar ataxias is approximately 3/100,000. In their compilation, which was highly biased toward studies in industrialized nations, SCA3 was the most common diagnosis, followed by SCA2 and SCA6. Presumably reflecting founder effects, there are regional and national differences in the proportions of polyQ SCAs. In Japan, for example, the data of Maruyama et al. (12) suggest that SCA3 is the most common dominantly inherited SCA (~27%), followed by SCA6 (~21%), and DRPLA (~10%). Leaving aside HD and SCA3, it is likely that the prevalence rates of individual polyQ diseases are very low. One estimate for SBMA is that its prevalence in the United States is approximately 1–2/100,000, although a higher rate was suggested to occur in Japan due to the founder effect (13).
In the case of HD, prevalence is maintained partly by new mutations. The de novo mutation rate may be in the range of 5–10% of patients presenting for diagnosis (8, 14–16). Apparent de novo mutations have two sources: repeat expansion from a parent with a reduced penetrance allele (36–39 repeats) and repeat expansion from a male parent with an intermediate-length allele (IA; 27–35 repeats). The latter are likely particularly important in generating de novo mutations. IAs are relatively common in populations of European descent, occurring in approximately 6% of individuals. The larger the repeat within an IA, the more likely it is that an expansion into the pathological range will occur. European-descended populations have an increased likelihood of repeat expansions above the normal range, higher average repeat numbers, and a higher prevalence of haplotypes with predispositions for repeat expansions. These types of phenomena may occur with other repeat expansion diseases.
Despite their rarity, polyQ disorders have considerable social impact. Again, HD is the best-documented example. Divino et al. (17) used administrative data to estimate the direct medical costs of HD. A crude summary of their analysis is that the annual per capita direct medical costs are ~$30,000 per year. There is no estimate of indirect costs due to HD, but an older estimate of annual income lost in working-age patients with Parkinson’s disease is $39,000 (in 1994 dollars) per patient per year (18). Indirect costs related to informal caregiving and earning losses of family members are likely to be substantial. As most people with polyQ diseases experience disease onset in middle age, the cumulative social costs are likely to be in the range of billions of dollars per year. These dominant disorders likely cause substantial intergenerational negative effects in unaffected family members. The general clinical impression is that these diseases push affected families toward the bottom of the social ladder.
INSIGHTS INTO DISEASE MECHANISMS
A number of excellent reviews over the last decade highlight what we have come to understand about individual polyQ disorders. Rather than focus on each disease separately, we instead emphasize important concepts that have emerged from the study of this group of disorders. For each concept highlighted below, we discuss critical investigations that brought these ideas to the attention of the field.
The CAG/PolyQ Tract Is Sufficient to Cause Neurological Dysfunction but not to Cause Preferential Neuronal Vulnerability
The lack of sequence similarity, excluding the polyQ domain, among genes causing HD, SBMA, DRPLA, and SCAs 1, 2, 3, 6, 7, and 17; the existence of polyQ repeat-length thresholds for disease penetrance; and the positive correlation between the age of disease onset and repeat number all point to the conclusion that expanded repeat polyQ domains are the primary driver of neurodegeneration in this disease family. Experimental confirmation of this inference came from several murine model experiments. Transgenic mice expressing a fragment of expanded repeat exon 1 huntingtin (HTT) exhibited an aggressive phenotype, with some lines developing marked motor dysfunction and premature death within weeks to months of birth (19, 20). These lines exhibit widely distributed neuronal inclusion pathology and, in the most aggressive lines, significant neuronal loss (21). In a particularly informative experiment, Ordway et al. (22) created a novel polyQ disease model by inserting a long CAG repeat into the gene encoding hypoxanthine phosphoribosyl transferase (Hprt), which does not contain a polyQ domain. In mice, inactivation of the X-linked Hprt gene does not produce a phenotype. Mice heterozygous and hemizygous for expanded polyQ repeat Hprt develop a phenotype strongly reminiscent of HTT expanded repeat exon 1–fragment mice, with marked decline of motor function, seizures, early mortality, and widespread neuronal inclusion pathology. In a complementary analysis, Yamamoto et al. (23) used an inducible system to control the expression of a transgenic expanded repeat HTT exon 1 fragment. Stopping transgenic fragment expression after the development of motor and histological abnormalities resulted in phenotypic remission, including clearance of neuronal inclusion pathology. Analyses of these mice, as well as models of SCA1 (24) and SBMA (25), provided important evidence that expanded polyQ tracts induced CNS pathology not only through neuronal loss but also as a consequence of reversible dysfunction.
While the human genetic data and these mouse model experiments point to the determinate role of expanded polyQ domains as the primary pathological driver in this disease family, the nature of human pathologies and aspects of these murine model experiments indicate that other important factors influence disease-specific features. Each one of these diseases has a characteristic and relatively unique regional pattern of neurodegeneration. HD is characterized by early, prominent striatal degeneration; SBMA exhibits motor neuron degeneration and skeletal muscle pathology; and SCA7 is distinguished by retinal photoreceptor degeneration. This is unlike the expanded repeat exon 1 fragment HTT transgenic models, which exhibit diffuse pathology. These facts suggest that the mutant proteins are not passive carriers of neurotoxic expanded polyQ domains. Rather, features of the host protein likely influence regional and subregional patterns of neurodegeneration. Differential gene expression patterns likely contribute to differences in patterns of neurodegeneration. AR, for example, is expressed at relatively high levels in motor neurons and muscle, cell types that are significantly affected in SBMA (26). Expression patterns, however, cannot be the whole answer as there is discordance between gene expression and patterns of pathology in most polyQ disorders. For example, HTT apparently is expressed in all neurons, including neuronal populations resistant or relatively resistant to neurodegeneration (27, 28). It is likely that other features of carrier proteins influence pathogenesis. For example, several of these proteins function via participation in multiprotein complexes, and expanded polyQ domains likely alter the composition and function of these complexes. Experiments with SCA1 transgenic models indicate that impacts on these multiprotein complexes modulate neurodegeneration in a cell type–specific manner (29). In the polyQ SCAs, the vulnerability of cerebellar Purkinje neurons may be related to their unique metabolic and electrophysiogical properties and the high intrinsic excitability of these neurons (30). Another possible contributing factor to differing regional patterns of neurodegeneration could be combinations of GOF and LOF effects. While GOF effects are generally considered to be the most important general aspect of the neurotoxicity of expanded polyQ domains, LOF effects may also contribute to neurodegeneration. Because each gene causing polyQ disease has unique normal functions, a combination of LOF and GOF effects could contribute to pathological heterogeneity. In summary, the basic concept is that expanded polyQ domains drive neurodegeneration, but the specific features of each polyQ disorder are a function of the biology of the carrier protein.
PolyQ Aggregates Sequester Toxic Species but Also Exhibit Neurotoxic Properties
PolyQ disorders were discovered to exhibit characteristic intraneuronal inclusions as a result of efforts to develop murine genetic HD models. In 1997, Davies et al. (31) described huntingtin immunoreactive neuronal intranuclear inclusions (NIIs) in the R6 expanded repeat exon 1 model of HD. The subsequent evaluation of HD postmortem material with appropriate antisera disclosed both NIIs and neuronal cytoplasmic inclusions (32). Similar observations were made in other polyQ diseases (33). In retrospect, NIIs were present in electron micrographs of cortical biopsy specimens obtained from HD patients more than 20 years earlier (34).
The discovery of polyQ inclusions triggered a persistent debate over the pathogenic significance of inclusion pathology in these disorders. One hypothesis is that polyQ inclusions are important mediators of neuronal dysfunction and death (31). One suggested mechanism of neuronal dysfunction that may be mediated by NIIs is the sequestration of proteins, particularly transcription factors, that are important for nuclear function (see below). Both NIIs and cytoplasmic polyQ inclusions might have other pathogenic effects, for example, the physical disruption of normal cellular functions. An alternative hypothesis is that polyQ inclusions themselves are nonpathogenic and exert a protective role by sequestering smaller, toxic polyQ protein species (35). It was suggested also that both of these hypotheses could be correct: PolyQ inclusions might be protective in early disease phases but pathogenic in later phases (36). Finally, intracellular neuronal polyQ inclusions might simply be epiphenomena.
An influential experiment by Arrasate et al. (37) addressed some of these hypotheses. These researchers used an automated microscopy system to longitudinally evaluate the survival of large numbers of cultured striatal neurons transiently transfected with constructs containing expanded repeat exon 1 HTT fused with green fluorescent protein. The formation of neuronal inclusions was correlated with improved neuronal survival and reduced diffuse expression of mutant HTT, results consistent with the protective sequestration hypothesis. Complementary data also supported the protective sequestration hypothesis. Gutekunst et al. (38) described a poor correlation between the distribution of HTT immunoreactive neuronal inclusions and the regional pattern of pathology in HD. Kuemmerle et al. (39) examined inclusion formation in HD postmortem striatal specimens. Inclusions were sparse in the striatal medium spiny neurons that are most affected in HD. Both cytoplasmic inclusions and NIIs were abundant in a striatal interneuron population spared in HD. Mastroberardino et al. (40) crossed transgenic HTT expanded repeat exon 1 (R6/1) mice with tissue transglutaminase (TG2) knockout mice. The resulting R6/1tgTG2−/− mice exhibited longer survival, less neuronal degeneration, and a higher percentage of neurons containing NIIs.
As a result of these experiments and other work, the pendulum of opinion swung toward the protective sequestration hypothesis. More recent experiments conducted over the last 5 years, however, have breathed new life into the toxic inclusions hypothesis. Liu et al. (41) presented data suggesting that perinuclear inclusions containing mutant HTT disrupt the nuclear membrane and act as a trigger for neuronal death. In both transgenic HTT expanded repeat exon 1 (R6/2) mice and primary neuronal cultures, perinuclear inclusions were associated with cell cycle re-entry and neuronal death. Woerner et al. (42) presented data suggesting that cytoplasmic mutant HTT aggregates interfere with nucleocytoplasmic transport both of proteins and of RNAs. Bauerlein et al. (43) reported that HTT expanded polyQ exon 1 cytoplasmic aggregates cause endoplasmic reticulum (ER) dysfunction by disrupting ER membranes. These studies describe the deleterious effects of cytoplasmic aggregates, but other data support the concept of pathogenic NIIs in HD. Using sophisticated cell imaging methods, Li et al. (44) reported that NIIs distort chromatin and, via sticky interactions with key proteins, interfere with transcriptional regulation. Finally, the concept of both protective and pathogenic effects of inclusions received support from the work of Ramdzan et al. (45). Using a system analogous to that of Arrasate et al. (37), although not with neurons in primary culture but mainly AD293 cells, Ramdzan et al. presented data showing that an early effect of mutant HTT inclusions is to sequester smaller toxic species and mitigate apoptotic cell death, while a longer-term effect is to sequester other proteins, leading to a slower process of delayed necrotic cell death.
