Abstract
PURPOSE:
The purpose of this study was to investigate if human-extracted MRI tumor phenotypes of breast cancer could predict receptor status and tumor molecular subtype using MRIs from The Cancer Genome Atlas project.
MATERIALS AND METHODS:
Our retrospective interpretation study utilized the analysis of HIPAA-compliant breast MRI data from The Cancer Imaging Archive. One hundred and seven preoperative breast MRIs of biopsy proven invasive breast cancers were analyzed by 3 fellowship-trained breast-imaging radiologists. Each study was scored according to the Breast Imaging Reporting and Data System lexicon for mass and nonmass features. The Spearman rank correlation was used for association analysis of continuous variables; the Kruskal-Wallis test was used for associating continuous outcomes with categorical variables. The Fisher-exact test was used to assess correlations between categorical image-derived features and receptor status. Prediction of estrogen receptor (ER), progesterone receptor, human epidermal growth factor receptor, and molecular subtype were performed using random forest classifiers.
RESULTS:
ER+ tumors were associated with the absence of rim enhancement (P = 0.019, odds ratio [OR] 5.5), heterogeneous internal enhancement (P = 0.02, OR 6.5), peritumoral edema (P = 0.0001, OR 10.0), and axillary adenopathy (P = 0.04, OR 4.4). ER+ tumors were smaller than ER− tumors (23.7 mm vs 29.2 mm, P = 0.02, OR 8.2). All of these variables except the lack of axillary adenopathy were also associated with progesterone receptor+ status. Luminal A tumors (n = 57) were smaller compared to nonLuminal A (21.8 mm vs 27.5 mm, P = 0.035, OR 7.3) and lacked peritumoral edema (P = 0.001, OR 6.8). Basal like tumors were associated with heterogeneous internal enhancement (P = 0.05, OR 10.1), rim enhancement (P = 0.05, OR6.9), and perituomral edema (P = 0.0001, OR 13.8).
CONCLUSIONS:
Human extracted MRI tumor phenotypes may be able to differentiate those tumors with a more favorable clinical prognosis from their more aggressive counterparts.
Introduction
Breast cancers are heterogeneous with different molecular profiles, clinical behaviors, responses to therapy, and outcomes. The diverse spectrum of breast cancers has prompted extensive research assessing the underpinnings of this complex disease. The importance of tumor biology impacting prognosis and directing treatment has spurred work to identify biomarkers in an effort to predict tumor biology.1–4 Several commercially available tumor assays are currently in utilized by clinicians treating breast cancer in order to best tailor specific treatments for differing tumor subtypes based on differing tumor biology, such as the PAM50 assay. The PAM 50 assay analyzes the activity of 58 genes to estimate the risk of distant recurrence of hormone receptor positive breast cancer up to 10 years after diagnosis.5 Comparing gene expression profiles from each patient’s tumor with the 4 PAM50 molecular profiles is used to identify clinically relevant molecular subtypes of breast cancer-Luminal A, Luminal B, HER2 enriched, and basal like (BL).5 The spectrum of molecular subtypes confers different patterns of disease expression, response to treatment, and prognosis.1–4,6
At present, MRI of the breast remains a sensitive technique for detecting and characterizing breast masses given its ability to provide both spatial and kinetic information. MRI is particularly reliable in detailing tumor volume, extent, associated lymphadenopathy, and involvement of the chest wall, skin, and the nipple-areolar complex.7,8 A recent trend toward identifying surrogate markers for genetic testing has resulted in numerous publications describing relationships between MRI based phenotypes and tumor genetics.9–13 Several groups have reported primarily on computer extracted imaging features and their ability to predict tumor genetics.9,11,12,14–34 Yamaguchi et al described the relative lack of contrast washout with Luminal A tumors compared to non-Luminal A tumors which they postulated was due to differential histological components (ie, Luminal A tumors were more likely to have associated ductal carcinoma in situ resulting in less washout).15 Blaschke et al described an association between rapid contrast uptake and rapid washout with Luminal B and human epidermal growth factor receptor (HER2) subtype cancers which they postulated could be due to the associations of these tumors with vascular endothelial growth factor and the propensity for neoangiogenesis and therefore early contrast enhancement.16
Grimm et al found that differing molecular subtypes had different disease distribution patterns (ie, extent of disease) on MRI with HER2 and Luminal B tumors associated with a higher likelihood of multifocal and/or multicentric disease and nodal metastases.17 Li et al described associations between computer-extracted MRI phenotypes and hormone receptor status and found that estrogen receptor (ER) negative tumors tended to be larger, more irregular, more heterogeneous in contrast uptake, and have faster contrast uptake than ER+ tumors.12 The results of these studies offer a possible framework in which to further understand image based phenotypes or biomarkers of clinically relevant histopathologic prognostic indicators.
