Abstract
Three vignettes exemplify the potential of combining EM and X-ray crystallographic data with molecular dynamics (MD) simulation to explore the architecture, dynamics and functional properties of multicomponent macromolecular complexes. The first two describe how EM and X-ray crystallography were used to solve structures of the ribosome and the ARP2/3-actin complex, which enabled MD simulations that elucidated functional dynamics. The third describes how EM, X-ray crystallography, and microsecond MD simulations of a GPCR:G protein complex were used to explore transmembrane signaling by the β-adrenergic receptor. Recent technical advancements in EM, X-ray crystallography and computational simulation create unprecedented synergies for integrative structural biology to reveal new insights into heretofore intractable biological systems.
Introduction
The synergy among structural studies using X-ray crystallography, Electron Microscopy (EM) and image reconstruction is especially powerful for inferring the design and functional properties of multicomponent macromolecular complexes. A common strategy is to fit high-resolution X-ray or NMR structures into lower-resolution EM-derived molecular boundaries of the entire complex. Another approach is to use low to moderate resolution EM maps or EM-derived models to obtain initial phases for higher resolution X-ray crystallographic structure determination, exemplified by determination of the 9 Å structure of the 50S ribosome from an electron cryomicroscopy (cryoEM) map [1]. To our knowledge, the first example of this approach was determination of the 28 Å X-ray structure of tomato bushy stunt virus by the Harrison laboratory [2] using a phasing model provided by an EM map of negatively stained particles determined in the Crowther laboratory [3].
In the last decade there have been significant advances in EM technology and image processing methods, including improvements in microscope and stage stability, coherence and intensity of the electron beam, and, most recently, the development of direct electron detectors with a sensitivity that competes with film [4–8]. There is an expanding number of subnanometer resolution cryoEM maps of macromolecular complexes derived by single particle analysis, some approaching atomic resolution, exemplified by large megadalton (MDa) structures such as icosahedral viruses [9] and ribosomes [10– 12] and smaller complexes such as the TRP channel [13,14] (reviewed in this issue [15]), the HIV ENV complex [16], and γ-secretase [17]. Bona fide atomic resolution structures have been derived from analysis of 2D crystals of membrane proteins reconstituted into lipid bilayers [18] and protein structures derived by electron crystallography of 3D crystals [19]. Meanwhile, X-ray crystallography has been used to solve structures of membrane proteins and their complexes in membrane mimetic environments such as bicelles [20] and lipidic cubic phases [21]. Most recently, X-ray crystallography using a free electron laser (XFEL) has been used to solve structures from sub-micron crystals of large, biologically active complexes [22] and membrane proteins in a lipid environment [23] (reviewed in this issue by Feld and Frank [24]). In addition to these impressive advances in experimental structural biology, there are increasingly accurate and accessible computational tools for modeling structures with limited or sparse experimental data and investigating the functional dynamics of complex biological systems [25–28].
There are excellent historical and methodological reviews on the marriage of EM and X-ray crystallography, and on the myriad integrative structural biology approaches that include spectroscopic methods (e.g., NMR, EPR, FRET [29,30]). Here we focus on structural studies using EM and X-ray crystallography, which were brought into the realm of integrative structural biology by applications of molecular dynamics (MD) simulations. Also powerful, but not covered here, are macromolecular structural modeling methods that are guided or restrained by experimental data. These computational methods include the Integrative Modeling Platform from the Sali laboratory [25] and Rosetta from the Baker laboratory [31].
We highlight three vignettes that exemplify integrative structural and computational biology. In the first, studies of the ribosome employed cryoEM and X-ray crystallography to independently solve the same structure, from which MD studies were then possible. The second example describes how X-ray crystallography and electron tomography were used in tandem to solve the structure of the ARP2/3-actin complex, which enabled further analysis by MD simulation. In the third, an X-ray structure of the β-adrenergic receptor in a complex with the heterotrimeric G protein Gs was used to interpret structural flexibility of the complex by single particle EM image analysis. Microsecond MD simulations of the β-adrenergic receptor revealed the possible pathway of ligands to the orthosteric binding site and a mechanism for signaling across the membrane.
Structure and conformational dynamics of ribosomes
Ribosomes are massive MDa-sized ribonucleoprotein complexes that serve as the universal translator of genetic information, responsible for the conversion of messenger RNA (mRNA) transcripts to the polypeptides they encode. Prokaryotic ribosomes are typically formed by two subunits, constituting the 2.3 MDa 70s assembly: the larger 50s subunit is composed of 34 proteins and 3,000 ribosomal RNA (rRNA) nucleotides, and the smaller 30s subunit is formed from 21 proteins and 1,500 rRNA nucleotides. The mRNA transcript travels through a channel in the small subunit, which mediates the interactions between the anticodon-tRNA and the codons of the transcript. Catalytic activity for peptide bond formation resides in the large subunit. Together, the 50s and 30s subunits form the three sites for binding unique transfer RNA (tRNA) molecules [32].
Ribosomes were first visualized in cells by Palade using EM of fixed and stained thin sections of cells [33]. The overall architecture of the prokaryotic ribosome and the quaternary structure of many of its component proteins were gleaned from EM and immunolabeling of negatively stained preparations [34,35]. These studies provided the first, albeit low resolution, images of isolated ribosomes. Significant advances were made possible by cryoEM and three-dimensional image reconstruction of two-dimensional crystalline sheets of the eukaryotic 80s ribosome, which revealed the polypeptide exit pathway [36,37]. CryoEM of 70s ribosome particles embedded in vitreous ice [38,39] yielded maps at 23–25 Å resolution, revealing potential pathways in both subunits for the movement of the mRNA and the growing polypeptide, thereby enabling the first simple models of translation (Figure 1a). Five years later, cryoEM and single particle analysis of 70s ribosomes showed conformational changes that accompany binding of elongation factor G and subsequent GTP hydrolysis that permit mRNA translocation in the active site: rotation of the 30s subunit with respect to the 50s subunit and a subsequent widening of the mRNA channel [40]. This study demonstrated the significant capability of EM to directly detect mechanistically relevant conformational intermediates that may not be amenable to X-ray crystallography.
