Abstract
RNA-binding proteins are important players in post-transcriptional regulation, such as modulating mRNA splicing, translation, and degradation under diverse biological settings. Identifying and characterizing the RNA substrates is a critical step in deciphering the function and molecular mechanisms of the target RNA-binding proteins. High-throughput sequencing of the RNA fragments isolated by crosslinking immunoprecipitation (CLIP-seq) is one of the standard techniques to identify the in vivo transcriptome-wide binding sites of the target RNA-binding protein. This method is widely used in functional and mechanistic characterizations of RNA-binding proteins. In this review, we provide several practical considerations on performing and analyzing CLIP-seq experiments. Particularly, we focus on how to perform CLIP-seq experiments on endogenous RNA-binding proteins. In addition, we provide a practical summary on how to choose and use computational pipelines from an increasing number of computational methods and packages that are available for analyzing the sequencing datasets from the CLIP-seq experiments. We hope these practical considerations will facilitate experimental biologists in performing and analyzing CLIP-seq experiment to obtain biologically relevant mechanistic insights.
1. Background
RNA-binding proteins (RBPs) are important players in post-transcriptional regulation of gene expression under diverse biological processes. Importantly, malfunctions of RBPs have been implicated in a wide variety of human diseases, such as neurological disorders (1) and cancers (2). Thus, characterizing the biological functions and molecular mechanisms of RBPs has significant implications in both basic biology and potential translational applications.
RBPs usually control gene expression by binding to their substrate mRNAs. Therefore, a critical step in the functional and mechanistic characterization of the target RBP is to identify and characterize the RNA substrates it binds. Multiple biochemical methods have been developed for this purpose. Over the past decade, high-throughput sequencing of the RNA fragments isolated by crosslinking and immunoprecipitation (CLIP-seq) has become one of the standard techniques to identify the in vivo transcriptome-wide binding sites of RBPs (3). Briefly, in this approach, the cells are first treated by UV crosslinking, which only introduces covalent bonds between the RNA and the protein that are in direct contact. Thereby, the in vivo RNA-protein interactions are preserved. Then the cells are lysed and treated with an RNase that degrades naked RNAs but not RNA regions bound by proteins. The target RBP and the RNA fragments it binds are isolated via immunoprecipitation. The 5’ and 3’ ends of the RNA fragments bound by the RBP are cued (e.g. prepare RNA fragments with 5’ end phosphorylated and 3’ end hydroxyl) so that they are compatible for downstream sequencing library preparation. The RNA:RBP complex is then resolved on an SDS-PAGE gel followed by transferring to a nitrocellulose membrane. The membrane areas containing the target RNA-protein complexes are surgically cut, and proteinase K digestion is used to remove the RBP from the RNA-protein complexes. The resulting RNA fragments are subject to library construction followed by high throughput sequencing. Mapping and analyzing the sequencing reads from the RNA fragments reveals where the target RBP binds in the transcriptome.
CLIP-seq offers several major advantages in identifying RNA substrates of the target RBP (3). First, the RNA-protein interactions identified by CLIP reflect in vivo interactions due to the nature of the UV crosslinking. Second, CLIP enables the identification of RNA fragments that are directly bound by the target RBP, but not those bound by other proteins interacting with the target RBP (4, 5). Since UV crosslinking introduces covalent bonds between the RNA and the protein that are in direct contact, high stringent washes (e.g., 1M NaCl in many protocols) can be applied during the immunoprecipitation step. These stringent washes not only reduce non-specific interactions but also, more importantly, can disassociate protein complexes, preventing immunoprecipitated RNAs bound to non-target RBPs. In addition, isolating the target RNA:RBP complex on the nitrocellulose resulting from the SDS-PAGE and transfer further ensures that the resulting RNA fragments are those directly bound by the target RBP. Third, in addition to identifying the RNA species that the target RBP binds to, CLIP-seq analysis can also reveal where the target RBP binds in each of the identified RNA species. This information provides critical insights into how the RBP recognize its substrates (e.g., sequence motifs, structural features, etc.) and how it regulates target RNA expression (e.g., intronic binding region may suggest splicing regulations, etc.). Collectively, these strengths make the CLIP-seq approach one of the major methods of characterizing in vivo global RNA-protein interactions.
Although there are tens of steps in performing the CLIP-seq experiment, the inventors of this method and many experts provide detailed and reproducible protocols and excellent explanations on performing this experiment (6, 7). In addition, multiple modifications and derivations have been applied to the original CLIP-seq protocol to make the whole process easier to undertake and more likely to succeed. For example, to enhance the UV crosslinking efficiency on RNA:RBP complexes, PAR-CLIP (photoactivatable ribonucleoside-enhanced CLIP) was developed (8). In this method, cells are first treated with photoactive ribonucleoside analogs, such as 4-thiouridine, which can be incorporated into nascent RNAs. Then crosslinking is performed at a specific wavelength that activates the photoactive ribonucleoside analog (e.g., UV365nm for 4-thiouridine). This approach significantly enhances the crosslinking efficiencies for several RBPs and facilitates the identification of their transcriptome-wide RNA targets. In addition, taking advantage of the observation that during the cDNA synthesis of CLIP-seq library construction, the amino acid residue(s) of the RBP crosslinked to the RNA fragment which cannot be completely removed by protease K digestion tend to inhibit the read-through of the reverse transcriptase, a cDNA-circularization based library construction strategy was developed (9). This approach enables the identification of the RNA-RBP interaction sites at high resolution. Recently, an enhanced CLIP (eCLIP) protocol was developed that significantly reduces PCR duplicate reads during sequencing library construction and enables CLIP-seq experiments to be performed in a high throughput fashion (10). Together with the original method, these modifications and many others make the CLIP-seq approach easily accessible to many investigators studying in vivo RNA:protein interactions. Since there are already many outstanding reviews and protocols on CLIP-seq (6-8, 11-13), we will not discuss the technical details of the CLIP-seq experiment.
In this review, we will focus on two issues in performing and analyzing CLIP-seq experiment. First, we will discuss how to perform CLIP-seq experiment on endogenous RBPs for which high-quality antibodies (e.g., immunoprecipitation grade) are not available. We believe focusing on endogenous RBPs is critical to obtain functional and mechanistic insights that are biologically relevant. Second, after obtaining the sequencing datasets from the CLIP-seq experiment, how to analyze them and extract relevant information regarding in vivo RNA:RBP interactions is still a challenge for most experimental biologists. Moreover, although there are many computational algorithms and packages that are developed for CLIP-seq data analysis, quite different from standard RNA-seq data analysis, currently there are no standardized computational pipelines that are widely used for analyzing CLIP-seq datasets. Thus, here we discuss a general procedure for computational analysis CLIP-seq datasets and share our opinions in using several different computational packages in analyzing CLIP-seq data. We hope that these two practical considerations can be of help to other investigators in performing and analyzing CLIP-seq experiments.
