Abstract
Introduction
Altered microbial diversity has been associated with gastrointestinal (GI) symptoms in persons with irritable bowel syndrome (IBS). Less is known about the relationship of microbiome with extra-intestinal pain and psychological distress symptoms and quality of life (QOL) in persons with IBS. We aimed to evaluate the relationship of fecal microbiota to GI symptoms, stool consistency, psychological distress, extra-intestinal pain, and QOL in participants meeting Rome III criteria for IBS.
Methods
Seventy-six women completed a 28-day diary that included GI, stool consistency, psychological distress, and extra-intestinal pain ratings. Participants completed the IBS-Specific-Quality of Life questionnaire. Stool samples were collected and analyzed by 16S rRNA gene sequencing. Principal component analysis was performed and the first two components (PC1, PC2) were used to test relationships among bacterial families and clinical measures.
Results
Participants were categorized as IBS-constipation (n=22), IBS-diarrhea (n=39), IBS-mixed (n=13), and IBS-unsubtyped (n=2). There was a significant group effect for the Firmicutes to Bacteroidetes (FB) ratio and PC1. Lower microbial diversity and richness were associated with increased urgency and extra-intestinal pain, worse QOL, and looser stools. Lower extra-intestinal pain was associated with increased Rikenellaceae, Christensenellaceae, Dehalobabacteriaceae, Oscillospiraceae, Mogibacteriaceae, Ruminococcaceae, Sutterellaceae, Desulfovibrionaceae, and Erysipelotrichaceae abundances. QOL was positively associated with many of these same bacterial families. Higher FB ratio was positively associated with loose stools. There were no statistically significant relationships between daily psychological distress or abdominal pain and bacterial families.
Conclusion
Stool microbial diversity and composition are linked to daily extra-intestinal symptoms, stool consistency, and QOL in women with IBS.
INTRODUCTION
Approximately 10–20% of adults experience chronic abdominal pain/discomfort symptoms associated with altered bowel habits (constipation and/or diarrhea) compatible with a diagnosis of irritable bowel syndrome (IBS).1, 2 The public health impact of IBS in the US is large with direct and indirect costs totaling approximately $1.35 billion/year.3–5 The management of patients with IBS is challenging in that it is a heterogeneous condition with multiple factors (e.g., stress, diet, dysmotility, altered brain-gut axis) likely contributing to its etiology. To date, there are few biomarkers available to help diagnose, identify relevant subgroups and/or provide guidance on the management of IBS symptoms.6–8
Recent studies support the idea that alterations in gut microbiome composition and function (dysbiosis) may play an important role in the pathophysiology and clinical symptoms of IBS.9–12 Emerging evidence suggests that IBS symptoms and severity may be linked to microbial signatures. Utilizing retrospective questionnaires regarding IBS symptoms and severity, both Pozuelo et al. (2015) and Tap et al. (2016) identified key microbial taxa associated with IBS characteristics. Pozuelo et al.13 found differences in microbial community composition, at various taxonomic levels, between 113 IBS patients and 66 healthy controls. These included lower microbial diversity in patients with IBS as well as higher average counts of phylum Bacteriodetes and lower counts of phylum Firmicutes in patients with IBS-D (diarrhea) and IBS-M (mixed) subtypes. Pozuelo et al.13 also correlated microbiota composition and subjective IBS symptoms in 44 patients who did not take any medications. Flatulence was positively correlated with family Lachnospiraceae from the phylum Firmicutes. Abdominal pain was positively correlated with the genera Bacteroides (family Bacteroidaceae) and genera Ruminoccocus (family Lachnospiraceae) and an unclassified member of the family Barneisiellaceae. In contrast, abdominal pain was negatively associated with the genera Prevotella (family Prevotellaceae) and Catenibacterium (family Erysipelotrichaceae). Using retrospective questionnaires and a two-week stool diary, Tap et al. found no differences in microbiota richness or composition between healthy controls and IBS participants or among IBS subtypes. However, using a machine-learning approach, Tap et al. identified a microbiota signature for those with severe IBS symptoms (IBS-Symptom Severity Score > 300) that included lower diversity and reduced abundances of genera Methanobacteriales (phylum Euryarchaeota) and genera Prevotella (family Prevotellaceae) and increased abundances of genera Bacteroides (family Bacteroidaceae).14
Gut microbiota appear to not only influence immune and inflammatory responses within the GI tract but also systemic inflammatory responses15 and, in some cases, behavior. A current hypothesis is that dysbiosis activates mucosal innate immune responses which increase epithelial permeability, activate nociceptive sensory pathways and dysregulate the enteric nervous system.16–18 Such changes then contribute to visceral hypersensitivity and possibly extra-intestinal or co-morbid symptoms (e.g., fatigue, joint/muscle pain, mood state) that patients with IBS frequently report.19 Yet the relationship of microbiota to extra-intestinal symptoms of IBS remains unexplored. Studies with rodents show the relationship of the gut microbiota to anxiety-like and depression-like behaviors.20–22 When compared to specific pathogen-free mice, germ-free (GF) mice display lower levels of anxiety-like behavior in response to water maze stress testing.23 In humans, healthy adults show improvement in anxiety after taking probiotics.24–29 These pieces of evidence suggest that gut microbiome characteristics may contribute to mood symptoms that are often associated with IBS, such as psychological distress. Assessing memory testing and social behaviors, animal studies provide some support that gut bacteria influence cognition. However, in humans the notion that gut bacteria is linked to quality of life remains unexplored.
Most studies to date relating subjective measures including symptoms and quality of life have relied on retrospective questionnaires, which lack resolution of a daily scale and are often hampered by recall bias 30. Prospectively collected daily diaries represent an improvement over this situation.31 When used in combination with retrospective measures, the two approaches to symptom capture provide insight regarding the patient’s daily experience and overall perception of condition. Understanding the linkages between microbiota and average daily IBS symptoms may offer unique therapeutic strategies. Therefore, we aimed to: 1) test for differences in gut microbiota measures and diversity among IBS subtypes, i.e., IBS-Constipation (IBS-C), IBS-Diarrhea (IBS-D), IBS-Mixed (IBS-M) and IBS-unsubtyped (IBS-U) ; and 2) test the relationships of gut microbiota measures to daily IBS symptoms (e.g., abdominal pain), stool consistency, psychological distress, extra-intestinal pain, and overall quality of life (QOL) measures.
METHODS
Stool samples from a baseline assessment of 76 women enrolled in a behavioral intervention trial were collected and microbiota analyses performed.32 All participants needed to have a diagnosis of IBS given by a health care provider and be between the ages of 18 and 70. The subject’s medical history was reviewed by an experienced gastroenterologist. In addition, subjects needed to report current IBS symptoms. Exclusion criteria included had a history of coexisting GI pathology (e.g., IBD, peptic ulcers) or surgery (e.g., open abdominal surgery or midline abdominal incisions), renal or reproductive pathology (e.g., endometriosis). Subjects were excluded if currently taking or had been taking antibiotics in the 2 months prior to study. In addition, we excluded women taking anticholinergics, cholestyramine, narcotics, docusate, enema preparations, iron supplements, or laxatives.32 Although both men and women were recruited into the larger study, microbiome analyses were performed only on the women’s samples. Subjects completed a 4-week daily diary recording of GI symptoms, stool consistency, psychological distress, and extra-intestinal pain symptoms. Additional measures included Rome III Questionnaire and IBS Quality of Life (IBS QOL).