In an effort to assess whether image-based phenotypes are associated with specific tumor genetics, and therefore able to predict tumor aggressiveness, The Cancer Imaging Archive (TCIA) was formed through multi-institutional participation providing breast imaging studies from those genetically fully sequenced tumors in The Cancer Genome Atlas (TCGA) database.35,36 Utilizing the TCIA database, we sought to assess whether human-extracted MRI features of known cancers in the TCGA database were associated with tumor receptor status (ie, ER, Progesterone Receptor [PR], and HER2) and the molecular subtypes of breast cancer based on the PAM50 assay.
Methods
Study Population
De-identified patient data were downloaded from the National Cancer Institute’s TCGA Breast Invasive Carcinoma and TCIA initiatives under IRB-approved HIPAA compliant protocols, this study was deemed exempt from institutional IRB approval and informed consent was waived. At the time of our study, 108 cases with breast MRI data, a subset of the entire TCGA breast cancer cases, had been collected and made available in the TCIA (http://www.cancerimagingarchive.net).35 One patient was excluded due to incomplete image sequences, leaving the total number of patients included in the study at 107 (Table 1), “entire study population.” Those patients with “research based” PAM50 assay results and complete MRI data available (“PAM 50 subgroup,” n = 88) were included in a subgroup analysis of imaging phenotype predictors of molecular subtypes (Table 2). Of note, for these TCGA samples, the clinical PAM50 assay results could not be obtained—only the “research-based” assay outputs as described by Parker et al were available.5 Cancer subtypes were determined by using the PAM50 classifier on TCGA mRNAseq data as described by Cirello et al.37 84–91 patients have been previously reported.9–13 The prior articles reported on the ability of computer extracted MR imaging phenotypes of the known breast cancers in the common database to predict stage, molecular classifications, and recurrence scores of the known breast cancers in the current study.9–13 Our study included all patients with available MRI and TCGA clinical data and included all patients with available PAM50 assays. Additionally, the current study focused exclusively on the ability of human extracted or radiologist scored MRI phenotypes to predict receptor status and molecular subtypes of breast cancer.
TABLE 1.
Patient demographics, entire study population (n = 107)
| Patient characteristics | |
|---|---|
| Sex | Women |
| Age (mean, range) | 53.7, (29–82) |
| Tumor characteristics | |
| Size (mean, range) | 24.6 mm, (7.8–87.6 mm) |
| Origin (frequency) | |
| Ductal | 91 (85%) |
| Lobular | 12 (11.2%) |
| Mixed | 4 (3.7%) |
| ER Status | |
| Positive | 90 (84.1%) |
| Negative | 17 (15.9%) |
| PR Status | |
| Positive | 81 (75.7%) |
| Negative | 26 (24.3%) |
| HER2 Status | |
| Positive | 22 (20.5%) |
| Negative | 85 (79.4%) |
Abbreviations: ER, estrogen receptor; PR, progesterone receptor, HER2, human epidermal growth factor receptor.
TABLE 2.
PAM50 tumor subtypes, PAM50 subgroup (n = 88)
| Luminal A | 57 (64.8%) |
| Luminal B | 10(11.4%) |
| HER2 Enriched | 5 (5.7%) |
| Basal Like | 11 (12.5%) |
| Normal Like | 5 (5.7%) |
Image Data
All MRIs were acquired utilizing a standard double breast coil and were performed on 1.5 T GE (Milwaukee, WI) whole body MRI systems. T1-weighted dynamic contrast-enhanced and T2-weighted images were analyzed for this study, including 1 pre and 3–5 postcontrast images obtained using a T1-weighted 3 dimensional spoiled gradient echo sequence with a gadolinium-based contrast agent. In-plane resolution on the dynamic series was 0.53–0.86 mm, and spacing between slices was 2–3 mm. Imaging was performed across 4 institutions including Memorial Sloan Kettering Cancer Center, Mayo Clinic, University of Pittsburgh Medical Center, and Roswell Park Cancer Institute. All 107 TCGA patients with biopsy proven invasive breast cancer and available breast MRI results were assessed by a panel of radiologists (TCGA Breast Phenotype Research Group) using Clear Canvas software (ClearCanvas, Toronto, Ontario, Canada).38 Imaging was scored based on a variety of mass and nonmass features according to the Breast Imaging Reporting and Data System 2013 edition39 (Table 3). Each breast MRI examination was reviewed and annotated independently by 3 of the 11 expert board-certified breast radiologists who were blinded to outcomes with final scoring reached by consensus. The breast imaging experience of radiologists in this study ranged from 4 to 29 years.
