Abstract
Production of fuels, therapeutic drugs, chemicals, and biomaterials using sustainable biological processes have received renewed attention due to increasing environmental concerns. Despite having high industrial output, most of the current chemical processes are associated with environmentally undesirable by-products which escalate the cost of downstream processing. Compared to chemical processes, whole cell biocatalysts offer several advantages including high selectivity, catalytic efficiency, milder operational conditions and low impact on the environment, making this approach the current choice for synthesis and manufacturing of different industrial products. In this review, we present the application of whole cell actinobacteria for the synthesis of biologically active compounds, biofuel production and conversion of harmful compounds to less toxic by-products. Actinobacteria alone are responsible for the production of nearly half of the documented biologically active metabolites and many enzymes; with the involvement of various species of whole cell actinobacteria such as Rhodococcus, Streptomyces, Nocardia and Corynebacterium for the production of useful industrial commodities.
Keywords: Actinobacteria, biocatalysts, nitriles, biotransformation, biofuel, ethylene glycol
Introduction
Biotransformation is the process by which substrates are converted into useful products using biocatalysts either in the form of whole cells or their enzymes (Ward and Köhler, 2015; Bezborodov and Zagustina, 2016).The classical chemical based transformation of substrates is prone to several disadvantages, including ecologically unfavorable conditions and associated undesirable by-products (Lin and Tao, 2017). Unlike chemical methods, biocatalysts provide several benefits such as their availability from renewable resources, they work at low temperature and pH, are easy to degrade biologically and are selective in both substrate and product stereochemistry (Jemli et al., 2016). Enzyme based biotransformations, however, are also not free of drawbacks including their high cost, higher susceptibility to changes in operating conditions and substrate or product toxicity (Garzón-Posse et al., 2018). In contrast to other catalytic reactions, whole cell biocatalysts allow transformation of substrates via multiple cascades of reactions, help generation of cofactors, have high regio- and stereo-selectivity, work under mild operational, environmentally-friendly conditions, and help selective hydroxylation of non-activated carbon atoms. The latter is not possible with chemical catalysts (de Carvalho, 2017). In addition to these advantages, compounds produced by microorganisms are considered to be safe, which attract many health-conscious consumers (de Carvalho, 2017).
To meet the growing call for efficient and economically feasible biocatalysts, researchers are testing different groups of microorganisms, including actinobacteria (Mukhtar et al., 2017), Escherichia coli (Lin et al., 2013; Ladkau et al., 2014; de Carvalho, 2017), Pseudomonas putida (Gehring et al., 2016), Bacillus cereus (Banerjee and Ghoshal, 2010), Enterococcus faecalis and Saccharomyces cerevisiae (Whited et al., 2010; Lin and Tao, 2017). Actinobacteria are widely distributed in nature, with several phenotypes including anaerobes, aerobes, spore formers, unicellular, and filamentous forms (Lewin et al., 2016). They are one of the most diverse, well characterized and metabolically versatile group of microorganisms. They play an essential role in maintaining soil structure and carbon recycling through decomposition of various organic matter such as cellulose, chitin, and pectin (Priyadharsini and Dhanasekaran, 2015; Kim et al., 2016). Furthermore, they produce several enzymes (amylases, cellulases, proteases, chitinases, xylanases, and pectinase) (Mukhtar et al., 2017), antibiotics, antitumor agents, plant growth regulators, and vitamins (Prakash et al., 2013; Kamjam et al., 2017).
Over 22,000 biologically active microbial metabolites reported and actinobacteria alone represented 45% of them which are followed by fungi (38%) and unicellular bacteria, especially Bacillus sp. and Pseudomonas sp. (17%) (Bérdy, 2005; Demain and Sanchez, 2009). Among the described 140 genera of actinobacteria, only few of them produce the majority of active compounds (Jensen et al., 2005; Bull and Stach, 2007; Pimentel-Elardo et al., 2010; Adegboye and Babalola, 2013). Streptomyces alone represents three fourth of the total active metabolites produced by actinobacteria (Lam, 2007; Solecka et al., 2012; Barka et al., 2015; Chater, 2016). Table 1 below highlights the approximate share of each microbial group for active metabolite production.
Table 1.
Microbial share of active bioactive metabolites (Bérdy, 2005).
| Source | Total bioactive metabolites | Antibiotics | Other bioactive metabolites |
|---|---|---|---|
| Bacteria | 3800 | 2900 | 900 |
| Eubacteriales | 2750 | ||
| Bacillus sp. | 860 | ||
| Pseudomonas sp. | 795 | ||
| Myxobacter | 410 | ||
| Cyanobacter | 640 | ||
| Actinobacteria | 10100 | 8700 | 1400 |
| Streptomyces sp. | 7630 | ||
| Other genera | 2470 | ||
| Fungi | 8600 | 4900 | 3700 |
| Microscopic fungi | 6450 | ||
| Penicillium/Aspergillus | 1950 | ||
| Basidiomycetes | 2000 | ||
| Yeasts | 140 | ||
| Slime molds | 60 | ||
| Total | 22500 | 16500 | 6000 |
Apart from the above contributions, actinobacteria play a vital part in the development of a sustainable bioenergy industry, predominantly through their cellulolytic enzymes which decompose plant biomass to produce simple sugars that serve as raw materials for biofuel production. Furthermore, their diverse biosynthetic capacity allow them to mediate various environmental interactions which lead to synthesis of various biologically active products (Lewin et al., 2016). Here, we review the application of whole cells actinobacteria for biotransformation of various substrates in a way to produce more active and less toxic compounds as well as biofuels.
Biotransformation of Harmful Compounds
Nitrile Biotransformation
Microbial or enzymatic biotransformation of nitriles result the conversion of these toxic compounds into industrially important compounds like acids and amides. Nitriles constitute a group of chemicals widely used in drugs, rubbers, and plastic industries. These compounds contain a cyano group in their backbone which is highly correlated with toxicity (Ramteke et al., 2013). Their high rate of manufacture and continuous usage make them an important source of environmental pollution and have been detected in different environmental samples including sediments of water-treatment plants, in marinas and beach areas (Baxter and Cummings, 2006). Clinically, nitrile toxicity has been associated with cancer and different health problems such as bronchial irritation, respiratory disorders, convulsions, coma, and skeletal deformities (Ramteke et al., 2013). Some researchers highlighted the link with psycho-behavioral abnormalities including learning, memory, motor nerve anomalies in rats treated with aliphatic nitriles such as acetonitrile, acrylonitrile and crotononitrile (Boadas-Vaello et al., 2007; Ramteke et al., 2013).