Eligibility for the study included the following process. Potential subjects who responded to posted advertisements were screened by telephone. If eligible, questionnaires were mailed to the subject’s home and then returned at the first visit. At the visit the subject met with the research nurse, consent was obtained, questionnaires reviewed, and instructions on the use of the daily diary and stool collection given.
Human participants institutional review approval was obtained prior to enrolling participants (February 2008) and renewed thereafter. Written informed consent was obtained from the participants. This study was registered with clinicaltrials.gov through the U.S. National Institutes of Health (Clinicaltrials.gov identifier: NCT00907790).
Microbiota Characterization
Fecal samples were collected at each participant’s home and immediately stored at −20°C. Frozen samples were transported to our laboratory and stored thereafter at −80°C. Fecal microbial communities were characterized using 16S rRNA gene sequencing as previously described.33 Briefly, community DNA was extracted from each sample using the PowerSoil DNA extraction kit (MoBIO, Carlsbad, CA) with modifications to the manufacturer’s protocol.34 Community DNA quantity and quality were assessed using fluorometric methods (Qubit, Waltham, MA), and amplicon libraries were prepared using the NEXTflex V4 Amplicon-Seq kit 2.0 (Bioo Scientific, Austin, TX). Sequence libraries were generated using the MiSeq platform (Illumina, San Diego, CA).
Sequence libraries were demultiplexed by barcode, and primer sequence were removed using cutadapt.35 PhiX reads were removed by mapping amplicon reads to the PhiX genome using bowtie2 (version 2.0.6)36 with the fast-local setting and retaining all unmapped read pairs. Reads were then further quality-filtered and clustered into operational taxonomic units (OTUs) using LOTUS (version 1.462).37 The LOTUS platform retained reads with minimum average quality scores >20 and minimum read length of 170 bp. Reads were clustered into OTUs at a 97% similarity threshold using the UPARSE algorithm (version 8.0.1623)38 and potentially chimeric sequences were identified among the OTU representative sequences using the VSEARCH algorithm (version 1.9.7).39 Non-chimeric representative sequences were classified using the following classifier – database pairs: Ribosomal Database Project (RDP) classifier and SILVA (version 123) database, RDP classifier and HitDB database, and UTAX classifier and RDP trainset 15 (https://www.drive5.com/utax/data/utax_rdp_16s_tainset15.tar.gz). A consensus taxonomy was derived in which classifications were supported by 2 or more calls at any taxonomic level. In cases of disagreement, the taxonomic call was limited to the finest level of resolution supported by two or more of the classifiers. For example, if a taxonomic call was supported by two or more classifiers at the family level but lacked majority consensus at the genus level, the family-level designation was retained and “unclassified” was used in place of the genus-level designation.
Descriptive Measures
ROME III Questionnaire
The participants completed the ROME III questionnaire for IBS, a diagnostic questionnaire developed by the Rome Foundation to identify patients with IBS. The development and validation of this questionnaire have been previously described.2, 40, 41
Daily Diary Measures
Every day for 4 weeks subjects completed a daily diary which included 26 symptoms rated on a scale of 0 (not present), 1 (mild), 2 (moderate), 3 (severe) or 4 (very severe).42 The score for each individual symptom was summarized as the mean rating over the 28 days. Symptoms analyzed for this report included the GI symptoms of abdominal pain or discomfort, bloating, constipation, diarrhea and urgency. A composite measure of extra-intestinal pain was computed as the average of headache, joint pain, muscle pain, backache, and headache. The psychological distress composite score included anxiety, depression, panic, and stressed.
Participants also rated the consistency of each stool using the following scale of 0 (no stool), 1 (very hard), 2 (hard), 3 (formed), 4 (loose) or 5 (watery) as previously described.43 These daily ratings were summarized as the mean consistency score over 28 days, with higher values indicating looser stools.
Quality of Life for IBS
The IBS-Quality of Life (IBSQOL) questionnaire is a 42-item questionnaire with nine scales: sleep, emotional, mental health beliefs, energy, physical functioning, diet, social role, physical role, and sexual relations.44 Example questions are “How often did your IBS make you feel fed up or frustrated?” e.g., 1 (always), 2 (often), 3 (sometimes), 4 (seldom), to 5 (never); and “My IBS affected my ability to succeed at work/main activity” e.g., 1 (strongly agree) to 5 (strongly disagree). The scales were converted to a standard 0 to 100 scale. A total score was computed by averaging all scales. Lower scores indicate worse IBS impact on QOL.
Statistical Analysis
The analysis strategy consisted of two steps. The first step evaluated associations of clinical measures with five microbial summary measures of microbiome composition applying the Benjamini-Hochberg procedure to control for multiple comparisons in these initial analyses. Only those clinical measures that showed significant association with microbiota summary measures in this first step were analyzed in the second step. The purpose of this second step was to understand which families and genera are associated with those clinical measures. No multiple comparisons adjustments were done in the second step, the protection against inflated type 2 error having been accomplished in the first step.
Five microbiome summary measures were computed in the initial analyses: Diversity measures (Shannon and operational taxonomic unit [out] richness) were computed on the OTU level. FB ratio was computed as the fractional abundance of phylum Firmicutes divided by fractional abundance of phylum Bacteriodetes. A small constant, 0.02, was added to the denominator so that a fractional abundance of phylum Bacteriodetes close to zero would not result in an extremely high ratio.
The remaining two summary measures were defined from principal component analyses (PCA) applied to the logit-transformed family data using only the 35 most abundant bacterial families. The logit transform was applied to the relative abundance of these families: log10 [ (p+.0001) / ((1-p)+.0001) ], where p is the read count associated with each bacterial family divided by the total read count. The constant 0.0001 was added to accommodate relative abundance being zero or one. This PCA was based on partial correlations controlling for total sequencing read count and subject age. Only the first two PCs were used in subsequent analyses.
Supplemental Figure 1 shows the distributions of these 35 families across subjects. Supplemental Figure 2 shows the correlation of each family to the two PCs, as a description of which families were important components of each of these two PCs.
Analysis of variance (ANOVA) was used to test differences in the five microbiota summary measures by IBS bowel pattern subtypes, controlling for read count and subject age. Partial correlations, controlling for read count and age, were computed between each of the five microbiome summary measures and the clinical measures of diary stool consistency and symptoms, and QOL. The clinical measures which showed significant correlations with one or more of the five summary measures after controlling for multiple comparisons, were included in analyses of individual families and genera. Statistical analyses were performed using with IBM SPSS Statistics for Windows, version 19 (SPSS, Inc., Armonk, NY: IBM Corp, USA).