2. Epitope-tagging endogenous RBPs via genomic editings for CLIP-seq
Immunoprecipitation of the target RBP and its associated RNA fragments is a critical step in the CLIP-seq experiment. Thus, a high-quality IP-grade antibody to the target RBP is essential for the success of the CLIP-seq experiment. Although a large number of antibodies for many RBPs are commercially available, a lot of these antibodies are not rigorously validated, let alone having the capacity to specifically immunoprecipitate the endogenous target RBPs. Therefore, an important technical challenge in performing the CLIP-seq analysis is the availability of high-quality antibodies to the target RBP(s).
One approach commonly used to circumvent this technical challenge is to ectopically express the target RBP in an epitope-tagged form from either a plasmid or a viral vector, and then perform the CLIP-seq experiment using high-quality antibodies against the epitope tag. Although some mechanistic insights regarding RNA:RBP interactions can be obtained (e.g., binding motif preference of the target RBP, etc.), an important caveat of this approach is that the RNA species identified may not be the real targets of the endogenous RBPs. This is mainly due to the following two reasons. First, the increased RBP expression level may lead to changes in the binding kinetics, resulting in identifying weak or non-optimal RNA:RBP-interactions that do not occur when the target RBP is expressed at its endogenous level. Second, the expressions of many RBPs are tightly regulated, and alterations in their expression levels may lead to transcriptomic and physiological changes of the cell. For instance, in terminal erythropoiesis, an RBP, Cpeb4, is transcriptionally induced during differentiation. Interestingly, however, Cpeb4 also translationally represses its own mRNA in differentiating erythroid cells. Thereby, the Cpeb4 protein level is maintained within a specific range during terminal erythropoiesis. Importantly, changing the expression level of Cpeb4 by either knock-down or over-expression inhibits the terminal differentiation of erythroid cells (14). Thus, if CLIP-seq is performed on the over-expressed Cpeb4 in erythroid cells, the target RNAs identified may not represent the real RNAs that endogenous Cpeb4 binds in differentiating erythroid cells. This example highlights that in order to obtain biologically relevant functional and mechanistic insights, it is important to study RBPs under their endogenous levels.
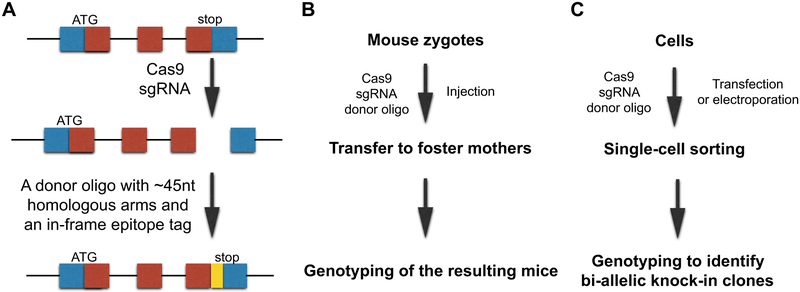
We recently used CRISPR/Cas9-based genomic editing techniques to generate epitope-tagged proteins for mechanistic studies on endogenous RBPs (15). In this approach (Fig. 1A), a DNA double-strand break is introduced to the genomic region of the C-terminus (or the N-terminus) of the target RBP by a small guide RNA (sgRNA) and the Cas9 protein. Then the double-strand break is repaired in the presence of a donor oligo, which contains two ~45nt homologous arms and the coding sequence of a small epitope tag (e.g., V5, FLAG, etc.). Through homologous recombination, the epitope tag sequence is integrated in-frame with the target RBP. The resulting tagged RBP is expressed at its endogenous level because the knocked-in tag sequence changes neither the promoter nor the 3’UTR sequences, which are two key elements in controlling the expression level of the RBP. We successfully generated a Zfp36-V5 knock-in mouse using this approach (Fig. 1B) (15), which enabled us to perform mechanistic studies on the endogenous Zfp36, which is an important RBP regulating inflammatory responses with no commercially available high-quality antibodies. Using a V5 antibody, we identified the proteins interacting with the endogenous Zfp36 and the target mRNAs it binds in primary mouse macrophages. This epitope-tag knock-in approach can also be used in cultured cells (Fig. 1C). Since there is no selection marker in this approach, after introducing the sgRNA, Cas9, and the donor oligo, the cells are subject to single cell sorting (using the GFP from the Cas9 expressing vector). The expanded clones are genotyped to identify bi-allelic knock-ins, and the expression of the tagged RBP will be examined by Western blot using an antibody against the knocked-in epitope tag. The resulting cell lines with bi-allelic epitope-tag knocked-in to the target RBP locus will be great resources for downstream functional and mechanistic studies on the endogenous RBP at the cell level.
Figure 1.
Epitope-tagging the endogenous RBP via genome editing. (A) Outline of the CRISPR/Cas9-based approach for epitope-tagging the endogenous RBP locus. (B) Outline of the procedures of using this approach to generate epitope-tag knock-in mouse. (C) Outline of the procedures of using this approach to generate bi-allelic knock-in cell lines.
There are several important considerations when applying this epitope-tag knock-in approach to study endogenous RBPs.
First, to study RBPs expressed at the endogenous levels, it is important to avoid using selection markers (e.g., puromycin) as described in some other genome-editing-mediated epitope-tag knock-in methods (16). Although facilitating the selection of correct knock-in alleles, the presence of the selection marker replaces the endogenous 3’UTR of the target RBP with an exogenous 3’UTR. This change may alter the expression level of the endogenous RBP because numerous RBPs (e.g., Cpeb4, Zfp36, etc.) can auto-regulate their own expression through binding to the regulatory elements in the 3’UTRs of their own mRNAs (14, 17). Moreover, many exogenous 3’UTRs (e.g., 3’UTRs derived from the SV40 virus or BGHR etc.) used in the selection marker approach can significantly stabilize the resulting mRNA and enhance the expression of the target RBP, leading an increased target RBP level. The selection-free donor oligo knock-in approach as described above, however, does not have this problem, as the potential regulatory elements in the endogenous 3’UTR are maintained in the mRNA expressed from the knock-in allele.
Second, like all the experiments using tagged proteins, it is important to examine whether the tagged protein is functional or not. One way to test this is to examine and compare the cellular or molecular phenotypes among the wild-type cell, the bi-allelic knock-in cell, and the target RBP knock-out or knock-down cells. Or alternatively, the tagged protein can be used to examine whether it can rescue the phenotype(s) of the target RBP knock-out or knock-down cells.
Third, in terms of obtaining bi-allelic knock-ins, although the donor oligo mediated epitope-tag knock-in approach is not as easy to perform as the knock-in methods using selection markers, due to the high efficiency of the CRISPR/Cas9 genome editing system in many types of cells, we still can obtain reasonable number of bi-allelic-edited cell clones for several RBPs tested. In addition, during the CRISPR/Cas9 mediated genomic editing, the non-homologous end joining pathway can be blocked by chemical inhibitors (e.g., SCR7 in mouse cells) (18), thereby promoting the donor oligo mediated homologous recombination and enhancing the chances of obtaining bi-allelic knock-in cells.
