Abstract
Ribosomes associated with nonsense-mediated decay factors Upf1, Upf2, or Upf3 were purified by immunoprecipitation, and enrichment and stoichiometry of Upf factors and ribosomal proteins were analyzed by western blot and mass spectrometry. Using a small RNA library preparation protocol that eliminates in-gel RNA and cDNA size selection and incorporates four random nucleotides on each side of the ribosome-protected RNA fragment allowed recovery, detection, and analysis of all size classes of protected fragments from a sample simultaneously.
Keywords: NMD, Selective ribosome profiling, Upf proteins
1. Introduction
Nonsense-mediated mRNA Decay (NMD) is a conserved translation-dependent mRNA decay pathway generally triggered when a premature termination codon (PTC) occupies the ribosomal A site [1–5]. NMD targets polysome-associated mRNAs [6–9] that account for as much as 15% of the complexity of an organism’s transcriptome [6, 10–18]. PTCs characteristic of NMD-targeted mRNAs can arise from mutations in genomic DNA, alternative pre-mRNA processing, or non-productive DNA rearrangements, but they are also inherent to a subset of normal transcripts, including those for which the ribosome fails to utilize or maintain the proper reading frame [19]. NMD substrates have substantial biological impact: not only does NMD ensure “junk” removal (e.g., byproducts of alternate splicing, cytoplasmic intron-containing transcripts, or pseudogene mRNAs [6, 20]), but it also effectively renders most nonsense alleles as null alleles [21, 22] and has been coopted to regulate the levels and locations of specific proteins [23, 24].
UPF1, UPF2, and UPF3 encode the key factors controlling NMD [1, 3, 4, 25–30] and their inactivation stabilizes nonsense-containing mRNAs while having no significant effects on most wild-type transcripts [6, 30, 31]. The Upf proteins are mostly cytoplasmic and interact with each other, the ribosome, and multiple mRNA decay and translation factors, but their exact roles in NMD, and their mechanism of association with a premature termination complex, remain to be clarified [1]. Upf1 is NMD’s key regulator. Overexpression of UPF1 compensates for the nonsense suppression phenotype of mutations in UPF2 and UPF3 (without an effect on NMD phenotypes), but not vice versa [32], and maximal in vitro activation of Upf1’s helicase and ATPase activities requires both Upf2 and Upf3 [33]. The Upf proteins associate with ribosomes [34–38], but there are conflicting views as to whether this occurs stochastically, only to be activated at a PTC, or whether it is premature termination per se that leads to Upf recruitment and function [5]. Very little is known about when in the translation cycle Upf factors bind to ribosomes, whether NMD is triggered by these binding events, and which Upf factors, if any, confer degradation specificity to an NMD substrate.
To address this conundrum, we have established methods for enriching and profiling yeast ribosomes that are associated with specific Upf factors. Most ribosome profiling libraries are prepared using some version of the protocol developed by Ingolia et al. [39] and require in-gel size-selection of ribosome-protected RNA fragments, size selection and circularization of cDNA, and some method of PCR product size selection. In addition to being time-consuming and difficult to prepare libraries from small amounts of input RNA, these protocols are seldom designed to recover the full spectrum of potential ribosome-protected RNA fragment size classes. Further, unlike many small or micro-RNA library preparation protocols, most ribosome profiling library preparation methods do not include random nucleotide bar codes to eliminate PCR duplicates from downstream analysis.
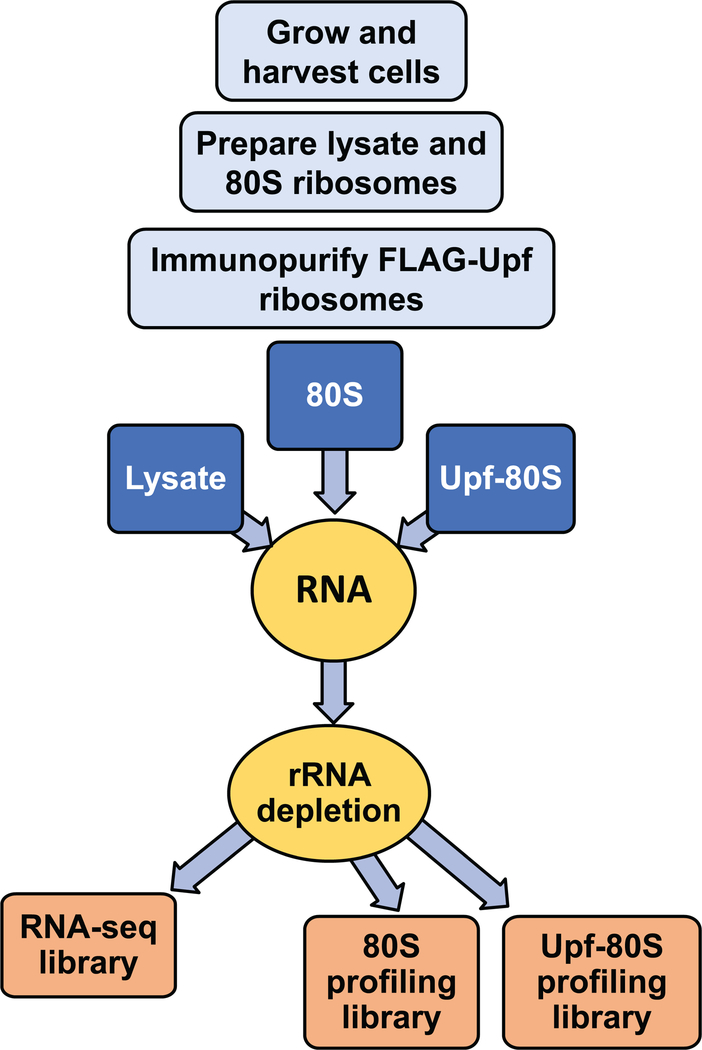
We have developed a workflow (summarized in Fig. 1) which not only allows efficient recovery of Upf-associated ribosomes by immunoprecipitation, but also enables a snapshot of the translational status and stoichiometry of Upf factors and ribosomes. In addition, by incorporating a library preparation method which places four random nucleotides at each end of the ribosome protected fragment and eliminates in-gel RNA and cDNA size selection, our workflow has allowed us to eliminate PCR duplicates from our analyses, recover a broad range of ribosome-protected fragment size classes, and compare the read length distributions of all ribosome-protected fragment size classes present in a sample across the transcriptome.
Figure 1.
Diagram of protocol workflow.
2. Methods
2.1. Equipment and reagents
2.1.1. Recommended equipment is listed in Table 1.
Table 1.
Equipment
| Name | Manufacturer |
|---|---|
| Shaking incubator | Thermo Scientific |
| 2.8liter Fernbach flasks | Pyrex |
| 1liter vacuum filtration device | Kontes |
| Cryomill | Retsch |
| Stainless steel jar | Retsch (01.462.0332) |
| 20mm stainless steel ball | Retsch (05.368.0062) |
| Tabletop refrigerated centrifuge | Thermo Fisher |
| Ultracentrifuge | Beckman Coulter |
| Spectrophotometer | Beckman |
| Thermomixer | Eppendorf |
| Microcentrifuge | Eppendorf |
| Platform rocker | Fisher scientific |
| Tabletop ultracentrifuge | Beckman Coulter |
| Mini-PROTEAN Tetra Cell | BioRad |
| Trans-Blot SD Semi-Dry Electrophoretic Transfer Cell | BioRad |
| Shaking water bath | VWR |
| Film processor | Kodak |
| Speed vac | Savant Instruments, Inc. |
| NanoAcquity UPLC | Waters |
| 75 µm I.D. analytical column | |
| Q Exactive hybrid mass spectrometer | Thermo |
| Mascot Distiller version 2.5 | Matrix Sciences, Ltd. |
| Mascot Server version 2.5 | Matrix Sciences, Ltd. |
| Scaffold Viewer | Proteome Software, Inc.) |
| Qubit 3.0 Fluorometer | Life Technologies |
| Thermocycler | Eppendorf |
| Fragment analyzer | Advanced Analytics |
Reagents and consumables necessary for performing this protocol are listed in Table 2.
Table 2.