RESULTS
Demographics
Most of participants were Caucasian (71%) and had a college degree (80%). The mean age was 39.0 years (SD 15.1) (Table 1). Based on Rome III diagnostic criteria, IBS-diarrhea was the predominant bowel subtype (IBS-D, n=39) followed by IBS-constipation (IBS-C, n=22), IBS-mixed (IBS-M, n=13) and 2 participants who did not meet the criteria for IBS-C, IBS-D, or IBS-M were categorized as IBS-unsubtyped (IBS-U).
Table 1.
Descriptive Characteristics Based on Rome III Irritable Bowel Syndrome (IBS) Bowel Pattern Predominance in Women with IBS
| Variable | IBS-D diarrhea (n = 39) |
IBS-C constipation (n=22) |
IBS-M mixed (n=13) |
IBS-U unsubtyped (n=2) |
|---|---|---|---|---|
| Demographics | ||||
| Age (mean [SD], yr) | 37.8 (14.3) | 41.9 (15.6) | 33.4 (13.1) | 67.5 (0.7) |
| Body mass index (kg/m2, mean [SD]) | 25.8 (6.0) | 25.4 (6.8) | 24.6 (6.4) | 22.8 (0.7) |
| Race (White, % [n]) | 69% (25) | 91% (20) | 67% (8) | 100% (2) |
| Married or partnered (% [n]) | 49% (19) | 55% (12) | 30% (4) | 0 |
| Education (Bachelors or above, % [n]) | 69% (27) | 76% (16) | 62% (8) | 100% (2) |
| Income (> $60 000/yr, (% [n]) | 44% (14) | 47% (9) | 46% (6) | 50% (1) |
| Professional job (% [n]) | 49% (18) | 10% (2) | 31% (4) | 50% (1) |
| Daily Symptoms (mean [SD]) | ||||
| Abdominal pain | 1.39 (0.66) | 1.30 (0.52) | 1.49 (0.57) | 1.11 (0.15) |
| Bloating | 1.10 (0.83) | 1.13 (0.61) | 1.48 (0.83) | 1.11 (0.05) |
| Constipation* | 0.48 (0.54) | 1.14 (0.63) | 0.70 (0.58) | 0.57 (0.25) |
| Diarrhea* | 0.91 (0.60) | 0.17 (0.19) | 0.55 (0.81) | 0 |
| Urgency* | 0.91 (0.70) | 0.30 (0.31) | 0.90 (1.07) | 0 |
| Stool consistency* | 3.30 (0.56) | 2.44 (0.35) | 3.18 (0.43) | 2.75 (0.11) |
| Psychological distressa | 0.59 (0.49) | 0.77 (0.51) | 0.59 (0.31) | 0.35 (0.28) |
| Extra-intestinal painb | 0.42 (0.42) | 0.60 (0.34) | 0.43 (0.48) | 0.25 (0.33) |
| IBS quality of life | 66.9 (15.8) | 68.1 (12.6) | 66.0 (18.5) | 68.4 (14.7) |
Psychological distress is a composite of daily reporting of anxiety, depression, panic, and stressed;
Extra-intestinal pain is a composite of daily reporting of heartburn, joint pain, muscle pain, backache, and headache.
p<0.05 by ANOVA
Microbiome and IBS Subtypes
Table 2 shows the microbiota summary measures by bowel pattern IBS subgroup, IBS-C, IBS-D, and IBS-M. FB ratio differed significantly by group, being lowest in the IBS-C group and highest in the IBS-D group. PC1 also differed signficantly across bowel pattern groups (Table 2).
Table 2.
Comparison of Microbiota Summary Measures Based on Irritable Bowel Syndrome (IBS) Rome III Bowel Pattern Subgroups
| Microbiome summary measures (mean, (SD)) |
IBS-C (n = 22) | IBS-D (n = 39) | IBS-M (n = 13) | p valuea |
|---|---|---|---|---|
| Number of OTUs | 324 (106) | 290 (84) | 256 (86) | NS |
| Shannon diversity | 3.33 (0.58) | 3.22 (0.56) | 3.00 (0.45) | NS |
| FB ratio | 9.92 (9.68) | 21.04 (13.9) | 19.04 (16.7) | 0.018* |
| PC1 | 0.44 (0.86) | −0.21 (0.93) | −0.26 (1.17) | 0.02* |
| PC2 | −0.20 (1.07) | 0.28 (0.96) | −0.39 (0.81) | 0.08 |
Unadjusted p-value from analysis of covariance controlling for read count and age.
Significant under Benjamini-Hochberg procedure to control false discovery rate at 0.05, across the 5 hypothesis tests in this table.
IBS-C = IBS-constipation; IBS-D = IBS-diarrhea; IBS-M = IBS-mixed; OTUs = operational taxonomic units; FB = Firmicutes/Bacteroidetes ratio; PC1 = principal component 1; PC2 = principal component 2.
Correlation of Microbiota Summary Indicators with Gastrointestinal Measures and Quality of Life
Table 3 shows the correlation of the five microbiota summary indicators with stool consistency, GI symptom measures, and IBS-specific QOL. Higher diversity (Shannon and OTU richness) was associated with lower values for stool consistency (i.e., harder stools) and better IBS-specific QOL. The OTU richness, but not Shannon diversity, was associated with less diarrhea and urgency. Increased FB ratio was associated with looser stools, but none of the other individual GI symptom measures. PC1 showed a correlation pattern similar to OTU richness, while PC2 was associated with better QOL. Also, note that none of the microbiome summary indicators were significantly associated with abdominal pain, bloating, or constipation. These correlations are illustrated in Figure 1.
Table 3.
Correlations of Microbiota Summary Measures with Stool Consistency, Individual Gastrointestinal Symptoms, Urgency, Psychological Distress and Extra-intestinal Summary Measures and Irritable Bowel Syndrome (IBS)-Specific Quality of Life in Women with IBSa
| Stool consistency |
Abdominal pain |
Bloating | Constipation | Diarrhea | Urgency | Psychological distressb |
Extra- intestinal painc |
Urgency | IBS- Specific Quality of life |
|
|---|---|---|---|---|---|---|---|---|---|---|
| Shannon diversity | −0.24* | 0.04 | −0.07 | 0.14 | −0.06 | −0.15 | −0.19 | −0.36*† | −0.15 | 0.31** |
| Number of OTUs | −0.29** | −0.12 | −0.15 | 0.11 | −0.25* | −0.29** | −0.24* | −0.33*† | −0.29** | 0.43** |
| FB ratio | 0.38** | −0.17 | −0.12 | −0.21 | 0.21 | 0.23 | 0.15 | 0.06 | 0.23 | −0.05 |
| PC1 | −0.33** | 0.05 | 0.01 | 0.18 | −0.19 | −0.31** | −0.18 | −0.25* | −0.31** | 0.25* |
| PC2 | 0.04 | −0.17 | −0.17 | −0.01 | −0.02 | −0.04 | −0.15 | −0.23 | −0.04 | 0.30** |
PC = principal component; OTUs = operational taxonomic units; FB ratio = Firmicutes/Bacteroidetes ratio
Partial correlations controlled for read count and age;
Psychological distress summary measure includes daily anxiety, depression, panic and stressed;
Extra-intestinal pain summary measure includes daily heartburn, joint pain, muscle pain, backache, and headache
unadjusted p<0.05;
significant under Benjamini-Hochberg procedure to control false discovery rate at 0.05, across the 35 correlations in this table.