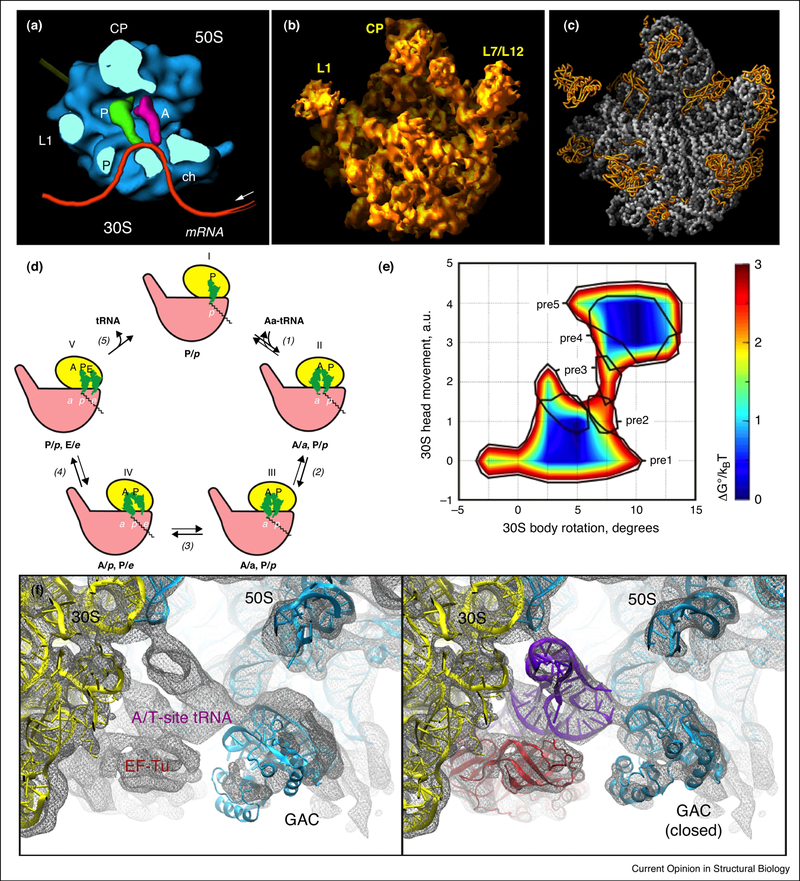
Figure 1. Structure and dynamics of the ribosome.
A more accurate mechanism for translation has emerged due to progressive improvements in resolution and the use of integrative structural methods. (a) A 25-Å resolution 3D reconstruction derived by cryoEM and single particle image analysis of E. coli 70s ribosomes, shown as a surface representation and in cross-section at the tRNA binding sites in the active site, provided a framework upon which to speculate the path of the mRNA transcript through the translation machinery [38]. (b) A surface representation of a 9-Å resolution electron density map of the H. marismortui 50s ribosomal subunit, derived by X-ray crystallography. Initial phasing, leading to location of heavy atoms, was made possible by molecular replacement using a 20-Å cryoEM map derived from the 25-Å reconstruction shown in (a) [1]. (c) The 2.4-Å X-ray structure of the H. marismortui 50s ribosomal subunit revealed the positions of all protein and nucleic acid components and showed that the active site is composed entirely of rRNA [43]. (d) Factor-free elongation cycle illustrating the conformational dynamics of the ribosome during translation. The small ribosomal subunit undergoes rigid body-type motions, shown here as tilting from an open or unlocked position (steps I, IV, and V) to a closed or locked conformation (steps II and III). In factor-assisted translation, EF-Tu –GTP catalyzes step 1 (docking of proper aminoacyl-tRNA) and EF-G-GTP catalyzes step 4 (translocation) [56]. (e) A plot of the free energy landscape of ribosome conformations during “hybrid” tRNA states. Each tRNA state is characterized by an ensemble of global conformational states of the 30S subunit, including the ratchet-like movement relative to the 50s subunit position, which fluctuate around a state with minimum free energy [50]. (f) All-atom molecular dynamics flexible fitting (MDFF) was used to model protein and rRNA components into the 6.7-Å resolution cryoEM map of the E. coli ribosome ternary complex EF-Tu-aminoacyl-tRNA-GDP stalled by the antibiotic kirromycin (TC-bound ribosome). Systems were typically composed of ~250,000 atoms. Structures were docked as rigid bodies into the corresponding maps and used as initial coordinates for flexible fitting (shown on the left). The resulting flexibly fitted structures are shown on the right [63].
Due to the heterogeneity, asymmetrical assembly, flexibility, and immense size of ribosomes, well-ordered, isotropic crystals were difficult to obtain. In the 1980s and 1990s the Yonath laboratory was able to obtain 3D crystals that exhibited diffraction to ~3 Å resolution [41,42]. However, pathological defects such as twinning effectively truncated the data sets to medium resolution. A major breakthrough came in the late 1990s when Steitz and colleagues determined a 9-Å resolution X-ray structure of the H. marismortui 50s ribosomal subunit (Figure 1b). Initial low-resolution phasing to 20-Å was performed by molecular replacement, with phases provided by a 20-Å cryoEM map [1]. The EM-derived phases allowed determination of the substructure of bound heavy atom clusters in the crystals and subsequent phase extension to 9-Å. The ability to phase the X-ray crystallographic data beginning only with the EM maps validated the accuracy of the EM reconstructions. Heretofore unseen features of the ribosome were revealed by the 9-Å electron density maps, yet the precise demarcation between the protein and RNA components was elusive.
In 2000, fundamental insight into the molecular basis of translation was first revealed in atomic detail by determination of the 2.4-Å X-ray structure of the 50s ribosomal subunit [43] (Figure 1c). This leap in resolution was made possible by modifications to crystal growth and harvesting conditions that significantly improved diffraction and prevented twinning. Additional prokaryotic ribosome subunit crystal structures followed quickly, permitting further understanding of the translation machinery [44,45]. These initial structures demonstrated that the active site was devoid of protein, and the enzymatic function of the ribosome was entirely due to RNA, a feature that had been previously proposed but not directly observed [46]. Crystal structures of complete ribosomes revealed the interfacial features of the large and small subunit [47] and, with higher resolution structures, the position of the mRNA transcript in the channel and how the tRNAs fit in each binding site [48]. We now have a treasure trove of crystal structures that elucidate every step of translation, from initiation to termination [32]. Utilizing newly developed direct electron detectors, a number of recent atomic resolution structures of bacterial, mammalian, and mitochondrial ribosomes have been determined via single particle cryoEM [10–12].
Recent studies employing single molecule fluorescence resonance energy transfer (smFRET; reviewed in [49]) and moderate resolution single particle cryoEM reconstructions (e.g., [50,51]) suggest that ribosomal function involves substantial conformational heterogeneity and stochastic dynamics [52] (Figure 1d,e). For example, during translocation, there is a low-energy threshold to interconversion between two conformations that appears to be facilitated by inherent structural features of the ribosome. Evidence of spontaneous transitions between these states [53] led to the hypothesis that passage from one step of translocation to the next is less dependent on external stimuli and more on conformational fluctuations intrinsic to the ribosome itself [54]. The major conformational states of the ribosome are distinguished by various intersubunit rotation angles and/or configurations of the tRNA in the active sites [50] (Figure 1e). Additional discrete conformational substates have been proposed recently based on smFRET and subsequent cryoEM studies of an rRNA mutant (reviewed in [55]).