TABLE 3.
Imaging phenotypes (n = 107)—distribution of radiologist scored BIRADS features in the entire study population
| Breast observations | Radiologist score (frequency) |
|---|---|
| Breast composition | Extreme (13)/Heterogeneous (43)/Scattered (42)/Fatty (9) |
| Background enhancement | Marked (5)/Moderate (23)/Mild (53)/Minimal (26) |
| Extent | Unicentric (58)/Multicentric (21)/Multifocal (28) |
| Mass observations | |
| Shape | Round-oval (38)/Irregular (69) |
| Margin | Circumscribed (8)/Spiculated (67)/Irregular (32) |
| T2 pattern | Homogeneous (56)/Heterogeneous (51) |
| T2 signal intensity | Hyperintense (25)/Isointense (78)/Hypointense (4) |
| T2 peritumoral edema | Present (29)/Absent (78) |
| Dark internal septations | Present (2)/Absent (105) |
| Internal enhancement | Heterogeneous (63)/Homogeneous (44) |
| Rim enhancement | Present (17)/Absent (90) |
| Lesion size | Maximum diameter |
| Axillary adenopathy | Present (27)/Absent (80) |
| Pectoralis invasion | Present (2)/Absent (105) |
| Chest wall invasion | Present (0)/Absent (107) |
| Edema | Present (1)/Absent (0) |
| Nipple Inversion | Present/Absent |
| Nipple retraction | Present (6)/Absent (101) |
| Nipple Invastion | Present (3)/Absent (104) |
| Skin thickening | Present/Absent |
| Skin invasion | Present (2)/Absent (105) |
| Skin retraction | Present/Absent |
| Nonmass observations | |
| Associated nonmass enhancement | Present (33)/Absent (74) |
| Nonmass distribution | Focal (16)/Linear (16)/Diffuse (11)/Segmental (0)/Regional (16)/Multiple Regions (19) |
| Nonmass internal enhancement | Heterogeneous (28)/Clumped (44)/Clustered Ring (14) |
Statistical Analysis
Batch corrected gene expression data was obtained from the TCGA Data Portal. The Spearman rank correlation was used for association analysis of continuous variables; the Kruskal-Wallis test was used for associating continuous outcomes with categorical variables.40 The Fisher-exact test was used to assess correlations between categorical image-derived features and ER, PR, and HER2 receptor status.41 Correction for multiple hypothesis testing was conducted using the Benjamini-Hochberg method to control the False Discovery Rate.42 Associations with adjusted P-values no larger than 0.05 were deemed as statistically significant and selected for interpretation. Prediction of ER, PR, HER2, and molecular subtype (from PAM50 data) were performed using random forest classifiers. The random forest classifier is an ensemble of decision-trees, can handle a mixture of numeric (eg, lesion size) and categorical data (eg, Breast Imaging Reporting and Data System margin descriptor) to predict class labels, and is capable of handling large numbers of features. The prediction performance is assessed via a bootstrap sampling (using an “out-of-bag” set for model assessment).43 Variable importance plots were obtained by measuring the improvement in the probability of correct classification induced by a specific variable.44 An adjusted P-value of 0.05 was selected as the threshold for determining statistical significance. For ER and/or PR status prediction, the difference of the random forest classifier’s performance using area under the receiver operating characteristic curve, area under the curve (AUC) relative to random classi-fication (AUC = 0.5) is assessed via P-value from a Mann-Whitney hypothesis test (using R-package, “verification”).45
Results
The entire study population of 107 biopsy proven invasive breast cancers included 91(85%) ductal, 12 (11.2%) lobular, and 4 (3.7%) mixed tumors, the majority of which were ER+ (n = 90). Thirteen cases were triple negative breast cancers (TNBCs) (Table 1). Of those cases with PAM50 assay data available, the PAM50 subgroup, (n = 88), 76 (86.4%) were ductal, 10 (11.4%) were lobular, and 2 (2.2%) were mixed, the majority of which, 64.8%, were classified as Luminal A (n = 57). Eleven of the PAM50 subgroup tumors were BL, of which 10 were TNBC with the final BL tumor being ER+/PR+/HER2+ (Table 2).