Reagents and Consumables
| Product | Manufacturer | Catalog number | |
|---|---|---|---|
| 1M Tris, pH7.5 | Corning | 46–030-CM | |
| 5M NaCl | Cellgro | 46–032-CV | |
| 1M MgCl2 | Ambion | AM9530G | |
| RNase-free water | Ambion | AM9922 | |
| Triton X-100 | Sigma | T8787 | |
| Dithiothreitol (DTT) | Sigma | D9779 | |
| Phenylmethylsulfonyl fluoride | Sigma | P7626 | |
| Igepal CA-630 | Sigma | I8896 | |
| 0.2M MES-NaOH, pH5.5 | Boston Bioproducts | BB-109 | |
| 10X PBS | Fisher Scientific | BP3994 | |
| Tween-20 | Sigma | P1379 | |
| Sodium dodecyl sulfate (SDS) | Sigma | L3771 | |
| β-mercaptoethanol | Life Technologies | 21985 | |
| 0.8μm cellulose acetate membrane filter | Sterlitech | CA089025 | |
| Protease inhibitors | Pierce | 88266 | |
| Cycloheximide | Calbiochem | 239764 | |
| RNaseI | Invitrogen | AM2294 | |
| Superase-In | Ambion | AM2694 | |
| anti-FLAG M2 affinity gel | Sigma | A2220 | |
| Amicon-Pro affinity exchange device | Millipore | ACS500024 | |
| 3/8” Tough-Spot | Diversified Biotech | SPRL-1100 | |
| 20g BD PrecisionGlide hyperdermic needle | BD | 305175 | |
| 3X FLAG peptide | Sigma | F4799 | |
| Amicon Ultra-2 100K centrifugal filter unit | Millipore | UFC210024 | |
| Sucrose | Sigma | S0389 | |
| Thick wall polyallomer 7X20mm centrifuge tubes | Beckman Coulter | 343621 | |
| 4X Laemmli sample buffer | BioRad | 161–0747 | |
| 10-well, 30 µl 4–20% Mini-PROTEAN® TGX Precast Protein Gel | BioRad | 4561093 | |
| Prosieve Quadcolor protein marker, 4.6– 300kDa | Lonza | 00193837 | |
| 10X Tris/glycine/SDS buffer | BioRad | 161–0732 | |
| Immobilon-P PVDF membrane | Millipore | IPVH00010 | |
| Nonfat dry milk | Grocery store | ||
| Bovine serum albumin | Sigma | A4503 | |
| anti-FLAG antibody | Sigma | F7425 | |
| ECL Rabbit IgG, HRP-linked whole Ab (from donkey) | GE Healthcare | NA934 | |
| Mouse IgG (H+L) secondary antibody | ThermoFisher Scientific | 625620 | |
| Hyperfilm ECL | Sigma | GE28–9068-35 | |
| Colloidal Blue staining kit | Invitrogen | LC6025 | |
| Ammonium bicarbonate | |||
| Dithiothreitol | |||
| Iodoacetamide | |||
| Acetonitrile | |||
| Trypsin | |||
| ProteaseMAX Surfactant | Promega | ||
| Formic acid | |||
| Trifluoroacetic acid | |||
| 5 µm (200Å) Magic C18AQ | Bruker-Michrom | ||
| 3 µm (100Å) Magic C18AQ | Bruker-Michrom | ||
| Turbo DNase | ThermoFisher Scientific | AM2238 | |
| miRNeasy mini kit | Qiagen | 217004 | |
| 1.5ml flat cap microcentrifuge tube | Laboratory Products Sales | L259901 | |
| Qubit RNA HS assay kit | ThermoFisher Scientific | Q32852 | |
| High sensitivity RNA analysis kit (15 nt) | Advanced Analytical | DNF-472 | |
| Baseline-ZERO DNase | Epicentre | DB0715K | |
| Phenol:Chloroform:IAA | Ambion | AM9732 | |
| 3M sodium acetate (NaOAc) | Sigma | S7899 | |
| Glycogen | Roche | 10901393001 | |
| Ethanol | Sigma | E7023 | |
| Ribo-Zero Gold rRNA removal kit (Yeast) | Illumina | MRZY1324 | |
| TruSeq Stranded mRNA sample prep kit | Illumina | RS-122–2101 | |
| NextSeq 500/550 High Output v2 kit (75 cycles) | Illumina | FC-404–2005 | |
| Qubit DNA HS assay kit | ThermoFisher Scientific | Q32851 | |
| High Sensitivity NGS fragment analysis kit | Advanced Analytical | DNF-474 | |
| T4 polynucleotide kinase | New England Biolabs | M0201 | |
| RNA Clean and Concentrator-5 | Zymo Research | R1016 | |
| 100mM ATP | Thermo Scientific | R0441 | |
| NEXTflex Small RNA-Seq Kit v3 | BIOO Scientific | NOVA-5132–06 | |
| 0.2ml 96-well PCR plate | USA Scientific | 1402–9600 | |
| Qubit dsDNA HS assay kit | ThermoFisher Scientific | Q32851 | |
| 5X -TRP dropout mix (grams): | |||
| Uracil | 2 | Sigma | U-0750 |
| Histidine | 4 | “ | H-8125 |
| Arginine | 4 | “ | A-5131 |
| Methionine | 4 | “ | M-9625 |
| Tyrosine | 4 | “ | T-3754 |
| Isoleucine | 4 | “ | I-2752 |
| Lysine | 4 | “ | L-5626 |
| Adenine | 2 | “ | A-8751 |
| Phenylalanine | 4 | “ | P-2126 |
| Leucine | 4 | “ | L-8000 |
| Aspartic acid | 4 | “ | A-6683 |
| Valine | 4 | “ | V-0500 |
| Threonine | 4 | “ | T-8625 |
| Serine | 4 | “ | S-4500 |
| -TRP liquid medium | |||
| Yeast nitrogen base | 1.7 g | ||
| (NH4)2SO4 | 5.0 g | ||
| Water | up to 850 ml | ||
| Autoclave, then add (per liter): | |||
| 40 % glucose | 50 ml | ||
| 5X-TRP (2g/100ml water) | 100 ml | ||
2.1.2. Buffers used in this protocol are:
Footprinting buffer (FB): 20mM Tris, pH7.4, 150mM NaCl, 5mM MgCl2
Immunoprecipitation (IP) buffer: 150mM Tris, pH7.5, 150mM NaCl, 20mM MgCl2, 0.1% Igepal
1.5X MES-NaOH buffer: 150 mM MES-NaOH, pH 5.5, 450 mM NaCl, 15 mM MgCl2, 15 mM β-mercaptoethanol
1X PBS-T: 1X PBS, 0.1% Tween 20
Western stripping buffer: 2% SDS, 62.5mM Tris, pH6.8, 0.8% β-mercaptoethanol
2.2. High-copy expression of tagged Upf proteins
Yeast Upf proteins were tagged by N-terminal fusion of a FLAG epitope (amino acid sequence DYKDDDDK) [40] to the open reading frame of each gene. N-terminal tagging was chosen because previous experiments have demonstrated that these alleles are stably expressed and functional in vivo [29, 41]. FLAG-UPF3 was expressed under the TPI promoter as this was determined to be necessary for high-level expression of this protein (data not shown).
2.2.1. Yeast strains are described in Table 3.
Table 3.
Yeast strains
| Name | Genotype |
|---|---|
| HFY114 | MAT a ade2–1 his3–11, 15 leu2–3, 112 trp1–1 ura3–1 can1–100 UPF1 UPF2 UPF3 |
| HFY871 | MAT a ade2–1 his3–11, 15 leu2–3, 112 trp1–1 ura3–1 can1–100 upf1::HIS3 UPF2 UPF3 |
| HFY115 | MAT a ade2–1 his3–11, 15 leu2–3, 112 trp1–1 ura3–1 can1–100 UPF1 upf2::HIS3 UPF3 |
| HFY861 | MAT a ade2–1 his3–11, 15 leu2–3, 112 trp1–1 ura3–1 can1–100 UPF1 UPF2 upf3::HIS3 |
Plasmids [29, 41] are described in Table 4.
Table 4.
Plasmids
| Name | Description |
|---|---|
| YEplac112 | Yeast episomal plasmid (https://www.addgene.org/vector-database/4892/) |
| pG1-FLAG-Upf1 | Yeast episomal plasmid containing entire UPF1 coding region as a 3.6kb BamHI fragment; FLAG-tag fused at the N-terminus; expressed under the TDH3 promoter |
| YEplac112-FLAG-Upf2 | YEplac112 containing entire UPF2 coding region, and the endogenous promoter; FLAG-tag fused at the N-terminus; same as YEplac112-F2-NMD2* (*NMD2=UPF2) |
| YEplac195-TPI-FLAG-Upf3 | Yeast episomal plasmid containing entire UPF3 coding region as a 2.2 kb SalI-XbaI fragment; FLAG-tag fused at the N-terminus; expressed under the TPI promoter |
2.2.2. Yeast strains were transformed with plasmids according to standard methods and grown on selective medium until a final shift to rich medium prior to harvest.