A recent hypothesis has emerged in which ribosomal function can be described by a processive Brownian ratchet model [54]. In a general sense, the model proposes that the driving force for a molecular machine is Brownian motion. The motion is biased along a particular trajectory (i.e., there is a pawl or barrier to prevent reversion) by free energy released from chemical reactions, thereby effecting a functional end [56]. The general idea in explanation of ribosome function was initially proposed by Bretscher [57] and Spirin [58] independently, supported by the cryoEM analyses of Frank and Agarwal [40], and brilliantly elaborated as a mechanistic explanation of translation, again by Spirin [56] (Figure 1d).
Unfortunately, models for protein synthesis have been limited by gaps between static structural snapshots from the translation process and spectroscopic data related to kinetics. Computational methods and simulation have provided a valuable tool to integrate these structural and kinetic data. Models have been derived by fitting high-resolution crystal structures into lower-resolution EM maps, and have enabled the modeling of the structural transitions between different conformational states. Notable MD-based tools employed for this purpose include real-space refinement [59], normal mode analysis flexible fitting [60], conformational space sampling constrained by low-resolution maps and restrained by a deformable elastic network (DEN) [61,62], and molecular dynamics flexible fitting (MDFF) [63] (Figure 1f). Most of these applications have dealt most directly with static structures and shape-based transitions between structures rather than trying to capture the physical dynamics of the transitions directly. Nonetheless, physical simulation, as well as probabilistic integration tools such as IMP [25], provide powerful and readily generalizable tools for combining multi-resolution experimental data and physical constraints such as the chemical composition of the macromolecular components.
Structure and conformational dynamics of the ARP2/3-Actin Complex
Directional motility in cells is achieved by the assembly of a dense, branched network of actin filaments, producing a physical force that results in the extension of a leading edge. Without branching, actin is too long and flexible to produce sufficient pushing force [64]. Formation of branches between two actin filaments is mediated by the actin-related protein Arp2/3 complex, which is comprised of seven proteins, of which five subunits are named ARPC1–5 and serve a supporting role for the Arp2 and Arp3 subunits. Interaction of the Arp2/3 complex with an actin filament in the presence of ATP and nucleation promoting factors catalyzes the initiation of a new daughter actin strand [65]. In 2001, an immense step forward in the field of cell motility was achieved by Pollard and colleagues, who determined the X-ray structure of the inactive, non-nucleating Arp2/3 (Figure 2a) [66–68].
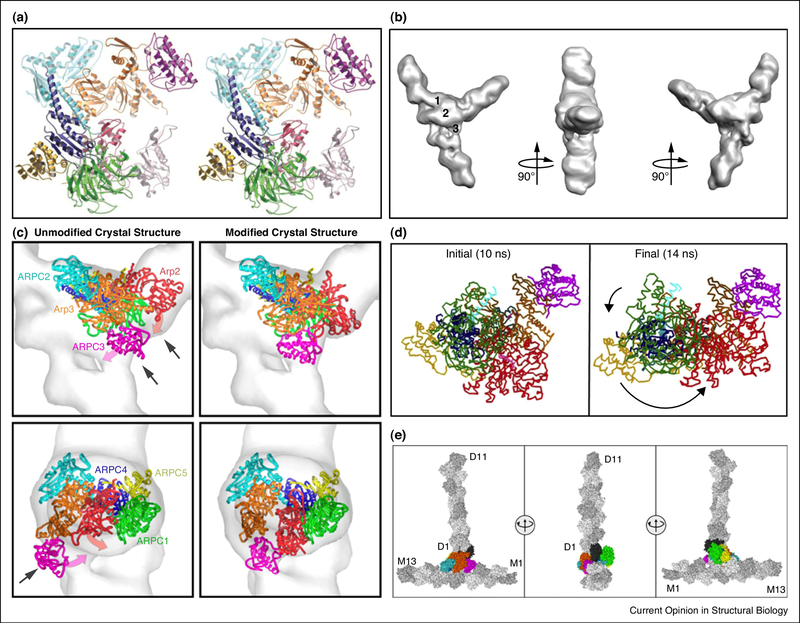
Figure 2. Structure and dynamics of the Arp2/3-actin complex.
(a) X-ray crystal structure of the inactive Arp2/3 complex viewed as a stereopair of ribbon diagrams [66]. (b) Reconstruction of a filament branch junction formed by the A. castellanii Arp2/3 complex. Three views are related by 90° clockwise rotations. Numbers indicate three bridges of density between the two branches [69]. (c) Fits of unmodified and modified crystal structures in the reconstruction of branch junctions. The “Unmodified” structure shows the best fit of the Arp2/3 complex crystal structure. Arrows indicate mismatches with the reconstruction. The magenta and red arrows indicate the movement of ARPC3 and Arp2 upon remodeling. “Modified” shows the best fit of the remodeled Arp2/3 complex [69]. (d) Complex in the initial and final conformations during MD simulations with application of directional forces between the Cα values of subdomains 3 and 4 of Arp2 in the inactive position and the target position next to Arp3, while restraining the movements of the Cα values of Arp3 subdomains 1, 2, and 4 [70]. (e) Starting structure for the simulation of the Arp2/3 complex branch junction. Three views related by 90° clockwise rotations, with protein subunits in surface representation and with mother (M1 to M13) and daughter (D1 to D11) filaments colored gray [65]. The common color code for subunits of the Arp2/3 complex is used throughout: Arp2, red; Arp3, orange; ARPC1, green; ARPC2, cyan; ARPC3, magenta; ARPC4, blue; and ARPC5, yellow. In (e) the Arp2/3 complex subunits are colored and labeled using the common coloring scheme except for Arp2 in black.
In 2008, Volkman and Hanein used dual-axis electron tomography followed by single-particle volume processing to generate a 3D reconstruction of a negatively stained A. castellanii actin branch junction at 26-Å resolution (Figure 2b). This was an essential contribution as it provided a basis to orient the crystal structures of the Arp2/3 complex in the context of an actin branch and to infer the active state conformation. Individual subunits of the Arp2/3 complex and actin filaments were docked by rigid body fitting into the tomogram. The ARPC1, ARPC2, ARPC4, ARPC5, and the daughter actin filament could be positioned within the molecular boundary of the reconstruction without modification. To fit the rest of the complex, Arp2 had to undergo a ~30-Å conformational shift that brought it next to Arp3 and ARPC3 (Figure 2c). In addition, actin monomers in the mother filament had to be modified from a filamentous to a monomeric conformation. This allowed the filament to untwist locally so that it could be positioned within the density map [69]. While the model gleaned from tomography provided insight regarding relative orientations of individual subunits and large conformational changes between inactive and active complex structures, the limited resolution precluded the understanding of atomistic details.