The removal of nitrile compounds from the environment is possible using microbial methods, due to their associated low cost and user-friendly approach (Fang et al., 2015). Microorganisms degrade nitrile compounds through the hydrolytic route, which comprises two enzymatic systems as indicated in Figure 1. In the first route, nitrile hydratase (NHase, EC 4.2.1.84) catalyzes the formation of amides from nitriles, which are later changed to ammonia and carboxylic acids by amidase (EC 3.5.1.4). Alternatively, nitrilase (EC 3.5.5.1) catalyzes for the direct conversion of nitriles into carboxylic acids and ammonia (Ramteke et al., 2013).
FIGURE 1.
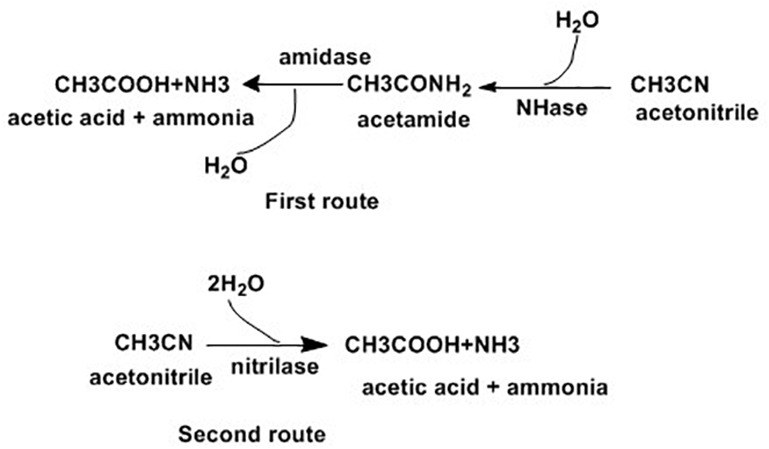
Enzymatic pathways for nitrile hydrolysis (Ramteke et al., 2013).
The production of acids and amides from nitriles is possible using chemical catalysts. However, this approach is only achieved under harsh conditions like extreme temperature, acidity or alkalinity (Nigam et al., 2009). Currently, many microorganisms having either of the two nitrile degradation enzymatic systems have been reported. These microorganisms can be categorized into two groups. The first is made up of bacteria like Mesorhizobium sp. F28 (Feng and Lee, 2009), Klebsiella oxytoca (Kao et al., 2006), Rhodococcus erythropolis A4 and Rhodococcus rhodochrous PA-34 (Vesela et al., 2012), which only contain a single enzyme system of NHase/amidase and Streptomyces sp. MTCC 7546 only contains nitrilase (Nigam et al., 2009). The other group contain bacteria such as Nocardia globerula NHB-2 (Bhalla and Kumar, 2005), Amycolatopsis sp. IITR215 (Babu et al., 2010), Bacillus subtilis ZJB-063 (Zheng et al., 2008) and R. rhodochrous BX2 (Fang et al., 2015), which have both NHase/amidase and nitrilase.
Variation among nitrile degrading microorganisms also exists in terms of the end products of nitrile degradation. Bacteria in the single enzyme system like R. rhodochrous PA-34 convert nitrile only into amides while K. oxytoca and Mesorhizobium sp. F28 convert nitriles into corresponding amides and carboxylic acids. Those bacteria which utilize the two enzyme system result in amides and carboxylic acids. Among these group of bacteria, R. rhodochrous BX2, B. subtilis AJB-063 and Paracoccus sp. SKG (Santoshkumar et al., 2011) displayed completed degradation of carboxylic acids with final end product of ammonia. As indicated above nitrile degradation systems vary among different bacterial genera as well as with in the same genus such as Rhodococcus. Unlike others, Streptomyces sp. MTCC 7546 in the immobilized as well as Free State biotransforms acrylonitrile into acrylic acid without the formation of amides. The authors suggested that due to several reasons such as operational stability (allow to reuse the system several times), and ease of production on a large scale, the conversion of acrylonitrile using immobilized cells is better than cells in the free state (Nigam et al., 2009).
Biotransformation of Aromatic Ring Containing Compounds
Phthalate esters and phenols are the two most common chemicals used in industry for stabilization and modification of the characteristics and performance of polymers (He Z. et al., 2014). Di-n-butyl phthalate (DBP), a type of phthalate ester, is a component of different merchandises including pesticides, wrapping materials, makeups, wrappers, wears, and insulators in electric disposals (Dargnat et al., 2009). Similarly, phenol can be applied for the manufacturing of drugs, rubbers, polycarbonate resins, and nylon (Christen et al., 2011).
Phthalates are major environmental pollutants which come into contact with humans and animals through contaminated water systems (He Z. et al., 2014). The European community listed these compounds among the 33 dangerous substances to be controlled in surface water (Dargnat et al., 2009). As they are a constituent of plastics which are now ubiquitous in diverse environments phthalates are now present almost everywhere (Fang et al., 2010). Phthalate toxicity is associated with endocrine system disruption in different species of fish and mammals. These compounds were also observed to interfere with the reproductive system and in human and animal development (Lottrup et al., 2006; Li et al., 2010). Concurrent observation of phenols and phthalate esters has been reported in the Selangor River basin in Malaysia (Santhi and Mustafa, 2013) and induction of lactate dehydrogenase release from Sertoli cells, which is associated with infertility, coexist compared to individual chemical effects (Li et al., 2010).