Despite more than two decades of work, much of it performed by highly accomplished investigators, the potential pathogenic role or roles of expanded polyQ inclusions is uncertain. As with other aspects of the polyQ field, the interpretation of experiments is clouded by the use of multiple differing systems, including both in vitro and in vivo models based on the overexpression of fragment constructs, and the frequent use of nonneuronal systems. As suggested originally by Tallaksen-Greene et al. and supported by the recent experiments by Ramdzan et al. (45), it is plausible that inclusions containing expanded polyQ HTT exhibit both pathogenic and protective qualities. There might also be multiple pathogenic mechanisms that are secondary to inclusions containing expanded polyQ. This concept is consistent with the general idea that expanded polyQ proteins injure neurons via pleiotropic effects on cell functions (see below).
The Nucleus Is an Important Site of PolyQ Toxicity
Despite robust evidence for the disruption of key cytoplasmic processes in polyQ disease, data from multiple diseases indicate that the nucleus is a critical site of toxicity. This notion was first established experimentally for SCA1. Building upon the observation that polyQ ataxin-1 (ATXN1) alters nuclear matrix structures (46), Klement et al. (47) characterized transgenic mouse lines expressing polyQ ATXN1, one of which contained an intact nuclear localization sequence (NLS) and the other with a disrupted NLS. NLS-mutant mice failed to develop the age-dependent Purkinje cell dendritic atrophy and ataxia that occurred in mice expressing polyQ ATXN1 with an intact NLS. Similar data demonstrating the importance of nuclear localization of the mutant protein subsequently emerged from the study of other polyQ disorders. In SBMA mouse and Drosophilia models, the presence of androgen is required to induce nuclear translocation of the polyQ AR and trigger cytotoxicity (48, 49). These observations were reproduced in cultured motor neurons, enabling genetic analyses that demonstrated both nuclear localization and ligand-dependent protein misfolding are required for neuronal toxicity (50). Further data supporting the importance of nuclear localization were generated from studies in a mouse model of polyQ toxicity in which gene targeting was used to insert a long CAG repeat into the mouse Hprt protein (51). The addition of an NLS hastened the occurrence of neuropathology and behavioral abnormalities in these mice, whereas the addition of a nuclear export sequence delayed these features. Collectively, these data indicate the importance of the nucleus as a site of toxicity mediated by polyQ proteins.
Posttranslational Modifications and Interacting Proteins Influence the Toxicity of PolyQ Proteins
Corroborating the notion that the biology of the carrier protein underlies specific features of each polyQ disorder are data indicating that posttranslational modifications of mutant proteins potently modulate toxicity. These posttranslational modifications occur on amino acids outside the polyQ tract and often influence protein–protein interactions or function. The first example of these modulatory effects was provided for ATXN1 (52), in which the phosphorylation of serine 776 of ATXN1 was critical for polyQ-induced toxicity in transgenic mice. Mice expressing polyQ ATXN1 with serine 776 mutated to alanine displayed substantially reduced Purkinje cell degeneration. Conversely, a phosphomimetic serine to aspartic acid mutation in wild-type ATXN1 induced motor dysfunction and Purkinje neuron degeneration in transgenic mice (53). Phosphorylation of ATXN1 at serine 776 facilitates the dissociation of ATXN1 from 14–3-3 in the cytosol, enabling the nuclear translocation of polyQ ATXN1 to mediate toxicity (54). Another important association of ATXN1 is with capicua (CIC), a protein that binds DNA and represses the expression of target genes. The importance of the association of ATXN1 with CIC in SCA1 was highlighted by the fact that reducing CIC levels in SCA1 knockin mice, either genetically or by exercise, improved survival and Purkinje neuron loss (55). Toxicity, stabilization, and the seeding of oligomers formed by polyQ ATXN1 also require interaction with CIC, demonstrating an important role for this protein complex in SCA1 toxicity. Notably, both LOF and toxic GOF with CIC coexist to trigger polyQ ATXN1-mediated toxicity (55). This is indicated by a study showing that disruption of the ATXN1–CIC complex during development is detrimental, resulting in a spectrum of neurobehavioral phenotypes both in mice and in humans that are a consequence of LOF (56).
Subsequent work has demonstrated that posttranslational modifications of disease-causing proteins modulate phenotypes in other polyQ disorders. In cellular models of SBMA, the phosphorylation of polyQ AR at serine residues 215 and 792 by Akt diminished ligand binding and reduced toxicity (57). Akt-mediated phosphorylation of polyQ AR was promoted by insulin-like growth factor-1 (IGF-1), and in mice, disease phenotypes were partially rescued either by transgenic overexpression of IGF-1 in muscle or the peripheral administration of IGF-1 (58, 59). Phosphorylation of polyQ AR at alternative sites also impacted protein stability and toxicity in models of disease (60), suggesting that several kinase pathways may offer therapeutic targets. Moreover, the transcriptional regulatory activity of ligand-bound polyQ AR was modulated by the covalent attachment of small ubiquitin-like modifier (SUMO) to lysine residues 385 and 518 (61, 62). In mice, the disruption of this modification recovered the partial LOF conferred by polyQ tract expansion and diminished disease phenotypes (62). Ligand-triggered conformational changes also modulated polyQ AR toxicity, as genetic disruption of the AR’s N-terminal and C-terminal interactions delayed disease onset in transgenic mice (63).
SUMOylation was first implicated as a disease-modifying posttranslational modification by Steffan et al. (64) in the study of HD models. Using exon 1 fragment models, they showed that SUMOylation stabilized the toxic HTT protein, promoted transcriptional repression, and exacerbated neurodegeneration in Drosophila. SUMOylation of HTT was mediated by the E3 SUMO ligase protein inhibitor of activated STAT 1 (PIAS1), the knockdown of which reduced mutant HTT accumulation and behavioral phenotypes in fragment R6/2 mice (65). That the N terminus of polyQ HTT modifies toxicity in a complex manner was further supported by characterization of bacterial artificial chromosome (BAC) transgenic mice in which the N-terminal 17 amino acids were deleted, resulting in more overt disease (66). Moreover, similar to polyQ AR, mutant HTT toxicity was modulated by phosphorylation at consensus Akt sites. Phosphorylation at serine 421 mitigated neurodegeneration in mice and neuronal cultures by increasing proteasome-dependent turnover of the mutant protein (67). As with the polyQ AR, it is likely that phosphorylation at additional residues by alternative kinases impacts toxicity and may thereby present further targets for therapeutic interventions.
Non–Cell Autonomous Mechanisms Contribute to Disease Phenotypes
The preferential vulnerability of distinct neuronal populations results partly from cell autonomous toxicity that is triggered by the expression of the mutant protein. Additionally, the toxic effects of polyQ proteins expressed in closely connected cells also contribute to disease pathogenesis. These non–cell autonomous contributions may arise either from glial cells or functionally associated target cells that are coupled by synaptic connections. Glial involvement in the pathogenesis of SCA7 was implicated through studies in which transgenic overexpression of polyQ ataxin-7 (ATXN7) in Bergmann glia was shown to disrupt glutamate transport and trigger behavioral impairment (68). Conversely, genetic deletion of polyQ ATXN7 in Bergmann glia partially rescued the behavior of SCA7 transgenic mice (69). Similarly, transgenic expression of an amino-terminal fragment of polyQ HTT in astrocytes diminished mouse motor performance, body weight, and survival (70). The notion that glia contribute to HD pathogenesis was further supported by studies in which human glial progenitor cells (hGPCs) were grafted into the brains of immunodeficient mice (71). When hGPCs expressing mutant HTT were grafted into wild-type mice, the animals developed progressive motor deficits. In contrast, the grafting of normal hGPCs into the brains of R6/2 fragment mice ameliorated behavioral phenotypes.
Non–cell autonomous mechanisms have been observed in SBMA as well, where the polyQ AR leads to progressive degeneration both of lower motor neurons and their synaptic target, skeletal muscle. Transgenic overexpression of wild-type or polyQ AR only in muscle was sufficient to cause muscle atrophy, diminished motor performance, and motor axon loss (72, 73). Moreover, the diminished expression of polyQ AR in peripheral tissues, but not in the spinal cord, following the administration of antisense oligonucleotides (ASOs) or genetic deletion of polyQ AR only from muscle rescued animals from disease (74, 75). Collectively, these studies have established the important contribution of skeletal muscle to SBMA pathogenesis and identified this peripheral site as a potential target for therapies. Similarly, it is likely that synaptically connected neurons contribute to non–cell autonomous toxicity in other polyQ diseases through transneuronal mechanisms. In support of this conclusion, the conditional deletion of mutant HTT in a BAC transgenic model of HD demonstrated that the reduced expression of mutant protein in cortical neurons improved postsynaptic marker protein expression and synaptic neurotransmission within medium spiny neurons (76).
Multiple Downstream Pathways Are Disrupted by Mutant Proteins
During the prior three decades, researchers studied polyQ diseases with a host of genetic models in organisms ranging from yeast to nonhuman primates, in patient-derived cells and tissues, and in living participants. This body of data demonstrates that polyQ mutations disrupt a large number of downstream pathways. For the most intensely studied polyQ disease, HD, more than a dozen proximate mechanisms of neurodegeneration receive support in the literature (Table 3), and analogous studies of other polyQ disorders are consistent with the pleiotropic pathogenic effects of expanded CAG/polyQ repeats in all polyQ diseases. Both GOF and LOF mechanisms are implicated.
Table 3.
Proximate mechanisms of neurodegeneration implicated in Huntington’s disease
| Processes | Mechanisms |
|---|---|
| Nucleus and gene expression | Transcriptional dysregulation |
| DNA damage and repair | |
| Toxic RNAs | |
| RAN translation | |
| Impaired nucleocytoplasmic transport | |
| Cytoplasmic processes | Excitotoxicity |
| Abnormal intracellular Ca2+ buffering | |
| Altered axonal transport | |
| Neurotrophic factor dysregulation | |
| Mitochondrial dysfunction | |
| Altered neuronal cholesterol metabolism | |
| Autophagy dysregulation | |
| Defective ciliogenesis | |
| Astrocyte dysfunction |
Abbreviation: RAN, repeat-associated non-ATG.