Fourth, even though there are many outstanding antibodies against the commonly used epitope tags (e.g., V5, FLAG, etc.), once the bi-allelic knock-in cells are obtained, it is still important to test several different antibodies against the same epitope tag to identify the one with the high specificity and IP efficiency in the target cell type. This is because different cell types have different proteomes. An antibody that works well in one type of the cell does not necessarily mean that it has the same specificity in a different type of cell. However, thanks to a large number of commercially available antibodies to each of the commonly used epitope tags, it is usually not difficult to find such a good antibody to the cell types under investigation.
3. Practical considerations on computationally analyzing CLIP-seq datasets
Several automatic or semi-automatic pipelines were developed for bioinformatic analysis of CLIP-seq (19-22). These pipelines take raw sequencing data, in most cases, Fastq files as input, and output the locations and statistical significance of binding sites. Some of the pipelines also output RBP motifs and bigwig/wig files for visualization. However, experimental biologists may need to customize pipelines for their specific datasets. In this review, we provide a practical guide on how to choose and use software packages as building blocks of a CLIP-seq analysis pipeline.
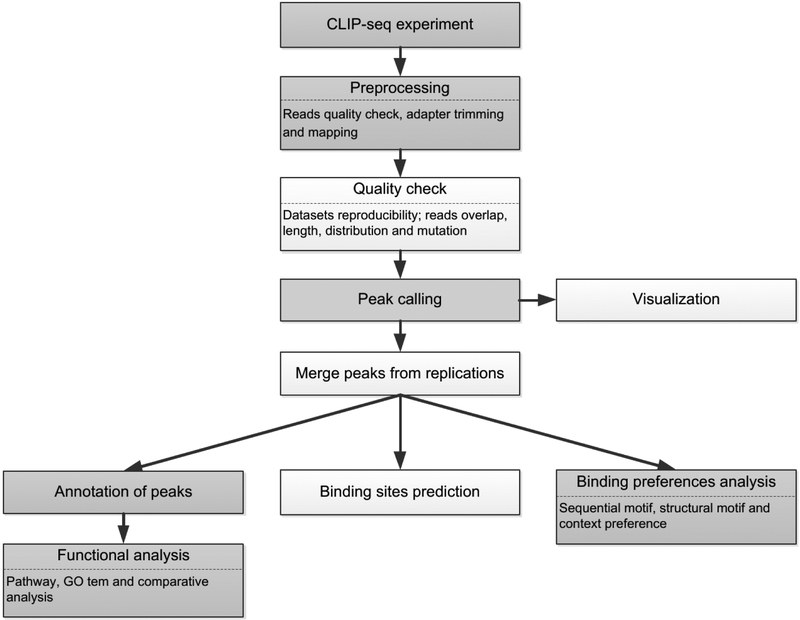
The workflows of the bioinformatic analysis for CLIP-seq data are similar to each other. The difference is in the choice of software and parameters. A typical pipeline is shown in Fig. 2. Data preprocessing includes trimming and quality check of the raw reads, read alignment and quality check of the mapping results. The next step of the analysis is peak calling, which is the core of bioinformatic analysis of CLIP-seq data. From the resulting peaks, post-processing usually includes merging the peaks from replicates, predicting or rescuing the peaks, binding preferences and functional analysis.
Figure 2.
The workflow of the bioinformatic analysis for CLIP-seq data. The shaded diagrams are the basic steps, and the remaining ones are the optional steps.
3.1. Preprocessing
The bioinformatic analysis usually starts with adapter trimming and quality check. Adapter and quality trimming can be performed by tools such as Cutadapt (23), Fastx-toolkit (24), Trimmomatch (25), Prinseq (26) and TrimGalore (27). For iCLIP/eCLIP dataset, the low-quality bases should only be trimmed at the 3' end of the fragment, since the 5' end shows the position of truncation. Reads shorter than 18nt are likely to be mapped to wrong locations or multiple locations and should be discarded after trimming. FastQC (28) is a widely used tool to check the RNA library by displaying statistical characteristics, such as read quality scores and read duplication levels. High level of read duplication often suggests contamination or improper preprocessing. After adapter trimming and quality check, the resulting reads are mapped to the genome or transcriptome. Mapping to the transcriptome increases sensitivity, while mapping to the genome preserves the information of the binding sites in precursor messenger RNA and unannotated regions. Since the experimental sensitivity is usually sufficient, mapping to the genome is the better choice in most cases (29). This also saves the hassle caused by different transcriptome annotations and is easier to visualize by IGV or genome browser. Mapping tools that can map reads across the junctions should be used for alternative splicing. HISAT (30) and STAR (31) are popular mapping tools using Burrows-Wheeler transform, which enable a fast search of reads locations. STAR is the choice in the eCLIP-seq bioinformatics pipeline developed by the inventors of eCLIP-seq, which searches in uncompressed suffix arrays. It takes more than 20 GB of RAM for aligning to human or mouse genome and usually runs on servers. HISAT is a more recent tool whose speed is comparable to STAR and requires only ~4 GB of RAM for human or mouse genome. While STAR should be used with existing pipelines for compatibility, HISAT is recommended in customized CLIP-seq analysis pipelines. Novoalign shows better mapping rate on some CLIP-seq datasets (32), but its full version is not free. The multi-mapped reads, usually the reads with MAPQ score less than 10, should be removed from the mapping result. Reverse transcription polymerase chain reaction (RT-PCR) is a technique widely used in CLIP-seq experiments, by which the RNA molecules are first converted into their complimentary DNA (cDNA), then a standard polymerase chain reaction (PCR) is performed to make multiple copies of the cDNAs (33). While RT-PCR facilitates the study for small samples, the amplification of cDNA is not evenly distributed for different regions, which introduces additional bias to the library. A commonly used method to correct this bias in the CLIP-seq protocol is to insert a random barcode or random-mer into the primer. Reads mapped to the same location with the same random barcode are collapsed into one copy. The random barcodes should also be removed before mapping. If random barcodes are not used, tools like fastx_collapser in the Fastx-toolkit (24), Rmdup in SAMtools packages (34) and MarkDuplicates in the Picard toolkit (35) can be used to remove the potential PCR duplicates. And fastuniq (36) is a tool designed to remove duplicates in paired-end reads.