Figure 1.
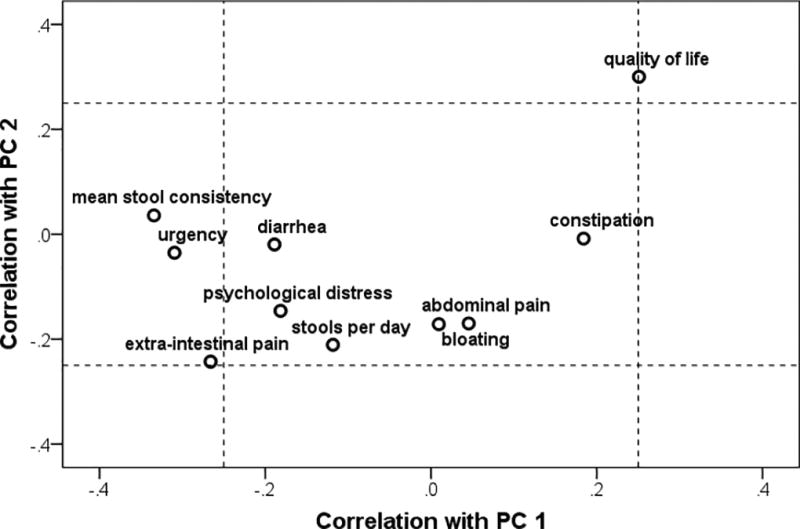
Clinical measures and their correlations with the first two principal components (PC1, PC2) defined based on family-level microbial community composition in the stool communities of women with irritable bowel syndrome.
Correlation of Microbiota Summary Indicators with Extra-intestinal Pain and Psychological Distress Measures
The summary measure of extra-intestinal pain includes heartburn, joint pain, muscle pain, backache, and headache. The extra-intestinal pain summary measure was inversely associated with Shannon diversity and number of OTUs (Table 4). PC1 was inversely related to extra-intestinal pain summary measure while no statistical relationships were noted in the individual pain symptoms. Microbiome summary measures were not significantly associated with the psychological distress summary measure (Table 4).
Table 4.
Correlations of Microbiota Summary Measures with Individual Daily Extra-Intestinal Symptoms in Women with Irritable Bowel Syndromea
| Heartburn | Joint pain | Muscle pain | Backache | Headache | |
|---|---|---|---|---|---|
| Shannon diversity | −0.28* | −0.39*† | −0.38*† | −0.20 | −0.26* |
| Number of OTUs | −0.32* | −0.25* | −0.33*† | −0.23 | −0.29* |
| FB ratio | 0.07 | 0.07 | 0.11 | −0.02 | 0.17 |
| PC1 | −0.18 | −0.21 | −0.22 | −0.18 | −0.18 |
| PC2 | −0.18 | −0.17 | −0.22 | −0.15 | −0.15 |
PC = principal component; OTUs = operational taxonomic units; FB ratio = Firmicutes/Bacteroidetes ratio
Partial correlations controlled for read count and age;
unadjusted p<0.05;
significant under Benjamini-Hochberg procedure to control false discovery rate at 0.05, across the 25 correlations in this table.
Correlation of Microbiota Families with Daily Clinical Measures and Quality of Life
Supplemental Table 1 shows partial correlations between the logit of relative abundance of phyla and families with mean stool consistency, urgency, psychological distress, extra-intestinal pain, and QOL. As seen in Supplemental Table 1, many correlations were significant at p<0.05. However, focusing on the stronger correlations with p<0.01 the following results were noted. Looser stool consistency was associated with lower abundance of the phylum Bacteroidetes and the families Bacteroidaceae, Christensenellaceae, Dehalobacteriaceae and Oscillospiraceae, and was associated with higher abundance of the family Erysipelotrichaceae. Better QOL was associated with higher abundance of the families Christensenellaceae, Dehalobacteriaceae, Mogibacteriaceae, Ruminococcaceae, Eubacteriaceae and Enterococcaceae. Higher extra-intestinal pain was associated with lower abundance of the families Christensenellaceae, Dehalobacteriaceae, Oscillospiraceae, Mogibacteriaceae and Ruminococcaceae. There was no significant relationship at p<0.01 between psychological distress and microbial families. Supplemental Table 1 also shows the bacterial families and their relationship with Shannon diversity. Examples of family relationships with stool consistency are shown in Figure 2.
Figure 2.
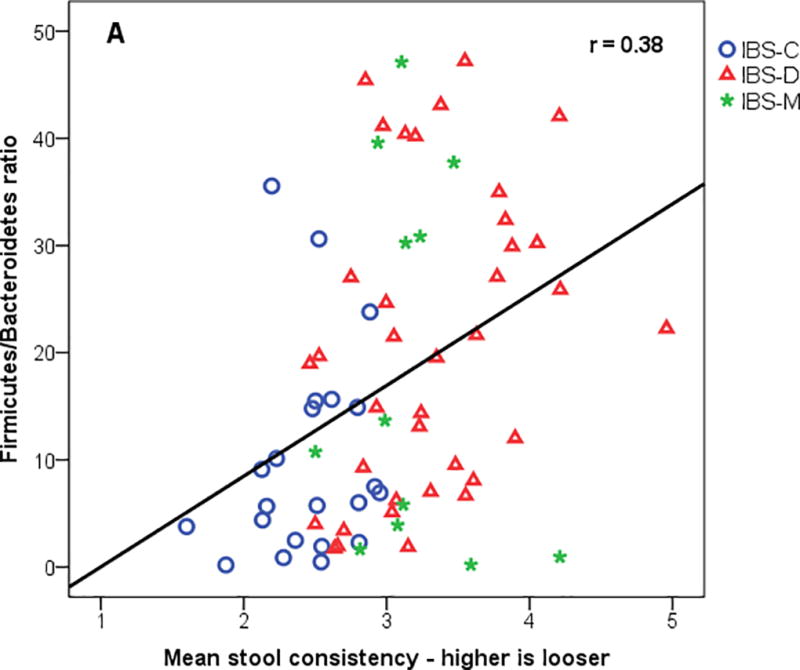
Mean stool consistency from daily diary versus (A) Firmicutes/Bacteroides (FB) ratio, (B) log of family Christensensellaceae, and (C) log of family Erysipelotrichaceae. IBS-C = IBS-constipation; IBS-D = IBS-diarrhea; IBS-M = IBS-mixed.