2.3. Preparation of lysate and 80S ribosomes
2.3.1. Growth and harvest of cells
Cells are expanded in selective medium to maintain the plasmid bearing the FLAG-tagged UPF allele. Growth for ~2 doublings in rich medium prior to cell harvest ensures maximal occupancy of translating ribosomes on mRNA.
Inoculate 100ml selective medium from a single colony or cell patch grown on selective medium, shake at 30°C, 210 rpm, overnight. In the morning, dilute the culture to A600=0.1 in 100ml selective medium and shake at 30°C, 210 rpm, for 6–8 hours. The starting cell density for the next step may vary depending on the growth rate of the strain, but generally dilute culture to A600=0.0015–0.003 into 2 liters prewarmed selective medium in a 6 liter flask, shake at 30°C, 120 rpm, 16 hours. In the morning, if the A600 has not exceeded 0.8, dilute the culture to A600=0.15–0.2 into individual 2.8 liter Fernbach flasks containing 1 liter prewarmed YAPD medium (YEPD medium supplemented with adenine; [42]), 2 to 6 liters total, depending on the application. For immunoprecipitation experiments, a total of 6 liters of culture are grown per strain and cycloheximide condition; otherwise, 1 liter of culture is grown. Shake at 30°C, 210rpm, to A600=0.6 to 0.8.
Recover each liter of culture individually by vacuum filtration onto a 0.8μm cellulose acetate membrane filter, quickly collect cells with a spatula, flash freeze by plunging into 50ml polyethylene centrifuge tubes filled with liquid nitrogen, and drip in 2.5ml footprinting buffer (FB) plus 1% TritonX-100, 0.5mM DTT, 1mM phenylmethylsulfonyl fluoride (PMSF), 1X protease inhibitors. When indicated, 100μg/ml final concentration cycloheximide is added to the FB and culture, with swirling to mix, immediately prior to vacuum filtration.
Pool frozen cells plus FB from 2 liters total of cell culture and lyse in a Cryomill (Retsch) in a 50ml stainless steel jar with a 20mm stainless steel ball; precool at 5Hz, 2 minutes, and grind at 10Hz, 15 minutes, precooling all tools and containers in liquid nitrogen and keeping all samples on dry ice. Pool frozen powder from up to 6 liters of culture from each culture condition into two new 50ml polyethylene centrifuge tubes and store at −80°C.
2.3.2. Purification of 80S ribosomes
The following ribosome preparation protocol is adapted from Ingolia, et al. [39]:
Thaw pooled frozen cell powder by immersing the tube in 30°C water and stirring with a sterile pipette until just thawed, and then place on ice. Centrifuge twice for 5 minutes, 5,000 rpm (4,696 g), in a 4°C tabletop centrifuge. Place supernatant into chilled, balanced 10.4ml polycarbonate tubes and spin in an ultracentrifuge at 18,000 rpm (29,321 g), type 50Ti rotor, 4°C, 10 minutes. Carefully collect the supernatant with a Pasteur pipette, avoiding the lipid layer and pellet, into new chilled 10.4ml polycarbonate tubes; rebalance and spin again at 18,000 rpm (29,321 g), type 50Ti rotor, 4°C, 15 minutes. Carefully collect the supernatant as before into a chilled 50ml centrifuge tube on ice.
Measure lysate A260 in a spectrophotometer. Reserve 0.5ml lysate in 100–200μl aliquots on dry ice and store at −80°C for later RNA extraction for use in making RNA-Seq libraries. Divide remaining lysate into 1ml aliquots in 1.7ml microcentrifuge tubes. Add 15U RNaseI per A260 unit of clarified lysate, and incubate for 1 hour at 25°C and 700rpm in a Thermomixer. Place digested lysate on ice and add Superase-In RNase Inhibitor to 0.8U per A260.
Layer 3–4ml of digested lysate onto 6–7ml 1M sucrose cushion in FB plus 0.5mM DTT in a 10.4ml polycarbonate tube and centrifuge in a 50Ti rotor, 50,000 rpm (226,240 g), 105 minutes, 4°C. Dissolve ribosome pellet on ice in 0.5ml FB plus 0.5mM DTT, 1X protease inhibitors, 10U/ml Superase-In per tube, or 2ml total for ribosomes from 6 liters of culture. Transfer dissolved ribosomes to microcentrifuge tubes, spin in a microcentrifuge at 15,000 rpm (21,130 g), 1 minute, to pellet insoluble material and then transfer supernatant to new microcentrifuge tubes. Measure A260 of ribosomes in a spectrophotometer and freeze 100U aliquots for immunoprecipitation reactions. Additionally, freeze four aliquots of ribosomes equivalent to 20μl A260=100 for later RNA extraction if performing ribosome profiling on ribosomes which have not undergone immunopurification.
2.4. Immunopurification of Upf-associated ribosomes
2.4.1. Prepare affinity gel
All steps in this section are performed at 4°C.
Thaw anti-FLAG M2 Affinity Gel on ice; one 5 ml bottle is enough for two immunoprecipitation experiments of 6 bindings per condition. Aliquot 800μl 50% anti-FLAG M2 Affinity resin into each Amicon-Pro Affinity exchange device for a total of 6 devices per experiment. Spin in a refrigerated benchtop centrifuge at 1000g, 1 minute.
Resuspend each aliquot of affinity gel in 8ml cold IP buffer and spin at 1000g, 2 minutes; repeat.
2.4.2. Binding of ribosomes
Resuspend each washed aliquot of affinity gel in 8ml cold IP buffer plus 0.5mM DTT, 1X protease inhibitors, 100U/ml Superase-In.
Save three 1μl aliquots of input ribosomes in 15μl IP buffer total from each condition on dry ice for later analysis by western blot and mass spectrometry; store aliquots at −80°C. Add 16 A260 of ribosomes per aliquot of resuspended affinity gel (96 A260 of ribosomes total per condition across 6 devices). Place any remaining ribosomes on dry ice and store at −80°C. Place a 3/8” Tough-Spot over the hole in the Amicon-Pro Affinity exchange device, rubbing firmly to secure. Close the cap tightly, and secure the cap with parafilm. Incubate for 90 minutes at 4°C with rocking at 12 rpm on a platform rocker.
After binding, remove parafilm, pierce the 3/8” Tough-Spot with a 20g hypodermic needle, and collect affinity gel by centrifugation at 4°C, 1000g, 2 min. Pool the flowthrough from each condition and save three aliquots of 15μl each as in step 2. Wash affinity gel 3 times with 8ml IP buffer plus 0.5mM DTT, 1X protease inhibitors, with centrifugation at 1000g for 2 minutes between each wash. Pool the final wash from each condition and save three 15μl aliquots as in step 2.
Place a new 3/8” Tough-Spot over the hole of the Amicon-Pro Affinity Exchange Device. Incubate each binding reaction with 2ml 0.15mg/ml 3X FLAG peptide in IP buffer plus 0.5mM DTT, 1X protease inhibitors, 100U/ml Superase-In for 30min, 4°C, rocking as in step 2. Pierce the 3/8” Tough-Spot and collect eluate by centrifugation as in step 3.
2.4.3. Concentration of ribosomes
Spin 2ml eluate from each elution through an Amicon Ultra-2 100K Centrifugal Filter Unit at 4°C, 4000g, until the volume is ~200μl. Recover the concentrate by inverting the filter unit into the tubes provided by the manufacturer and spin at 1000g, 3 minutes. Pool the concentrate from each condition (6 tubes) into one Amicon Ultra-2 100K Centrifugal Filter Unit and spin at 4000g until the total volume is ~160μl; recover the concentrate as with the previous spin.
In thick wall polyallomer 7X20mm centrifuge tubes, layer 160μl concentrate on top of a 40μl 1M sucrose cushion in IP buffer plus 1X protease inhibitors, 10U/ml Superase-In, and spin in a tabletop ultracentrifuge at 66,000 rpm (194,173 g),100 minutes, 4°C, in a TLA100 rotor. Carefully and completely remove the supernatant and resuspend the pellet in 60μl IP buffer plus 0.5mM DTT, 1X protease inhibitors, 20U/ml Superase-In. Determine A260 with a spectrophotometer. Save three 1μl aliquots of the ribosome pellet in 15μl IP buffer total from each condition on dry ice for later analysis by western blot; save one 2μl aliquot of ribosomes in 15μl IP buffer total from each condition for mass spectrometry. Store remaining immunopurified sample as one aliquot for RNA preparation. Freeze ribosomes and aliquots on dry ice and store at −80°C.