To confirm that the CLIP-seq experiment is successfully performed and save time from interpreting bewildering peak calling results, it is recommended to check the statistical characteristics of the mapping result. Read mutation, length and location distribution, reproducibility, and read overlap should be checked for both CLIP-seq data and size-matched input (SMInput) data. Most of the reads should be mapped to a small portion of the genome. A straightforward way to check the read location distribution is to divide the genome into bins of the same length and count the read percentage in the top ranked bins (37). If the reads scatter all over the genome instead of clustering in a small portion of the genome, the datasets may contain too much noise from non-specific RNA:RBP interactions. The Pearson’s correlation coefficients of TPM (Transcripts Per Million) between replicates of CLIP-seq data and SMInput data can be used to check the reproducibility of the datasets. Compared to RPKM (Reads Per Kilobase of transcript, per Million mapped reads) or FPKM (Fragments Per Kilobase of transcript, per Million mapped reads), whose sums in different samples may vary, the sums of TPM are the same across samples, which facilitates the comparison among samples. For iCLIP/eCLIP, both complete and truncated reads are agglomerated at the RNA:RBP crosslink sites. Therefore, the datasets contain both shorter and longer reads. The starting position of truncated reads should overlap at binding sites. ICLIPro (37) can be used to visualize and check the overlapping of read starts.
3.2. Peak calling
Many tools were developed for CLIP-seq peak calling (38). A typical peak calling process can be divided into two tasks (39). The first task is to determine the regions enriched with reads. The second task is to calculate the statistical significance of the selected regions. Table 1 summarizes a list of popular peaking calling tools.
Table 1.
Software packages for peak calling
| Name | CLIP-seq protocol | Pre- process |
Input | Control | Statistical model | Feature | Resolution |
|---|---|---|---|---|---|---|---|
| Piranha | All | No | BED and BAM | Yes | Zero-truncated negative binomial distribution (default) | Choices of statistical model and covariates | Bins |
| ASPeak | CLIP-seq and RIP-seq | No | SAM, BAM, BOWTIE and BED | Yes | Negative binomial distribution | Expression sensitive | Peaks |
| Pyicoclip | All | Yes | Eland, SAM, BAM and BED | No | Background estimation | Complete pipeline | Predefined regions |
| iCount | iCLIP | Yes | BAM | No | Background estimation | Complete pipeline | Peaks |
| CLIPper | eCLIP | Yes | BAM | Yes | Poisson distribution | Cubic splines fit of reads profile | Peaks |
| PARalyzer | PAR-CLIP | Yes | SAM, BAM and BOWTIE | No | Gaussian kernel density estimator | N/A | Reads clusters |
| Bmix | PAR-CLIP | No | BAM | No | Constrained three-component binomial mixture model | Explicitly accounts for the sources of noise | Reads clusters |
| WavClusteR | PAR-CLIP | No | BAM | No | Non-parametric, two component mixture model | Complete pipeline | Reads clusters |
| MiClip | CLIP-seq, HITS-CLIP and PAR-CLIP | No | SAM | No | Zero inflated binomial distribution | Online interface | Reads clusters |
| pyCRAC | HITS-CLIP, PAR-CLIP and CRAC | Yes | SAM, BAM and Novoalign | No | Background estimation | Complete pipeline | Reads clusters |
| PureCLIP | eCLIP | No | BAM | Yes | Zero-truncated negative binomial distribution | Integrates motif information | Single nucleotide |
| PIPECLIP | HITS-CLIP, PAR-CLIP and iCLIP | Yes | BAM | Yes | Zero-truncated negative binomial distribution | Online interface | Reads clusters |
| CTK | All | Yes | BED | No | Background estimation | Complete pipeline | Single nucleotide |
In all variations of CLIP-seq data, reads are agglomerated around the crosslinking sites. To detect the unexpectedly enriched reads clusters, Piranha (40) divides the genome into bins of equal length and counts read starts in each bin. By default setting of Piranha, the read counts are modeled by zero-truncated negative binomial distribution, and then a p-value is assigned to each bin, which shows the likelihood that the bin is in the background. The zero-truncated negative binomial distribution was justified to be a good fit for read counts distribution by using over 100 CLIP-seq datasets (40) and is widely used by CLIP-seq peak callers. Modeling background from the whole genome may lead to a loss of sensitivity in lowly expressed genes. ASPeak (41) uses RIP-input or RNA-seq to estimate expression levels in user-defined genomic intervals, most commonly, genome annotations in GTF or BED format. A tradeoff of calling peaks in user-defined genomic intervals is that intergenic peaks will be ignored. Instead of assigning p-values to bins of equal length, CLIPper (42) interpolates the reads heights, centers and width through the pre-mRNA using cubic splines. A Poisson distribution is used to calculate the p-value of the peaks. CLIPper pipeline includes a script to normalize each peak against the SMInput data by using Χ2 test or Fisher's exact test. Instead of fitting a specific distribution, Pyicoclip (43) and iCount (44) generate background by randomly distributing the read counts in user-defined genomic regions.
In addition to reads coverage, mutations and truncations can be used in peak calling. HITS-CLIP introduces mutations depending on the RBPs. PAR-CLIP induces T to C transitions at the crosslinking sites. Instead of mutations, eCLIP and iCLIP generate truncated reads at the crosslinking sites. The T to C transitions in PAR-CLIP are modeled by a kernel-density-based classifier in PARalyzer (45), to distinguish the regions of crosslinking and non-crosslinking. To further reduce the false positives, BMix (46) and wavClusteR (47, 48) filter out the non-experimentally induced T to C conversion before performing the peak calling procedure. MiClip (49) handles the CLIP clusters and binding sites with mutations by a two-pass Hidden Markov Model (HMM). Python package pyCRAC (50) provides scripts as well as classes and functions to customize the analysis of HITS-CLIP, PAR-CLIP and CRAC data. To utilize the truncated reads information, PureCLIP (51) models the combination of truncation patterns and reads enrichment as four states in HMM. PureCLIP also provides an option to incorporate background crosslink-associated (CL) motif learned from SMInput data, which is useful when the target RPBs recognize different CL motifs.
Comprehensive tools were developed to utilize both the mutation and the truncation information. In addition to identifying CLIP tag clusters, they use experiment induced mutations or truncations to report reliable crosslink sites from CLIP tag clusters, depending on which CLIP-seq protocol is used. PIPE-CLIP allows the user to specify the types of mutations. CTK package (20) uses a valley seeking algorithm for peak calling to distinguish a broad peak and two neighboring peaks. Then crosslink-induced mutation sites (CIMS) (52) or crosslink-induced truncation sites (CITS) are identified for PAR-CLIP/HITS-CLIP or iCLIP/eCLIP. In CITS analysis, CTK package also takes consideration of deletions presenting in readthrough reads.