A detailed review of all of these potential mechanisms is beyond the scope of this overview; instead, we focus on some of the best-supported proximate disease mechanisms. These alterations target nuclear processes that regulate gene expression at transcriptional or posttranscriptional steps, as well as pathways that are critical to maintaining cellular homeostasis, including the regulation of energy metabolism and ions, protein quality-control pathways, and intracellular trafficking.
Gene expression.
Nuclear processes that contribute to disease pathogenesis have attracted considerable attention, spurred in part by the observation that NIIs are frequently pathological hallmarks of polyQ diseases. Several genes affected in polyQ disorders are regulators of gene expression (Table 1). PolyQ domains are common among transcriptionally active proteins, and expanded polyQ domains could plausibly promote abnormal interactions among transcriptional regulators and disease-causing mutant proteins. The observation that transcriptional coregulators—many of which are present in limiting quantities within the nucleus—are sequestered in NIIs led to the notion that their impaired function underlies transcriptional dysregulation in disease. Among the factors implicated in pathogenesis are the cAMP response element-binding (CREB) protein, specific protein-1 (Sp1), and TAFII130 (77–81), all of which are components of the transcriptional regulatory apparatus and impact histone acetylation. Attempts to compensate for the diminished function of these and related factors using histone deacetylase (HDAC) inhibitors abrogated disease phenotypes in models of SBMA and HD (82, 83); subsequent work in models of other polyQ disorders showed similar beneficial effects. Translating this approach to patients proved challenging, in part because of the broad effects of HDAC inhibitors on gene expression that can lead to toxicity with the long-term administration required to treat these chronic disorders.
Other fundamental aspects of gene expression are also disrupted in polyQ disease at the RNA level. In vitro studies suggest that CAG repeat RNAs induce splicing changes by sequestering the muscleblind-like-1 (Mbnl1) splicing factor through a mechanism analogous to that implicated in the myotonic dystrophies (84). Similarly, in yeast, the expression of mutant HTT protein disrupts preRNA splicing (85). The generation of an exon 1 protein fragment in HD mice by aberrant HTT gene splicing suggests that these mechanisms may contribute to aberrant gene regulation in mammals (86). In addition to splicing dysregulation, evidence suggests that long CAG-containing RNAs may trigger the occurrence of repeat-associated non-ATG (RAN) translation, a process wherein aberrant RNA structures, such as hairpins, trigger the production of neurotoxic polypeptides through noncanonical translation (87). Homopolymeric proteins generated by bidirectional RAN translation have been shown to occur in HD brain tissue (88). These alterations in gene expression are associated with abnormalities in nuclear envelope integrity, the disruption of nucleocytoplasmic transport (42, 89), and evidence implicating DNA damage and repair (90), further indications that nuclear functions are disrupted in polyQ diseases.
Axonal transport.
In addition to effects on critical nuclear processes, substantial data indicate that polyQ disorders are associated with significant disruptions of cytoplasmic processes that are essential for neuronal homeostasis. Abnormalities of fast axonal transport (FAT), which may contribute to neurodegeneration in numerous genetic disorders (91), have been demonstrated in several polyQ disorders, including HD and SBMA (92–94). These abnormalities are good examples of the pleiotropic effects of expanded polyQ HTT and perhaps serve as examples of LOF effects. HTT normally functions, at least partly, as a scaffold protein in several multiprotein complexes (95). HTT interacts with molecular motor machinery, probably directly with dynein and indirectly via other members of this complex (96, 97). The reduced expression of wild-type HTT slows FAT, while the overexpression of wild-type HTT enhances FAT. The expansion of HTT’s polyQ tract appears to have some of the same effects on FAT as reducing expression of the wild-type protein. Zala et al. (98) suggested a particularly interesting role for huntingtin in FAT. They demonstrated that wild-type HTT ties important glycolytic enzymes, such as GAPDH, to fast-moving vesicles, with these enzymes being responsible for local production of the ATP needed to fuel FAT.
Reduced FAT is likely to have multiple consequences. One specific consequence in HD that has attracted interest is the reduced trophic support of striatal neurons. Medium spiny projection neurons depend on brain-derived neurotrophic factor (BDNF) secreted by corticostriate neurons for normal function and integrity. Postmortem analyses indicate reduced striatal BDNF levels in HD, and experimentally induced BDNF deficiency in mice reproduces some of the features of HD striatal pathology (99–101). Reduced corticostriate transport of BDNF due to defects in FAT induced by mutant huntingtin is a plausible mechanism of BDNF deficiency and non–cell autonomous toxicity in HD. In addition, there are also suggestions that altered FAT reduces tropomyosin receptor kinase B (TrkB; a BDNF receptor) transport in striatal neuron dendrites, further impairing trophic support (102).
Mitochondrial function.
Additional evidence indicates that the expression of mitochondrial proteins involved in oxidative phosphorylation is altered in polyQ disorders. A potential role for mitochondrial dysfunction in HD was suspected because subacute systemic inhibition of oxidative phosphorylation with 3-nitropropionic acid in nonhuman primates reproduced the striatal neuropathological changes of HD (103). Spectrophotometric assays in postmortem brain tissue demonstrated that impaired oxidative phosphorylation was restricted to the basal ganglia in HD brain tissue (104). Other data—such as magnetic resonance spectroscopy (MRS) evidence of increased lactate production in the brains of HD patients and MRS studies of muscle bioenergetics in HD patients—also suggest mitochondrial dysfunction in HD (105, 106). Transcriptional dysregulation by mutant HTT could explain mitochondrial dysfunction in HD (107). However, a methodologically sophisticated effort to assess regional brain respiratory quotients in HD patients produced results inconsistent with oxidative phosphorylation deficits (108). Similarly, normal levels of jugular bulb lactate are reported in HD patients, and recent measurements of muscle respiratory activity were normal in biopsy specimens from premanifest carriers of HD mutant alleles (109, 110). These results are inconsistent with major mitochondrial defects occurring in HD. Although not a feature of early disease in SCA1, studies in a mouse model demonstrated reduced mitochondrial transcripts and protein during the course of disease (111). In transgenic mouse models of SCA3, mitochondrial DNA depletion is seen in affected neuronal regions (112). There is no clear evidence for mitochondrial dysfunction in relevant neurons in SCA2 and SCA7. In contrast, alterations in mitochondrial membrane potential, mitochondrial number, and gene expression occur in skeletal muscle of knockin SBMA mice (113, 114).
Excitotoxicity and ion homeostasis.
Excitotoxicity is a well-established mechanism of neuronal death in acute neurological disorders, such as hypoxia–ischemia, hypoglycemia, and status epilepticus. Excitotoxicity may also be involved in neurodegenerative disorders, particularly HD. Approximately 30 years ago, several groups identified that acute intrastriatal administration of N-methyl-d-aspartate (NMDA) glutamate receptor agonists produced neuronal death with histopathological features resembling HD pathology (115). Partly because neurotransmitter receptors were regarded as promising targets for small molecule drug development, the investigation of potential excitotoxic mechanisms was pursued vigorously. Thorough work by Raymond (116) in transgenic murine models strongly supported the concept of excitotoxic injury in HD. Raymond demonstrated that expanded polyQ HTT preferentially enhanced the activity of the NR2B subtype of NMDA receptors (116). Enhanced ion flux, particularly Ca2+ flux, through this receptor was associated with neuronal dysfunction and death. These results were supported by experiments in a well-characterized knockin line in which double mutant mice were bred expressing both the expanded polyQ repeat murine homolog of huntingtin (Hdh) and an NR2B NMDA receptor transgene causing elevated NR2B NMDA receptor activity (117). The transgene had no deleterious effects. The double mutant mice exhibited marked potentiation of striatal neuron dysfunction and degeneration. Unfortunately, despite strong evidence for excitotoxicity as an important mechanism of neurodegeneration in HD, this approach turned out to be therapeutically barren. NMDA receptors were a logical target and like other heteromeric, ionotropic neurotransmitter receptors, have a rich pharmacology. NMDA receptors, however, also mediate trophic effects on neurons. While overactivation of these receptors produces neuronal death, inadequate activation also results in neuronal death, and there is no clear way to determine the appropriate therapeutic index for treatment with an NMDA receptor antagonist (118, 119).
Other aspects of neuronal calcium homeostasis are affected by expanded polyQ HTT (116). In addition to providing evidence for direct interactions between expanded polyQ HTT and NMDA receptors, Bezprozvanny’s group (120, 121) presented evidence that expanded polyQ HTT enhances the interactions of inositol triphosphate and its receptor. This apparently lowers the threshold for Ca2+ release from ER stores, potentially compromising the many cellular processes influenced by cytosolic Ca2+ levels.
Altered membrane excitability in polyQ ataxia.
Changes in Purkinje neuron membrane excitability tied to altered ion channel expression or function, or both, are widespread in models of polyQ ataxia. These changes are important because they are likely the proximate cause of motor dysfunction at stages of disease preceding overt neurodegeneration (30, 122–125). Altered membrane excitability in cerebellar Purkinje neurons manifests as alterations in spiking. Purkinje neurons normally exhibit autonomous spiking, even in the absence of synaptic input. In mouse models at early stages of SCA1, SCA2, and SCA3, increased membrane excitability results in membrane depolarization and a lack of spiking (30, 122, 123), which is attributed to depolarization block. At different disease stages, a reduction in the frequency of Purkinje neuron firing is consistently seen in models of SCA1 (126), SCA2 (123, 124), and SCA6 (125, 127). In some other mouse models or at other stages of disease, irregular spiking is present (125, 127, 128). Notably, improving Purkinje neuron membrane excitability through activating K+ channels in models of SCA1 (30) and SCA2 (128) improves motor dysfunction and slows degeneration. Currently approved K+ channel activators are tolerated in patients with SCAs and may provide symptomatic benefits (129). Interestingly, other pathways, such as protein kinase C signaling, also appear to converge on these alterations in intrinsic membrane excitability to modify neurodegeneration (130).
Consequences for therapy development.
From a therapeutic standpoint, the collective impact of work on polyQ disease pathogenesis led to a disappointing conclusion: There is no one single pathway disrupted by polyQ proteins in any of these disorders that is both necessary and sufficient to cause disease pathogenesis. The existence of multiple downstream pathways that are dysfunctional suggests that therapeutically targeting any one of them will lead to inadequate responses. This notion has guided more recent attempts to target the ultimate mediator of pathogenesis, the mutant protein or gene, for therapeutic benefit. These approaches have shown promise in experimental models of disease, and several are advancing toward pivotal clinical trials.
TRANSLATIONAL ADVANCES
Leveraging scientific advances for the benefit of patients suffering from polyQ disorders has proven to be a significant challenge. Much current effort is focused on discovering disease biomarkers to facilitate clinical trials and on to evaluating therapeutic strategies in preclinical models and patients with these diseases.