2.5. Analysis of immunopurified ribosomes by western blot
2.5.1. Western blotting
Add 5μl 4X Laemmli Sample Buffer plus β-mercaptoethanol to each 15μl aliquot of input ribosomes, flowthrough, final wash, and ribosome pellet from the immunopurification steps. Incubate at 100°C for 5 minutes, cool briefly, and vortex for 2–3 minutes to completely disrupt the ribosomes. Pulse spin very briefly to collect the liquid.
Load the samples onto a 10-well, 30 µl 4–20% precast polyacrylamide gel; include protein markers (4.6–300kDa) and run in 1X Tris/glycine/SDS buffer at 40mA per gel for 30 minutes. Transfer samples to a PVDF Membrane using a semi-dry electrophoretic transfer apparatus at 15V, 45 minutes.
If the FLAG-tagged protein of interest is larger than 55kD, cut each membrane with a razor blade at the 55kD protein size marker, block with 5% nonfat dry milk, 1X PBS-T, and probe the top membrane portion with 1:10,000 diluted anti-FLAG antibody in 3% BSA, 1X PBS-T followed by 1:5000 diluted ECL Rabbit IgG, HRP-linked whole Ab (from donkey) secondary antibody; probe the bottom membrane portion with 1:5000 diluted anti-Tcm1 antibody (gift of Jonathan Warner, [43]) followed by 1:5000 diluted Mouse IgG (H+L) secondary antibody. If the FLAG-tagged protein of interest is smaller than 55kD, it will be necessary to probe first with anti-FLAG antibody, followed by stripping in western stripping buffer at 50°C for 45 minutes with shaking and washing extensively with water, followed by probing with anti-Tcm1 antibody. Detect antibody signal with ECL homebrew reagents (The Coller Lab Protocol Book, https://case.edu/med/coller/Coller%20Protocol%20Book.pdf) using Hyperfilm ECL film.
2.6. Analysis of immunopurified ribosomes by mass spectrometry
2.6.1. Preparation of samples for mass spectrometry
Prepare input and pellet samples for mass spectrometry as for western blotting by incubating 1μl input ribosomes or 2μl pellet ribosomes in 15μl IP buffer total with 5μl 4X Laemmli Sample Buffer plus β-mercaptoethanol at 100°C for 5 minutes followed by vortexing for 2–3 minutes.
Load samples onto a 10-well, 30 µl 4–20% precast polyacrylamide gel and run at 40mA for 5 minutes. Do not exceed 5 minutes of run time.
Fix and stain gel with Colloidal Blue Staining Kit for 3 hours; decant stain and wash with distilled water just until sample bands are visible. Place gel onto plastic wrap and cut out the entire stained region of each sample with a razor blade, not more than 1cm x 1cm pieces. Place gel slice into a microcentrifuge tube and add 500μl water. Store at 4°C until the samples are sent to a mass spectrometry core facility, within a few days.
In the mass spectrometry facility, remove water and add 200µl of 250 mM ammonium bicarbonate. For reduction, add 20µl of a 45mM solution of 1, 4 dithiothreitol (DTT) and incubate at 50ºC for 30 min. Cool samples to room temperature and then, for alkylation, add 20µl of a 100mM iodoacetamide solution and allow to react for 30 min. Wash gel slices 2 X with 1 ml water aliquots. Remove water and place 1ml of 50:50 (50 mM ammonium bicarbonate: acetonitrile) in each tube and incubate samples at room temperature for 1hr. Remove the solution and add 200 µl of acetonitrile to each tube at which point the gels slices will turn opaque white. Remove acetonitrile and further dry gel slices in a Speed Vac. Rehydrate gel slices in 100µl of 4ng/µl of sequencing grade trypsin (Sigma) in 0.01% ProteaseMAX Surfactant (Promega): 50 mM ammonium bicarbonate. Add additional bicarbonate buffer to ensure complete submersion of the gel slices. Incubate samples at 37ºC for 18 hrs. Remove the supernatant of each sample and place in a separate 1.5ml Eppendorf tube. Further extract gel slices with 200µl of 80:20 (acetonitrile: 1% formic acid). Combine the extracts with the supernatants of each sample, then dry the samples completely in a Speed Vac.
2.6.2. LC/MS/MS Analysis
Reconstitute tryptic peptide digests in 25µl 5% acetonitrile containing 0.1% (v/v) trifluoroacetic acid and separate on a NanoAcquity (Waters) UPLC. In brief, a 2.5µl injection is loaded in 5% acetonitrile containing 0.1% formic acid at 4.0 µl/min for 4.0 min onto a 100 µm I.D. fused-silica pre-column packed with 2 cm of 5µm (200Å) Magic C18AQ (Bruker-Michrom) and eluted using a gradient at 300nl/min onto a 75µm I.D. analytical column packed with 25cm of 3µm (100Å) Magic C18AQ particles to a gravity-pulled tip. The solvents are A, water (0.1% formic acid) and B, acetonitrile (0.1% formic acid). A linear gradient is developed from 5% solvent A to 35% solvent B in 60 minutes. Ions are introduced by positive electrospray ionization via liquid junction into a Q Exactive hybrid mass spectrometer (Thermo). Mass spectra are acquired over m/z 300–1750 at 70,000 resolution (m/z 200) and data-dependent acquisition selects the top 10 most abundant precursor ions for tandem mass spectrometry by HCD fragmentation using an isolation width of 1.6 Da, collision energy of 27, and a resolution of 17,500
2.6.3. Data analysis
Raw data files are peak processed with Mascot Distiller (version 2.5, Matrix Sciences, Ltd.) prior to database searching with Mascot Server (version 2.5) against the S. cerevisiae index of the SwissProt database. Search parameters include trypsin specificity with two missed cleavages. The variable modifications of oxidized methionine, pyroglutamic acid for N-terminal glutamine, N-terminal acetylation of the protein, and a fixed modification for carbamidomethyl cysteine are considered. The mass tolerances are 10 ppm for the precursor and 0.05Da for the fragments. Extracted ion chromatograms are generated for the precursor ions and quantitated using a variation of the average method described by Silva et al. [44] in Mascot Distiller. Search results and precursor intensity data are also loaded into the Scaffold Viewer (Proteome Software, Inc.) for peptide/ protein validation and label free quantitation.
2.7. Preparation of RNA and removal of ribosomal RNA
2.7.1. RNA preparation from lysate
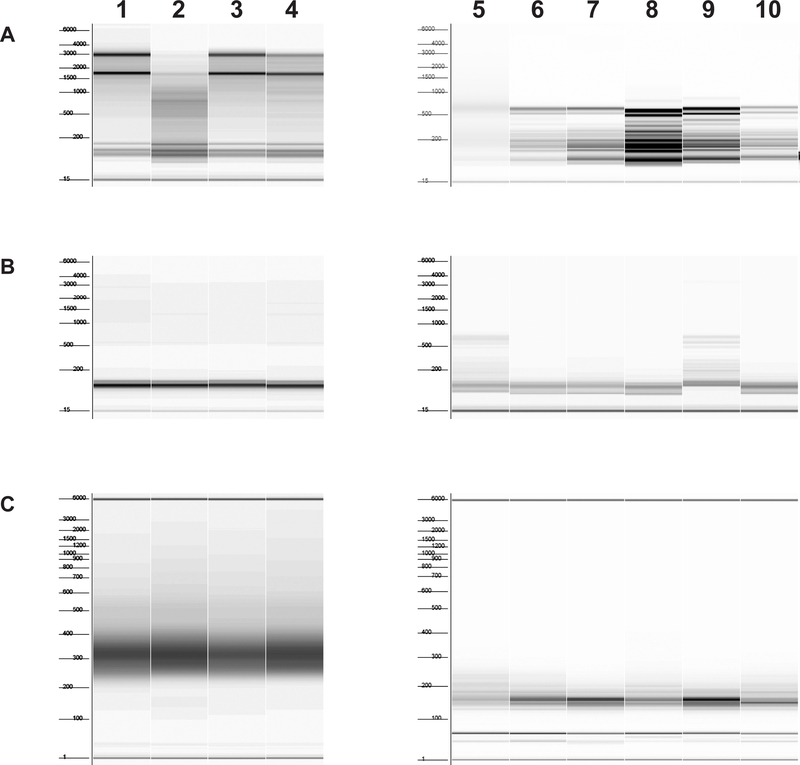
Prepare RNA from the equivalent of 60μl A260 = 100 undigested clarified lysate from step 3.2.2. with the miRNeasy mini kit as in 2.7.2, step 3. Quantitate on a spectrophotometer. Analyze RNA on a Fragment Analyzer with a High Sensitivity RNA Analysis Kit (15 nt) to check quality of RNA. (Fig. 2A, left panel)
Bring 30μg total RNA to 17μl in water. Add 2μl 10X Baseline-ZERO DNase Reaction Buffer, 1μl Baseline-ZERO DNase. Incubate at 37°C, 15–30 minutes in a thermocycler.