MRI Phenotype Association with Histopathologic Variables
Based on the entire study population (n = 107), the absence of all of the following were associated with ER+ tumors: rim enhancement (P = 0.019, odds ratio [OR] 5.5), heterogeneous internal enhancement (P = 0.02, OR 6.5), peritumoral edema (P = 0.0001, OR 10.0), and axillary adenopathy (P = 0.04, OR 4.4). ER+ tumors were smaller than ER+ tumors (23.7 mm vs 29.2 mm, P = 0.02, OR 8.2). All of these variables except the lack of axillary adenopathy also maintained their statistically significant associations with PR status (Table 4).
TABLE 4.
Imaging phenotype associations—Receptor status and molecular subtype associations with the three imaging phenotypes which emerged as main features associated with specific tumor profile. Of note, the 5 normal like tumors from the PAM50 subgroup (n = 88) were not included as no imaging phenotypes were associated with this molecular subtype
| Entire study population (n = 107) | Average size (mm) | Rim enhancement (%) | Heterogeneous internal enhancement (%) | Peritumoral edema (%) |
|---|---|---|---|---|
| ER+ | 23.7 | 11 (10/90) | 53 (48/90) | 19 (17/90) |
| ER- | 29.2 | 41 (7/17) | 88 (15/17) | 70 (12/17) |
| PR+ | 23.8 | 11 (9/81) | 50 (41/81) | 58 (15/81) |
| PR- | 27.1 | 31 (8/26) | 85 (22/26) | 54 (14/26) |
| HER2+ | 23.5 | 18(4/22) | 59(13/22) | 32 (7/22) |
| HER2- | 24.8 | 15 (13/85) | 58 (50/85) | 26 (22/85) |
| TNBC | 27.9 | 46 (6/13) | 92 (12/13) | 77 (10/13) |
| PAM50 subgroup (n = 88) | ||||
| Lum A | 21.8 | 9 (5/57) | 47 (27/57) | 10 (6/57) |
| Lum B | 25.5 | 10 (1/10) | 70 (7/10) | 20 (2/10) |
| HER2 | 27.8 | 40 (2/5) | 40 (2/5) | 60 (3/5) |
| Basal Like | 29.2 | 45 (5/11) | 91 (10/11) | 62 (8/11) |
Abbreviations: ER, estrogen receptor; PR, progesterone receptor, HER2, human epidermal growth factor receptor; TNBC, triple negative breast cancer; Lum A, Luminal A; Lum B, Luminal B.
Based on the PAM50 subgroup (n = 88), when looking at the Luminal A tumors compared to non-Luminal A (n = 57), Luminal A tumors were smaller (21.8 mm vs 27.5 mm for non-Luminal A, P = 0.035, OR 7.3) and they lacked peritumoral edema (P = 0.001, OR 6.8). When comparing BL tumors (n = 13) to non-BL (n = 75), BL tumors were associated with heterogeneous internal enhancement (P = 0.05, OR 10.1), rim enhancement (P = 0.05, OR 6.9), and perituomral edema (P = 0.0001, OR 13.8) (Table 5).
TABLE 5.
Imaging phenotype associations—associations between receptor status of tumors in entire the study population (n=107) as well as in the PAM50 subgroup and imaging phenotypes which reached statistical significance
| Entire study population (n = 107) | Imaging phenotype | BH corrected P value | Odds ratio |
|---|---|---|---|
| Receptor status: | |||
| ER+ | T2 Peritumoral edema absent | 0.0002 | 10.0 |
| Lesion Size | 0.02 | 8.2 | |
| Heterogeneous internal enhancement absent | 0.02 | 6.5 | |
| Rim enhancement absent | 0.02 | 5.5 | |
| Axillary LAD absent | 0.04 | 4.4 | |
| PR+ | T2 Peritumoral edema absent | 0.001 | 5.0 |
| Lesion Size | 0.05 | 4.7 | |
| Heterogeneous internal enhancement absent | 0.02 | 3.9 | |
| Rim enhancement absent | 0.05 | 3.5 | |
| PAM50 subgroup (n=88) | |||
| Molecular subtype | |||
| Luminal A | Lesion size | 0.04 | 7.3 |
| T2 peritumoral edema absent | 0.001 | 6.8 | |
| Basal Like | T2 peritumoral edema present | 0.001 | 13.8 |
| Rim enhancement | 0.05 | 6.9 |
Abbreviations: ER, estrogen receptor; PR, progesterone receptor.