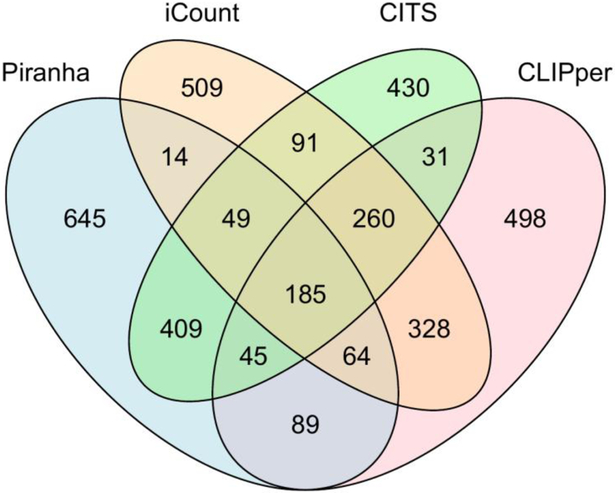
Because various features and models are used, peak calling tools may report different binding sites from the same datasets. In one study (39), only 15.5% of the peaks were output by at least one peak caller in Piranha, CLIPper, and Ext. blockbuster (53), were reported by all three peak callers. Various methods were used to benchmark the performance of peak callers. S. Bottini et al. compared the percentage of peaks called from Ago2 HITS-CLIP datasets containing canonical miRNA binding sites (54). A. M. Chakrabarti et al. compared the peak coverage of RNA splice sites in an iCLIP-seq PTBP1 data set (29). For most of the CLIP-seq datasets, sequence motifs can be used for benchmarking. Here we use an eCLIP-seq dataset of Zfp36 (15) whose sequence motif is known to evaluate the specificity. The sequence motif of Zfp36 determined from in vitro RNA-binding studies (55) is consistent with the one defined from our eCLIP-seq data. Therefore, this dataset is suitable as a benchmark. A binding site determined by peak callers will be considered as a true positive if it is close to the location of the sequence motif. Sequence motif was used to evaluate the specificity of peak callers in many previous studies (20, 51). Peak callers developed specifically for PAR-CLIP require specific mutation patterns, such as PARalyzer, were not compared in this review. We chose two popular peak callers, Piranha and CLIPper and two recently developed tools, iCount and CTK package for comparison. CTK package provides crosslinking induced mutation site (CIMS) and crosslinking induced truncation site (CITS) analysis, which call peaks at single nucleotide resolution. For eCLIP-seq data, CITS pipeline in CTK package was used. CITS and iCount do not have functions to normalize by SMInput data. To be fair, the peak calling results from CITS and iCount were normalized with SMInput data by using scripts in CLIPper pipeline. Peak callers generated different numbers of peaks, and the top 1500 peaks from each peak caller were used for comparison (Fig. 3). Different peak widths were generated by different peak callers. Therefore, instead of peak boundary, peak center was used for comparative analysis.
Figure 3.
Venn diagram of overlapped genomic locations of peak regions in Zfp36 eCLIP-seq datasets called by the four peak calling methods.
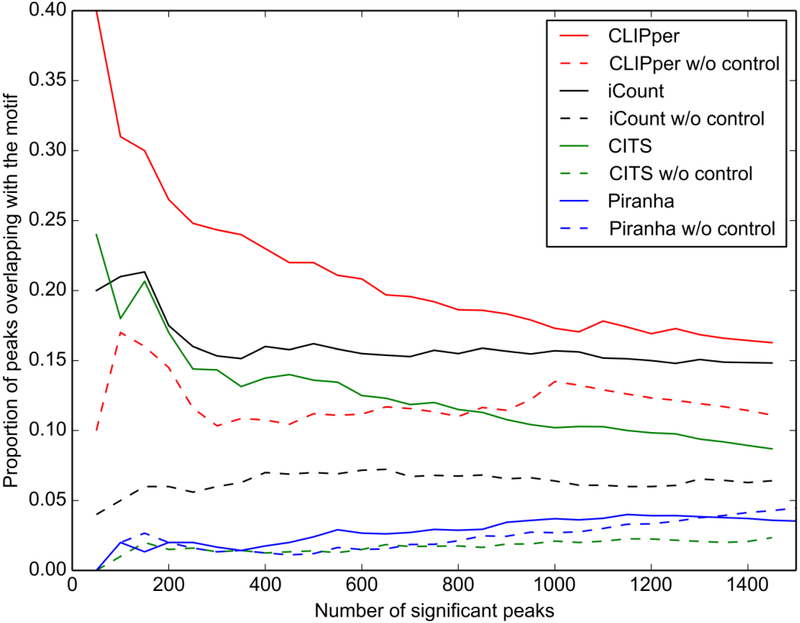
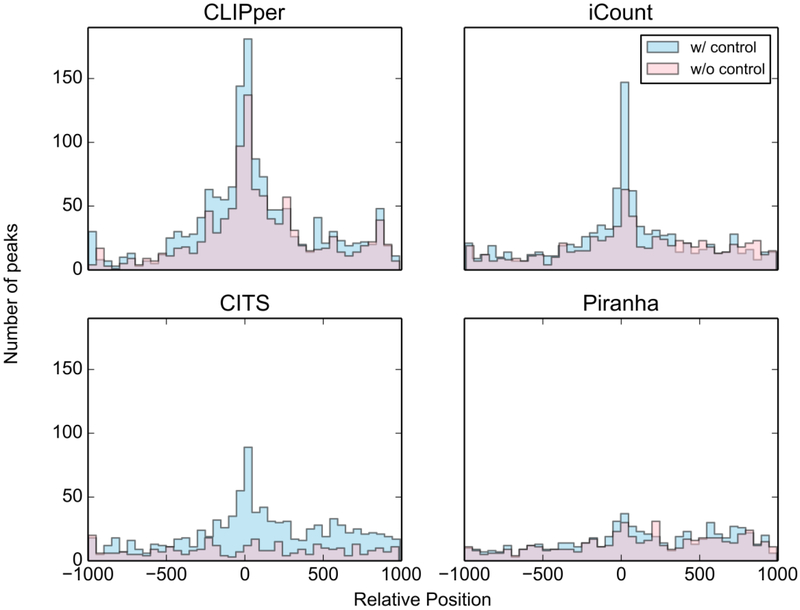
As shown in Fig. 4 and Fig. 5, CLIPper identified more binding sites overlapping with the motif, implying higher specificity than other tools on this dataset, while the peak center in iCount's result shows better coincidence with motif start positions, indicating more accurate peak positions. The performance of peak callers may vary in different datasets. The results from different peak callers can be intersected for specificity or combined for sensitivity. When sensitivity is sufficient (for example, peaks number > 10k), intersection of different peak callers’ results is recommended. Take this Zfp36 eCLIP-seq data set as an example, peaks called by iCount that are overlapped with calls of CLIPper can be used as peak calling result for high specificity and accuracy.
Figure 4.
Proportion of peaks called by the four peak calling methods in the Zfp36 eCLIP-seq datasets overlapping with motif “UAUUUAUU”.
Figure 5.
The position of top 1500 peaks called by the four methods in the Zfp36 eCLIP-seq datasets relative to motif “UAUUUAUU”.