Steered MD simulations of the ARP2/3 complex provided insight into conformational changes that Arp2 undergoes from its inactive conformation to its position in the branched junction. Force was applied to Arp2 while restraining Arp3 to bring the two structures together as in the branched junction. The simulation showed that one block of the complex (Arp2, ARPC1, the globular domain of ARPC4 and 5) rotated 30° counterclockwise around a pivot point in the α-helix of ARPC4 to align Arp2 next to Arp3 (Figure 2d). The final structure of the junction buried more surface area than the inactive conformation [70]. MD in combination with protein-protein docking was also performed on the crystal structure of the ARPC2/ARPC4 heterodimer from the inactive Arp2/3 complex and an eight-monomer actin filament. These simulations generated an independent model similar to the EM model (Cα RMSD = 5.9 Å) and predicted key actin-binding residues on ARPC2 and ARPC4 that were confirmed by polymerization assays on the mutant constructs [71].
Subsequently, a large-scale MD study was performed by Voth and co-workers to test the stability and validity of the branched junction model. A 3 million atom simulation of the complex of 31 protein subunits (the mother filament with 13 ADP-actin subunits, the Arp2/3 complex, and the daughter filament with 11 ADP-actin subunits) (Figure 2e) in explicit solvent was performed for an aggregate time of 175 ns (one 75 ns simulation of the original EM model and two 50 ns simulations with alternate derived structures of actin or Arp2/3 complex). The simulations suggested that the branch structure was indeed very stable and that the interface between the Arp2/3 complex and the mother actin filament contains a large number of dynamic salt bridges and hydrophobic contacts that can form or break during the timescale of these simulations. Notably, these results improved the fits of individual complex subunits to the EM density maps [65]. As a test of the MD-based models, 1000 representative poses of each individual subunit from the last 25 ns of one of the simulations were docked by rigid body fitting into the EM map. Even though the MD was not constrained by the EM density, the simulations improved the fits of Arp3, ARPC2, the daughter filament and the mother filament. Cumulatively, these studies demonstrate the power of MD in providing atomistic detail beginning with low-resolution EM-based models and docked X-ray structures, and insights into dynamics that cannot be determined by static structures. MD simulations on large complexes such as the Arp2/3 complex are computationally demanding, but advances in computational resources are making such calculations feasible to a wider community.
Flexible α-helical Domain of Gsα in a β2-adrenergic receptor Gs complex
G protein coupled receptors (GPCRs) represent 1–2% of the human genome [72,73] and comprise about a third of all pharmaceutical targets. Extracellular signals such as light, odorants, or neurotransmitters are transduced by GPCRs to heterotrimeric G proteins, composed of an α subunit, and a βγ dimer, which reside on the cytoplasmic side of the plasma membrane. In the inactive state of the receptor-G protein complex, GDP is bound to Gα [74]. Upon activation, GDP is exchanged for GTP, and GTP-bound Gα can dissociate from both the GPCR and Gβγ. Until the GTP on Gα is hydrolyzed back to GDP, both Gα and Gβγ can regulate downstream effector proteins.
GPCRs exhibit multiple conformational states and are notoriously unstable when solubilized in detergents, greatly impeding structural studies. Important advances that have enabled structural analysis of GPCRs include identification of high-yield expression systems and a number of factors that stabilize the complexes: high-affinity ligands, new detergents, additives such as cholesterol hemisuccinate, thermostabilizing mutations, fusion proteins such as T4 lysozyme, and the addition of stabilizing nanobodies [75–79]. With this foundation of technical advances, Kobilka and colleagues achieved a seminal discovery in 2011 by solving the 3.2 Å X-ray crystal structure of an active-state β2-adrenergic receptor (β2AR) in complex with the heterotrimeric G protein Gsαβ1γ2 (Fig. 3a) [80]. This structure captured the agonist-bound receptor in complex with a nucleotide-free G protein, poised to bind activating GTP. When compared to isolated Gα structures, the most surprising aspect of the crystal structure was the relationship between the α-helical and Ras-like domains of Gsα. When nucleotide is bound, a cleft between these two domains forms the nucleotide binding site [81]. The β2AR-Gs structure represents a snapshot in the signaling cascade from receptor to G protein: the empty state of Gs subsequent to receptor dependent GDP release. The crystal structure showed that in the absence of bound nucleotide, there is a large rotational displacement (127°) of the α-helical domain of Gsα away from the Gsα Ras-like domain, relative to the X-ray structure of the nucleotide-bound G protein [80]. This rigid-body motion of the Gs α-helical domain, which was proposed previously using other techniques [82], exposes the nucleotide binding pocket, providing an elegantly simple mechanism for GTP/GDP exchange, the raison d’être of GPCRs.
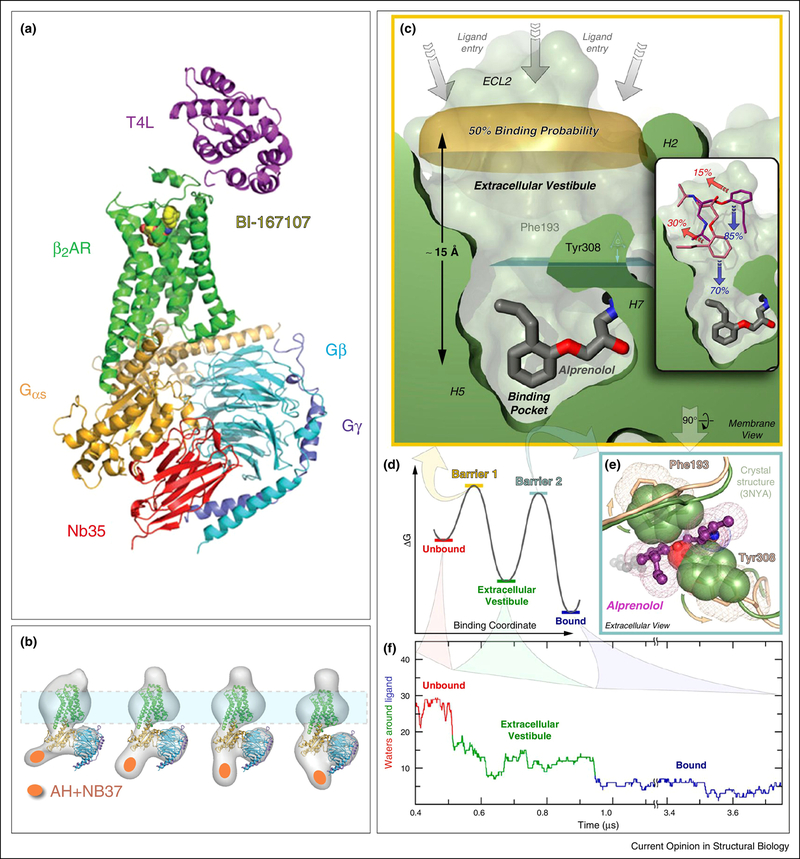
Figure 3. Structure and dynamics of a GPCR-G protein complex.