Add 2μl 10X Baseline-ZERO DNase Stop solution, incubate at 65°C, 5 min. Add 180μl water, extract 1X with Phenol:Chloroform:IAA, pH6.6. Reserve the aqueous layer and add 20μl 3M NaOAc, 2μl 10mg/ml glycogen, 600μl ethanol; place on dry ice 30 minutes; spin in a benchtop centrifuge at 12000 rpm (12,638 g), 4°C, 30 minutes; and wash the pellet twice with 70% ethanol. Decant ethanol, dry the pellet and dissolve in 30μl water. Measure the concentration on a spectrophotometer.
Figure 2. Fragment analyzer examples.
Starting material lanes 1–4, yeast lysate, untreated with RNase I; lanes 5–6, immunopurified ribosomes; lanes 7–10, total ribosomes. Numbers to the left of each figure indicate marker sizes in nt. A. RNA before removal of ribosomal RNA. Samples in lanes 2 and 5 are degraded and should be re-prepared; if the problem persists, it is necessary to make another preparation of lysate and ribosomes. Samples in lanes 5–10 have fragmented ribosomal RNA due to incubation of the lysate from which they were prepared with RNase I; this is expected. B. RNA after removal of ribosomal RNA. Samples in lanes 5 and 9 have incomplete removal of ribosomal RNA; these RNAs should be subjected to another round of ribosomal RNA removal. C. Finished libraries after PCR and cleanup. Sample in lane 5 is a library with a broad size range. This library should be prepared again if sequencing results indicate poor quality.
2.7.2. RNA preparation from ribosomes
Prepare RNA from the entire sample of immunoprecipitated ribosomes. If preparing a library from ribosomes which have not undergone immunoprecipitation, dilute the ribosomes in IP buffer to the same concentration as the immunoprecipitated ribosomes in 60μl IP buffer plus 0.5mM DTT, 1X protease inhibitors, 20U/ml Superase-In.
Add 2.5 U Turbo DNase to 60μl ribosomes; incubate on ice, 10 min.
Immediately extract RNA from ribosomes using the miRNeasy mini kit, following the manufacturer’s protocol for the preparation of total RNA including small RNA, eluting into 30μl water in a 1.5ml flat cap microcentrifuge tube. Quantitate on a Qubit 3.0 Fluorometer with the RNA HS assay kit. For immunopurified ribosomal RNA, dilute 1:10 in water; for total ribosomes, dilute 1:50 prior to quantitation. Freeze purified RNA samples on dry ice and store at −80°C until library preparation, up to 1 week. Analyze RNA on a Fragment Analyzer with a High Sensitivity RNA Analysis Kit (15 nt) to check quality of RNA (Fig. 2A, right panel).
2.7.3. Removal of ribosomal RNA
Ribosomal RNA is removed from RNA purified from both ribosomes and lysate using the Ribo-Zero Gold rRNA Removal Kit (Yeast).
For RNA from lysate, perform ribosomal RNA removal on one aliquot of 5μg RNA. Perform the rRNA removal using the manufacturer’s protocol for 2.5–5μg input RNA. Transfer the supernatant to a fresh 1.5ml flat cap microcentrifuge tube and bring the volume up to 180μl with water.
For RNA from ribosomes, divide all the RNA from one sample and perform ribosomal RNA removal on two separate aliquots of ~14μl each. Perform the rRNA removal using the manufacturer’s protocol for 1–2.5μg input RNA. Pool the supernatants from one sample for a combined total of 180μl and transfer to a 1.5ml flat cap microcentrifuge tube.
Add to 180μl RNA: 18μl 3M NaOAc, 2μl 1M MgCl2 (to ribosome protected fragment (RPF) RNA only), 2μl 10mg/ml glycogen, 600μl cold 100% ethanol; place on dry ice, 30 minutes; spin in a benchtop centrifuge at 12,000 rpm (12,638 g), 4°C, 30 minutes; and wash the pellet twice with 80% ethanol. Carefully decant ethanol and dry the pellet. Dissolve RPF RNA in 23μl water. Dissolve RNA from lysate in 20μl Fragment/Prime/Finish mix from the TruSeq Stranded mRNA Sample Prep kit. Analyze RNA on a Fragment Analyzer using the High Sensitivity RNA Analysis Kit (15 nt) to verify removal of ribosomal RNA (Fig. 2B).
2.8. Preparation of RNA-Seq libraries
Libraries are prepared for RNA-Seq using the TruSeq Stranded mRNA Sample Prep kit following manufacturer’s directions starting from the RNA fragmentation step (addition of Fragment/Prime/Finish Mix), without preceding steps to isolate polyA-containing mRNA.
2.9. Preparation of ribosome profiling libraries
2.9.1. 3’-Dephosphorylation and 5’-phosphorylation of ribosome protected fragments
The following protocol has been adapted from Guo, et al. [45].
Heat 20μl RPF RNA in a thermocycler at 80°C, 2 minutes. To 20μl RPF RNA, add 50μl 1.5X MES-NaOH buffer, 1μl SuperaseIn, and 1.2μl T4 polynucleotide kinase (T4 PNK). Incubate at 37°C, 2 hours; then 65°C, 20min. Add 72.2μl H2O.
Clean up with RNA Clean and Concentrator-5 kit but adjust RNA Binding Buffer (total volume as needed) by mixing an equal volume of buffer and ethanol. Add 290μl adjusted RNA Binding Buffer to each 145μl sample, mix. Proceed according to manufacturer’s instructions, eluting into 16μl water.
Add to 15.6μl RNA: 2μl 10X T4PNK buffer, 2μl 10mM ATP, 0.4μl T4 PNK, 1μl SuperasIn. Incubate in a thermocycler 37°C, 1 hour, then 60°C, 10 minutes.
Clean up with RNA Clean and Concentrator-5, adjusting RNA Binding Buffer (total volume as needed) by mixing an equal volume of buffer and ethanol. Add 40μl adjusted RNA Binding Buffer to each 20μl sample, mix. Proceed according to manufacturer’s instructions, eluting into 12μl water.
2.9.2. Ribosome profiling libraries
Ribosome profiling libraries are made using the NEXTflex Small RNA-Seq Kit v3 with the following modifications:
All steps can be performed in a 0.2ml 96-well PCR plate in a thermocycler.
Use 1:4 dilutions of both the NEXTflex 3’ 4N Adenylated Adapter and NEXTflex 5’ 4N Adapter.
Libraries will require 22–25 cycles of PCR amplification depending on the starting amount of RPF RNA. Check 1μl on a Fragment Analyzer after 22 cycles using the High Sensitivity NGS Fragment Analysis Kit, holding PCR reactions at −20°C during analysis. Thaw samples and perform 2–3 more PCR cycles if needed and check on a Fragment Analyzer again before gel-free size selection and cleanup. All samples for one study ideally should have the same number of PCR cycles.
Perform gel-free size selection and cleanup of the cDNA and final PCR products according to the manufacturer’s supplemental protocol. For the PCR bead cleanup step, the protocol was altered by bringing the PCR reaction to 25μl with water, using 65μl beads, and eluting into 13.5μl Resuspension Buffer.
2.9.3. Quantitate RNA-Seq and ribosome profiling libraries with a Qubit using the dsDNA HS Assay Kit.
Check the quality of the libraries on a Fragment Analyzer using the High Sensitivity NGS Fragment Analysis Kit (Fig. 2C), prior to sequencing on a NextSeq500 using a NextSeq 500/550 High Output v2 kit (75 cycles) to generate single-end reads. Careful pipetting and dilution of samples and reagents for Qubit quantitation can eliminate the need for subsequent qPCR quantitation.
2.10. Important considerations
For efficient cell rupture, do not grind more than 15ml cell volume in the Cryomill at one time. Harvested cells from 2 liters of medium fulfill this requirement if the final A600 is less than 0.8.
Work quickly and efficiently when harvesting cells by vacuum filtration to ensure quality ribosome profiling libraries. Rapid transfer of cells to liquid nitrogen immediately after the growth medium is removed by filtration results in libraries with excellent three-nucleotide periodicity even in the absence of cycloheximide.
Pierce the caps of any centrifuge tubes used to hold liquid nitrogen prior to use to prevent rupture.