Biomarkers
Evaluating potential therapies for polyQ disorders presents interesting opportunities but also challenges beyond the usual difficulties of therapeutic clinical research for neurodegeneration. Unlike many neurodegenerative disorders, which are clinically or pathologically defined syndromes, or both, and likely of heterogeneous origin, polyQ disorders are monogenic, highly penetrant illnesses. Even though there is phenotypic variation within each polyQ disorder, straightforward genetic testing identifies carriers of mutant alleles with virtually perfect accuracy. The relative rarity of these disorders, however, is an obstacle to efficient clinical research, particularly trials. The most prevalent polyQ disorder in developed nations, HD, is rare, and the other polyQ disorders are generally less prevalent (see above). For most polyQ disorders, even small trials require multicenter, frequently international, collaborations, with the attendant regulatory, administrative, and data gathering and storage complications. Collaborative groups have arisen to address these obstacles, including the North America–based Huntington Study Group, the European Huntington’s Disease Network, the European Integrated Project on Spinocerebellar Ataxias, and the Clinical Research Consortium for Spinocerebellar Ataxia.
Clinical trials for polyQ disorders also suffer from the absence of easily obtained and interpreted outcome measures, which is common in neurodegenerative disorder research. Typical outcome measures are clinical rating scales based on functional performance, such as the HD Total Functional Capacity scale, and examination-based scales, such as the Unified Huntington’s Disease Rating Scale, the Scale for Assessment and Rating of Ataxia, and the Adult Myopathy Assessment Tool. Such scales provide neither clear end points nor truly continuously varying measures, can be confounded by placebo effects and other forms of bias, and exhibit nonlinearity over the total course of disease. The ultimate goal of clinical research into polyQ disease is to develop therapies that delay or prevent the onset of manifest disease. At this point, there are no clear clinical standards for assessing transitions from premanifest to manifest disease. In particular, longitudinal studies of premanifest HD populations have documented a long prodromal phase of insidious alterations in behavior, cognitive functions, and motor function, but did not identify any overt milestones suitable for assessing phenotypic conversion (131, 132). The declaration of manifest disease remains a clinical judgment with a significant subjective component.
Biomarkers are proposed to be the solution to these and other problems in clinical research into polyQ disease and other neurodegeneration disorders. Biomarker is a much-used (perhaps abused) term that can refer to different entities. Potentially useful biomarkers include measurements that provide objective evidence of disease activity and might be suitable as surrogate end points in trials. Other biomarkers might reflect intervention activity (target engagement) without necessarily reflecting disease activity. A useful example of the potential impact of biomarkers comes from multiple sclerosis research. Biomarkers identified by magnetic resonance imaging (MRI) of intermittent disease activity and cumulative brain injury were crucial to demonstrating the relevant effects of immunomodulatory therapies in early-phase trials. These findings set the stage for pivotal Phase III trials using conventional clinical end points. Imaging biomarkers have drawn the most attention in HD in the context of interventions in premanifest carriers of mutant alleles. Longitudinal MRI studies indicate that changes in striatal volumes are an early and quantifiable abnormality in premanifest carriers of mutant alleles (133). Striatal volume changes are inferred to occur many years, perhaps decades, prior to the obvious onset of manifest HD. These studies indicate that striatal volume changes have the statistical properties suitable to be used as a biomarker of disease activity and could plausibly serve as a rational surrogate end point. An example of a potentially useful target engagement biomarker comes from a recent trial of an antisense oligonucleotide (ASO) for treating HD. This ASO was administered intrathecally, allowing analysis of HTT levels in participants’ cerebrospinal fluid (CSF). Target engagement was demonstrated, with ASO dose-related reductions in CSF HTT concentrations. As shown by these examples, the development and application of biomarkers should be guided by clear, specifically defined concepts of their use.
Therapeutics
The goal of delaying or preventing the onset of manifest disease poses additional problems in developing therapies for polyQ diseases. As suggested by research with premanifest carriers of HD mutant alleles, neurodegeneration may begin many years prior to the onset of manifest disease. One possible implication is that early, prolonged treatment may be needed to delay or prevent disease onset. Prior to manifest disease, however it is defined, carriers of mutant alleles are clinically normal. Successful preventive therapies will require a high therapeutic index. Side effects producing significant impairments in daily function undermine the goal of maintaining normalcy. In addition, even minor or cosmetic side effects tend to reduce compliance significantly. Individuals taking antihypertensive agents, for example, discontinue treatment at the rate of approximately 10% per year. Successful treatments may need to be both highly efficacious and largely free of side effects, a very demanding standard.
Another potential problem with preventive treatment is that partially effective treatments that delay the onset of manifest disease may also slow disease progression. As disease prevalence is a function of disease duration, this would have the superficially paradoxical consequence of effective therapies increasing disease prevalence. The specific consequences of slowing disease progression would likely vary between specific polyQ diseases. SBMA and SCA6, for example, are distinguished by relatively selective motor deficits. Slowing the progression of SBMA or SCA6 might prolong independent ambulation in manifest patients, and even a modest extension of the period of independent ambulation would have significant benefits. Treating more diffuse polyQ neurodegenerations, such as HD, might be more problematic. A significant fraction of early HD patients have psychiatric problems. These patients can be extremely difficult to manage for their families and physicians. A recent modeling study suggested that gains in delaying the onset of HD would be largely offset by extensions in the duration of manifest disease (134). It is not clear that families of HD patients would regard a treatment that extended the duration of manifest HD as a positive development. It is also plausible that prolonging the duration of polyQ diseases will result in the emergence of previously undocumented disease features. While it is recognized that clinical disease features are largely restricted to the CNS (with the exception of muscle involvement in SBMA), disease-causing genes are expressed in other tissues. In Parkinson’s disease dopamine replacement therapy has altered the natural history of the disease, with significant life extension and the consequent emergence and recognition of important disease features, such as dementia, that were not appreciated in historic descriptions.
The possibility of developing partially effective treatments points to a need for therapies that target important symptomatic disease features. Barring the unlikely event of the development of highly effective treatments that completely prevent manifest disease, the development of disease-modifying therapies may actually increase the need for therapies targeting particularly problematic symptoms. Given the complexity of many clinical features of polyQ diseases, this is arguably as great a challenge as developing disease-modifying therapies.
Given the likely pleiotropic effects of expanded repeat polyQ–containing proteins or mRNAs, or both, strategies to target the mutant protein or gene have garnered increased attention. Three general strategies to accomplish this have emerged. The first is to harness endogenous pathways that degrade the misfolded mutant proteins and aggregates to clear cells of toxic species. These efforts have focused on activating either macroautophagy or chaperone-dependent pathways to enhance cell-clearance mechanisms. Robust evidence indicates that macroautophagy clears both cytosolic protein aggregates and dysfunctional mitochondria, and it is essential for maintaining neuronal homeostasis. The induction of this pathway was first shown to occur in models of HD (135), followed by evidence that autophagy degrades aggregate-prone polyQ proteins (136). Subsequent work showed that the activation of autophagy ameliorates disease in models of HD and other polyQ disorders (137). Efforts are underway to identify small molecules that stimulate this clearance mechanism in neurons and are safe for chronic administration.
Chaperone-targeted approaches have focused partly on heat shock protein 90 (Hsp90) and Hsp70, which function together in a multiprotein complex to regulate the proteostasis of client proteins (138). Notably, this machinery regulates both HTT and AR. Interaction with Hsp90 stabilizes client proteins, and small-molecule inhibitors of Hsp90 promote polyQ HTT and AR degradation to ameliorate phenotypes in disease models. Interaction with Hsp90 is lost as client proteins misfold or following treatment with an Hsp90 inhibitor. Client-bound Hsp70 then recruits chaperone-dependent E3 ubiquitin ligases, such as C terminus of HSC70–interacting protein (CHIP), to ubiquitinate target proteins and facilitate their degradation. There are several significant challenges to targeting this machinery for therapeutic benefit. Hundreds of proteins are clients of the Hsp90 chaperone machinery, including critical regulators of signal transduction and gene expression: Treatment with Hsp90 inhibitors leads to the degradation of many of these substrates, not simply those that are misfolded. An alternative approach to more selectively target misfolded clients is to activate Hsp70-dependent ubiquitination, a strategy that has been shown to work in disease models, but for which small molecules that are safe and active in humans are lacking (139). Additionally, because the overexpression of Hsp70 is beneficial in several models, efforts to target its transcriptional regulator, heat shock factor protein-1 (HSF-1), have garnered attention (140). These efforts complement evidence of a diminished HSF-1-dependent stress response in HD models (141), and thereby may address the impaired activity of protein quality control in disease. One related point is noteworthy. The chaperone machinery regulates client proteins by interacting with them at hydrophobic clefts, sites of inherent instability that include sites of ligand binding (138). Truncated fragments of polyQ proteins may lack the binding sites present in full-length proteins and, therefore, exhibit a differential response to proteostasis manipulation. Such is the case for the polyQ AR, where the ligand-binding domain is at the C terminus and the polyQ tract is at the N terminus. After cells are treated with an Hsp70 inhibitor, truncated N-terminal fragments of the polyQ AR respond differently from how the full-length protein does(142).
A second approach, along the general lines of enhancing cellular capacity to deal with the stress of polyQ mutations, is to modulate the critical pathways implicated in aging. This concept gained traction as a one-size-fits-all approach to treatments for neurodegeneration. While polyQ disorders exhibit an earlier age of onset than common neurodegenerations, such as Alzheimer’s and Parkinson’s diseases, disease expression is age related. Several groups applied interventions that retard aging in rodents, such as caloric restriction, to HD genetic models in the hope of producing therapeutic effects. Results have been equivocal (143).
A third emerging therapeutic strategy is to employ approaches to suppress the expression of the mutant gene or to correct the mutation by genome editing. Two technologies that suppress gene expression are being actively pursued in preclinical and early-phase clinical studies: ASOs and adeno-associated viral vector delivery of small-interfering RNAs (AAV-siRNAs). Both approaches reduce translation via the induction of mRNA degradation. Expression suppression approaches have the major theoretical advantage of apparently striking at the root cause of neurodegeneration, thereby sidestepping the difficulties of targeting what are likely to be multiple proximate causes of neuronal dysfunction and death. ASOs containing chemically modified backbones that enhance their stability and tolerability have been used successfully to ameliorate aspects of the disease phenotype in models of several polyQ disorders, including HD, SBMA, and SCA3 (74, 144, 145), and an ongoing Phase I–IIa trial of this approach is being conducted in early-stage HD patients. Similarly, AAV-siRNAs have shown benefit in animal models of several polyQ disorders (146, 147). In the last 2–3 years, efforts have also explored the use of CRISPR/Cas9 genome editing to delete the mutant gene in HD mice (148). Although all of these approaches show promise in rodent models of disease, the extent to which they will sufficiently target cells in the much larger human CNS is an important unresolved question. Moreover, there are both theoretical and practical obstacles to ASO and AAV-siRNA treatments for polyQ diseases.