Correlation of Microbiota Genera with Clinical Measures
Ninety-five genera were identified that met the criteria of being present in at least 10% of the samples. Genus-level data, along with correlations with clinical measures, are shown in Supplemental Table 2. As expected, many of the specific genera from families in which significant relationships were found also demonstrated significant (p<0.05) relationships with clinical measures. For example, genus Dehalobacterium from the family Dehalobacteriaceae was inversely related (r = −0.32) to stool consistency and positively associated with QOL (r = 0.34). However, when adjusted for multiple comparisons, none of these relationships remained significant. Supplemental Table 2 also shows the bacterial genera and their relationships with Shannon diversity.
DISCUSSION
In this sample of 76 women with Rome III-diagnosed IBS, we sought to determine if differences in microbial communities existed across IBS predominant bowel pattern groups and the relationships of stool consistency, urgency, psychological distress, extra-intestinal pain, and QOL to specific bacterial taxa. Using Shannon diversity, OTU richness, FB ratio, PC1, and PC2 as microbial summary indicators, we determined that the FB ratio is markedly higher in the IBS-D group and PC1 is higher in the IBS-C group. When the five microbial summary indicators were correlated with clinical measures, greater diversity was associated with harder stools, fewer extra-intestinal pain symptoms, and higher QOL. Further examination of individual families identified a cluster of three families within the phylum Bacteroidetes and eight families within the phylum Firmicutes that showed positive associations with stool microbial diversity. Bowel pattern group was significantly associated with the abundances of seven families, i.e., Bacteroidaceae, Porphyromonadaceae, Christensellaceae, Leuconostocaceae, Gemellaceae, Methanobacteriaceae, and Erysipelotrichaceae. A summary extra-intestinal pain measure that included daily heartburn, joint pain, muscle pain, backache, and headache was inversely related to microbial summary indicators. We found no statistically significant relationships of microbiota summary indicators with daily abdominal pain, bloating, intestinal gas, and psychological distress ratings.
This is the first study to examine the relationship of bacterial families with both daily GI symptoms as well as extra-intestinal pain, i.e., heartburn, joint pain, muscle pain, backache, and headache. Several Firmicutes families (Dehalobacteriaceae, Oscillospriraceae, Mogibacteriaceae, Ruminococcaceae) were associated with lower extra-intestinal pain severity. The lack of association with diary abdominal pain symptoms with bacterial families was likely related to the fact that all participants reported abdominal pain on the diary. However, not all women reported extra-intestinal pain symptoms on the daily diary. Such variability in extra-intestinal pain measures may have contributed to the stronger associations with bacterial families. Without healthy controls in this study, we do not know if these relationships are unique to women with IBS, but they provide intriguing targets for future study.
While not statistically significant, we did observe a trend towards decreased microbial diversity for IBS-D compared to IBS-C patients. This could indicate that an altered gut microbiome may be a differentiating feature and potential underlying mechanism for the development of IBS-D versus IBS-C symptoms.
Somewhat similar to our results, other investigators have demonstrated reduced diversity is associated with diarrhea or loose stools45–47 as well as taxonomic differences between controls and persons with constipation. Comparing mucosal microbiota from women with constipation to healthy women using a single taxon approach, Parthasarathy et al.48 found the phylum Bacteroides and the genus Feacalibacterium were higher in constipated women. Several other genera were associated with reduced colonic transit measured with scintigraphy. Roager47 also studied colonic transit time in 98 healthy individuals using radio-opaque markers. Fecal Shannon index and OTU richness were positively associated with slower colonic transit time. Slower colon passage time may contribute to a greater abundance of methanogens (seen more predominantly in harder stools).46 While another group has shown no IBS subgroup differences in methane breadth testing49, our IBS-C group had a greater abundance of family Methanobacteriaceae relative to IBS-D and their abundance showed an inverse trend (Supplemental Table 1) with stool consistency. The role of this bacterial family in symptoms in individuals with IBS remains to be determined. Our observation of an inverse relationship of stool consistency with family Christensenellaceae (r=-0.37) is consistent with Parthasarathy’s association findings related to this family and methane production. However, they found no significant association of this family with colonic transit. To date, little is known about the physiological significance of family Christensenellaceae in persons with IBS-C. Whether the presence of increased fecal diversity combined with higher levels of specific families such as family Christensenella influence motility could be an impetus for further studies. It is unlikely that variations in the water content from stool samples (i.e., less water in harder stools, more water in looser stools) affect microbial diversity measures.50
Although the utility of the FB ratio has been questioned in the context of obesity51, it appears to be meaningful in the context of IBS. Jeffery et al.18 found that a high FB ratio was characteristic of IBS and was closely associated with predominant stool form. Likewise, our data support that a relationship between the FB ratio and predominant stool type exists in IBS. The IBS-C group had a lower FB ratio as compared to the IBS-D group (Table 1). Parthasarathy et al. (2016) in a study of mucosal microbiota found that women with chronic constipation and/or IBS-constipation and -mixed had elevated levels of genera from the phylum Bacteroidetes.48 At this time, it is not known whether a higher ratio of FB contributes to diarrhea and/or is a consequence of rapid transit and diarrhea.
In the current study, the family Erysipelotrichaceae, which belongs to the phylum Firmicutes, was positively associated with loose stools. There is a growing literature about family Erysipelotrichaceae, its taxa, and their potential roles in infection/immune activation.52–54 Dietary factors, specifically the dietary non-fermentable fiber hydroxypropyl methylcellulose, increased the abundance of family Erysipelotrichaceae in a murine model.55
This study is the first to address the relationship among prospectively collected daily extra-intestinal pain symptoms and fecal microbiota composition in the context of IBS. A summary measure of daily heartburn, joint pain, muscle pain, backache, and headache was inversely associated with microbial diversity measures. The literature on musculoskeletal pain and microbiome is scarce. Whether these pain-related symptoms are linked to overall heightened somatic pain sensitivity due to central pain pathway activation, a product of immune system activation, and/or a behavioral tendency to report multiple symptoms cannot be discerned.
In the current study only fecal microbiota were examined. Multiple studies in the published literature indicate that the mucosal and fecal microbiota differ from one another. However, they are not completely distinct and bacteria with host-modulating properties can be detected in both locations. For example, this is the case with Fusobacterium nucleatum in colorectal cancer.56 Despite the imperfect (mucosal) resolution, fecal microbial community profiles do have the potential to provide insight into pathophysiology. This is substantiated further by the number of studies which show relationships between fecal microbial composition and clinical symptoms (e.g., stooling characteristics).45
This study has several limitations. A single fecal sample was collected from each woman enrolled in an intervention trial for IBS and as such, there was no control group. We excluded women who had used antibiotics for two months prior to study entry, whereas other microbiome studies, including the Human Microbiome Project, have required longer antibiotic-free periods (e.g., 6 months) prior to microbiome testing.34 All women in the study were between ages 18 and 70 with approximately two-thirds of those under age 45 receiving some form of hormonal contraception. No effort was made to control for menstrual cycle phase or menopausal status, which may be important when describing the microbiota composition.57 In addition, we did not have dietary intake information. Because the date of stool collection was not available on a third of our sample, we chose to use the 28-day average for stool consistency and reports of diarrhea and constipation. On the other hand, this study has several strengths including the use of a daily diary to collect symptoms and stool consistency markers and the measurement of quality of life.