Imaging Phenotype Importance in Predicting Receptor Status
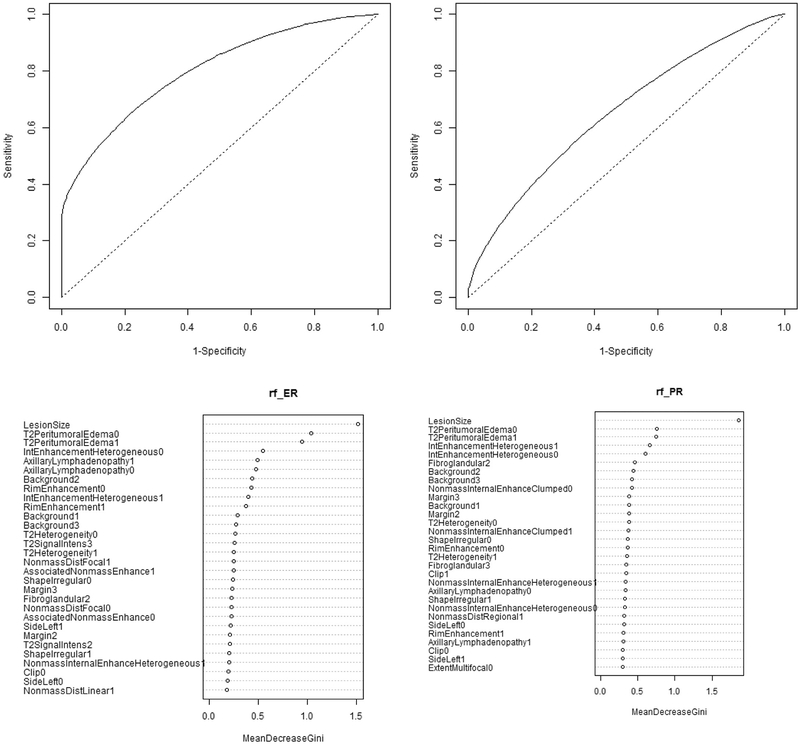
In the entire study population (n = 107), the following variables ranked at the top of the variable importance plots using the random forest classifier to predict receptor status (Fig 1A–B): ER status: lesion size, peritumoral edema, and internal enhancement (AUC 0.8, 95% confidence interval [CI] 0.654–0.88, P = 0.0001); PR status: lesion size, peritumoral edema, and internal enhancement (AUC 0.633, 95% CI 0.505–0.776, P = 0.02). MRI tumor phenotypes did not reliably predict HER2 status (AUC 0.48, P = 0.4). Variable importance plots rank order those features most important in predicting a given receptor status, for both ER and PR tasks, a sharp drop-off was noted after the first few features making those at the top most important in predicting receptor status in this population.
FIG 1.
(A, B) Receptor status prediction tasks and associated variable importance plots from full study population (n = 107). AUC for all MRI phenotypes used to predict receptor status and corresponding variable importance plots. The AUC for the ER status prediction task is 0.8 with confidence intervals [0.654, 0.88], and P-value 0. For PR status prediction the AUC is 0.633 with confidence interval [0.505, 0.776], P-value = 0.02. For both ER and PR status, imaging features performed well at predicting receptor status. Variable importance plots rank order those features most important in predicting a given receptor status, for both ER and PR tasks, a sharp drop-off was noted after the first few features making those at the top most important in predicting receptor status in this population.