A) X-ray structure [80] of agonist (yellow spheres) bound β2AR (green) interacting with the heterotrimeric G protein Gs, composed of Gαs (orange), which has an extensive binding surface with the β2AR, and Gβ (cyan) and Gγ (purple). The Nb35 nanobody (red) binds between Gα and Gβ, and T4 lysozyme (magenta) was expressed as a fusion protein at the β2AR N-terminus. Nb35 and T4 lysozyme facilitated crystallization. B) Fitting the X-ray crystal structure of T4L-β2AR in A) into 2D projections of particle class averages imaged by EM [83]. The Nb37 nanobody (orange oval) binds to the α-helical (AH) domain of Gsα and was used in the EM experiments to visualize the conformational flexibility of the AH domain. (C-F) MD simulation of antagonist binding [89]. C) Extracellular vestibule and orthosteric binding pocket of β2AR in relation to the antagonist alprenolol. D) Energy barriers of binding alprenolol to the extracellular vestibule and the orthosteric binding site E) Residues impeding access to the orthosteric binding site F) Microsecond simulation of dehydration of alprenolol during binding to the β2AR in a ~50,000 atom system.
In a companion EM study, Skiniotis led a team that examined the same β2-adrenergic receptor-Gs complex (β2AR-Gs) [83]. A similar open orientation of the α-helical domain of Gsα was observed in the β2AR-Gs complex imaged by single particle EM [83]; the map was interpreted by fitting the β2AR-Gs X-ray structure into the EM density using rigid body docking (Fig. 3b). Binding of a nanobody (Nb37) specifically engineered to the α-helical domain also aided the identification of the α-helical domain in the EM reconstructions (Fig. 3b). Interestingly, addition of nucleotide or nucleotide mimetic caused the α-helical domain to swing closer to the Ras-like domain in the β2AR-Gs complex [83], approximating a closed conformation. Modeling of the α-helical domain in the closed state was performed manually to match the EM densities that resulted from the presence of nucleotide.
Although EM and X-ray crystallography provided structures of different conformational states of a GPCR-G protein complex, MD simulations add another perspective on transient conformations that may not be amenable to static structural analysis. For example, biochemical studies have suggested the importance of an “ionic lock” in GPCRs that binds the intracellular ends of helix 3 and 6 together in the inactive state. Disruption of this ionic lock, which occurs during rhodopsin activation [84], was thought to be a prerequisite to the active state of a GPCR. However, crystal structures of both the β1 and β2 adrenergic receptors bound to antagonist displayed no evidence of the ionic lock [85,86]. This enigma was reconciled by Dror et al., who used microsecond timescale MD simulations of the β2AR to discover that either in the apo state or bound to antagonist, the ionic lock frequently formed, but was in equilibrium with an unlocked inactive state of the receptor [87,88].
Another example of an intermediate conformation revealed by MD simulation involved in silico ligand binding studies with the β1and β2 adrenergic receptors, in which transitory binding of both agonist and antagonist was demonstrated at an extracellular vestibule, framed by ECL2 and 3 and transmembrane helices 5–7 [89]. Figure 3c illustrates the relationship between the extracellular vestibule and the orthosteric binding pocket of the β2AR. The energy barrier to the final, orthosteric ligand binding is indicated in Fig. 3d. One of the components of the energy barrier to ligand binding in the orthosteric site is dehydration of the ligand, as well as the orthosteric binding site; another is the conformation of certain residues, such as Phe 193 and Tyr 308 (Fig. 3e), which need to move apart before ligand can enter the binding pocket. The importance of longer MD simulations is apparent in Fig. 3F, which only reveals the orthosteric ligand binding at approximately one microsecond; this length of time was necessary to overcome the deep energy barrier introduced by the extracellular binding site. Thus, MD has proven a useful tool in the elucidation of transitory conformational states not typically amenable to EM or X-ray crystallography.
Conclusion
Historically, molecular simulation has played an important role in determining the likely conformation of large biomolecular assemblies given maps derived by EM and X-ray crystallography. In addition, simulation methods have been particularly useful to integrate data provided by a variety of methods and when the experimental data are insufficient to uniquely determine an atomic resolution conformation in a straightforward manner. The formal basis for such work was elegantly expressed in the context of NMR by Nilges and co-workers [90]. We expect that the use of such approaches will continue to increase and that new technical improvements such as direct electron detectors and free-electron-laser nanocrystallography will bring new opportunities and challenges in this regard.
As EM, crystallographic and spectroscopic approaches yield more high-resolution information regarding multiple conformational states of important macromolecules and complexes, one particular use of MD simulation is to “connect the dots”. The goal in such a cases is to obtain models not only of biomolecular structure but also of conformational transitions and dynamics in a manner based on experimental data and physical principles. This has long been done in a geometry-based fashion, from simple “morphing” [59] to more sophisticated methods such as geometry-based normal mode analysis [91,92]. However, high-fidelity prediction of dynamics in a manner that might be quantitatively predictive of experimental dynamics has been challenging, particularly for large complexes and slow transitions. The vignettes described above highlight important representative successes. Although many problems are indeed moderately tractable using current techniques, further progress in this area is not simply a matter of larger and faster computers. One key area for methodological development is how to incorporate experimental data most efficiently to construct dynamic models, while not biasing the dynamics themselves in potentially unphysical ways. Many approaches achieve one but not the other: morphing provides an extreme example of the first, while unconstrained MD simulation provides an example of the second. Many other approaches can yield thermodynamic information efficiently but not dynamics or dynamics only in the case where one can appropriately choose collective variables. Much more can be said on the trade-offs involved and how many promising approaches become challenging for complex biomolecular systems. Another important challenge for the future is efficiently incorporating experimental kinetic data. Needless to say, we believe the combination of data provided by intermediate and high-resolution structural methods and MD simulation will be a fruitful area in the years to come and that such integrative approaches will be key to understanding the structure and dynamics of macromolecular complexes.
Acknowledgments
We gratefully acknowledge support from NIH grants R01 HL48908, R01 GM066087, R01 GM084545, and P50 GM082545.
Footnotes
Frank and Agarwal 2000: CryoEM analysis of 70s ribosomes in various states due to the presence of different substrate analogs and effectors revealed two distinct rotations of the small subunit with respect to the large subunit during the translocation cycle.