Different approaches have been documented for removal of DBP from natural environments. These are hydrolysis, photo degradation and biodegradation (Lau et al., 2005; Jonsson et al., 2006; Chen et al., 2009). Two of the former approaches were not effective due to the structural nature of DBP and microbial mediated metabolic transformation of DBP is the current choice. Microbial mediated degradation of DBP involves initial conversion of DBP into phthalic acid and which is further transformed with the help of two dioxygenase pathways into 4,5-dihydroxyphalate and 4,5-dihydroxyphalate in gram negative and gram positive bacteria, respectively. Finally, these two compounds are transformed into a common intermediate protocatechuate under aerobic conditions (Wu et al., 2011). For the degradation of phenols the first step is hydroxylation of phenol to catechol followed by ring cleavage of catechol into 2-hydroxymuconic semialdehyde for the meta-pathway aided by catechol-2, 3-dioxygenase and into cis, cis-muconate with the help of catechol-1, 2-dioxygenase for ortho pathway. Finally 2-hydroxymuconic semialdehyde oxidized 4-oxalocrotonate or hydrolyzed it to 2-oxopent-4-enoate and cis, cis-muconate into muconolactone (Banerjee and Ghoshal, 2010).
Several bacteria strains have the ability to degrade DBP, such as Rhodococcus sp. (Yu et al., 2009; Jin et al., 2010), Gordonia sp. (Wu et al., 2011), Agrobacterium sp. (Wu et al., 2011) and Enterobacter sp. (Fang et al., 2010). Members of the Rhodococcus genus, have been demonstrated for degradation of Phenol in addition to DBP (Saa et al., 2010), individually as well as via simultaneously mineralization of DBP and phenol (Lu et al., 2009; He et al., 2013). Individual or synchronous biodegradation of DBP and phenol by Rhodococcus ruber strain DP-2 was also reported in similar study (He Y.C. et al., 2014).
Chlorophenols (CPs) are aromatic compounds which contain a minimum of one chlorine and a hydroxyl group on the benzene rings. Five types of CPs, as indicated in Figure 2 below, based on chemical structures include monochlorophenols, polychlorophenols, chloronitrophenols, chloroaminophenols, and chloromethylphenols (Arora and Bae, 2014). They are largely used as fungicidal, germicidal and wood preservatives agents. They are also important for synthesis of dyes and drugs (Arora and Bae, 2014). CPs and their derivatives rank among the top environmental pollutants where industrial wastes, pesticides, herbicides, and complex chlorinated hydrocarbons are major sources of contamination (Olaniran and Igbinosa, 2011). Direct skin contact and eating or drinking of contaminated substances are major sources of people exposure (Arora and Bae, 2014). Cellular exposure to CPs are associated with cytotoxic, mutagenic and carcinogenic properties, with several types of polychlorophenols labeled as potential human carcinogens by the World Health Organization and the International Agency for Research on Cancers (Igbinosa et al., 2013).
FIGURE 2.
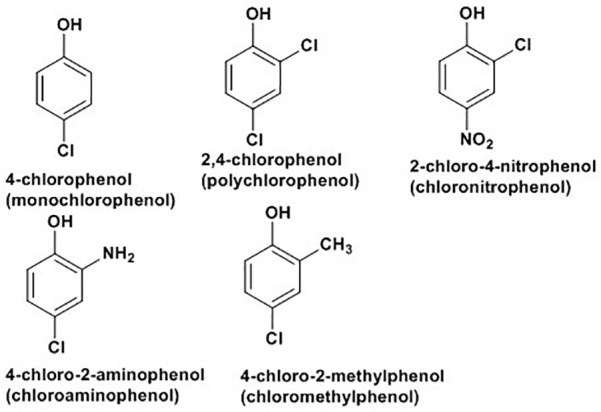
Some examples of chlorophenols (Arora and Bae, 2014).
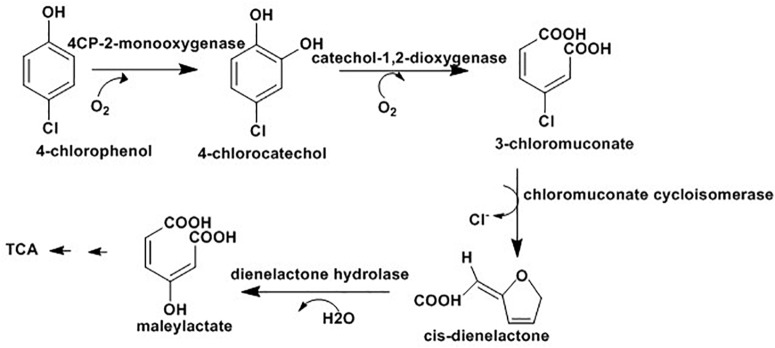
Different possible mechanisms are reported for bacterial degradation of CPs and their derivatives. In the first mechanism, hydroxylation of chlorophenolic rings at ortho-positions with the help of monooxygenases results in formation of chlorocatechols which are further degraded (Hollender et al., 1997; Solyanikova and Golovleva, 2011) or hydroxylated prior to ring cleavage (Nordin et al., 2005). In the second mechanism, with the same enzyme, hydroxylation of chlorophenolic rings at meta position results in chlorocatechols which degrade via hydroxylation (Nordin et al., 2005) or dehalogenation (Xun et al., 1992) prior to ring cleavage. The third mechanism applicable for degradation of chloronitrophenols where the degradation may be initiated by hydroxylation (Arora and Jain, 2012), reductive dehalogenation (Pandey et al., 2011), or reduction of the nitro group (Arora and Jain, 2012). Finally, in case of chloroaminophenols degradation, the pathway may start with removal of ammonium ions by the enzyme deaminase followed by the ring cleavage (Arora and Bae, 2014) or the dehalogenation (Arora and Bae, 2014). The detail mechanisms of different routes of biodegradation of chlrophenols and its derivatives with various bacteria, such as Pseudomonas knackmussii B-13, Rhodococcus opacus 1G, Arthrobacter chlorophenolicus A6, Streptomyces rochei 303, Pseudomonas sp. NCIB9340, Bacillus insolitus, Nocardioides sp. K44, Mycobacterium chlorophenolicum PCP1 and Mycobacterium fortuitum CG-2 are well documented in a recent review by Arora and Bae (2014); and the following Figure 3 presents degradation of 4-chlorophenol as an example of the process.
FIGURE 3.
4-chlorophenol degradation via ortho postion where the final intermidate, maleylactate, inter for TCA cycle for complet mineralizatio (Arora and Bae, 2014).