Control samples or SMInput are used as negative controls in CLIP-seq, which are crucial for the normalization in peak calling. Although there is no gold standard for how to use these negative controls, a widely used method is to use statistical tests to estimate the significance of the peaks against the control samples or SMInput and output a p-value for each peak. Fig. 4 and Fig. 5 show that all four peak callers including CITS and iCount, which are designed to generate a random background instead of using a control sample or SMInput, have better specificity after normalizing against SMInput. When control samples and SMInput are not available, RNA-seq data can be used as a negative control to correct the bias from transcripts abundance. But nonspecific binding sites will not be filtered when using RNA-seq data instead of SMInput. For peak callers without the function to normalize by using the control dataset, some publicly available scripts can be used to do this. Typically, these scripts take peaks' locations, mapping results of both CLIP-seq dataset and the SMInput as input, and output a new file containing a statistical significance value for each peak. GetDifferentialPeaks function from Homer (56) is one of the options. This function was developed for Chip-seq data analysis. It filters out the insignificant peaks by read counts fold enrichment and Poisson enrichment p-value over the background, at user set cutoffs. For eCLIP-seq datasets, a script (Peak_input_normalization_wrapper.pl) from Yeo Lab (57) can be used.
Some of the peak callers model replicates explicitly, such as OmniCLIP (58). But for most of the peak callers, users must merge the peaks from replicates manually. In most cases, the intersection of peaks is used for specificity. The intersect function in BEDTools package (59, 60), clusters function in iCount package and mergePeaks function in Homer package can be used for this purpose.
3.3. Downstream analysis
A typical downstream analysis includes binding preferences analysis, binding site prediction and functional analysis of binding targets.
3.3.1. Binding preference analysis
RBPs specify binding sites through recognizing sequence and structural features in their binding targets. Motifs of many RBPs are collected into databases such as the CISBP-RNA Database (61). Many tools were also developed to discover motifs from CLIP-seq data. Since the motifs may not be in the binding sites but close to them, peaks identified from CLIP-seq data are extended in both directions for motif discovery. The sequences of extended peaks can be generated by getfasta function in BEDTools package in a strand-specific manner. Motif searching software packages take these sequences in fasta format as input, and output one or more sequence motifs that are enriched in the input sequences, as well as their statistical significance and locations. MEME (62, 63) outputs position weight matrices (PWM) of sequence motifs, by using expectation maximization (EM) algorithm to fit a two-component finite mixture model to the input sequences. With the PWM output by MEME, FIMO (64) reports the occurrences of motifs whose false discovery rate (FDR) is lower than a user-defined threshold. To take secondary structure into consideration, motif analysis software packages often take secondary structures that were folded by computation programs as an additional input. S. J. Lange et al. indicates that extending binding sites by 150nt is suitable for computational folding of local structures, because more than 85% of the bases pairs span less than this distance (65). With the secondary structure annotation, MEMERIS (66) searches RBPs motifs only in single strand regions. Zagros (67) models sequence, pairness and diagnostic event by using a mixture model. RNAcontext (68) extends the structural states from pairness to a collection of pairness, unstructured region, hairpin loop and others. CapR (69) calculates a secondary structural profile around RBP binding sites. Graphprot (70) models sequence and structural information as hypergraph (71) instead of calculating them separately. A new tool ssHMM (72) incorporates sequence and structural information of RBP motifs as a set of symbol-emitting states in HMM, in which the symbols are the four nucleotides and the states are structural context. The model of ssHMM is trained by input sequences and their structural annotations to output both the motif starts and binding preferred structures. Beside the k-mer sequence and structural feature, binding preferences include core spacing motif and flanking nucleotide composition (73). While the question of finding sequence motif is solved by software packages such as MEME Suite, finding structural motif from CLIP-seq datasets is still an open question. The noise introduced by peak calling as well as secondary structure prediction makes it difficult to detect the signal of structural motif.
3.3.2. Binding site prediction
Binding site prediction is a process in CLIP-seq data analysis to minimize the false negative of peak calling. Peak calling tools may not be able to identify the binding sites with insufficient mapped reads, such as in lowly expressed genes and regions with low mappability. Peak prediction is typically modeled as a classification problem to divide given genomic regions into two classes, binding and non-binding sites. Traditional machine learning models start from binding preference analysis. Graphprot stores the encoded binding preference information into feature vectors and uses a support vector machine for classification. Deboost (74) uses the bag-of-words model to encode sequence features, then applies a deep-boosting based method for classification. Instead of interpretable features like sequence and structural motif, recent deep learning models iDeep (75) and iDeepS (76) train for optimal parameters and weights in neuron networks, and lightly improve the prediction. CLAM (77) uses a different strategy to reduce the false negative by re-assigning multi-mapped reads, from which more peaks can be identified.
3.3.3. Functional analysis of binding targets
After the determination of the peak locations through peak calling and optional peak prediction, Gene Ontology (10) term and pathway analyses are often performed as the last step of the bioinformatics pipeline (15, 78). The cellular processes that the RBP may relate to can be obtained by calculating the enrichment of genes in GO term and pathway databases (79, 80). For CLIP-seq data, the results of functional analysis provide clues to understand post-transcriptional regulation, such as modulating mRNA splicing, translation, and degradation under diverse biological settings. When prior knowledge of the RBP’s function is available, the result of GO term and pathway analyses can be used to verify the peak calling result. If large fraction of binding targets shares same GO terms and is consistent with the prior knowledge, it is very likely that the peak calling was performed successfully. To perform the GO term and pathway analyses, binding sites need to be annotated to transcripts. The redundant items in genome annotations can be manually removed, then transcripts that contain binding sites can be obtained as binding targets. Some tools were developed for this task, such as Annotatepeaks.pl in Homer package, annotate function in iCount package and bed2annotation.pl in CTK package. Functional analysis tools such as DAVID (81) and Enrichr (82) take binding transcripts' symbols or IDs as input, and output enriched biological themes and visualize genes on the pathways. To analyze the interaction between RBPs, binding sites from different CLIP-seq datasets are often compared. Databases such as StarBase (83, 84), CLIPdb (85), RBPDB (86), DoRiNA (87, 88) and POSTAR (89) provide binding sites information of hundreds of RBPs. To compare binding sites of a RBP under different conditions, dCLIP (90) uses HMM to detect common and differential binding sites. A comprehensive web server Seten was developed specifically for RBP function enrichment analysis. In addition to traditional GO term and pathway information, comparative analysis with preprocessed CLIP-seq datasets is also provided by Seten as bubble charts (91).
4. Concluding remarks
In summary, CLIP-seq is a powerful tool for examining in vivo transcriptome-wide RNA:protein interactions. Here we discussed two practical aspects of performing and analyzing CLIP-seq experiments. First, using the epitope-tag knock-in approach as described above, CLIP-seq experiments can be performed on the endogenous RBPs that do not have high-quality antibodies, so that biologically relevant mechanistic insights can be obtained. We believe future developments in the genomic editing technologies will make it easier and more efficient to epitope-tag endogenous proteins, which will greatly facilitate mechanistic studies on endogenous RBPs. Second, to reduce the computational challenges faced by many molecular biologists in analyzing the CLIP-seq datasets, we compared the computational tools that are currently available for analyzing the CLIP-seq data and provided our opinions on how to choose among existing computational tools. We feel that further developing standardized computational pipelines or software packages in processing and analyzing CLIP-seq datasets will be of great help for maximally extracting meaningful information from the CLIP-seq experiments.