The theoretical concern is based on the possibility that haploinsufficiency contributes to neurodegeneration in polyQ diseases. Existing technologies do not permit mutation-specific knockdowns, although there has been some work with haplotype-related expression suppression. In this scenario, expression reductions may actually exacerbate disease progression. The prevailing impression is that GOF effects dominate pathogenesis in polyQ diseases, but judging the balance of GOF versus LOF effects in preclinical experiments is often quite difficult. In several cases, we do not possess complete information about the normal function of these proteins. Murine knockouts of target genes are used to assess haploinsufficiency effects. The absence of an overt phenotype in heterozygous animals provides some reassurance that LOF effects are not important. Among other issues, the relatively short life span and limited behavioral repertoire of mice confounds the use of these genetic models to definitively assess mutation-nonspecific expression suppression interventions. Reassuring AAV-siRNA experiments for HTT suppression were performed in nonhuman primates, but these experiments are necessarily limited by the small number of animals and short follow-up periods (149). Complete LOF mutations in AR cause testicular feminization and not neuromuscular disease, providing strong support that a GOF mechanism predominates in SBMA. However, even in this case, the loss of anabolic support conferred by the polyQ expansion potentially contributes to the neuromuscular phenotype of SBMA patients and may confound the effects of expression suppression in them.
Major practical obstacles are related to ASO and AAV delivery to the CNS. ASOs do not pass the blood–brain barrier. This approach requires intrathecal or intracisternal delivery. Repeated lumbar puncture for intrathecal delivery is the approach used in an early-phase trial of HTT ASOs. Mitigating this problem somewhat is the fact that ASOs appear to have relatively durable effects, reducing HTT expression significantly for periods of weeks and raising the possibility that infrequent but regular treatment might be adequate. Intrathecal delivery, however, has other drawbacks. The normal pattern of CSF flow is from the ventricles to the subarachnoid space and up over the hemispheres. Intrathecal administration primarily delivers ASOs to the cortex, with relatively little penetration into subcortical structures. For HD, which exhibits prominent subcortical and cortical pathology, treatment effects may be only partial. This may be less of an issue for other polyQ disorders, particularly SCAs with their prominent brainstem and cerebellar pathology, as these structures are bathed in CSF, and SBMA, in which skeletal muscle is a potentially important peripheral target.
Present AAV approaches involve a different set of challenges. AAV administration is necessarily highly invasive and its effects are limited by the regional and subregional spread of virus. For a disease such as HD, which causes widespread neurodegeneration, treating the large human brain is a major challenge. Other polyQ disorders, such as SCA6, exhibit more restricted pathology, and focused AAV injection may be more efficacious. AAV strains that exhibit CNS penetration and neuronal transfection after intravenous administration may obviate these delivery problems (150). Regardless of the delivery issues, another potential drawback of AAV-mediated treatments is irreversibility. While the persistence of effects is attractive from the perspective of producing durable treatment effects, it also means that significant side effects are likely to be permanent.
As with any clinical trial, participants’ safety will be a paramount concern. Because of potential haploinsufficiency effects and the functionally irreversible consequences of AAV-mediated treatments, long-term follow-up will be particularly important in trials of gene expression suppression for polyQ diseases.
RECOMMENDATIONS
Symptomatic Therapies
Two particularly salient features have emerged from research on polyQ diseases: the pleiotropic pathogenic effects of mutated genes and proteins and the difficulty in identifying transitions to manifest disease. As discussed above, several factors favor expression suppression approaches as emerging therapies, but there are both practical challenges and theoretical obstacles that raise concern that suppression may be only partially successful. These factors emphasize the importance of developing therapies for symptomatic features of polyQ diseases. It makes little sense to improve disease survival without concomitantly improving the quality of life. Developing useful symptomatic therapies will likely require different research approaches. The complex features of major disease symptoms reflect circuit-level dysfunctions that are secondary consequences of primary neuronal dysfunction and neurodegeneration. Understanding how neuronal dysfunction, including alterations in neuronal spiking and synaptic signaling, contributes to circuit dysfunctions requires investigations of pathological neurobiology that are different from the molecular- and cellular-based approaches used to understand pathogenesis that dominate the field.
Rigorous and Conceptually Clear Biomarker Studies
The lack of clear milestones identifying manifest disease emphasizes the need for appropriate biomarkers to delineate intervention trial outcomes in premanifest carriers of mutations. Such biomarker work has to be conceptually and technically rigorous. Careful attention has to be paid to the utility of the proposed biomarkers. Are they aimed at demonstrating target engagement? Are they surrogate markers for disease activity? Do they reflect basic pathogenic features or useful epiphenomena? Do they monitor intervention activity? The criteria for developing and evaluating these distinct types of biomarkers are different. All biomarkers must be statistically robust and ultimately validated against relevant clinical outcomes.
Developing a Standard for What Constitutes a Successful Therapy
The possibilities of partially effective therapies and trade-offs between delaying the onset of manifest disease versus extending the duration of manifest disease raise the issue of what constitutes a successful therapy. Although it is informed by data, this is not a scientific issue. The preferences of patients, at-risk individuals, and family members are essential in exploring this difficult issue. The criteria for what constitutes an acceptable therapy should be decided in advance of major intervention trials. Efforts should be made to clarify patients’, at-risk individuals’, and family members’ understandings of and preferences about potential therapies. This information should be the basis for formulating criteria to evaluate an intervention’s success.
Despite encouraging results using expression suppression in preclinical models and an early-phase trial in HD, we are clearly a long way away from having successful therapies for polyQ diseases. The only existing effective intervention is genetic screening and embryo selection. This intervention is complex because almost all polyQ diseases are dominant. The experience with HD indicates that at-risk individuals are generally not interested in knowing whether they carry the mutation. Ethical genetic screening in this context requires blinded in vitro fertilization and preimplantation genetic diagnosis. This approach is technically demanding and relatively expensive but could be made widely available as it presently represents a viable option for mitigating the considerable burden of polyQ diseases.
ACKNOWLEDGMENTS
Work in the authors’ laboratories is supported by the USs National Institutes of Health (grants R01 NS055746 and R21 NS101030 to A.P.L., R01 NS085054 to V.G.S., P50 NS091856 and R21 NS088302 to R.L.A.) and the Muscular Dystrophy Association (MDA 513702 to A.P.L.).
Footnotes
DISCLOSURE STATEMENT
Dr. Albin receives compensation for service on the Data Safety and Monitoring Boards of the LEGATO-HD and IONIS HTTRX studies. Dr. Shakkottai serves as a member of the Research Advisory Board for Cadent Therapeutics. Dr. Lieberman is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Paulson H 2018. Repeat expansion diseases. Handb. Clin. Neurol 147:105–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stoyas CA, La Spada AR. 2018. The CAG-polyglutamine repeat diseases: a clinical, molecular, genetic, and pathophysiologic nosology. Handb. Clin. Neurol 147:143–70 [DOI] [PubMed] [Google Scholar]
- 3.Giorgetti E, Lieberman AP. 2016. Polyglutamine androgen receptor–mediated neuromuscular disease. Cell. Mol. Life Sci 73:3991–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, et al. 2010. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466:1069–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee T, Li YR, Ingre C, Weber M, Grehl T, et al. 2011. Ataxin-2 intermediate-length polyglutamine expansions in European ALS patients. Hum. Mol. Genet 20:1697–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Damme P, Veldink JH, van Blitterswijk M, Corveleyn A, van Vught PW, et al. 2011. Expanded ATXN2 CAG repeat size in ALS identifies genetic overlap between ALS and SCA2. Neurology 76:2066–72 [DOI] [PubMed] [Google Scholar]
- 7.Pringsheim T, Wiltshire K, Day L, Dykeman J, Steeves T, Jette N. 2012. The incidence and prevalence of Huntington’s disease: a systematic review and meta-analysis. Mov. Disord 27:1083–91 [DOI] [PubMed] [Google Scholar]
- 8.Kay C, Hayden MR, Leavitt BR. 2017. Epidemiology of Huntington disease. Handb. Clin. Neurol 144:31–46 [DOI] [PubMed] [Google Scholar]
- 9.Evans SJ, Douglas I, Rawlins MD, Wexler NS, Tabrizi SJ, Smeeth L. 2013. Prevalence of adult Huntington’s disease in the UK based on diagnoses recorded in general practice records. J. Neurol. Neurosurg. Psychiatry 84:1156–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher ER, Hayden MR. 2014. Multisource ascertainment of Huntington disease in Canada: prevalence and population at risk. Mov. Disord 29:105–14 [DOI] [PubMed] [Google Scholar]
- 11.Ruano L, Melo C, Silva MC, Coutinho P. 2014. The global epidemiology of hereditary ataxia and spastic paraplegia: a systematic review of prevalence studies. Neuroepidemiology 42:174–83 [DOI] [PubMed] [Google Scholar]
- 12.Maruyama H, Izumi Y, Morino H, Oda M, Toji H, Nakamura S, Kawakami H. 2002. Difference in disease-free survival curve and regional distribution according to subtype of spinocerebellar ataxia: a study of 1,286 Japanese patients. Am. J. Med. Genet 114:578–83 [DOI] [PubMed] [Google Scholar]