CONCLUSION
Stool microbial diversity and composition are linked to greater extra-intestinal pain symptoms and reduced QOL.
Supplementary Material
Figure 3.
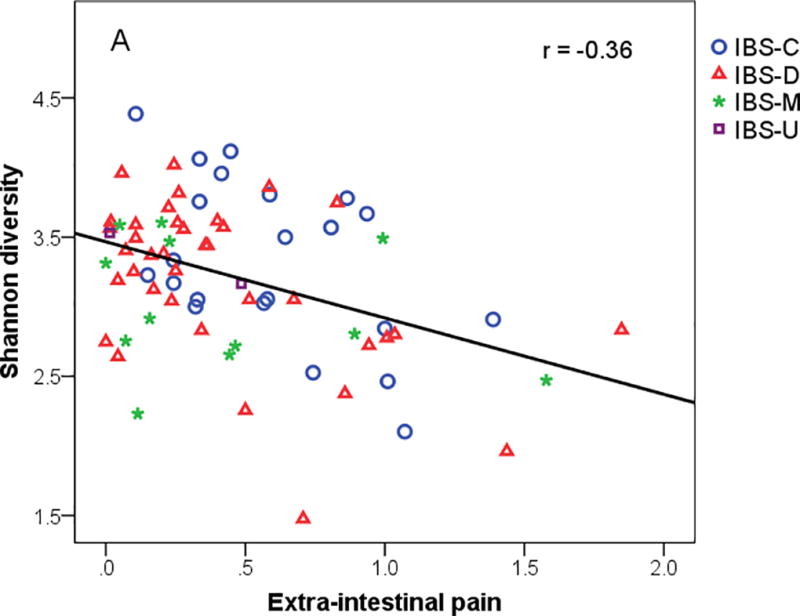
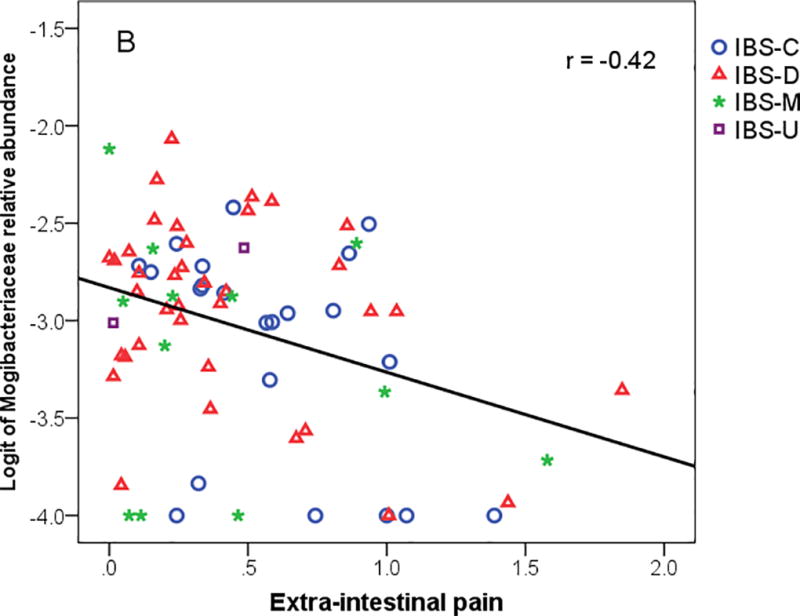
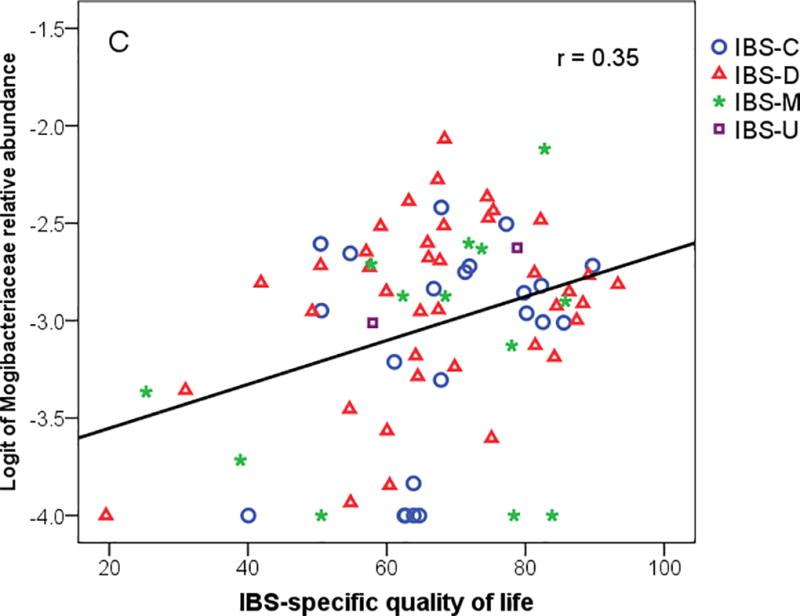
(A) Extra-intestinal pain rating from daily dairy versus Shannon diversity, (B) Extra-intestinal pain summary measure versus logit of family Mogibacteriaceae, and (C) IBS-Specific Quality of life with logit of family Mogibacteriaceae. IBS-C = IBS-constipation; IBS-D = IBS-diarrhea; IBS-M = IBS-mixed.
Acknowledgments
Supported by a grants from the NIH NINRR21NR014331 and NR01004142.
The corresponding author takes responsibility for the integrity of the work from the initial manuscript inception to the final edits and subsequent publication (if accepted).
Footnotes
- Emily B. Hollister, PhD. - Led the data analyses and interpretation. Also provided input regarding the data collection protocol.
- Kevin C. Cain, PhD. - Biostatistician – took the lead on all data analyses and co-investigator on the grant, assisted with design of the study
- Robert J. Shulman, MD. - Co-PI of the study. Co-wrote the grant and contributed to data analytic strategy and interpretation.
- Monica E. Jarrett, PhD. - Co-PI of the study. Directly involved in all aspects of the study from its inception to completion.
- Robert L. Burr, PhD. - Biostatistician, worked with Dr. Cain on the data analytic strategy.
- Cynthia Ko, MD. - Study gastroenterologist. She played a key role in subject recruitment and screening and contributed to interpretation of data.
- Jasmine Zia, MD. - Study gastroenterologist and played a key role in subject recruitment and screening. She also contributed to the writing and interpretation of symptom data.
- Claire J. Han, PhD. - Postdoctoral fellow assisted with technical writing and visualization of data. Also contributed to data interpretation.