Finally, like many other great techniques, CLIP-seq analysis also has its own limitations. For example, although CLIP-seq analysis can reveal the in vivo physical interactions between RNA and the target proteins, whether or not the identified interactions have a functional consequence on gene expression, however, cannot be inferred from the CLIP-seq results alone. In addition, due to the complexity of the crosslinking reaction, whether the CLIP-seq signal intensity can be used as an indication of in vivo binding strength of the target RBP is debatable. Thus, care should be taken in interpreting results from CLIP-seq experiments. Nonetheless, with technology advancement in both genomic editing and bioinformatics, we believe CLIP-seq analysis in combination with functional assays will reveal more exciting gene-expression regulatory networks mediated by RBPs.
Highlights.
Epitope-tagged endogenous RNA-binding proteins using CRISPR/Cas9-based genome editing
General consideration of the computational analysis on CLIP-seq datasets
Comparison of several widely used computational programs for peak calling on CLIP-seq datasets
Acknowledgement
This work is supported by grants from the NIH (R01 HL141112, R01 GM102515), the DOD (BM170039), and the Mayo Foundation for Education and Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brinegar AE, Cooper TA, Roles for RNA-binding proteins in development and disease. Brain research 1647, 1–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pereira B, Billaud M, Almeida R, RNA-Binding Proteins in Cancer: Old Players and New Actors. Trends in cancer 3, 506–528 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Ule J, Jensen K, Mele A, Darnell RB, CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods 37, 376–386 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Darnell R, CLIP (cross-linking and immunoprecipitation) identification of RNAs bound by a specific protein. Cold Spring Harb Protoc 2012, 1146–1160 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Zhang C, Darnell RB, Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol 29, 607–614 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darnell RB, HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA 1, 266–286 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee FCY, Ule J, Advances in CLIP Technologies for Studies of Protein-RNA Interactions. Mol Cell 69, 354–369 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Ascano M, Hafner M, Cekan P, Gerstberger S, Tuschl T, Identification of RNA-protein interaction networks using PAR-CLIP. Wiley Interdiscip Rev RNA 3, 159–177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huppertz I et al. , iCLIP: protein-RNA interactions at nucleotide resolution. Methods 65, 274–287 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Nostrand EL et al. , Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 13, 508–514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wheeler EC, Van Nostrand EL, Yeo GW, Advances and challenges in the detection of transcriptome-wide protein-RNA interactions. Wiley Interdiscip Rev RNA 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garzia A, Meyer C, Morozov P, Sajek M, Tuschl T, Optimization of PAR-CLIP for transcriptome-wide identification of binding sites of RNA-binding proteins. Methods 118-119, 24–40 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Nostrand EL, Huelga SC, Yeo GW, Experimental and Computational Considerations in the Study of RNA-Binding Protein-RNA Interactions. Adv Exp Med Biol 907, 1–28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu W, Yuan B, Lodish HF, Cpeb4-mediated translational regulatory circuitry controls terminal erythroid differentiation. Dev Cell 30, 660–672 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Chen X, Liu Q, Zhang S, Hu W, Translation repression via modulation of the cytoplasmic poly(A)-binding protein in the inflammatory response. Elife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Nostrand EL et al. , CRISPR/Cas9-mediated integration enables TAG-eCLIP of endogenously tagged RNA binding proteins. Methods 118-119, 50–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brooks SA, Connolly JE, Rigby WF, The role of mRNA turnover in the regulation of tristetraprolin expression: evidence for an extracellular signal-regulated kinase-specific, AU-rich element-dependent, autoregulatory pathway. J Immunol 172, 7263–7271 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Maruyama T et al. , Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol 33, 538–542 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maragkakis M, Alexiou P, Nakaya T, Mourelatos Z, CLIPSeqTools—a novel bioinformatics CLIP-seq analysis suite. RNA 22, 1–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shah A, Qian Y, Weyn-Vanhentenryck SM, Zhang C, CLIP Tool Kit (CTK): a flexible and robust pipeline to analyze CLIP sequencing data. Bioinformatics 33, 566–567 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen B, Yun J, Kim MS, Mendell JT, Xie Y, PIPE-CLIP: a comprehensive online tool for CLIP-seq data analysis. Genome Biol 15, R18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khorshid M, Rodak C, Zavolan M, CLIPZ: a database and analysis environment for experimentally determined binding sites of RNA-binding proteins. Nucleic Acids Res 39, D245–D252 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin M, Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal 17, pp. 10–12 (2011). [Google Scholar]
- 24.Gordon A, Hannon G, Fastx-toolkit. FASTQ/A short-reads preprocessing tools (unpublished) http://hannonlab cshl. edu/fastx_toolkit 5, (2010). [Google Scholar]
- 25.Bolger AM, Lohse M, Usadel B, Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmieder R, Edwards R, Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krueger F, Trim galore. A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files, (2015). [Google Scholar]
- 28.B. Bioinformatics, FastQC: a quality control tool for high throughput sequence data. Cambridge, UK: Babraham Institute, (2011). [Google Scholar]
- 29.Chakrabarti AM, Haberman N, Praznik A, Luscombe NM, Ule J, Data Science Issues in Studying Protein–RNA Interactions with CLIP Technologies. (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim D, Langmead B, Salzberg SL, HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobin A et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29,15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bottini S, Pratella D, Grandjean V, Repetto E, Trabucchi M, Recent computational developments on CLIP-seq data analysis and microRNA targeting implications. Brief Bioinform, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farrell RE Jr, RNA Methodologies: laboratory guide for isolation and characterization. (Academic Press, 2009). [Google Scholar]
- 34.Li H et al. , The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKenna A et al. , The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu H et al. , FastUniq: a fast de novo duplicates removal tool for paired short reads. PLoS One 7, e52249 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hauer C et al. , Improved binding site assignment by high-resolution mapping of RNA-protein interactions using iCLIP. Nat Commun 6, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De S, Gorospe M, Bioinformatic tools for analysis of CLIP ribonucleoprotein data. Wiley Interdiscip Rev RNA 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uhl M, Houwaart T, Corrado G, Wright PR, Backofen R, Computational analysis of CLIP-seq data. Methods 118-119, 60–72 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Uren PJ et al. , Site identification in high-throughput RNA-protein interaction data. Bioinformatics 28, 3013–3020 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kucukural A, Ozadam H, Singh G, Moore MJ, Cenik C, ASPeak: an abundance sensitive peak detection algorithm for RIP-Seq. Bioinformatics 29, 2485–2486 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lovci MT et al. , Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nature Structural & Molecular Biology 20, 1434–1442 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Althammer S, Gonzalez-Vallinas J, Ballare C, Beato M, Eyras E, Pyicos: a versatile toolkit for the analysis of high-throughput sequencing data. Bioinformatics 27, 3333–3340 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Konig J et al. , iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17, 909–915 