Hou and his colleagues reported for the first time magnetically immobilized R. rhodochrous cells for biodegradation of CPs. Their study demonstrated that R. rhodochrous DSM6263 depends on constitutively expressed enzymes for hydroxylation of CPs resulting in chlorocatechol formation and complete degradation. Their observation was consistent with another study where Rhodococcus sp. AN-22 (an aniline-assimilating bacterium) produced cis, cis-muconic acid from catechol (Matsumura et al., 2006) and highlighted how these compounds could also be metabolized with immobilized cells of R. rhodochrous DSM6263 (Hou et al., 2016). Researchers advocated the use of immobilized cells, over free cells, to degrade toxic chemicals due to many reasons such as long-term stability of the catalyst and the immobilization also protects cells from harmful effects of toxic pollutants (Jianlong et al., 2002). Immobilization of cells can be done using a number of techniques including surface adsorption, natural or artificial flocculation, covalent or electrostatic binding to carriers, and encapsulation in a polymer-gel (Hou et al., 2016).
Atrazine (2-chloro-4-(ethylamino)-6-(isopropylamino)-1, 3, 5-triazine) was first introduced in 1950s as an emergent herbicide and they are among widely used pesticides in different countries such as United States, Canada, Africa, and Asia Pacific region (Huang et al., 2003; Jablonowski et al., 2011). Usage of atrazine banned in European countries in 2004 as atrazine concentrations in water surpassed or were estimated to surpass allowable limits (Jablonowski et al., 2011). Due to factors like its widespread utilization as a herbicide and its persistence in the environment, it is common to observe traces of atrazine both in surface and ground water bodies (Gilliom et al., 2006). Traces of atrazine were detected in widely dispersed areas which are far from urban and agricultural areas such as in rainwater in different places (Brun et al., 2008) in fog, arctic ice and seawater.
Contact with atrazine is associated with a serious threat to human and ecosystem health. One of the most notable effects of exposure is endocrine (Solomon et al., 2008; Rohr and McCoy, 2010). Many studies link atrazine with harmful effects on the health of animals and humans, such as sexual abnormalities (demasculinization) in frogs, low testosterone production in rats and higher levels of prostate cancer in workers at an atrazine manufacturing factory (Hecker et al., 2005; Liu and Parales, 2009), and it is also categorized as a group 3 carcinogen according to the International Agency for research on cancer1. The above observations indicated that there is cause for concern regarding atrazine residues in soil, groundwater, and surface waters.
Due to its persistence in the environment and being highly toxic, it is very important to develop approaches to degrade and remove atrazine deposits from the environment. Microbial-degradation is one of the methods for elimination of atrazine from soil (Tappe et al., 2002). Different species of microorganisms associated with degradation of atrazine with various degree of biodegradation where some undergo complete mineralization while others produce various intermediates including hydroxyatrazine, deethylatrazine, deisopropylatrazine, n-isopropylammilide, n-ethylammilide, and cyanuric acid (Mandelbaum et al., 1995; Ralebits et al., 2002; Ghosh and Philip, 2004; Getenga et al., 2009). Specifically, atrazine degradation was documented with the help of Rhodococcus sp. BCH2 (Kolekar et al., 2014), Arthrobacter sp. (Getenga et al., 2009), Nocardioides sp. (Topp et al., 2000). Pseudomonas sp. strain ADP (Rousseaux et al., 2001; Liu and Parales, 2009) was the first bacterium reported that could completely mineralize atrazine; with most of the degradation studies based on study this strain. As indicated in Figure 4 (below) atrazine degradation is achieved because of the presence of the genes, atzA, atzB and atzC, which code for enzymes such as chlorohydrolase (AtzA), hydroxyatrazine ethylaminohydrolase (AtzB) and N-isopropylammelide isopropylaminohydrolase (AtzC), respectively which convert atrazine sequentially to cyanuric acid (Neumann et al., 2004). Some strains of Pseudomonas can further degrade cyanuric acid into CO2 and NH3.
FIGURE 4.
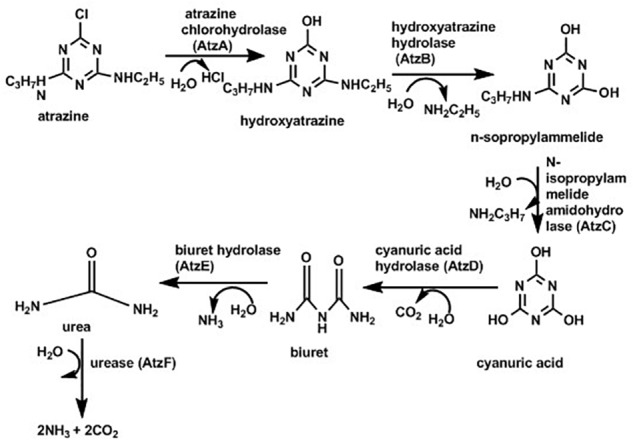
Pathway for atrazine degradation (Liu and Parales, 2009).
1, 4-Dioxane is a cyclic ether with many applications including components of deodorants, detergents and various types of paints. The process for manufacturing of polyesters also results 1,4-dioxane production. Various factors including illegal dumping of industrial wastes contribute to 1, 4-dioxane associated water contamination. High level of 1, 4-Dioxane can cause liver and nasal cancers in rats module (Dourson et al., 2014) and are listed as group 2B human carcinogen (Inoue et al., 2016). These compounds are soluble in water with low volatility and have a lower chance of absorbance in solids (Stepien et al., 2014). Therefore, once 1, 4-dioxane appears in the environment it can persist for many days and a high degree of 1, 4-dioxane pollution was observed in surface water, groundwater, and landfill leachatewa (Stepien et al., 2014).