- 13.Tanaka F, Doyu M, Ito Y, Matsumoto M, Mitsuma T, et al. 1996. Founder effect in spinal and bulbar muscular atrophy (SBMA). Hum. Mol. Genet 5:1253–57 [DOI] [PubMed] [Google Scholar]
- 14.Semaka A, Hayden MR. 2014. Evidence-based genetic counselling implications for Huntington disease intermediate allele predictive test results. Clin. Genet 85:303–11 [DOI] [PubMed] [Google Scholar]
- 15.Semaka A, Kay C, Doty CN, Collins JA, Tam N, Hayden MR. 2013. High frequency of intermediate alleles on Huntington disease–associated haplotypes in British Columbia’s general population. Am. J. Med. Genet. B Neuropsychiatr. Genet 162B:864–71 [DOI] [PubMed] [Google Scholar]
- 16.Sequeiros J, Ramos EM, Cerqueira J, Costa MC, Sousa A, et al. 2010. Large normal and reduced penetrance alleles in Huntington disease: instability in families and frequency at the laboratory, at the clinic and in the population. Clin. Genet 78:381–87 [DOI] [PubMed] [Google Scholar]
- 17.Divino V, Dekoven M, Warner JH, Giuliano J, Anderson KE, et al. 2013. The direct medical costs of Huntington’s disease by stage: a retrospective commercial and Medicaid claims data analysis. J. Med. Econ 16:1043–50 [DOI] [PubMed] [Google Scholar]
- 18.Whetten-Goldstein K, Sloan F, Kulas E, Cutson T, Schenkman M. 1997. The burden of Parkinson’s disease on society, family, and the individual. J. Am. Geriatr. Soc 45:844–49 [DOI] [PubMed] [Google Scholar]
- 19.Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, et al. 1996. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87:493–506 [DOI] [PubMed] [Google Scholar]
- 20.Schilling G, Becher MW, Sharp AH, Jinnah HA, Duan K, et al. 1999. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum. Mol. Genet 8:397–407 [DOI] [PubMed] [Google Scholar]
- 21.Stack EC, Kubilus JK, Smith K, Cormier K, Del Signore SJ, et al. 2005. Chronology of behavioral symptoms and neuropathological sequela in R6/2 Huntington’s disease transgenic mice. J. Comp. Neurol 490:354–70 [DOI] [PubMed] [Google Scholar]
- 22.Ordway JM, Tallaksen-Greene S, Gutekunst CA, Bernstein EM, Cearley JA, et al. 1997. Ectopically expressed CAG repeats cause intranuclear inclusions and a progressive late onset neurological phenotype in the mouse. Cell 91:753–63 [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto A, Lucas JJ, Hen R. 2000. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington’s disease. Cell 101:57–66 [DOI] [PubMed] [Google Scholar]
- 24.Zu T, Duvick LA, Kaytor MD, Berlinger MS, Zoghbi HY, et al. 2004. Recovery from polyglutamine-induced neurodegeneration in conditional SCA1 transgenic mice. J. Neurosci 24:8853–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chevalier-Larsen ES, O’Brien CJ, Wang H, Jenkins SC, Holder L, et al. 2004. Castration restores function and neurofilament alterations of aged symptomatic males in a transgenic mouse model of spinal and bulbar muscular atrophy. J. Neurosci 24:4778–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simerly RB, Chang C, Muramatsu M, Swanson LW. 1990. Distribution of androgen and estrogen receptor mRNA-containing cells in the rat brain: an in situ hybridization study. J. Comp. Neurol 294:76–95 [DOI] [PubMed] [Google Scholar]
- 27.Strong TV, Tagle DA, Valdes JM, Elmer LW, Boehm K, et al. 1993. Widespread expression of the human and rat Huntington’s disease gene in brain and nonneural tissues. Nat. Genet 5:259–65 [DOI] [PubMed] [Google Scholar]
- 28.Li SH, Schilling G, Young WS 3rd, Li XJ, Margolis RL, et al. 1993. Huntington’s disease gene (IT15) is widely expressed in human and rat tissues. Neuron 11:985–93 [DOI] [PubMed] [Google Scholar]
- 29.Jafar-Nejad P, Ward CS, Richman R, Orr HT, Zoghbi HY. 2011. Regional rescue of spinocerebellar ataxia type 1 phenotypes by 14–3–3ε haploinsufficiency in mice underscores complex pathogenicity in neurodegeneration. PNAS 108:2142–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dell’Orco JM, Wasserman AH, Chopra R, Ingram MA, Hu YS, et al. 2015. Neuronal atrophy early in degenerative ataxia is a compensatory mechanism to regulate membrane excitability. J. Neurosci 35:11292–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, et al. 1997. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell 90:537–48 [DOI] [PubMed] [Google Scholar]
- 32.DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, et al. 1997. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277:1990–93 [DOI] [PubMed] [Google Scholar]
- 33.Paulson HL, Perez MK, Trottier Y, Trojanowski JQ, Subramony SH, et al. 1997. Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron 19:333–44 [DOI] [PubMed] [Google Scholar]
- 34.Tellez-Nagel I, Johnson AB, Terry RD. 1974. Studies on brain biopsies of patients with Huntington’s chorea. J. Neuropathol. Exp. Neurol 33:308–32 [DOI] [PubMed] [Google Scholar]
- 35.Saudou F, Finkbeiner S, Devys D, Greenberg ME. 1998. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell 95:55–66 [DOI] [PubMed] [Google Scholar]
- 36.Tallaksen-Greene SJ, Ordway JM, Crouse AB, Jackson WS, Detloff PJ, Albin RL. 2003. Hprt(CAG)146 mice: age of onset of behavioral abnormalities, time course of neuronal intranuclear inclusion accumulation, neurotransmitter marker alterations, mitochondrial function markers, and susceptibility to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine. J. Comp. Neurol 465:205–19 [DOI] [PubMed] [Google Scholar]
- 37.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. 2004. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431:805–10 [DOI] [PubMed] [Google Scholar]
- 38.Gutekunst CA, Li SH, Yi H, Mulroy JS, Kuemmerle S, et al. 1999. Nuclear and neuropil aggregates in Huntington’s disease: relationship to neuropathology. J. Neurosci 19:2522–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuemmerle S, Gutekunst CA, Klein AM, Li XJ, Li SH, et al. 1999. Huntington aggregates may not predict neuronal death in Huntington’s disease. Ann. Neurol 46:842–49 [PubMed] [Google Scholar]
- 40.Mastroberardino PG, Iannicola C, Nardacci R, Bernassola F, De Laurenzi V, et al. 2002. ‘Tissue’ transglutaminase ablation reduces neuronal death and prolongs survival in a mouse model of Huntington’s disease. Cell Death Differ. 9:873–80 [DOI] [PubMed] [Google Scholar]
- 41.Liu KY, Shyu YC, Barbaro BA, Lin YT, Chern Y, et al. 2015. Disruption of the nuclear membrane by perinuclear inclusions of mutant huntingtin causes cell-cycle re-entry and striatal cell death in mouse and cell models of Huntington’s disease. Hum. Mol. Genet 24:1602–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woerner AC, Frottin F, Hornburg D, Feng LR, Meissner F, et al. 2016. Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science 351:173–76 [DOI] [PubMed] [Google Scholar]
- 43.Bauerlein FJB, Saha I, Mishra A, Kalemanov M, Martinez-Sanchez A, et al. 2017. In situ architecture and cellular interactions of polyQ inclusions. Cell 171:179–87 [DOI] [PubMed] [Google Scholar]
- 44.Li L, Liu H, Dong P, Li D, Legant WR, et al. 2016. Real-time imaging of Huntingtin aggregates diverting target search and gene transcription. eLife 5:e17056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramdzan YM, Trubetskov MM, Ormsby AR, Newcombe EA, Sui X, et al. 2017. Huntingtin inclusions trigger cellular quiescence, deactivate apoptosis, and lead to delayed necrosis. Cell Rep. 19:919–27 [DOI] [PubMed] [Google Scholar]
- 46.Skinner PJ, Koshy BT, Cummings CJ, Klement IA, Helin K, et al. 1997. Ataxin-1 with an expanded glutamine tract alters nuclear matrix–associated structures. Nature 389:971–74 [DOI] [PubMed] [Google Scholar]
- 47.Klement IA, Skinner PJ, Kaytor MD, Yi H, Hersch SM, et al. 1998. Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell 95:41–53 [DOI] [PubMed] [Google Scholar]
- 48.Katsuno M, Adachi H, Kume A, Li M, Nakagomi Y, et al. 2002. Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron 35:843–54 [DOI] [PubMed] [Google Scholar]
- 49.Takeyama K, Ito S, Yamamoto A, Tanimoto H, Furutani T, et al. 2002. Androgen-dependent neurodegeneration by polyglutamine-expanded human androgen receptor in Drosophila. Neuron 35:855–64 [DOI] [PubMed] [Google Scholar]
- 50.Montie HL, Cho MS, Holder L, Liu Y, Tsvetkov AS, et al. 2009. Cytoplasmic retention of polyglutamine-expanded androgen receptor ameliorates disease via autophagy in a mouse model of spinal and bulbar muscular atrophy. Hum. Mol. Genet 18:1937–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jackson WS, Tallaksen-Greene SJ, Albin RL, Detloff PJ. 2003. Nucleocytoplasmic transport signals affect the age at onset of abnormalities in knock-in mice expressing polyglutamine within an ectopic protein context. Hum. Mol. Genet 12:1621–29 [DOI] [PubMed] [Google Scholar]
- 52.Emamian ES, Kaytor MD, Duvick LA, Zu T, Tousey SK, et al. 2003. Serine 776 of ataxin-1 is critical for polyglutamine-induced disease in SCA1 transgenic mice. Neuron 38:375–87 [DOI] [PubMed] [Google Scholar]
- 53.Duvick L, Barnes J, Ebner B, Agrawal S, Andresen M, et al. 2010. SCA1-like disease in mice expressing wild-type ataxin-1 with a serine to aspartic acid replacement at residue 776. Neuron 67:929–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lai S, O’Callaghan B, Zoghbi HY, Orr HT. 2011. 14–3-3 binding to ataxin-1 (ATXN1) regulates its dephosphorylation at Ser-776 and transport to the nucleus. J. Biol. Chem 286:34606–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fryer JD, Yu P, Kang H, Mandel-Brehm C, Carter AN, et al. 2011. Exercise and genetic rescue of SCA1 via the transcriptional repressor Capicua. Science 334:690–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu HC, Tan Q, Rousseaux MW, Wang W, Kim JY, et al. 2017. Disruption of the ATXN1–CIC complex causes a spectrum of neurobehavioral phenotypes in mice and humans. Nat. Genet 49:527–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Palazzolo I, Burnett BG, Young JE, Brenne PL, La Spada AR, et al. 2007. Akt blocks ligand binding and protects against expanded polyglutamine androgen receptor toxicity. Hum. Mol. Genet 16:1593–603 [DOI] [PubMed] [Google Scholar]