Selmer et al. (2006): A 2.8 Å crystal structure of a prokaryotic 70s ribosome with partial to full occupancy of tRNAs at the A-, P-, and E-sites and mRNA bound in the translation center persists as one of the most complete ribosome structures.
Frank and Gonzalez (2010): An exhaustive review of the multiple structural methods and associated experiments that have been employed to probe the mechanism of translation, with a thesis put forward by the authors that the ribosome behaves as a Brownian ratchet.
Moore (2012): A fascinating assessment of the status of ribosome research that provides a concise historical account but, more importantly, lays out a “post-structural” roadmap where a combination of static structures, kinetic, energetic and thermodynamic data will be required for an accurate and complete movie of translation.
Trabuco et al. (2008): The development of the molecular dynamics flexible fitting (MDFF), described in this paper, was spearheaded by the need to model and morph ribosome crystal structures into lower resolution EM maps in order to understand the nature and extent of conformational changes in the subunits.
Robinson et al. (2001): Crystal structure of the Arp 2/3 complex (ARPC1, ARPC2, ARPC3, ARPC4, ARPC5, Arp2 and Arp3) solved to 2.0 Å resolution.
Rouiller et al. (2008): 3D reconstruction of the actin branch junction containing the Arp2/3 complex and both the mother and daughter actin filament solved to 26 Å resolution via electron tomography followed by single-particle volume processing.
Pfaendtner et al. (2011): Molecular dynamics simulations of the actin branch junction demonstrating the stability of the branch junction, the dynamic salt bridges and hydrophobic contacts of the structure, and the ability of molecular dynamics to improves fits to low resolution electron densities.
Rasmussen et al. (2011): Kobilka and colleagues were the first to publish the purification of a stable receptor G protein complex, the β2-adrenergic receptor and Gs, and solve the crystal structure at 3.2 Å.
Westfield et al. (2011): This companion paper to Rasmussen et al. used single particle EM analysis to visualize the β2-adrenergic receptor Gs complex, and identify flexibility in the α-helical domain of Gα.
Dror et al. (2009): Dror used molecular dynamics simulations to reconcile the GPCR ionic lock hypothesis with biochemical and crystallographic data.
Dror et al. (2011): Dror used molecular dynamics simulations to reveal transient ligand binding sites in β-adrenergic receptors that could be targets for drug development.
Rieping et al. (2005): This paper outlines how a Bayesian formalism can be used for structure determination and the implementation for Inferential Structure Determination (ISD). Technical details and major applications are given in other manuscripts by the authors, but this serves as a concise and clear introduction to the concept.
References and recommended reading
- 1.Ban N, Freeborn B, Nissen P, Penczek P, Grassucci RA, Sweet R, Frank J, Moore PB, Steitz TA: A 9 A resolution X-ray crystallographic map of the large ribosomal subunit. Cell 1998, 93:1105–1115. [DOI] [PubMed] [Google Scholar]
- 2.Harrison SC, Jack A: Structure of tomato bushy stunt virus: three-dimensional x-ray diffraction analysis at 16 A resolution. J Mol Biol 1975, 97:173–191. [DOI] [PubMed] [Google Scholar]
- 3.Crowther RA, Amos LA: Three-dimensional image reconstructions of some small spherical viruses. Cold Spring Harb Symp Quant Biol 1972, 36:489–494. [DOI] [PubMed] [Google Scholar]
- 4.Scheres SH: A Bayesian view on cryo-EM structure determination. J Mol Biol 2012, 415:406–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheres SH: RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 2012, 180:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henderson R, McMullan G: Problems in obtaining perfect images by single-particle electron cryomicroscopy of biological structures in amorphous ice. Microscopy (Oxf) 2013, 62:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhlbrandt W: Introduction to Electron Crystallography. In Electron Crystallography of Soluble and Membrane Proteins Edited by Schmidt-Krey I, Cheng Y: Humana Press; 2013:1–16. Methods in Molecular Biology, vol 955.] [DOI] [PubMed] [Google Scholar]
- 8.Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y: Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 2013, 10:584–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burnett RM: The structure of the adenovirus capsid. II. The packing symmetry of hexon and its implications for viral architecture. J Mol Biol 1985, 185:125–143. [DOI] [PubMed] [Google Scholar]
- 10.Wong W, Bai XC, Brown A, Fernandez IS, Hanssen E, Condron M, Tan YH, Baum J, Scheres SH: Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine. Elife (Cambridge) 2014:e03080. [DOI] [PMC free article] [PubMed]
- 11.Voorhees RM, Fernandez IS, Scheres SH, Hegde RS: Structure of the Mammalian ribosome-sec61 complex to 3.4 a resolution. Cell 2014, 157:1632–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amunts A, Brown A, Bai XC, Llacer JL, Hussain T, Emsley P, Long F, Murshudov G, Scheres SH, Ramakrishnan V: Structure of the yeast mitochondrial large ribosomal subunit. Science 2014, 343:1485–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao M, Cao E, Julius D, Cheng Y: Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 2013, 504:107–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao E, Liao M, Cheng Y, Julius D: TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 2013, 504:113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao M, Cao E, Julius D, Cheng Y: Single particle electron cryo-microscopy of a mammalian ion channel. Curr Opin Struct Biol 2014, 27C:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, et al. : Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 2013, 342:1484–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu P, Bai XC, Ma D, Xie T, Yan C, Sun L, Yang G, Zhao Y, Zhou R, Scheres SH, et al. : Three-dimensional structure of human gamma-secretase. Nature 2014. [DOI] [PMC free article] [PubMed]
- 18.Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T: Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature 2005, 438:633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi D, Nannenga BL, Iadanza MG, Gonen T: Three-dimensional electron crystallography of protein microcrystals. Elife 2:e01345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morgan JL, McNamara JT, Zimmer J: Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat Struct Mol Biol 2014, 21:489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pebay-Peyroula E, Rummel G, Rosenbusch JP, Landau EM: X-ray structure of bacteriorhodopsin at 2.5 angstroms from microcrystals grown in lipidic cubic phases. Science 1997, 277:1676–1681. [DOI] [PubMed] [Google Scholar]