Cell pellets from 2 liters of each culture condition and ground cell powder can be pooled and stored at −80°C for several weeks until needed. Be certain to prevent thawing or warming of the cell pellets or frozen cell powder until use.
When thawing the cell powder, only stir the tube in tepid water until most, but not all, of the frozen material is thawed; place the sample on ice and continue gently stirring until remaining ice is thawed. Keep cell lysate at 4°C until the RNaseI digestion step.
Do not exceed 0.5mM DTT in the IP buffer to avoid damaging the affinity gel.
Do not exceed 5 minutes of run time on a polyacrylamide gel when preparing samples for mass spectrometry, to minimize the amount of gel entering the mass spectrometry apparatus.
Do not use an old (>2 months) previously opened miRNeasy kit for the RNA preparation steps, as we have found this to produce RNA of variable quality.
Small RNA fragments are susceptible to loss by adhesion to the sides of the microcentrifuge tube, or loss of pellet during ethanol precipitation steps. Always resuspend RNA pellets by pipetting, never vortex, and very carefully decant ethanol to avoid loss. Save all decanted ethanol until the final washed pellet is verified visually. After the final ethanol wash, pulse spin and carefully remove remaining ethanol with a round-tip gel loading pipette tip and dry before dissolving in buffer, being careful not to lose the pellet due to static charge. We have found that the microcentrifuge tubes used in this protocol are the least susceptible to RNA loss. Addition of MgCl2 to the RPF RNA ethanol precipitation step enhances recovery of small (<30nt) RNA [46].
Four RNA-Seq or four ribosome profiling libraries can be pooled per sequencing run.
The first four and last four bases of each ribosome profiling library read following adapter trimming are random barcodes, thereby allowing removal of PCR duplicates from bioinformatic analysis if desired.
3. Results and Discussion
3.1. Overexpressed N-terminally FLAG-tagged Upf proteins complement nonsense-mediated mRNA decay and growth phenotypes.
Preliminary co-immunoprecipitation experiments determined that expression of tagged Upf1 on a single-copy yeast plasmid and subsequent pulldown of purified ribosomes did not produce sufficient material or enrichment of Upf1 relative to ribosomal proteins to perform ribosome profiling analysis of Upf1-associated ribosomes; however, expression of FLAG-Upf1 under the TDH3 [47] promoter on an episomal yeast plasmid and subsequent pulldown resulted in near stoichiometric relative amounts of Upf1 and ribosomal proteins as determined by mass spectrometry (data not shown). Therefore, FLAG-tagged Upf proteins expressed from yeast episomal plasmids were introduced into their respective yeast deletion strains. In addition, Upf3 was placed under control of the TPI1 promoter for efficient expression.
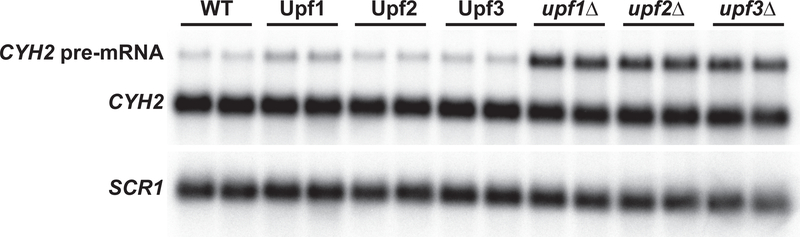
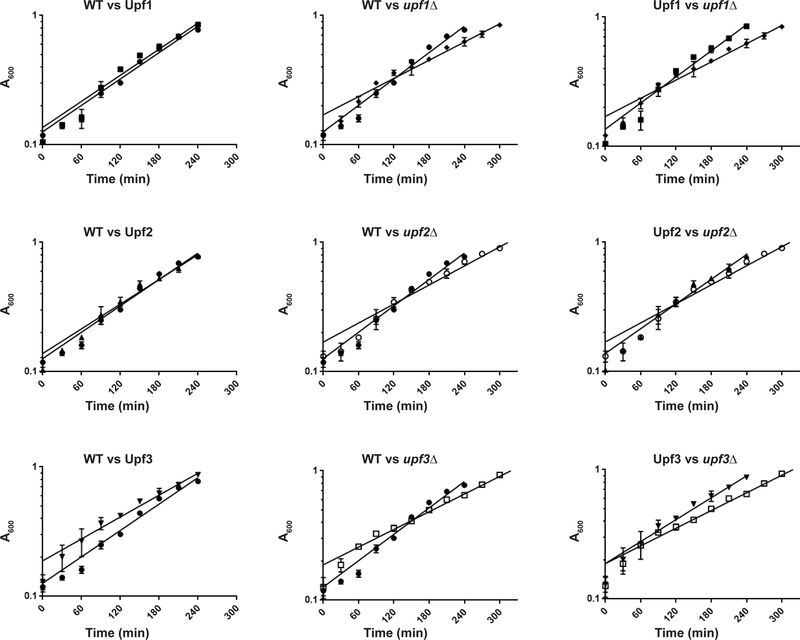
Overexpression of Upf proteins has been demonstrated to rescue mutant NMD as well as nonsense suppression phenotypes in yeast and human cells [32, 48–53]. To ascertain whether the increased expression levels of these proteins were likely to yield biologically relevant results in downstream experiments, we measured the growth rate and NMD phenotype of strains bearing those plasmids. All three proteins were able to complement the NMD (Fig. 3) and growth phenotypes (Fig. 4) of their respective deletion strains. The growth curves of both FLAG-Upf1 and FLAG-Upf2 overexpression strains were indistinguishable from the wild-type strain (Fig 4, left panel), while the FLAG-Upf3 overexpression strain exhibited a growth curve intermediate between wild-type and upf3Δ (Fig. 4, bottom row). That said, we recognize that overexpression of the three Upfs probably skews the equilibrium of ribosome association in favor of binding.
Figure 3. Overexpressed N-terminally FLAG-tagged Upf proteins complement nonsense-mediated mRNA decay phenotypes.
Northern blot of total RNA from yeast strains expressing: empty vector (WT); high-copy FLAG-tagged Upf1, Upf2, or Upf3 in the respective deletion strains; or bearing a deletion of the UPF1, UPF2 or UPF3 coding region (designated upf1Δ, upf2Δ, or upf3Δ). NMD phenotype was determined by hybridization with a random-primed labeled probe for CYH2. SCR1 was used as a loading control.
Figure 4. Growth curve comparisons.
Left panel, between WT and FLAG-tagged Upf strains; center panel, between WT and deletion strains; right panel, between FLAG-tagged Upf strains and corresponding deletion strains. WT, ●; FLAG-Upf1, ■; FLAG-Upf2, ▲; FLAG-Upf3, ▼; upf1Δ, ♦; upf2Δ, ○; upf3Δ, □. Timepoints as mean A600 from cultures of two independent isolates. Error bars=range of values.
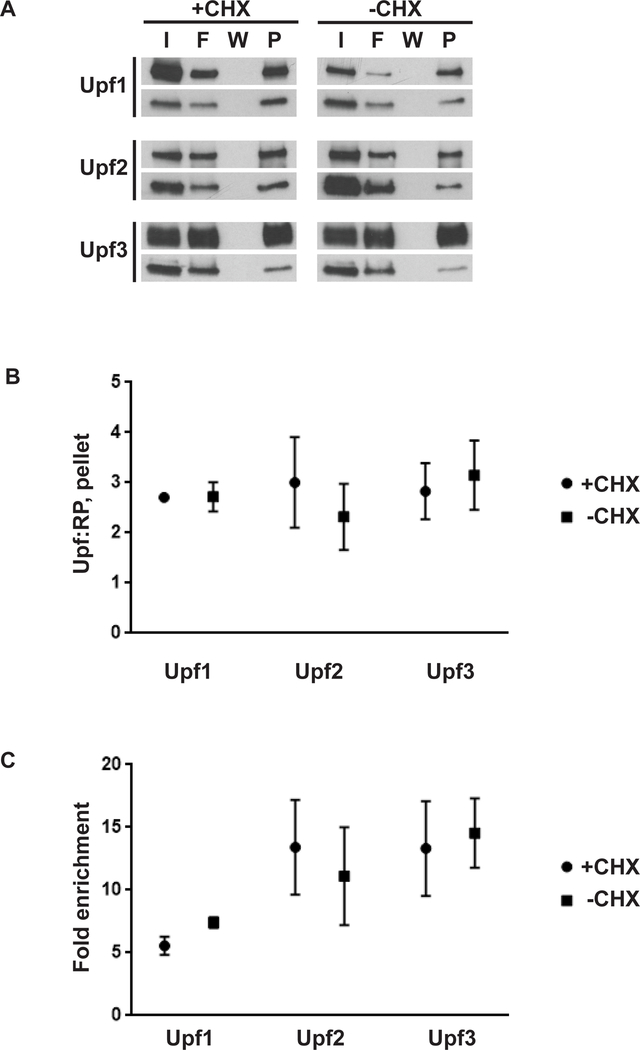
3.2. Immunopurification selects for and enriches Upf-associated ribosomes as determined by western blotting and mass spectrometry.