Their removal from water bodies is an important public concern, especially as the routine physical and chemical methods for water decontamination are not effective for removal of 1, 4-dioxane (Adams et al., 1994). Advanced procedures such as the combination of ozone and hydrogen peroxide treatments are expensive to use (Adams et al., 1994; Kishimoto et al., 2008). Thus, cost effective as well as reliable methods for cleaning of 1, 4-dioxane from water lead to nocardioform actinobacteria such as Pseudonocardia (Matsui et al., 2016) and Rhodococcus (Deeb and Alvarez-Cohen, 1999) which account for the major portion of capable microorganisms. Inoue et al. (2016) tested various species of Pseudonocardia and Rhodococcus for their ability to degrade 1, 4-dioxane. Their findings indicated P. dioxanivorans JCM 13855T (also known as P. dioxanivorans CB1190) (Parales et al., 1994) was the only Pseudonocardia sp. tested that used 1,4-dioxane as a carbon source and degraded it. In contrast, they observed the inability of R. ruber JCM 3205T to degrade 1, 4-dioxane. However, there are reports on the ability of Rhodococcus spp. such as R. ruber 219 (Bernhardt and Diekmann, 1991) and R. ruber T1 and T5 (Oyama et al., 2013) to biodegrade 1,4-dioxane. This highlighted species variation among Rhodococcus in relation to effective degradation and utilization of 1, 4-dioxane.
Production of Important Compounds
Ethylene Glycol Synthesis
Due to the continuing concern about climate change and depletion of fossil energy, expansion of biological processes using renewable biological resources to produce different chemical feed stocks and energy has become an attractive approach for the chemical industry. Ethylene glycol is a feedstock which serves as a starting material for the manufacture of several items including polymers, anti-freeze agents, and coolants (Zhang and Yu, 2013). Routinely, ethylene glycol is produced through a costly chemical process using ethylene derived from the petrochemical industry as a starting material. Therefore, there is a preference for biological synthesis of ethylene glycol over chemical methods as the former has a low impact on the environment and the reaction is selective (Mattam et al., 2013).
Fermentation of carbohydrates is an economical process for production of ethylene glycol from biofeed stock. Corynebacterium glutamicum, an actinobacterium, has been designed for manufacture of ethylene glycol directly from glucose via extension of the serine synthesis pathway. Serine is produced by most microorganisms from 3-phosphoglycerate (a glycolysis intermediate) using three enzymatic steps: 3-phosphoglycerate converted into P-hydroxypyruvate with the help of 3-phosphoglycerate dehydrogenase (PGDH; serA), followed by conversion of P-hydroxypyruvate to P-serine by phosphoserine aminotransferase (PSAT; serC) and P-serine to serine with the help of phosphoserine phosphatase (PSP; serB) (Peters-Wendisch et al., 2005). Chen et al. (2016) proposed two ways for ethylene glycol synthesis via the extension of the serine synthesis pathway where serine is the starting material for both systems with the end product glycoaldehyde (Chen et al., 2016). The first route involves deamination of serine with aminotransferase or amino acid dehydrogenase resulting in hydroxypyruvate. Finally, glycoaldehyde is produce from hydroxypyruvate with the help of α-ketoacid decarboxylase. Similarly, the other route also has two steps where ethanolamide produced from serine with decarboxylation and then converted to glycoaldehyde following oxidation by monoamine oxidase. Finally, glycoaldehyde can be reduce to ethylene glycol by alcohol dehydrogenase such as yqhD as indicated in Figure 5.
FIGURE 5.
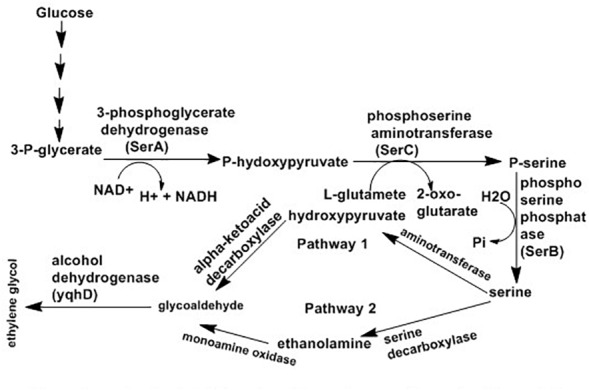
Proposed pathways for synthesis of ethylene glycol from glucose using serine intermediate (Chen et al., 2016).
In addition to glucose, C. glutamicum uses several substrates such as sugars present in molasses (sucrose and fructose), pentose sugars present in lignocellulosic (Zahoor et al., 2012), n-acetyl-D-glucosamine and n-acetyl-D-muramic acid (Sgobba et al., 2018) for manufacturing of various amino acids (Bommareddy et al., 2014). Production of other compounds such as isobutanol, cadaverine, and succinate are possible with C. glutamicum (Blombach et al., 2011; Buschke et al., 2011; Litsanov et al., 2012).
Rhodococcus sp. are also associated with production of Ethylene glycol. Rhodococcus sp. CGMCC 4911 converted 1, 3-propanediol cyclic sulfate and its derivatives into corresponding diols. The growing cells of Rhodococcus successfully hydrolyzed ethylene sulfate, glycol sulfide, 1, 3-propanediol cyclic sulfate, and 1, 2-propanediol cyclic sulfate with different conversion rates (He et al., 2015).
Cyclic sulfates and its derivatives are very important compounds that serve as starting materials for the synthesis of various useful intermediates (Steinmann et al., 2001). Sulfatases hydrolyse organic sulfate esters into primary or secondary alkyl alcohols (Gadler and Faber, 2007). Microbial transformation of cyclic sulfates into diols can be achieved under mild reaction conditions. He et al. (2013) reported for the first time that Rhodococcus sp. CCZU10-1 can convert 1, 3-propanediol cyclic sulfate and its derivatives into diols, where factors like pH, temperature, and cells dose affect rate of biotransformation.
Production of Less Toxic and Biologically Active Drugs
The absolute configuration of chiral centers in molecules determine the biological activities of the compounds as these molecules bind with receptors made of enantiomerically pure protein (Kato et al., 2003). Most of the available chiral carbon containing drugs are racemic which contain equal concentration of S (+) and R (+) enantiomers. Since only one of the two is active and the other associated with toxicity, the current drug development gives priority to compounds only with a single enantiomer.
Ibuprofen [(R,S)-2-(4′-isobutylphenyl)propionic acid] are a group of effective, orally active, nonsteroidal, anti-inflammatory agents which include drugs such as naproxen, fenoprofen, and flurbiprofen (Sen and Anliker, 1996). Even though the S-(+)- ibuprofen form is more than 100 times more effective than the R-(-)-ibuprofen, the racemic form of ibuprofen is widely used (Kumaresan, 2010). The (R)-ibuprofen form can be converted to its enantiomer in the livers and kidneys of pigs and rats, though this process is not free of toxicity (Liu et al., 2009). Figure 6 below highlighted the application of whole cells N. corallina to biocatalyse the enantioselective hydrolysis of racemic ibuprofen nitrile [2-(4-isobutylphenyl) propanenitrile, 1] to optically active ibuprofen amide [2-(4-isobutylphenyl) propanamide, 2], a prodrug using nitrile hydratase (NHase) and hydrolyse this amide into optically active ibuprofen (3) using amidase.