- 22.Johansson LC, Arnlund D, Katona G, White TA, Barty A, DePonte DP, Shoeman RL, Wickstrand C, Sharma A, Williams GJ, et al. : Structure of a photosynthetic reaction centre determined by serial femtosecond crystallography. Nat Commun 2013, 4:2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu W, Wacker D, Gati C, Han GW, James D, Wang D, Nelson G, Weierstall U, Katritch V, Barty A, et al. : Serial femtosecond crystallography of G protein-coupled receptors. Science 2014, 342:1521–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feld GK, Frank M: Enabling membrane protein structure and dynamics with X-ray free electron lasers. Curr Opin Struct Biol 2014, 27C:69–78. [DOI] [PubMed] [Google Scholar]
- 25.Russel D, Lasker K, Webb B, Velazquez-Muriel J, Tjioe E, Schneidman-Duhovny D, Peterson B, Sali A: Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol 2012, 10:e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward AB, Sali A, Wilson IA: Biochemistry. Integrative structural biology. Science 2013, 339:913–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schroder GF, Levitt M, Brunger AT: Super-resolution biomolecular crystallography with low-resolution data. Nature 2009, 464:1218–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raman S, Lange OF, Rossi P, Tyka M, Wang X, Aramini J, Liu G, Ramelot TA, Eletsky A, Szyperski T, et al. : NMR structure determination for larger proteins using backbone-only data. Science 2010, 327:1014–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cowieson NP, Kobe B, Martin JL: United we stand: combining structural methods. Curr Opin Struct Biol 2008, 18:617–622. [DOI] [PubMed] [Google Scholar]
- 30.Webb B, Lasker K, Schneidman-Duhovny D, Tjioe E, Phillips J, Kim SJ, Velazquez-Muriel J, Russel D, Sali A: Modeling of proteins and their assemblies with the integrative modeling platform. Methods Mol Biol 781:377–397. [DOI] [PubMed] [Google Scholar]
- 31.van der Schot G, Zhang Z, Vernon R, Shen Y, Vranken WF, Baker D, Bonvin AM, Lange OF: Improving 3D structure prediction from chemical shift data. J Biomol NMR 2013, 57:27–35. [DOI] [PubMed] [Google Scholar]
- 32.Steitz TA: A structural understanding of the dynamic ribosome machine. Nat Rev Mol Cell Biol 2008, 9:242–253. [DOI] [PubMed] [Google Scholar]
- 33.Palade GE: A small particulate component of the cytoplasm. J Biophys Biochem Cytol 1955, 1:59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lake JA: Ribosome structure determined by electron microscopy of Escherichia coli small subunits, large subunits and monomeric ribosomes. J Mol Biol 1976, 105:131–139. [DOI] [PubMed] [Google Scholar]
- 35.Wittmann HG: Architecture of prokaryotic ribosomes. Annu Rev Biochem 1983, 52:35–65. [DOI] [PubMed] [Google Scholar]
- 36.Milligan RA, Unwin PN: Location of exit channel for nascent protein in 80S ribosome. Nature 1986, 319:693–695. [DOI] [PubMed] [Google Scholar]
- 37.Yonath A, Leonard KR, Wittmann HG: A tunnel in the large ribosomal subunit revealed by three-dimensional image reconstruction. Science 1987, 236:813–816. [DOI] [PubMed] [Google Scholar]
- 38.Frank J, Zhu J, Penczek P, Li Y, Srivastava S, Verschoor A, Radermacher M, Grassucci R, Lata RK, Agrawal RK: A model of protein synthesis based on cryo-electron microscopy of the E. coli ribosome. Nature 1995, 376:441–444. [DOI] [PubMed] [Google Scholar]
- 39.Stark H, Mueller F, Orlova EV, Schatz M, Dube P, Erdemir T, Zemlin F, Brimacombe R, van Heel M: The 70S Escherichia coli ribosome at 23 A resolution: fitting the ribosomal RNA. Structure 1995, 3:815–821. [DOI] [PubMed] [Google Scholar]
- 40.Frank J, Agrawal RK: A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 2000, 406:318–322. [DOI] [PubMed] [Google Scholar]
- 41.von Bohlen K, Makowski I, Hansen HA, Bartels H, Berkovitch-Yellin Z, Zaytzev-Bashan A, Meyer S, Paulke C, Franceschi F, Yonath A: Characterization and preliminary attempts for derivatization of crystals of large ribosomal subunits from Haloarcula marismortui diffracting to 3 A resolution. J Mol Biol 1991, 222:11–15. [DOI] [PubMed] [Google Scholar]
- 42.Yonath A, Harms J, Hansen HA, Bashan A, Schlunzen F, Levin I, Koelln I, Tocilj A, Agmon I, Peretz M, et al. : Crystallographic studies on the ribosome, a large macromolecular assembly exhibiting severe nonisomorphism, extreme beam sensitivity and no internal symmetry. Acta Crystallogr A 1998, 54:945–955. [DOI] [PubMed] [Google Scholar]
- 43.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA: The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 2000, 289:905–920. [DOI] [PubMed] [Google Scholar]
- 44.Wimberly BT, Brodersen DE, Clemons WM Jr., Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V: Structure of the 30S ribosomal subunit. Nature 2000, 407:327–339. [DOI] [PubMed] [Google Scholar]
- 45.Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A: High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 2001, 107:679–688. [DOI] [PubMed] [Google Scholar]
- 46.Steitz TA, Moore PB: RNA, the first macromolecular catalyst: the ribosome is a ribozyme. Trends Biochem Sci 2003, 28:411–418. [DOI] [PubMed] [Google Scholar]
- 47.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF: Crystal structure of the ribosome at 5.5 A resolution. Science 2001, 292:883–896. [DOI] [PubMed] [Google Scholar]
- 48.Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V: Structure of the 70S ribosome complexed with mRNA and tRNA. Science 2006, 313:1935–1942. [DOI] [PubMed] [Google Scholar]
- 49.Munro JB, Vaiana A, Sanbonmatsu KY, Blanchard SC: A new view of protein synthesis: mapping the free energy landscape of the ribosome using single-molecule FRET. Biopolymers 2008, 89:565–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H: Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature 2010, 466:329–333. [DOI] [PubMed] [Google Scholar]
- 51.Budkevich T, Giesebrecht J, Altman RB, Munro JB, Mielke T, Nierhaus KH, Blanchard SC, Spahn CM: Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol Cell 2011, 44:214–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moore PB: How should we think about the ribosome? Annu Rev Biophys 2012, 41:1–19. [DOI] [PubMed] [Google Scholar]
- 53.Shoji S, Walker SE, Fredrick K: Reverse translocation of tRNA in the ribosome. Mol Cell 2006, 24:931–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frank J, Gonzalez RL Jr.: Structure and dynamics of a processive Brownian motor: the translating ribosome. Annu Rev Biochem 2010, 79:381–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frank J: Intermediate states during mRNA-tRNA translocation. Curr Opin Struct Biol 2012, 22:778–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spirin AS: The ribosome as a conveying thermal ratchet machine. J Biol Chem 2009, 284:21103–21119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bretscher MS: Translocation in protein synthesis: a hybrid structure model. Nature 1968, 218:675–677. [DOI] [PubMed] [Google Scholar]
- 58.Spirin AS: How does the ribosome work? A hypothesis based on the two subunit construction of the ribosome. Curr Mod Biol 1968, 2:115–127. [DOI] [PubMed] [Google Scholar]