Ribosomes (80S) were purified from RNaseI-treated lysates prepared either in the presence or absence of cycloheximide and two biological replicates were generated for each combination of strain and cycloheximide condition. Yeast strains bearing FLAG-tagged Upf1, Upf2, or Upf3 were subjected to immunoprecipitation and elution from anti-FLAG M2 affinity gel and analyzed by western blot and mass spectrometry. Western blot analysis confirmed that FLAG-Upf-associated ribosomes were recovered from these preparations (Fig. 5A) and quantitation of mass spectrometry data by a variation of the average protocol [44] in Mascot Distiller was performed to determine the relative abundance of the Upf protein relative to all other proteins detected in the analysis (Supplementary Table 1) The ratio of the relative abundance of the Upf protein to the average relative abundance of the core ribosomal proteins (Upf:RP) present in the sample (Supplementary Table 2) was used to evaluate recovery and enrichment of Upf-associated ribosomes.
Figure 5. Overexpressed N-terminally FLAG-tagged Upf proteins allow efficient recovery of Upf-associated ribosomes.
A. Western blot of input (I), flowthrough (F), final wash (W), or pelleted ribosomes (P) from anti-FLAG immunoprecipitation reactions. Cultures were cycloheximide treated (+CHX) or untreated (-CHX) prior to cell collection. Western blots were probed with anti-FLAG (top panel per set) or anti-TCM1/RPL3 antibodies (bottom panel per set). B. Relative abundance of Upf protein per average relative abundance of core ribosomal proteins present in a sample (Upf:RP) in pellet as determined by mass spectrometry; mean and range of two biological replicates per strain and condition. C. Fold enrichment of Upf protein in pellet vs input; mean and range of two biological replicates per strain and condition. FLAG-tagged proteins are indicated, all panels.
For all three proteins examined here, Upf:RP was greater than 1:1 after immunopurification (Fig. 5B, Pellet fraction). Enrichment of Upf protein in immunopurified ribosomes was evaluated by dividing the Upf:RP of immunopurified ribosomes by that of their corresponding input ribosomes (Fig. 5C). Inclusion of cycloheximide had no significant effect on enrichment of Upf proteins or Upf:RP after immunoprecipitation (Figs. 5C and 5B). In addition to ribosomal proteins and the Upf factors, spectra apparently from nascent peptides were represented in all samples. Because the Upf proteins are N-terminally tagged, it is likely that a proportion of the Upf spectra also represent pulldown of their nascent peptides.
3.3. Deep sequencing analyses
The ribosome profiling library preparation method used here eliminates gel size selection and has fewer precipitation steps than conventional methods. These changes should result in less sample loss and more efficient recovery of ribosome protected fragments compared to standard methods. Due to the limited number of ribosomes recovered after immunopurification, efficient recovery of ribosome protected fragments was a particular concern. RNA-Seq and ribosome profiling libraries were generated from all samples, sequenced on a NextSeq500, and subjected to conventional bioinformatics analysis [19, 39, 54]. Inspection of read alignments from ribosome profiling libraries revealed that approximately 21% (Upf1), 6% (Upf2), and 9% (Upf3) of ribosomes were recovered subsequent to binding of the FLAG-tagged nascent peptide to the affinity gel (data not shown). While it is trivial to eliminate reads arising from these mRNAs from downstream analyses, it could be advantageous to use a C-terminal tag in future experiments, provided that such tagged proteins are biologically indistinguishable from their untagged cohort and that their associated ribosomes are recoverable by immunoprecipitation.
The incorporation of four random nucleotides on both sides of our ribosome protected fragments during library preparation allowed us to remove PCR duplicates from downstream analyses, thereby eliminating artificial read density “spikes” from the ribosome profiling data. Ribosome profiling libraries prepared with or without cycloheximide recovered a wide range of ribosome-protected RNA fragment sizes and allowed simultaneous observation of different fragment size classes within a given sample, fragment size class comparisons between total and immunopurified ribosome fractions, and read distribution across the transcriptome (manuscript in preparation).
In conclusion, high-level expression of epitope-tagged Upf proteins enabled purification and recovery of Upf factor-associated ribosomes, subsequent analysis of their protein content by mass spectrometry and, ultimately, identification of their bound mRNA fragments. The ribosome profiling library preparation method described here eliminates some unaddressed pitfalls of conventional methods and allows recovery and selective ribosome profiling analysis [55] of reads from purified, factor-bound ribosomes.
Supplementary Material
Example of Mascot Distiller quantitation of a sample of Upf1-associated ribosomes by mass spectrometry. Column “REF” indicates the reference protein, in this case Upf1 (Nam7). Column “Amount” calculates the amount of a given protein relative to the reference protein in the sample. A minimum of three spectra (column “#”) are required for quantitation by this method; therefore, some “Amount” cells are empty.
Core ribosomal proteins detected by mass spectrometry were quantitated in Mascot Distiller as described. For each sample, the ribosomal protein and its corresponding amount relative to the reference protein (indicated on the sheet label, bottom) are represented as paired columns. The average of ribosomal proteins quantitated (excluding those with no value for amount) was calculated and used to determine Upf:RP for a sample.
Highlights.
Immunopurification enables detailed analysis of ribosomes associated with factors essential for nonsense-mediated mRNA decay (NMD).
Stoichiometry and enrichment of NMD factors relative to ribosomal proteins pre-and post-purification can be estimated by mass spectrometry.
Eliminating in-gel RNA and cDNA size selection and including random barcodes in the preparation of ribosome profiling libraries enables a translational snapshot from a wide range of ribosome-protected fragment sizes.