FIGURE 6.
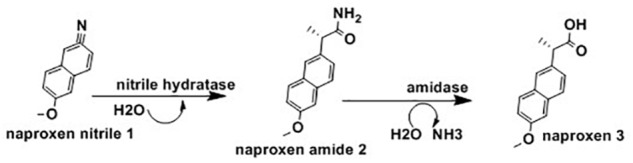
Biotransformation of ibuprofen nitrile 1 to ibuprofen amide 2 and ibuprofen 3 using Nocardia corallina B-276 (Lievano et al., 2012).
The results of the above observation indicated N. corallina B-276 displayed nitrile hydratase and amidase activities having low stereo-selectivity (Figure 6). This finding also revealed that N. corallina catalyzed deracemisation of racemic ibuprofen to (R)-ibuprofen enantiomer with an efficiency of >99%. This is the first observation where N. corallina B-276 catalyzed deracemisation processes. Currently, the (R)-enantiomer form of non-steroidal anti-inflammatory agents are the center of studies and research focus on resolving rac-ibuprofen (Pignatello et al., 2008); and due to this, recent efforts have been directed at resolving rac-ibuprofen (Trung et al., 2006).
A high percentage of optical active S-(+)-ibuprofen was reported from hydrolysis of ibuprofen amide and four related 2-phenylpropionamides using whole cell Rhodococcus AJ270 (Snell and Colby, 1999). This high purity was achieved through partial hydrolysis as complete hydrolysis of (R, S)-(+)-2-(4′-isobutylphenyl) propionamide (ibuprofen amide) results in a racemic mixture of ibuprofen enantiomers. The findings of this study suggested that prolonged hydrolysis of ibuprofen amide resulted in both ibuprofen amide and an optical purity of 90-94% of s-(+)-ibuprofen if the reaction is stopped before completion.
Due to its huge production and wide usage, ibuprofen is one of the most commonly detected compounds in wastewater (Buser et al., 1999). In addition to production of active ibuprofen, different species of actinobacteria including Patulibacter sp. strain I11 (Almeida et al., 2013) and Nocardia sp. NRRL 5646 (Chen and Rosazza, 1994; Cy et al., 2018) are involved in its biodegradation to the level where there is no more risk the community.
Carvedilol is a non-selective, β-adrenergic receptor antagonist and α1-adrenoceptor blocker, and it exists in two enantiomeric forms (Gagyi et al., 2008). The overall cardio-protective action of Carvedilol is due to its (S)-(-)-enantiomer, which is less hepatotoxic than the racemic mixture, and (R)-(+)- enantiomer (Hao and Kim, 2010). Ettireddy et al. (2017) tested Streptomyces halstedii and other bacteria for their ability to biotransform racemic carvedilol to its (S)-(-)-enantiomer. The result indicated some bacteria including S. halstedii exhibited incubation time dependence enantioselective conversion of carvedilol where the conversion rate increased up to 10 days of incubation and then after the rate is reduced and finally become zero.
Chiral amines have been widely used for manufacturing of several therapeutics such as codeine (pain relief), zoloft / sertraline (anti-depression), lariam (anti-malaria) and ethambuto (anti tuberculosis) and agrochemical intermediates including insecticides (imiprothrin, nornicotine), herbicides (imazapyr, imazapic) and fungicides (cyprofuram, fenbuconazole) (Ulrich et al., 2012). Several syntheses of optically active amines have been studied for many years. Asymmetric synthesis of chiral cyclic amine from cyclic imine achieved using whole-cell Streptomyces sp. GF3587 and 3546. As showed in Figure 7, these strains produced novel imine reductase enzymes which facilitate enantioselective redaction of cyclic 2-methyl-1-pyrroline into R-2-methylpyrrolidine and S-2-methylpyrrolidine in the presence of glucose.
FIGURE 7.
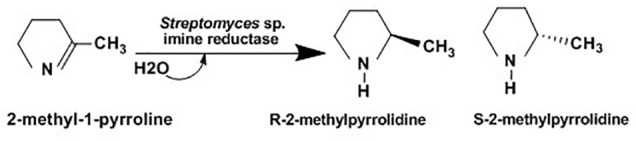
Asymmetric reduction of 2-methyl-1-pyrroline using Streptomyces sp. (Mitsukura et al., 2010).
In addition to enantioselective production of active drugs, regioselective addition or substitution of functional groups can also produce active drugs. Actinobacteria play a role in regioselective hydroxylation which mediates the conversion into more active forms. The hydroxylated form of isoflavones such as daidzein (4′, 7-dihydroxyisoflavone) and genistein (4′,5,7-trihydroxyisoflavone) are associated with lowering blood cholesterol and preventing cardiovascular diseases and cancer (Foti et al., 2005). As presented in Figure 8 below, some cytochrome P450 monooxygenases of Streptomyces avermitilis MA-4680 catalyze 3′-specific hydroxylation of daidzein and genistein to 3′,4′,7-trihydroxyisoflavone and 3′,4′,5,7-tetrahydroxyisoflavone, respectively (Roh et al., 2009).
FIGURE 8.
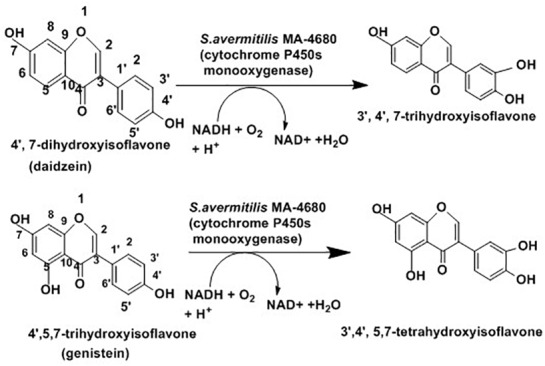
Schematic presentation for isoflavones hydroxylation with the help of Streptomyces avermitilis (Roh et al., 2009).