Discussion
The results from this study demonstrate that human extracted MRI features may be able to differentiate tumors with favorable prognosis from their more aggressive counterparts. Lesion size, internal enhancement characteristics, and the presence or absence of peritu-moral edema emerged as potential predictors of tumor receptor pro-files and molecular subtypes. From our results, it is apparent that those tumors which carry a more favorable prognosis and have known better response to treatment (ie, ER+, PR+, and Luminal A tumors) tended to be smaller, lacked heterogeneous internal enhancement, rim enhancement, peritumoral edema, and, with ER+ tumors, lacked axillary adenopathy. Those tumors which demonstrated heterogeneous internal enhancement, rim enhancement, and peritumoral edema conferred a worse prognosis as they were associated with triple negative and BL tumors which have a known resistance to treatment and poorer outcomes.46–48
The relationship between imaging phenotypes and tumor biology has been suggested in recent publications assessing breast MRI phenotypes and tumor genetics. Several published studies describe rim enhancement and heterogeneous internal enhancement associated with TNBC.49–53 Given the known crossover between TNBC and BL subtypes, our results suggest that these features when present may indicate the more aggressive TNBC and BL subtypes. Authors have hypothesized these features may be related to differential greater microvessel density, expression levels of vascular endothelial growth factor, and overall higher cellular proliferation markers leading to larger tumors with heterogeneous enhancement patterns.49–53 Lesion size was also an important variable in predicting receptor status and ranked at or near the top of all variable importance plots for each receptor subtype, with ER+ and PR+ tumors tending to be smaller than their receptor negative counterparts (Table 4, Fig 1A–B). These results are in keeping with recent publications highlighting the importance of size in predicting overall pathologic stage,9 risk of recurrence,11 and in the aforementioned reports of the tendency of more aggressive BL and TNBC subtypes to be larger than their Luminal A/B counterparts. In Burnside and Drukker et al’s study evaluating computer extracted image phenotypes from breast MRI and their ability to predict stage, size was the most powerful predictor of overall pathologic stage.9 Li et al recently reported the ability of computer extracted imaging features to predict risk of recurrence and found that heterogeneous enhancement (enhancement texture) and larger size correlated with a higher risk of recurrence based on various gene assay models, including PAM 50.11
There are several limitations of this study. First, the relatively small sample size resulted from the relatively limited number of patients with breast MRIs in the TCGA and/or TCIA databases with even fewer having PAM 50 assay data available. Given that the majority of tumors in this dataset were ER+, PR+, HER2−, and Luminal A, the receptor and molecular subtype analyses may have been skewed given relative fewer numbers of receptor negative and nonLuminal A tumors. The inability to identify any meaningful associations between image phenotypes and HER2 status is likely due to the relative small number of HER2+ tumors in our dataset (n = 22). Future studies will hopefully allow for a data set with more balanced molecular and receptor subtypes. The MRIs included in this study were acquired more than 10 years ago at 4 different institutions, each with variations in image acquisition including differing equipment and protocols, and therefore may not be representative of current MR practice standards with newer advanced technology and protocols. Despite such limitations, the TCGA and/or TCIA databases remain the largest publicly available resource for radiogenomics research. Future directions include expanding the study to a larger data set, including more current imaging technology and standardized protocols, with hopes to identify reproducible imaging biomarkers of clinically relevant prognostic indicators.
Conclusions
Human extracted MRI-based tumor phenotypes may be able to differentiate those tumors with a more favorable clinical prognosis from their more aggressive counterparts. The use of such MRI-based tumor phenotypes has the potential to provide useful prognostic information, and lead to a better understanding of tumor biology and aid in tailoring treatments for more precise breast cancer care.
Supplementary Material
Acknowledgment
This work was completed with the TCGA Breast Phenotype Research Group.
Grant Support: EJS and EAM were funded in part through the NIH/NCI Grant, P30 CA008748. MLG was funded in part through the NIH/NCI Grant, U01CA195564. AR was supported by CCSG Bioinformatics Shared Resource, P30 CA01667, an Institutional Research Grant from the University of Texas MD Anderson Cancer Center (MD Anderson) and a Career Development Award from the MD Anderson Brain Tumor SPORE. ESB was supported by the NIH grant, K24CA194251.
Footnotes
Supplementary materials
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1067/j.cpradiol.2018.08.003.