- 58.Palazzolo I, Stack C, Kong L, Musaro A, Adachi H, et al. 2009. Overexpression of IGF-1 in muscle attenuates disease in a mouse model of spinal and bulbar muscular atrophy. Neuron 63:316–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rinaldi C, Bott LC, Chen KL, Harmison GG, Katsuno M, et al. 2012. Insulinlike growth factor (IGF)-1 administration ameliorates disease manifestations in a mouse model of spinal and bulbar muscular atrophy. Mol. Med 18:1261–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Polanco MJ, Parodi S, Piol D, Stack C, Chivet M, et al. 2016. Adenylyl cyclase activating polypeptide reduces phosphorylation and toxicity of the polyglutamine-expanded androgen receptor in spinobulbar muscular atrophy. Sci. Transl. Med 8:370ra181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mukherjee S, Thomas M, Dadgar N, Lieberman AP, Iniguez-Lluhi JA. 2009. Small ubiquitin-like modifier (SUMO) modification of the androgen receptor attenuates polyglutamine-mediated aggregation. J. Biol. Chem 284:21296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chua JP, Reddy SL, Yu Z, Giorgetti E, Montie HL, et al. 2015. Disrupting SUMOylation enhances transcriptional function and ameliorates polyglutamine androgen receptor–mediated disease. J. Clin. Investig 125:831–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zboray L, Pluciennik A, Curtis D, Liu Y, Berman-Booty LD, et al. 2015. Preventing the androgen receptor N/C interaction delays disease onset in a mouse model of SBMA. Cell Rep. 13:2312–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC, et al. 2004. SUMO modification of Huntingtin and Huntington’s disease pathology. Science 304:100–4 [DOI] [PubMed] [Google Scholar]
- 65.O’Rourke JG, Gareau JR, Ochaba J, Song W, Rasko T, et al. 2013. SUMO-2 and PIAS1 modulate insoluble mutant huntingtin protein accumulation. Cell Rep. 4:362–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gu X, Cantle JP, Greiner ER, Lee CY, Barth AM, et al. 2015. N17 Modifies mutant Huntingtin nuclear pathogenesis and severity of disease in HD BAC transgenic mice. Neuron 85:726–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kratter IH, Zahed H, Lau A, Tsvetkov AS, Daub AC, et al. 2016. Serine 421 regulates mutant huntingtin toxicity and clearance in mice. J. Clin. Investig 126:3585–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Custer SK, Garden GA, Gill N, Rueb U, Libby RT, et al. 2006. Bergmann glia expression of polyglutamine-expanded ataxin-7 produces neurodegeneration by impairing glutamate transport. Nat. Neurosci 9:1302–11 [DOI] [PubMed] [Google Scholar]
- 69.Furrer SA, Mohanachandran MS, Waldherr SM, Chang C, Damian VA, et al. 2011. Spinocerebellar ataxia type 7 cerebellar disease requires the coordinated action of mutant ataxin-7 in neurons and glia, and displays non-cell-autonomous Bergmann glia degeneration. J. Neurosci 31:16269–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bradford J, Shin JY, Roberts M, Wang CE, Li XJ, Li S. 2009. Expression of mutant huntingtin in mouse brain astrocytes causes age-dependent neurological symptoms. PNAS 106:22480–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Benraiss A, Wang S, Herrlinger S, Li X, Chandler-Militello D, et al. 2016. Human glia can both induce and rescue aspects of disease phenotype in Huntington disease. Nat. Commun 7:11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Monks DA, Johansen JA, Mo K, Rao P, Eagleson B, et al. 2007. Overexpressioin of wild-type androgen receptor in muscle recapitulates polyglutamine disease. PNAS 104:113–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramzan F, McPhail M, Rao P, Mo K, Halievski K, et al. 2015. Distinct etiological roles for myocytes and motor neurons in a mouse model of Kennedy’s disease/spinobulbar muscular atrophy. J. Neurosci 35:6444–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lieberman AP, Yu Z, Murray S, Peralta R, Low A, et al. 2014. Peripheral androgen receptor gene suppression rescues disease in mouse models of spinal and bulbar muscular atrophy. Cell Rep. 7:774–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cortes CJ, Ling SC, Guo LT, Hung G, Tsunemi T, et al. 2014. Muscle expression of mutant androgen receptor accounts for systemic and motor neuron disease phenotypes in spinal and bulbar muscular atrophy. Neuron 82:295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang N, Gray M, Lu XH, Cantle JP, Holley SM, et al. 2014. Neuronal targets for reducing mutant huntingtin expression to ameliorate disease in a mouse model of Huntington’s disease. Nat. Med 20:536–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dunah AW, Jeong H, Griffin A, Kim YM, Standaert DG, et al. 2002. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington’s disease. Science 296:2238–43 [DOI] [PubMed] [Google Scholar]
- 78.Shimohata T, Nakajima T, Yamada M, Uchida C, Onodera O, et al. 2000. Expanded polyglutamine stretches interact with TAFII130, interfering with CREB-dependent transcription. Nat. Genet 26:29–36 [DOI] [PubMed] [Google Scholar]
- 79.Steffan JS, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu YZ, et al. 2000. The Huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription. PNAS 97:6763–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McCampbell A, Taylor JP, Taye AA, Robitschek J, Li M, et al. 2000. CREB-binding protein sequestration by expanded polyglutamine. Hum. Mol. Genet 9:2197–202 [DOI] [PubMed] [Google Scholar]
- 81.Nucifora FC Jr., Sasaki M, Peters MF, Huang H, Cooper JK, et al. 2001. Interference by huntingtin and atrophin-1 with CBP-mediated transcription leading to cellular toxicity. Science 291:2423–28 [DOI] [PubMed] [Google Scholar]
- 82.McCampbell A, Taye AA, Whitty L, Penney E, Steffan JS, Fischbeck KH. 2001. Histone deacetylase inhibitors reduce polyglutamine toxicity. PNAS 98:15179–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, et al. 2001. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature 413:739–43 [DOI] [PubMed] [Google Scholar]
- 84.Mykowska A, Sobczak K, Wojciechowska M, Kozlowski P, Krzyzosiak WJ. 2011. CAG repeats mimic CUG repeats in the misregulation of alternative splicing. Nucleic Acids Res. 39:8938–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jiang YJ, Che MX, Yuan JQ, Xie YY, Yan XZ, Hu HY. 2011. Interaction with polyglutamine-expanded huntingtin alters cellular distribution and RNA processing of huntingtin yeast two-hybrid protein A (HYPA). J. Biol. Chem 286:25236–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sathasivam K, Neueder A, Gipson TA, Landles C, Benjamin AC, et al. 2013. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. PNAS 110:2366–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cleary JD, Ranum LP. 2013. Repeat-associated non-ATG (RAN) translation in neurological disease. Hum. Mol. Genet 22:R45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Banez-Coronel M, Ayhan F, Tarabochia AD, Zu T, Perez BA, et al. 2015. RAN translation in Huntington disease. Neuron 88:667–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gasset-Rosa F, Chillon-Marinas C, Goginashvili A, Atwal RS, Artates JW, et al. 2017. Polyglutamine-expanded huntingtin exacerbates age-related disruption of nuclear integrity and nucleocytoplasmic transport. Neuron 94:48–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jones L, Houlden H, Tabrizi SJ. 2017. DNA repair in the trinucleotide repeat disorders. Lancet Neurol. 16:88–96 [DOI] [PubMed] [Google Scholar]
- 91.Hinckelmann MV, Zala D, Saudou F. 2013. Releasing the brake: restoring fast axonal transport in neurodegenerative disorders. Trends Cell Biol. 23:634–43 [DOI] [PubMed] [Google Scholar]
- 92.Morfini G, Pigino G, Szebenyi G, You Y, Pollema S, Brady ST. 2006. JNK mediates pathogenic effects of polyglutamine-expanded androgen receptor on fast axonal transport. Nat. Neurosci 9:907–16 [DOI] [PubMed] [Google Scholar]
- 93.Katsuno M, Adachi H, Minamiyama M, Waza M, Tokui K, et al. 2006. Reversible disruption of dynactin 1–mediated retrograde axonal transport in polyglutamine-induced motor neuron degeneration. J. Neurosci 26:12106–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kemp MQ, Poort JL, Baqri RM, Lieberman AP, Breedlove SM, et al. 2011. Impaired motoneuronal retrograde transport in two models of SBMA implicates two sites of androgen action. Hum. Mol. Genet 20:4475–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Saudou F, Humbert S. 2016. The biology of huntingtin. Neuron 89:910–26 [DOI] [PubMed] [Google Scholar]
- 96.Engelender S, Sharp AH, Colomer V, Tokito MK, Lanahan A, et al. 1997. Huntingtin-associated protein 1 (HAP1) interacts with the p150Glued subunit of dynactin. Hum. Mol. Genet 6:2205–12 [DOI] [PubMed] [Google Scholar]
- 97.Gunawardena S, Her LS, Brusch RG, Laymon RA, Niesman IR, et al. 2003. Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron 40:25–40 [DOI] [PubMed] [Google Scholar]
- 98.Zala D, Hinckelmann MV, Yu H, Lyra da Cunha MM, Liot G, et al. 2013. Vesicular glycolysis provides on-board energy for fast axonal transport. Cell 152:479–91 [DOI] [PubMed] [Google Scholar]
- 99.Ferrer I, Goutan E, Marin C, Rey MJ, Ribalta T. 2000. Brain-derived neurotrophic factor in Huntington disease. Brain Res. 866:257–61 [DOI] [PubMed] [Google Scholar]
- 100.Zuccato C, Marullo M, Conforti P, MacDonald ME, Tartari M, Cattaneo E. 2008. Systematic assessment of BDNF and its receptor levels in human cortices affected by Huntington’s disease. Brain Pathol. 18:225–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Strand AD, Baquet ZC, Aragaki AK, Holmans P, Yang L, et al. 2007. Expression profiling of Huntington’s disease models suggests that brain-derived neurotrophic factor depletion plays a major role in striatal degeneration. J. Neurosci 27:11758–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Virlogeux A, Moutaux E, Christaller W, Genoux A, Bruyere J, et al. 2018. Reconstituting corticostriatal network on-a-chip reveals the contribution of the presynaptic compartment to Huntington’s disease. Cell Rep. 22:110–22 [DOI] [PubMed] [Google Scholar]
- 103.Brouillet E, Hantraye P, Ferrante RJ, Dolan R, Leroy-Willig A, et al. 1995. Chronic mitochondrial energy impairment produces selective striatal degeneration and abnormal choreiform movements in primates. PNAS 92:7105–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Browne SE, Bowling AC, MacGarvey U, Baik MJ, Berger SC, et al. 1997. Oxidative damage and metabolic dysfunction in Huntington’s disease: selective vulnerability of the basal ganglia. Ann. Neurol 41:646–53 [DOI] [PubMed] [Google Scholar]
- 105.Jenkins BG, Rosas HD, Chen YC, Makabe T, Myers R, et al. 1998. 1H NMR spectroscopy studies of Huntington’s disease: correlations with CAG repeat numbers. Neurology 50:1357–65 [DOI] [PubMed] [Google Scholar]
- 106.Saft C, Zange J, Andrich J, Muller K, Lindenberg K, et al. 2005. Mitochondrial impairment in patients and asymptomatic mutation carriers of Huntington’s disease. Mov. Disord 20:674–79 [DOI] [PubMed] [Google Scholar]