Microbial biotransformation of natural steroids into pharmaceutically active intermediates has been practiced for many years (Fernandes et al., 2003). These active intermediates play a role in the manufacturing of all type I aromatase inhibitors (Lombardi, 2002) and several high-value steroidal drugs (Wadhwa and Smith, 2000).The bioconversion reaction proceeds with removal of the C-17 side chain of the steroid without any modification of the steroid nucleus (Szentirmai, 1990; Murohisa and Iida, 1993). As indicated in Figure 9, Mycobacterium sp. NRRL B-3683 and Mycobacterium sp. NRRL B-3805 facilitate a single step C-17 side chain cleavage of sitosterol, cholesterol, stigmasterol and ergosterol to produce C-19 steroids such as 1-androstene-3,17-dione and 1,4-androstadiene-3,17-dione (Sripalakit et al., 2006). The initial step of the side-chain oxidation of sterols is hydroxylation at C-17. The reaction is catalyzed by cytochrome P450 monooxygenase, then several dehydrogenase including 3-β-hydroxysterol dehydrogenase remove hydrogen to introduced a double bound at various points (Donova and Egorova, 2012).
FIGURE 9.
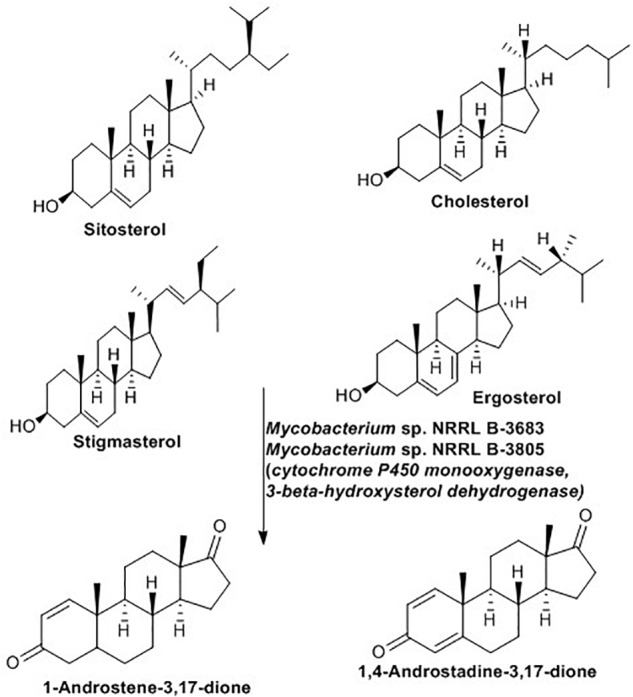
Sterol side-chain cleavage reaction mediated by Mycobacterium spp. (Sripalakit et al., 2006).
Amino Acid Production
Amino acids alone or in combination with other molecules can be used as drugs, food supplements, agriculture chemicals, and polymers. Ever year the annual production of amino acids has increased and is currently estimated to be about 3.7 million metric tons (Ikeda and Takeno, 2013). The global market for amino acids is estimated to be about US$6.6 billion and expected to increase by 8-10% each year (Leuchtenberger et al., 2005; Ikeda and Takeno, 2013). Amino acids including L-lysine, DL-methionine, L-threonine, and L-tryptophan account for the largest share in terms of production followed by L-glutamate, L-aspartate, and L-phenylalanine (Ikeda and Takeno, 2013).
Microbial fermentation is the primary source of most of the available amino acids in the market. The L-glutamate-producing bacterium, C. glutamicum, has been the cornerstone for the introduction of fermentation in industrial manufacturing of amino acids (Udaka, 1960). This bacterium is still the most widely utilized strain for manufacturing of important amino acids, mainly L-glutamate and L-lysine (Leuchtenberger et al., 2005). Manipulation of metabolic pathways of this bacterium allowed for the an improved product range including L-phenylalanine, L-aspartate, L-tryptophan, L-arginine, L-valine, nucleic acids such as purines, vitamins such as riboflavin and pantothenic acid (Burkovski, 2008; Gopinath et al., 2012) and significant amounts of organic acids such as acetic, lactic and succinic acid (Inui et al., 2004). Figure 10 highlights the biotechnological role of C. glutamicum. As indicated, C. glutamicum utilizes different starting materials for manufacturing of amino acids and other chemical commodities.
FIGURE 10.
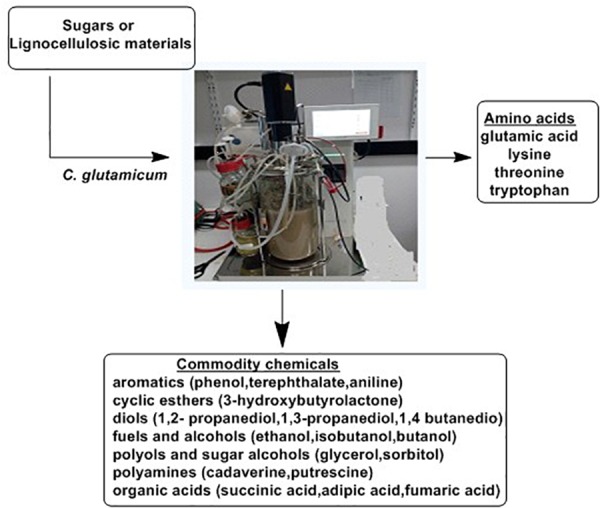
The Biotechnological potential of Corynebacterium glutamicum (Ikeda and Takeno, 2013).
Apart from the above mentioned biologically active compounds, different species of actinobacteria are involved in the synthesis of active compounds. Table 2 below summarizes some of these compounds.
Table 2.