References
- 1.Bagaria SP, et al. Personalizing breast cancer staging by the inclusion of ER, PR, and HER2. JAMA Surg 2014;149 125–9. [DOI] [PubMed] [Google Scholar]
- 2.Huber KE, Carey LA, Wazer DE. Breast cancer molecular subtypes in patients with locally advanced disease: impact on prognosis, patterns of recurrence, and response to therapy Seminars in Radiation Oncology. Elsevier; 2009. [DOI] [PubMed] [Google Scholar]
- 3.Wiechmann L, et al. Presenting features of breast cancer differ by molecular sub-type. Ann Surg Oncol 2009;16:2705–10. [DOI] [PubMed] [Google Scholar]
- 4.Nguyen PL, et al. Breast cancer subtype approximated by estrogen receptor, progesterone receptor, and HER-2 is associated with local and distant recurrence after breast-conserving therapy. J Clin Oncol 2008;26:2373–8. [DOI] [PubMed] [Google Scholar]
- 5.Parker JS, et al. Supervised risk predictor of breast cancer based on intrinsic sub-types. J Clin Oncol 2009;27:1160–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ko ES, et al. Apparent diffusion coefficient in estrogen receptor positive invasive ductal breast carcinoma: correlations with tumor-stroma[C0]ratio. Radiology 2013;271:30–7. [DOI] [PubMed] [Google Scholar]
- 7.Morris EA. Diagnostic breast MR imaging: current status and future directions. Radiol Clin N Am 2007;45:863–80. [DOI] [PubMed] [Google Scholar]
- 8.Morrow M, Waters J, Morris E. MRI for breast cancer screening, diagnosis, and treatment. Lancet 2011;378:1804–11. [DOI] [PubMed] [Google Scholar]
- 9.Burnside ES, et al. Using computer[C0]extracted image phenotypes from tumors on breast magnetic resonance imaging to predict breast cancer pathologic stage. Cancer 2016;122:748–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo W, et al. Prediction of clinical phenotypes in invasive breast carcinomas from the integration of radiomics and genomics data. J Med Imaging 2015;2 041007–041007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, et al. MR imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of MammaPrint, Oncotype DX, and PAM50 gene assays. Radiology 2016;281:152110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer 2016;2:16012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Y, et al. Deciphering genomic underpinnings of quantitative MRI-based radio-mic phenotypes of invasive breast carcinoma. Scientific Reports 2015;5:17787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto S, et al. Radiogenomic analysis of breast cancer using MRI: a preliminary study to define the landscape. Am J Roentgenol 2012;199:654–63. [DOI] [PubMed] [Google Scholar]
- 15.Yamaguchi K, et al. Intratumoral heterogeneity of the distribution of kinetic parameters in breast cancer: comparison based on the molecular subtypes of invasive breast cancer. Breast Cancer 2015;22:496–502. [DOI] [PubMed] [Google Scholar]
- 16.Blaschke E, Abe H. MRI phenotype of breast cancer: kinetic assessment for molecular subtypes. J Magn Reson Imaging 2015;42:920–4. [DOI] [PubMed] [Google Scholar]
- 17.Grimm LJ, et al. Can breast cancer molecular subtype help to select patients for preoperative MR imaging? Radiology 2014;274:352–8. [DOI] [PubMed] [Google Scholar]
- 18.Giger ML, Karssemeijer N, Schnabel JA. Breast image analysis for risk assessment, detection, diagnosis, and treatment of cancer. Annu Rev Biomed Eng 2013;15:327–57. [DOI] [PubMed] [Google Scholar]
- 19.Mazurowski MA, et al. Radiogenomic analysis of breast cancer: luminal B molecular subtype is associated with enhancement dynamics at MR imaging. Radiology 2014;273:365–72. [DOI] [PubMed] [Google Scholar]
- 20.Bhooshan N, et al. Computerized three-class classification of MRI-based prognostic markers for breast cancer. Phys Med Biol 2011;56:5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agner SC, et al. Computerized image analysis for identifying triple-negative breast cancers and differentiating them from other molecular subtypes of breast cancer on dynamic contrast-enhanced MR images: a feasibility study. Radiology 2014;272:91–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashraf AB, et al. Identification of intrinsic imaging phenotypes for breast cancer tumors: preliminary associations with gene expression profiles. Radiology 2014;272:374–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhooshan N, et al. Cancerous breast lesions on dynamic contrast-enhanced MR images: computerized characterization for image-based prognostic markers. Radiology 2010;254:680–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen W, et al. Automatic identification and classification of characteristic kinetic curves of breast lesions on DCE-MRI. Med Phys 2006;33:2878–87. [DOI] [PubMed] [Google Scholar]
- 25.Chen W, et al. Computerized interpretation of breast MRI: investigation of enhancement-variance dynamics. Med Phys 2004;31:1076–82. [DOI] [PubMed] [Google Scholar]
- 26.Chen W, et al. Volumetric texture analysis of breast lesions on contrast enhanced magnetic resonance images. Magn Reson Med 2007;58:562–71. [DOI] [PubMed] [Google Scholar]