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corcoran DL et al. , PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol 12, R79 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Golumbeanu M, Mohammadi P, Beerenwinkel N, BMix: probabilistic modeling of occurring substitutions in PAR-CLIP data. Bioinformatics 32, 976–983 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Comoglio F, Sievers C, Paro R, Sensitive and highly resolved identification of RNA-protein interaction sites in PAR-CLIP data. BMC Bioinformatics 16, 32 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sievers C, Schlumpf T, Sawarkar R, Comoglio F, Paro R, Mixture models and wavelet transforms reveal high confidence RNA-protein interaction sites in MOV10 PAR-CLIP data. Nucleic Acids Res 40, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang T, Chen B, Kim M, Xie Y, Xiao G, A model-based approach to identify binding sites in CLIP-Seq data. PloS one 9, e93248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Webb S, Hector RD, Kudla G, Granneman S, PAR-CLIP data indicate that Nrd1- Nab3-dependent transcription termination regulates expression of hundreds of protein coding genes in yeast. Genome biology 15, R8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krakau S, Richard H, Marsico A, PureCLIP: capturing target-specific protein-RNA interaction footprints from single-nucleotide CLIP-seq data. Genome Biol 18, 240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moore MJ et al. , Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nature protocols 9, 263 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langenberger D et al. , Evidence for human microRNA-offset RNAs in small RNA sequencing data. Bioinformatics 25, 2298–2301 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Bottini S et al. , From benchmarking HITS-CLIP peak detection programs to a new method for identification of miRNA-binding sites from Ago2-CLIP data. Nucleic Acids Res 45, e71–e71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brooks SA, Blackshear PJ, Tristetraprolin (TTP): interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochimica Et Biophysica Acta (BBA)-Gene Regulatory Mechanisms 1829, 666–679 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heinz S et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Nostrand EL et al. , Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nature methods 13, 508 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Drewe-Boss P, Wessels H-H, Ohler U, omniCLIP: Bayesian identification of protein-RNA interactions from CLIP-Seq data. bioRxiv, 161877 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quinlan AR, Hall IM, BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quinlan AR, BEDTools: the Swiss - army tool for genome feature analysis. Current protocols in bioinformatics 47, 11.12. 11–11.12. 34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ray D et al. , A compendium of RNA-binding motifs for decoding gene regulation. Nature 499, 172 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bailey TL, Williams N, Misleh C, Li WW, MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34, W369–W373 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bailey TL et al. , MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37, W202–W208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grant CE, Bailey TL, Noble WS, FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lange SJ et al. , Global or local? Predicting secondary structure and accessibility in mRNAs. Nucleic Acids Res 40, 5215–5226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hiller M, Pudimat R, Busch A, Backofen R, Using RNA secondary structures to guide sequence motif finding towards single-stranded regions. Nucleic Acids Res 34, e117–e117 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bahrami-Samani E, Penalva LO, Smith AD, Uren PJ, Leveraging cross-link modification events in CLIP-seq for motif discovery. Nucleic Acids Res 43, 95–103 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kazan H, Ray D, Chan ET, Hughes TR, Morris Q, RNAcontext: a new method for learning the sequence and structure binding preferences of RNA-binding proteins. PLoS computational biology 6, e1000832 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fukunaga T et al. , CapR: revealing structural specificities of RNA-binding protein target recognition using CLIP-seq data. Genome biology 15, R16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maticzka D, Lange SJ, Costa F, Backofen R, GraphProt: modeling binding preferences of RNA-binding proteins. Genome biology 15, R17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heyne S, Costa F, Rose D, Backofen R, GraphClust: alignment-free structural clustering of local RNA secondary structures. Bioinformatics 28, i224–i232 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heller D, Krestel R, Ohler U, Vingron M, Marsico A, ssHMM: extracting intuitive sequence-structure motifs from high-throughput RNA-binding protein data. Nucleic Acids Res 45, 11004–11018 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dominguez D et al. , Sequence, Structure, and Context Preferences of Human RNA Binding Proteins. Molecular cell 70, 854–867. e859 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li S et al. , A deep boosting based approach for capturing the sequence binding preferences of RNA-binding proteins from high-throughput CLIP-seq data. Nucleic Acids Res 45, e129–e129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pan X, Shen H-B, RNA-protein binding motifs mining with a new hybrid deep learning based cross-domain knowledge integration approach. BMC bioinformatics 18, 136(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pan X, Rijnbeek P, Yan J, Shen H-B, Prediction of RNA-protein sequence and structure binding preferences using deep convolutional and recurrent neural networks. BMC genomics 19, 511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Z, Xing Y, CLIP-seq analysis of multi-mapped reads discovers novel functional RNA regulatory sites in the human transcriptome. Nucleic Acids Res 45, 9260–9271 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saulière J et al. , CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nature structural & molecular biology 19, 1124 (2012). [DOI] [PubMed] [Google Scholar]
- 79.Kanehisa M, Goto S, KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ashburner M et al. , Gene Ontology: tool for the unification of biology. Nature genetics 25, 25 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dennis G et al. , DAVID: database for annotation, visualization, and integrated discovery. Genome biology 4, R60 (2003). [PubMed] [Google Scholar]
- 82.Chen EY et al. , Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li J-H, Liu S, Zhou H, Qu L-H, Yang J-H, starBase v2. 0: decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 42, D92–D97 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang J-H et al. , starBase: a database for exploring microRNA–mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res 39, D202–D209 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang Y-CT et al. , CLIPdb: a CLIP-seq database for protein-RNA interactions. BMC genomics 16, 51 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cook KB, Kazan H, Zuberi K, Morris Q, Hughes TR, RBPDB: a database of RNA-binding specificities. Nucleic Acids Res 39, D301–D308 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Anders G et al. , doRiNA: a database of RNA interactions in post-transcriptional regulation. Nucleic Acids Res 40, D180–D186 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blin K et al. , DoRiNA 2.0—upgrading the doRiNA database of RNA interactions in post-transcriptional regulation. Nucleic Acids Res 43, D160–D167 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hu B, Yang Y-CT, Huang Y, Zhu Y, Lu ZJ, POSTAR: a platform for exploring post-transcriptional regulation coordinated by RNA-binding proteins. Nucleic Acids Res 45, D104–D114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang T, Xie Y, Xiao G, dCLIP: a computational approach for comparative CLIP-seq analyses. Genome biology 15, R11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Budak G, Srivastava R, Janga SC, Seten: A tool for systematic identification and comparison of processes, phenotypes and diseases associated with RNA-binding proteins from condition-specific CLIP-seq profiles. RNA, rna. 059089.059116 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]