- 59.Gao H, Sengupta J, Valle M, Korostelev A, Eswar N, Stagg SM, Van Roey P, Agrawal RK, Harvey SC, Sali A, et al. : Study of the structural dynamics of the E coli 70S ribosome using real-space refinement. Cell 2003, 113:789–801. [DOI] [PubMed] [Google Scholar]
- 60.Tama F, Miyashita O, Brooks CL 3rd, : Normal mode based flexible fitting of high-resolution structure into low-resolution experimental data from cryo-EM. J Struct Biol 2004, 147:315–326. [DOI] [PubMed] [Google Scholar]
- 61.Schroder GF, Brunger AT, Levitt M: Combining efficient conformational sampling with a deformable elastic network model facilitates structure refinement at low resolution. Structure 2007, 15:1630–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bock LV, Blau C, Schroder GF, Davydov II, Fischer N, Stark H, Rodnina MV, Vaiana AC, Grubmuller H: Energy barriers and driving forces in tRNA translocation through the ribosome. Nat Struct Mol Biol 2013, 20:1390–1396. [DOI] [PubMed] [Google Scholar]
- 63.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K: Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 2008, 16:673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pollard TD, Borisy GG: Cellular motility driven by assembly and disassembly of actin filaments. Cell 2003, 112:453–465. [DOI] [PubMed] [Google Scholar]
- 65.Pfaendtner J, Volkmann N, Hanein D, Dalhaimer P, Pollard TD, Voth GA: Key structural features of the actin filament Arp2/3 complex branch junction revealed by molecular simulation. J Mol Biol 2012, 416:148–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robinson RC, Turbedsky K, Kaiser DA, Marchand JB, Higgs HN, Choe S, Pollard TD: Crystal structure of Arp2/3 complex. Science 2001, 294:1679–1684. [DOI] [PubMed] [Google Scholar]
- 67.Nolen BJ, Littlefield RS, Pollard TD: Crystal structures of actin-related protein 2/3 complex with bound ATP or ADP. Proc Natl Acad Sci U S A 2004, 101:15627–15632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nolen BJ, Pollard TD: Insights into the influence of nucleotides on actin family proteins from seven structures of Arp2/3 complex. Mol Cell 2007, 26:449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rouiller I, Xu XP, Amann KJ, Egile C, Nickell S, Nicastro D, Li R, Pollard TD, Volkmann N, Hanein D: The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol 2008, 180:887–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dalhaimer P, Pollard TD: Molecular dynamics simulations of Arp2/3 complex activation. Biophys J 2010, 99:2568–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goley ED, Rammohan A, Znameroski EA, Firat-Karalar EN, Sept D, Welch MD: An actin-filament-binding interface on the Arp2/3 complex is critical for nucleation and branch stability. Proc Natl Acad Sci U S A 2010, 107:8159–8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. : Initial sequencing and analysis of the human genome. Nature 2001, 409:860–921. [DOI] [PubMed] [Google Scholar]
- 73.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. : The sequence of the human genome. Science 2001, 291:1304–1351. [DOI] [PubMed] [Google Scholar]
- 74.Gilman AG: Nobel Lecture. G proteins and regulation of adenylyl cyclase. Biosci Rep 1995, 15:65–97. [DOI] [PubMed] [Google Scholar]
- 75.Xu F, Wu H, Katritch V, Han GW, Jacobson KA, Gao ZG, Cherezov V, Stevens RC: Structure of an agonist-bound human A2A adenosine receptor. Science 2011, 332:322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EY, Velasquez J, Kuhn P, Stevens RC: A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor. Structure 2008, 16:897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lebon G, Warne T, Edwards PC, Bennett K, Langmead CJ, Leslie AG, Tate CG: Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature 2011, 474:521–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chun E, Thompson AA, Liu W, Roth CB, Griffith MT, Katritch V, Kunken J, Xu F, Cherezov V, Hanson MA, et al. : Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure 2012, 20:967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, et al. : Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature 2011, 469:175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, et al. : Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature 2011, 477:549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sunahara RK, Tesmer JJ, Gilman AG, Sprang SR: Crystal structure of the adenylyl cyclase activator Gsalpha. Science 1997, 278:1943–1947. [DOI] [PubMed] [Google Scholar]
- 82.Van Eps N, Preininger AM, Alexander N, Kaya AI, Meier S, Meiler J, Hamm HE, Hubbell WL: Interaction of a G protein with an activated receptor opens the interdomain interface in the alpha subunit. Proc Natl Acad Sci U S A 2011, 108:9420–9424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Westfield GH, Rasmussen SG, Su M, Dutta S, DeVree BT, Chung KY, Calinski D, Velez-Ruiz G, Oleskie AN, Pardon E, et al. : Structural flexibility of the G alpha s alpha-helical domain in the beta2-adrenoceptor Gs complex. Proc Natl Acad Sci U S A 2011, 108:16086–16091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Standfuss J, Edwards PC, D’Antona A, Fransen M, Xie G, Oprian DD, Schertler GF: The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature 2011, 471:656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AG, Tate CG, Schertler GF: Structure of a beta1-adrenergic G-protein-coupled receptor. Nature 2008, 454:486–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, et al. : High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 2007, 318:1258–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dror RO, Arlow DH, Maragakis P, Mildorf TJ, Pan AC, Xu H, Borhani DW, Shaw DE: Activation mechanism of the beta2-adrenergic receptor. Proc Natl Acad Sci U S A 2011, 108:18684–18689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dror RO, Arlow DH, Borhani DW, Jensen MO, Piana S, Shaw DE: Identification of two distinct inactive conformations of the beta2-adrenergic receptor reconciles structural and biochemical observations. Proc Natl Acad Sci U S A 2009, 106:4689–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dror RO, Pan AC, Arlow DH, Borhani DW, Maragakis P, Shan Y, Xu H, Shaw DE: Pathway and mechanism of drug binding to G-protein-coupled receptors. Proc Natl Acad Sci U S A 2011, 108:13118–13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rieping W, Habeck M, Nilges M: Inferential structure determination. Science 2005, 309:303–306. [DOI] [PubMed] [Google Scholar]
- 91.Bahar I, Rader AJ: Coarse-grained normal mode analysis in structural biology. Curr Opin Struct Biol 2005, 15:586–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tama F, Valle M, Frank J, Brooks CL 3rd, : Dynamic reorganization of the functionally active ribosome explored by normal mode analysis and cryo-electron microscopy. Proc Natl Acad Sci U S A 2003, 100:9319–9323. [DOI] [PMC free article] [PubMed] [Google Scholar]