- 107.Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. 2006. Transcriptional repression of PGC-1α by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell 127:59–69 [DOI] [PubMed] [Google Scholar]
- 108.Powers WJ, Videen TO, Markham J, McGee-Minnich L, Antenor-Dorsey JV, et al. 2007. Selective defect of in vivo glycolysis in early Huntington’s disease striatum. PNAS 104:2945–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Buck E, Zugel M, Schumann U, Merz T, Gumpp AM, et al. 2017. High-resolution respirometry of fine-needle muscle biopsies in pre-manifest Huntington’s disease expansion mutation carriers shows normal mitochondrial respiratory function. PLOS ONE 12:e0175248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Josefsen K, Nielsen SM, Campos A, Seifert T, Hasholt L, et al. 2010. Reduced gluconeogenesis and lactate clearance in Huntington’s disease. Neurobiol. Dis 40:656–62 [DOI] [PubMed] [Google Scholar]
- 111.Ferro A, Carbone E, Zhang J, Marzouk E, Villegas M, et al. 2017. Short-term succinic acid treatment mitigates cerebellar mitochondrial OXPHOS dysfunction, neurodegeneration and ataxia in a Purkinje-specific spinocerebellar ataxia type 1 (SCA1) mouse model. PLOS ONE 12:e0188425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kazachkova N, Raposo M, Montiel R, Cymbron T, Bettencourt C, et al. 2013. Patterns of mitochondrial DNA damage in blood and brain tissues of a transgenic mouse model of Machado–Joseph disease. Neurodegener. Dis 11:206–14 [DOI] [PubMed] [Google Scholar]
- 113.Giorgetti E, Yu Z, Chua JP, Shimamura R, Zhao L, et al. 2016. Rescue of metabolic alterations in AR113Q skeletal muscle by peripheral androgen receptor gene silencing. Cell Rep. 17:125–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rocchi A, Milioto C, Parodi S, Armirotti A, Borgia D, et al. 2016. Glycolytic-to-oxidative fiber-type switch and mTOR signaling activation are early-onset features of SBMA muscle modified by high-fat diet. Acta Neuropathol. 132:127–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Beal MF, Kowall NW, Ellison DW, Mazurek MF, Swartz KJ, Martin JB. 1986. Replication of the neurochemical characteristics of Huntington’s disease by quinolinic acid. Nature 321:168–71 [DOI] [PubMed] [Google Scholar]
- 116.Raymond LA. 2017. Striatal synaptic dysfunction and altered calcium regulation in Huntington disease. Biochem. Biophys. Res. Commun 483:1051–62 [DOI] [PubMed] [Google Scholar]
- 117.Heng MY, Detloff PJ, Wang PL, Tsien JZ, Albin RL. 2009. In vivo evidence for NMDA receptor–mediated excitotoxicity in a murine genetic model of Huntington disease. J. Neurosci 29:3200–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Okamoto S, Pouladi MA, Talantova M, Yao D, Xia P, et al. 2009. Balance between synaptic versus extrasynaptic NMDA receptor activity influences inclusions and neurotoxicity of mutant huntingtin. Nat. Med 15:1407–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Papadia S, Hardingham GE. 2007. The dichotomy of NMDA receptor signaling. Neuroscientist 13:572–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tang TS, Tu H, Chan EY, Maximov A, Wang Z, et al. 2003. Huntingtin and huntingtin-associated protein 1 influence neuronal calcium signaling mediated by inositol-(1,4,5) triphosphate receptor type 1. Neuron 39:227–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pchitskaya E, Popugaeva E, Bezprozvanny I. 2018. Calcium signaling and molecular mechanisms underlying neurodegenerative diseases. Cell Calcium 70:87–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shakkottai VG, do Carmo Costa M, Dell’Orco JM, Sankaranarayanan A, Wulff H, Paulson HL. 2011. Early changes in cerebellar physiology accompany motor dysfunction in the polyglutamine disease spinocerebellar ataxia type 3. J. Neurosci 31:13002–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dell’Orco JM, Pulst SM, Shakkottai VG. 2017. Potassium channel dysfunction underlies Purkinje neuron spiking abnormalities in spinocerebellar ataxia type 2. Hum. Mol. Genet 26:3935–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hansen ST, Meera P, Otis TS, Pulst SM. 2013. Changes in Purkinje cell firing and gene expression precede behavioral pathology in a mouse model of SCA2. Hum. Mol. Genet 22:271–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jayabal S, Chang HH, Cullen KE, Watt AJ. 2016. 4-aminopyridine reverses ataxia and cerebellar firing deficiency in a mouse model of spinocerebellar ataxia type 6. Sci. Rep 6:29489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hourez R, Servais L, Orduz D, Gall D, Millard I, et al. 2011. Aminopyridines correct early dysfunction and delay neurodegeneration in a mouse model of spinocerebellar ataxia type 1. J. Neurosci 31:11795–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mark MD, Krause M, Boele HJ, Kruse W, Pollok S, et al. 2015. Spinocerebellar ataxia type 6 protein aggregates cause deficits in motor learning and cerebellar plasticity. J. Neurosci 35:8882–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kasumu AW, Hougaard C, Rode F, Jacobsen TA, Sabatier JM, et al. 2012. Selective positive modulator of calcium-activated potassium channels exerts beneficial effects in a mouse model of spinocerebellar ataxia type 2. Chem. Biol 19:1340–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bushart DD, Chopra R, Singh V, Murphy GG, Wulff H, Shakkottai VG. 2018. Targeting potassium channels to treat cerebellar ataxia. Ann. Clin. Transl. Neurol 5:297–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chopra R, Wasserman AH, Pulst SM, De Zeeuw CI, Shakkottai VG. 2018. Protein kinase C activity is a protective modifier of Purkinje neuron degeneration in cerebellar ataxia. Hum. Mol. Genet 27:1396–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Paulsen JS, Long JD, Ross CA, Harrington DL, Erwin CJ, et al. 2014. Prediction of manifest Huntington’s disease with clinical and imaging measures: a prospective observational study. Lancet Neurol. 13:1193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tabrizi SJ, Scahill RI, Owen G, Durr A, Leavitt BR, et al. 2013. Predictors of phenotypic progression and disease onset in premanifest and early-stage Huntington’s disease in the TRACK-HD study: analysis of 36-month observational data. Lancet Neurol. 12:637–49 [DOI] [PubMed] [Google Scholar]
- 133.Ross CA, Aylward EH, Wild EJ, Langbehn DR, Long JD, et al. 2014. Huntington disease: natural history, biomarkers and prospects for therapeutics. Nat. Rev. Neurol 10:204–16 [DOI] [PubMed] [Google Scholar]
- 134.Albin RL, Burke JF. 2015. Potential trade-offs in treatment of premanifest Huntington’s disease. Mov. Disord 30:1319–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kegel KB, Kim M, Sapp E, McIntyre C, Castano JG, et al. 2000. Huntingtin expression stimulates endosomal–lysosomal activity, endosome tubulation, and autophagy. J. Neurosci 20:7268–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ravikumar B, Duden R, Rubinsztein DC. 2002. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet 11:1107–17 [DOI] [PubMed] [Google Scholar]
- 137.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, et al. 2004. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet 36:585–95 [DOI] [PubMed] [Google Scholar]
- 138.Pratt WB, Gestwicki JE, Osawa Y, Lieberman AP. 2015. Targeting Hsp90/Hsp70-based protein quality control for treatment of adult onset neurodegenerative diseases. Annu. Rev. Pharmacol. Toxicol 55:353–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang AM, Miyata Y, Klinedinst S, Peng HM, Chua JP, et al. 2013. Activation of Hsp70 reduces neurotoxicity by promoting polyglutamine protein degradation. Nat. Chem. Biol 9:112–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Neef DW, Jaeger AM, Thiele DJ. 2011. Heat shock transcription factor 1 as a therapeutic target in neurodegenerative diseases. Nat. Rev. Drug Discov 10:930–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Labbadia J, Cunliffe H, Weiss A, Katsyuba E, Sathasivam K, et al. 2011. Altered chromatin architecture underlies progressive impairment of the heat shock response in mouse models of Huntington disease. J. Clin. Investig 121:3306–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang AM, Morishima Y, Clapp KM, Peng HM, Pratt WB, et al. 2010. Inhibition of hsp70 by methylene blue affects signaling protein function and ubiquitination and modulates polyglutamine protein degradation. J. Biol. Chem 285:15714–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Albin RL, Miller RA. 2016. Mini-review: retarding aging in murine genetic models of neurodegeneration. Neurobiol. Dis 85:73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kordasiewicz HB, Stanek LM, Wancewicz EV, Mazur C, McAlonis MM, et al. 2012. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron 74:1031–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Moore LR, Rajpal G, Dillingham IT, Qutob M, Blumenstein KG, et al. 2017. Evaluation of antisense oligonucleotides targeting ATXN3 in SCA3 mouse models. Mol. Ther. Nucleic Acids 7:200–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Harper SQ, Staber PD, He X, Eliason SL, Martins IH, et al. 2005. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. PNAS 102:5820–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xia H, Mao Q, Eliason SL, Harper SQ, Martins IH, et al. 2004. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat. Med 10:816–20 [DOI] [PubMed] [Google Scholar]
- 148.Yang S, Chang R, Yang H, Zhao T, Hong Y, et al. 2017. CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington’s disease. J. Clin. Investig 127:2719–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.McBride JL, Pitzer MR, Boudreau RL, Dufour B, Hobbs T, et al. 2011. Preclinical safety of RNAi-mediated HTT suppression in the rhesus macaque as a potential therapy for Huntington’s disease. Mol. Ther 19:2152–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Dufour BD, Smith CA, Clark RL, Walker TR, McBride JL. 2014. Intrajugular vein delivery of AAV9-RNAi prevents neuropathological changes and weight loss in Huntington’s disease mice. Mol. Ther 22:797–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Taylor JP, Lieberman AP, Fischbeck KH. Repeat expansion and neurological disease In Diseases of the Nervous System, 3rd Ed. (Asbury AK, McKhann GM, McDonald WI, Goadsby P, McArthur J, eds). Cambridge University Press, Cambridge, 2002, pp 32–54. [Google Scholar]