- 27.Chen W, et al. Computerized assessment of breast lesion malignancy using DCEMRI: robustness study on two independent clinical datasets from two manufacturers. Acad Radiol 2010;17:822–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Golden DI, et al. Dynamic contrast-enhanced MRI-based biomarkers of therapeutic response in triple-negative breast cancer. J Am Med Inf Assoc 2013;20:1059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grimm LJ, Zhang J, Mazurowski MA. Computational approach to radiogenomics of breast cancer: luminal A and luminal B molecular subtypes are associated with imaging features on routine breast MRI extracted using computer vision algorithms. J Magn Reson Imaging 2015;42:902–7. [DOI] [PubMed] [Google Scholar]
- 30.Lehmann BD, Pietenpol JA. Identification and use of biomarkers in treatment strategies for triple[C0]negative breast cancer subtypes. J Pathol 2014;232: 142–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makkat S, et al. Deconvolution-based dynamic contrast-enhanced MR imaging of breast tumors: correlation of tumor blood flow with human epidermal growth factor receptor 2 status and clinicopathologic findings—preliminary results 1. Radiology 2008;249:471–82. [DOI] [PubMed] [Google Scholar]
- 32.Mussurakis S, Buckley DL, Horsman A. Prediction of axillary lymph node status in invasive breast cancer with dynamic contrast-enhanced MR imaging. Radiology 1997;203:317–21. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto S, et al. Breast cancer: radiogenomic biomarker reveals associations among dynamic contrast-enhanced MR imaging, long noncoding RNA, and metastasis. Radiology 2015;275:384–92. [DOI] [PubMed] [Google Scholar]
- 34.Youk JH, et al. Triple-negative invasive breast cancer on dynamic contrast-enhanced and diffusion-weighted MR imaging: comparison with other breast cancer subtypes. Eur Radiol 2012;22:1724–34. [DOI] [PubMed] [Google Scholar]
- 35.Clark K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 2013;26:1045–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Network CGA. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ciriello G, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 2015;163:506–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mongkolwat P, et al. The National Cancer Informatics Program (NCIP) Annotation and Image Markup (AIM) Foundation Model. J Digit Imaging 2014;27:692–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris EA CC, Lee CH. ACR BI-RADS® Magnetic Resonance Imaging ACR BI-RADS® Atlas, Breast Imaging Reporting and Data System. Reston, VA: American College of Radiology; 2013. [Google Scholar]
- 40.Kruskal WH, Wallis WA. Use of ranks in one-criterion variance analysis. J Am Stat Assoc 1952;47:583–621. [Google Scholar]
- 41.Fisher RA. On the interpretation of χ2 from contingency tables, and the calculation of P. J R Stat Soc 1922;85:87–94. [Google Scholar]
- 42.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. Ser B (Methodol) 1995;57:289–300. [Google Scholar]
- 43.Breiman L Random forests. Mach Learn 2001;45:5–32. [Google Scholar]
- 44.Breiman L Manual on setting up, using, and understanding random forests v3.1. 2002. URL http://oz.berkeley.edu/users/breiman/Using_random_forests_V3; 2002 1.
- 45.Mason SJ, Graham NE. Areas beneath the relative operating characteristics (ROC) and relative operating levels (ROL) curves: Statistical significance and interpretation. Q R Meteorol Soc 2002;128:2145–66. [Google Scholar]
- 46.Voduc KD, et al. Breast cancer subtypes and the risk of local and regional relapse. J Clin Oncol 2010;28:1684–91. [DOI] [PubMed] [Google Scholar]
- 47.Millar EK, et al. Prediction of local recurrence, distant metastases, and death after breast-conserving therapy in early-stage invasive breast cancer using a five-bio-marker panel. J Clin Oncol 2009;27:4701–8. [DOI] [PubMed] [Google Scholar]
- 48.Carey LA, Cheang MCU, Perou CM. Genomics, Prognosis, and Therapeutic Interventions. Wolters Kluwer Health Adis (ESP); 2014. [Google Scholar]
- 49.Wang Y, et al. Estrogen receptor—negative invasive breast cancer: imaging features of tumors with and without human epidermal growth factor receptor type 2 overexpression 1. Radiology 2008;246:367–75. [DOI] [PubMed] [Google Scholar]
- 50.Teifke A, et al. Dynamic MR imaging of breast lesions: Correlation with microvessel distribution pattern and histologic characteristics of prognosis 1. Radiology 2006;239:351–60. [DOI] [PubMed] [Google Scholar]
- 51.Kobayashi M, et al. Two different types of ring[C0]like enhancement on dynamic MR imaging in breast cancer: correlation with the histopathologic findings. J Magn Reson Imaging 2008;28:1435–43. [DOI] [PubMed] [Google Scholar]
- 52.Jinguji M, et al. Rim enhancement of breast cancers on contrast-enhanced MR imaging: relationship with prognostic factors. Breast Cancer 2006;13:64–73. [DOI] [PubMed] [Google Scholar]
- 53.Costantini M, et al. Magnetic resonance imaging features in triple-negative breast cancer: comparison with luminal and HER2-overexpressing tumors. Clin Breast Cancer 2012;12:331–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.