Acknowledgments
We would like to thank Dr. Jon Warner for his kind gift of anti-L3 (Tcm1) monoclonal antibody [43]. This work was supported by Grants to A.J. (5R01 GM27757-37 and 1R35GM122468-02) from the U.S. National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCE LIST
- [1].Kervestin S, Jacobson A, NMD: a multifaceted response to premature translational termination, Nat Rev Mol Cell Biol 13(11) (2012) 700–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Popp MW, Maquat LE, Organizing principles of mammalian nonsense-mediated mRNA decay, Ann. Rev. Genet 47 (2013) 139–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nicholson P, Joncourt R, Muhlemann O, Analysis of nonsense-mediated mRNA decay in mammalian cells, Current protocols in cell biology editorial board, Juan S. Bonifacino [et al.] Chapter 27 (2012) Unit27 4. [DOI] [PubMed]
- [4].Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O, Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors, Cell Mol Life Sci 67(5) (2010) 677–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].He F, Jacobson A, Nonsense-mediated mRNA decay: Degradation of defective transcripts is only part of the story, Ann. Rev. Genet 49 (2015) 339–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A, Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5’ to 3’ mRNA decay pathways in yeast, Mol Cell 12(6) (2003) 1439–52. [DOI] [PubMed] [Google Scholar]
- [7].Atkin AL, Schenkman LR, Eastham M, Dahlseid JN, Lelivelt MJ, Culbertson MR, Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay, J. Biol. Chem 272(35) (1997) 22163–22172. [DOI] [PubMed] [Google Scholar]
- [8].Zhang S, Welch EM, Hogan K, Brown AH, Peltz SW, Jacobson A, Polysome-associated mRNAs are substrates for the nonsense-mediated mRNA decay pathway in Saccharomyces cerevisiae, RNA 3(3) (1997) 234–44. [PMC free article] [PubMed] [Google Scholar]
- [9].Hu W, Petzold C, Coller J, Baker KE, Nonsense-mediated mRNA decapping occurs on polyribosomes in Saccharomyces cerevisiae, Nat Struct Mol Biol 17(2) (2010) 244–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Johansson MJ, He F, Spatrick P, Li C, Jacobson A, Association of yeast Upf1p with direct substrates of the NMD pathway, Proc Natl Acad Sci U S A 104(52) (2007) 20872–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E, Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets, RNA 11(10) (2005) 1530–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC, Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise, Nat Genet 36(10) (2004) 1073–8. [DOI] [PubMed] [Google Scholar]
- [13].Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Monch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT, NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements, Genes Dev 22(10) (2008) 1381–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ramani AK, Nelson AC, Kapranov P, Bell I, Gingeras TR, Fraser AG, High resolution transcriptome maps for wild-type and nonsense-mediated decay-defective Caenorhabditis elegans, Genome Biol 10(9) (2009) R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Metzstein MM, Krasnow MA, Functions of the nonsense-mediated mRNA decay pathway in Drosophila development, PLoS Genet 2(12) (2006) e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wittkopp N, Huntzinger E, Weiler C, Sauliere J, Schmidt S, Sonawane M, Izaurralde E, Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival, Mol Cell Biol 29(13) (2009) 3517–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yepiskoposyan H, Aeschimann F, Nilsson D, Okoniewski M, Muhlemann O, Autoregulation of the nonsense-mediated mRNA decay pathway in human cells, RNA 17(12) (2011) 2108–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rayson S, Arciga-Reyes L, Wootton L, De Torres Zabala M, Truman W, Graham N, Grant M, Davies B, A role for nonsense-mediated mRNA decay in plants: pathogen responses are induced in Arabidopsis thaliana NMD mutants, PLoS One 7(2) (2012) e31917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Celik A, Baker R, He F, Jacobson A, High-resolution profiling of NMD targets in yeast reveals translational fidelity as a basis for substrate selection, RNA 23(5) (2017) 735–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].McGlincy NJ, Smith CW, Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense?, Trends Biochem Sci 33(8) (2008) 385–93. [DOI] [PubMed] [Google Scholar]
- [21].Culbertson MR, RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer, Trends Genet 15(2) (1999) 74–80. [DOI] [PubMed] [Google Scholar]
- [22].Miller JN, Pearce DA, Nonsense-mediated decay in genetic disease: Friend or foe?, Mutat. Res 762 (2014) 52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Johansson MJ, Jacobson A, Nonsense-mediated mRNA decay maintains translational fidelity by limiting magnesium uptake, Genes Dev 24(14) (2010) 1491–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lee MH, Schedl T, Translation repression by GLD-1 protects its mRNA targets from nonsense-mediated mRNA decay in C. elegans, Genes Dev 18(9) (2004) 1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cui Y, Hagan KW, Zhang S, Peltz SW, Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon, Genes Dev 9(4) (1995) 423–436. [DOI] [PubMed] [Google Scholar]
- [26].Perlick HA, Medghalchi SM, Spencer FA, Kendzior RJ Jr., Dietz HC, Mammalian orthologues of a yeast regulator of nonsense transcript stability, Proc Natl Acad Sci U S A 93(20) (1996) 10928–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Leeds P, Peltz SW, Jacobson A, Culbertson MR, The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon, Genes Dev 5(12A) (1991) 2303–2314. [DOI] [PubMed] [Google Scholar]
- [28].He F, Brown AH, Jacobson A, Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway, Mol Cell Biol 17(3) (1997) 1580–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].He F, Jacobson A, Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen, Genes Dev 9(4) (1995) 437–454. [DOI] [PubMed] [Google Scholar]
- [30].Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E, Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways, EMBO J 22(15) (2003) 3960–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lykke-Andersen J, Shu MD, Steitz JA, Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon, Cell 103(7) (2000) 1121–31. [DOI] [PubMed] [Google Scholar]
- [32].Maderazo AB, He F, Mangus DA, Jacobson A, Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p, Mol. Cell. Biol 20(13) (2000) 4591–4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chamieh H, Ballut L, Bonneau F, Le Hir H, NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity, Nat Struct Mol Biol 15(1) (2008) 85–93. [DOI] [PubMed] [Google Scholar]
- [34].Atkin AL, Altamura N, Leeds P, Culbertson MR, The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm, Mol Biol Cell 6(5) (1995) 611–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE, Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways, EMBO J 27(5) (2008) 736–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S, Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay, Genes Dev 20(3) (2006) 355–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Min EE, Roy B, Amrani N, He F, Jacobson A, Yeast Upf1 CH domain interacts with Rps26 of the 40S ribosomal subunit, RNA 19(8) (2013) 1105–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, Dong R, Guarani V, Vaites LP, Ordureau A, Rad R, Erickson BK, Wuhr M, Chick J, Zhai B, Kolippakkam D, Mintseris J, Obar RA, Harris T, Artavanis-Tsakonas S, Sowa ME, De Camilli P, Paulo JA, Harper JW, Gygi SP, The BioPlex Network: A Systematic Exploration of the Human Interactome, Cell 162(2) (2015) 425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS, The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments, Nat. Protoc 7(8) (2012) 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hopp KSP, T.P. Price VL, Libby RT, March CJ, Ceretti P, Urdal DL, and Conlon PJ, A short polypeptide marker sequence useful for recombinant protein identification and purification, Bio/Technology 6 (1988) 1204. [Google Scholar]
- [41].Czaplinski K, Weng Y, Hagan KW, Peltz SW, Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation, RNA 1(6) (1995) 610–23. [PMC free article] [PubMed] [Google Scholar]
- [42].Guthrie C, Fink GR, Methods in Enzymology: Molecular Biology of Saccharomyces cerevisiae., Academic Press, NY, 1991. [Google Scholar]
- [43].Vilardell J, Warner JR, Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA, Mol. Cell. Biol 17(4) (1997) 1959–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ, Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition, Mol Cell Proteomics 5(1) (2006) 144–56. [DOI] [PubMed] [Google Scholar]
- [45].Guo H, Ingolia NT, Weissman JS, Bartel DP, Mammalian microRNAs predominantly act to decrease target mRNA levels, Nature 466(7308) (2010) 835–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Razzell WE, The Precipitation of Polyribonucleotides with Magnesium Salts and Ethanol, J Biol Chem 238 (1963) 3053–7. [PubMed] [Google Scholar]
- [47].McAlister L, Holland MJ, Isolation and characterization of yeast strains carrying mutations in the glyceraldehyde-3-phosphate dehydrogenase genes, J Biol Chem 260(28) (1985) 15013–8. [PubMed] [Google Scholar]
- [48].de Pinto B, Lippolis R, Castaldo R, Altamura N, Overexpression of Upf1p compensates for mitochondrial splicing deficiency independently of its role in mRNA surveillance, Mol Microbiol 51(4) (2004) 1129–42. [DOI] [PubMed] [Google Scholar]
- [49].Maderazo AB, Belk JP, He F, Jacobson A, Nonsense-containing mRNAs that accumulate in the absence of a functional nonsense-mediated mRNA decay pathway are destabilized rapidly upon its restitution, Mol Cell Biol 23(3) (2003) 842–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ajamian L, Abel K, Rao S, Vyboh K, Garcia-de-Gracia F, Soto-Rifo R, Kulozik AE, Gehring NH, Mouland AJ, HIV-1 Recruits UPF1 but Excludes UPF2 to Promote Nucleocytoplasmic Export of the Genomic RNA, Biomolecules 5(4) (2015) 2808–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ajamian L, Abrahamyan L, Milev M, Ivanov PV, Kulozik AE, Gehring NH, Mouland AJ, Unexpected roles for UPF1 in HIV-1 RNA metabolism and translation, RNA 14(5) (2008) 914–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Shirley RL, Ford AS, Richards MR, Albertini M, Culbertson MR, Nuclear import of Upf3p is mediated by importin-alpha/-beta and export to the cytoplasm is required for a functional nonsense-mediated mRNA decay pathway in yeast, Genetics 161(4) (2002) 1465–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].He F, Ganesan R, Jacobson A, Intra- and intermolecular regulatory interactions in Upf1, the RNA helicase central to nonsense-mediated mRNA decay in yeast, Mol Cell Biol 33(23) (2013) 4672–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS, Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling, Science 324(5924) (2009) 218–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Becker AH, Oh E, Weissman JS, Kramer G, Bukau B, Selective ribosome profiling as a tool for studying the interaction of chaperones and targeting factors with nascent polypeptide chains and ribosomes, Nature protocols 8(11) (2013) 2212–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Example of Mascot Distiller quantitation of a sample of Upf1-associated ribosomes by mass spectrometry. Column “REF” indicates the reference protein, in this case Upf1 (Nam7). Column “Amount” calculates the amount of a given protein relative to the reference protein in the sample. A minimum of three spectra (column “#”) are required for quantitation by this method; therefore, some “Amount” cells are empty.
Core ribosomal proteins detected by mass spectrometry were quantitated in Mascot Distiller as described. For each sample, the ribosomal protein and its corresponding amount relative to the reference protein (indicated on the sheet label, bottom) are represented as paired columns. The average of ribosomal proteins quantitated (excluding those with no value for amount) was calculated and used to determine Upf:RP for a sample.