- Margaret M. Heitkemper, PhD. - PI of the study and responsible for the conduct of the study and the integrity of the data presented.
All authors reviewed and approved the final version.
Contributor Information
Emily B. Hollister, Email: holliste@bcm.edu.
Kevin C. Cain, Email: cain@uw.edu.
Robert J. Shulman, Email: rshulman@bcm.edu.
Monica E. Jarrett, Email: jarrett@uw.edu.
Robert L. Burr, Email: bobburr@uw.edu.
Cynthia Ko, Email: cwko@uw.edu.
Jasmine Zia, Email: jzia@medicine.washington.edu.
Claire J. Han, Email: jyh0908@uw.edu.
Margaret M. Heitkemper, Email: heit@uw.edu.
References
- 1.Rajilic-Stojanovic M, Smidt H, de Vos WM. Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol. 2007;9:2125–36. doi: 10.1111/j.1462-2920.2007.01369.x. [DOI] [PubMed] [Google Scholar]
- 2.Drossman D. Rome III: The Functional Gastrointestinal Disroders. 3. McClean, VA: Degnon Associates, Inc; 2006. [Google Scholar]
- 3.Mitra D, Davis KL, Baran RW. All-cause health care charges among managed care patients with constipation and comorbid irritable bowel syndrome. Postgrad Med. 2011;123:122–32. doi: 10.3810/pgm.2011.05.2290. [DOI] [PubMed] [Google Scholar]
- 4.Ladabaum U, Boyd E, Zhao WK, et al. Diagnosis, comorbidities, and management of irritable bowel syndrome in patients in a large health maintenance organization. Clin Gastroenterol Hepatol. 2012;10:37–45. doi: 10.1016/j.cgh.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Longstreth GF, Wolde-Tsadik G. Irritable bowel-type symptoms in HMO examinees. Prevalence, demographics, and clinical correlates. Dig Dis Sci. 1993;38:1581–9. doi: 10.1007/BF01303163. [DOI] [PubMed] [Google Scholar]
- 6.Pimentel M, Morales W, Rezaie A, et al. Development and validation of a biomarker for diarrhea-predominant irritable bowel syndrome in human subjects. PLoS One. 2015;10:e0126438. doi: 10.1371/journal.pone.0126438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jarrett ME, Cain KC, Barney PG, et al. Balance of Autonomic Nervous System Predicts Who Benefits from a Self-management Intervention Program for Irritable Bowel Syndrome. J Neurogastroenterol Motil. 2016;22:102–11. doi: 10.5056/jnm15067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim JH, Lin E, Pimentel M. Biomarkers of Irritable Bowel Syndrome. J Neurogastroenterol Motil. 2017;23:20–26. doi: 10.5056/jnm16135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajilic-Stojanovic M, Biagi E, Heilig HG, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792–801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 10.Saulnier DM, Riehle K, Mistretta TA, et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology. 2011;141:1782–91. doi: 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li G, Yang M, Jin Y, et al. Involvement of shared mucosal-associated microbiota in the duodenum and rectum in diarrhea-predominant irritable bowel syndrome. J Gastroenterol Hepatol. 2017 doi: 10.1111/jgh.14059. [DOI] [PubMed] [Google Scholar]
- 12.Rodino-Janeiro BK, Vicario M, Alonso-Cotoner C, et al. A Review of Microbiota and Irritable Bowel Syndrome: Future in Therapies. Adv Ther. 2018;35:289–310. doi: 10.1007/s12325-018-0673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pozuelo M, Panda S, Santiago A, et al. Reduction of butyrate- and methane-producing microorganisms in patients with Irritable Bowel Syndrome. Sci Rep. 2015;5:12693. doi: 10.1038/srep12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tap J, Derrien M, Tornblom H, et al. Identification of an Intestinal Microbiota Signature Associated With Severity of Irritable Bowel Syndrome. Gastroenterology. 2017;152:111–123. e8. doi: 10.1053/j.gastro.2016.09.049. [DOI] [PubMed] [Google Scholar]
- 15.Downs IA, Aroniadis OC, Kelly L, et al. Postinfection Irritable Bowel Syndrome: The Links Between Gastroenteritis, Inflammation, the Microbiome, and Functional Disease. J Clin Gastroenterol. 2017 doi: 10.1097/MCG.0000000000000924. [DOI] [PubMed] [Google Scholar]
- 16.O'Malley D. Immunomodulation of enteric neural function in irritable bowel syndrome. World J Gastroenterol. 2015;21:7362–6. doi: 10.3748/wjg.v21.i24.7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeffery IB, O'Toole PW, Ohman L, et al. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut. 2012;61:997–1006. doi: 10.1136/gutjnl-2011-301501. [DOI] [PubMed] [Google Scholar]
- 19.Costantini R, Affaitati G, Wesselmann U, et al. Visceral pain as a triggering factor for fibromyalgia symptoms in comorbid patients. Pain. 2017;158:1925–1937. doi: 10.1097/j.pain.0000000000000992. [DOI] [PubMed] [Google Scholar]
- 20.Bercik P, Denou E, Collins J, et al. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology. 2011;141:599–609. 609.e1–3. doi: 10.1053/j.gastro.2011.04.052. [DOI] [PubMed] [Google Scholar]
- 21.Bercik P, Park AJ, Sinclair D, et al. The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut-brain communication. Neurogastroenterol Motil. 2011;23:1132–9. doi: 10.1111/j.1365-2982.2011.01796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dinan TG, Cryan JF. Regulation of the stress response by the gut microbiota: implications for psychoneuroendocrinology. Psychoneuroendocrinology. 2012;37:1369–78. doi: 10.1016/j.psyneuen.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Neufeld KM, Kang N, Bienenstock J, et al. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol Motil. 2011;23:255–64. e119. doi: 10.1111/j.1365-2982.2010.01620.x. [DOI] [PubMed] [Google Scholar]
- 24.Benton D, Williams C, Brown A. Impact of consuming a milk drink containing a probiotic on mood and cognition. Eur J Clin Nutr. 2007;61:355–61. doi: 10.1038/sj.ejcn.1602546. [DOI] [PubMed] [Google Scholar]
- 25.Ki Cha B, Mun Jung S, Hwan Choi C, et al. The effect of a multispecies probiotic mixture on the symptoms and fecal microbiota in diarrhea-dominant irritable bowel syndrome: a randomized, double-blind, placebo-controlled trial. J Clin Gastroenterol. 2012;46:220–7. doi: 10.1097/MCG.0b013e31823712b1. [DOI] [PubMed] [Google Scholar]
- 26.Messaoudi M, Lalonde R, Violle N, et al. Assessment of psychotropic-like properties of a probiotic formulation (Lactobacillus helveticus R0052 and Bifidobacterium longum R0175) in rats and human subjects. Br J Nutr. 2011;105:755–64. doi: 10.1017/S0007114510004319. [DOI] [PubMed] [Google Scholar]
- 27.Messaoudi M, Violle N, Bisson JF, et al. Beneficial psychological effects of a probiotic formulation (Lactobacillus helveticus R0052 and Bifidobacterium longum R0175) in healthy human volunteers. Gut Microbes. 2011;2:256–61. doi: 10.4161/gmic.2.4.16108. [DOI] [PubMed] [Google Scholar]
- 28.Whorwell PJ, Altringer L, Morel J, et al. Efficacy of an encapsulated probiotic Bifidobacterium infantis 35624 in women with irritable bowel syndrome. Am J Gastroenterol. 2006;101:1581–90. doi: 10.1111/j.1572-0241.2006.00734.x. [DOI] [PubMed] [Google Scholar]
- 29.Logan AC, Venket Rao A, Irani D. Chronic fatigue syndrome: lactic acid bacteria may be of therapeutic value. Medical Hypotheses. 2003;60:915–923. doi: 10.1016/s0306-9877(03)00096-3. [DOI] [PubMed] [Google Scholar]
- 30.Mujagic Z, Leue C, Vork L, et al. The Experience Sampling Method--a new digital tool for momentary symptom assessment in IBS: an exploratory study. Neurogastroenterol Motil. 2015;27:1295–302. doi: 10.1111/nmo.12624. [DOI] [PubMed] [Google Scholar]
- 31.Palsson OS, Baggish JS, Turner MJ, et al. IBS patients show frequent fluctuations between loose/watery and hard/lumpy stools: implications for treatment. Am J Gastroenterol. 2012;107:286–95. doi: 10.1038/ajg.2011.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shulman RJ, Jarrett ME, Cain KC, et al. Associations among gut permeability, inflammatory markers, and symptoms in patients with irritable bowel syndrome. J Gastroenterol. 2014;49:1467–76. doi: 10.1007/s00535-013-0919-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luna RA, Oezguen N, Balderas M, et al. Distinct Microbiome-Neuroimmune Signatures Correlate With Functional Abdominal Pain in Children With Autism Spectrum Disorder. Cell Mol Gastroenterol Hepatol. 2017;3:218–230. doi: 10.1016/j.jcmgh.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aagaard K, Petrosino J, Keitel W, et al. The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. Faseb j. 2013;27:1012–22. doi: 10.1096/fj.12-220806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin M. Cutadapt removes adapter sequences from high-throughout sequencing reads. EMBnet.journal 17.1. 2011;17:10–12. [Google Scholar]
- 36.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hildebrand F, Tadeo R, Voigt AY, et al. LotuS: an efficient and user-friendly OTU processing pipeline. Microbiome. 2014;2:30. doi: 10.1186/2049-2618-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–8. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 39.Rognes T, Flouri T, Nichols B, et al. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitehead WE. Development and validation of the Rome III diagnostic questionnaire. McLean, VA: Degnon Associates; 2006. [Google Scholar]
- 41.Thompson W, Drossman D, Talley NJ. Rome III diagnostic questionnaire for the adult functional GI disorders (including alarm questions) and scoring algorithm. McLean, CA: Degnon Associates Inc; 2006. [Google Scholar]
- 42.Jarrett ME, Cain KC, Burr RL, et al. Comprehensive self-management for irritable bowel syndrome: randomized trial of in-person vs. combined in-person and telephone sessions. Am J Gastroenterol. 2009;104:3004–14. doi: 10.1038/ajg.2009.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shulman R, Schanler R, Lau C, et al. Early Feeding, Antenatal Glucocorticoids, and Human Milk Decrease Intestinal Permeability in Preterm Infants. Pediatric Research. 1998;44:519–523. doi: 10.1203/00006450-199810000-00009. [DOI] [PubMed] [Google Scholar]
- 44.Hahn B, Kirchdoerfer L, Fullerton S, et al. Evaluation of a new quality of life questionnaire for patients with irritable bowel syndrome. Aliment Pharmacol Ther. 1997;11:547–552. doi: 10.1046/j.1365-2036.1997.00168.x. [DOI] [PubMed] [Google Scholar]
- 45.Carroll IM, Ringel-Kulka T, Siddle JP, et al. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil. 2012;24:521–30. e248. doi: 10.1111/j.1365-2982.2012.01891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vandeputte D, Falony G, Vieira-Silva S, et al. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. 2016;65:57–62. doi: 10.1136/gutjnl-2015-309618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roager HM, Hansen LB, Bahl MI, et al. Colonic transit time is related to bacterial metabolism and mucosal turnover in the gut. Nat Microbiol. 2016;1:16093. doi: 10.1038/nmicrobiol.2016.93. [DOI] [PubMed] [Google Scholar]
- 48.Parthasarathy G, Chen J, Chen X, et al. Relationship Between Microbiota of the Colonic Mucosa vs Feces and Symptoms, Colonic Transit, and Methane Production in Female Patients With Chronic Constipation. Gastroenterology. 2016;150:367–79. e1. doi: 10.1053/j.gastro.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Di Stefano M, Mengoli C, Bergonzi M, et al. Breath Methane Excretion Is not An Accurate Marker of Colonic Methane Production in Irritable Bowel Syndrome. Am J Gastroenterol. 2015;110:891–8. doi: 10.1038/ajg.2015.47. [DOI] [PubMed] [Google Scholar]
- 50.Santiago A, Panda S, Mengels G, et al. Processing faecal samples: a step forward for standards in microbial community analysis. BMC Microbiol. 2014;14:112. doi: 10.1186/1471-2180-14-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sze MA, Schloss PD. Looking for a Signal in the Noise: Revisiting Obesity and the Microbiome. MBio. 2016;7 doi: 10.1128/mBio.01018-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaakoush NO. Insights into the Role of Erysipelotrichaceae in the Human Host. Front Cell Infect Microbiol. 2015;5:84. doi: 10.3389/fcimb.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen L, Wilson JE, Koenigsknecht MJ, et al. NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth. Nat Immunol. 2017;18:541–551. doi: 10.1038/ni.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mancabelli L, Milani C, Lugli GA, et al. Identification of universal gut microbial biomarkers of common human intestinal diseases by meta-analysis. FEMS Microbiol Ecol. 2017;93 doi: 10.1093/femsec/fix153. [DOI] [PubMed] [Google Scholar]
- 55.Cox LM, Cho I, Young SA, et al. The nonfermentable dietary fiber hydroxypropyl methylcellulose modulates intestinal microbiota. Faseb j. 2013;27:692–702. doi: 10.1096/fj.12-219477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Russo E, Bacci G, Chiellini C, et al. Preliminary Comparison of Oral and Intestinal Human Microbiota in Patients with Colorectal Cancer: A Pilot Study. Front Microbiol. 2017;8:2699. doi: 10.3389/fmicb.2017.02699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen KL, Madak-Erdogan Z. Estrogen and Microbiota Crosstalk: Should We Pay Attention? Trends Endocrinol Metab. 2016;27:752–755. doi: 10.1016/j.tem.2016.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