Industrial production of bioactive molecules using whole cells as biocatalysts.
| Compound | Microorganism | Biological activity | Reference |
|---|---|---|---|
| nicotinamide | Rhodococcus rhodochrous J1 | Meyer and Ruesing, 2008 | |
| L-lysine | Corynebacterium glutamicum | Amino acids | Burkovski, 2015 |
| L-glutamate | Corynebacterium glutamicum | Amino acids | Burkovski, 2015 |
| carboxylic acids | Rhodococcus sp. MTB5 | Various application | Ismailsab et al., 2017 |
| phenylpropanoic acid | Nocardia diaphanozonaria JCM3208 | Non-steroidal anti-inflammatory drugs | Mitsukura et al., 2002 |
| aromatic dicarboxylic acids | Rhodococcus jostii RHA1 | Aromatic chemicals synthesis | Mycroft et al., 2015 |
| ammonium acrylate | Rhodococcus ruber NCIMB 40757 | Raw materials for water-soluble polymers | Webster et al., 2001 |
| L-malic Acid | Nocardia sp. | Metabolites | Hronska et al., 2015 |
| butyramide | Rhodococcus rhodochrous | Drugs | Raj et al., 2007 |
| daptomycin | Streptomyces roseosporus | Antibiotics | Boeck et al., 1988 |
| hydroxylated adamantine (1-adamantanol) | Streptomyces griseoplanus | Pharmaceutical intermediate | Mitsukura et al., 2006 |
Production of Biofuels
A recent report indicated 80% of the world’s energy is obtained from fossil fuels such as petroleum, coal, and natural gas (Birol, 2017) with huge consequences for all living systems. Therefore, the search for environmentally benign sustainable energy from renewable sources continues. Plant biomass is considered the most promising feedstock to meet the global demand for sustainable energy and chemicals (Jojima et al., 2013). Lignocellulosic biomass hydrolysates are the primary polymers of plant biomass ideal for biofuel production. Glucose and xylose are the major components of this biomass followed by minor sugars, such as arabinose, and galactose (Elander et al., 2009).
Microbial conversion of plant biomass to sugars and their transformation into a wide array of important compounds, contribute in a major way to the development of a sustainable biofuel industry (Van Hamme et al., 2003; Fulton et al., 2015). Unlike other bacteria phyla, actinobacteria are equipped with the necessary enzymes for degradation of plant biomass (Berlemont and Martiny, 2013) and form the choice of biocatalysts in the biofuel industry (Lewin et al., 2016). Several species of actinobacteria including Streptomyces, Cellulomonas, Mycobacterium, Propionibacterium, Nocardia, Corynebacterium, Rhodococcus, and Micromonospora are rich in carbohydrate-degrading enzymes (glycoside hydrolase, endo/exo glucanases, cellulases, esterases) (Lombard et al., 2013).
The above mentioned enzymes are used to produce several types of simple sugars, which are further converted into biofuels and other compounds. The widespread utilization of bioethanol from corn and sugarcane and biodiesel from plant oil suffers from several disadvantages (Atsumi et al., 2008; Atabani et al., 2012; Caspeta and Nielsen, 2013). Therefore, alternative biofuels including microbially produced specialty biofuels with similar properties to traditional fuels have increased (Atsumi et al., 2008). These speciality biofuels which encompasses higher-alcohol biofuels, fatty acid alkyl esters and various isoprenoid compounds can be used directly as an energy source or fuel precursors (Peralta-Yahya et al., 2012; Beller et al., 2015). C. glutamicum is used for the production of specialty biofuels as this bacterium inherently resists the effect of isobutanol and synthesizes several amino acids like glutamate which are important for production of branched-chain alcohols. Genetic engineering of C. glutamicum was employed to produce similar amount of isobutanol as E. coli strains, the known producers of isobutanol (Smith et al., 2010; Blombach et al., 2011; Yamamoto et al., 2013). For the past few years, R. opacus PD630 has been the center of studies as they were capable of depositing up to 80% lipid in their biomass (Alvarez et al., 2000). The substrates used were alkanes, phenylalkanes, or non-hydrocarbons as the only source of carbon (Hong et al., 2011). Moreover, production of long chain fatty acid alkyl esters (biodiesel) were observed using a Streptomyces strain isolated from sheep feces (Lu et al., 2013); bisabolene, another alternative to diesel fuel (Phelan et al., 2014) produced by Streptomyces venezuelae; also produced 1-Propanol, an industrially relevant solvent with good fuel properties from Thermobifida fusca (Deng and Fong, 2011).
In addition to direct involvement in biofuels production, actinobacteria play a significant role in detoxifying fuel-associated toxic compounds. Starting from formation to maturation, microorganisms have been in contact with crude oil in different reservoirs which contribute for their adaptation to use and modify nearly all chemical categories in crude oil (Mohamed Mel et al., 2015). 85% of fossil fuels contain polycyclic aromatic hydrocarbons (PAHs) and about 13% of these PAHs contain nitrogen, oxygen or sulfur (hetero-PAHs) (Brinkmann et al., 2014). Burning of these fuels results different pollutants such as the oxides of carbon (COx), nitrogen (NOx), and sulfur (SOx) (Ma et al., 2006). Oxides of nitrogen and sulfur combine with water vapor in cloud and result acid rain of sulfuric and nitric acids, which become part of rain and snow (Gupta et al., 2005).
Among PAHs, sulfur (hetero-PAHs) in the form of thiophenic compounds such as benzothiophene (BT), dibenzothiophene (DBT), and their alkylated homologs (Khedkar and Shanker, 2015) have attracted increasingly stringent regulations due to their harmful effects on the environment and human health (Brinkmann et al., 2014; Sousa et al., 2016). The harmful effects of PAHs, particularly hetero-PAHs well documented (Brinkmann et al., 2014) and associated with mutagenic effects, chromosome aberrations, toxicity to daphnia and green algae, embryotoxicity of the zebrafish, and respiratory system irritations.
The concentration of sulfur compounds in crude oil ranges from 0.1 to 8% (w/w) (Müller et al., 2012) and their combustion associated with toxic pollution, most countries developed legislation which demands low level of sulfur in oils, and this in turn forces companies to produce ultra-low sulfur oil, which is currently a challenge (Kawaguchi et al., 2012; Khedkar and Shanker, 2015). Traditionally, reduction of sulfur from crude oil has been achieved with hydrodesulfurization (Gupta et al., 2005), a method that depends on high energy and pressure, which mostly reduces the quality of fuel in terms of energy (Shafi and Hutchings, 2000).
Biodesulfurization depends on whole microbes or their enzymes to eliminate sulfur atom selectively from various refractory compounds present in the fossil fuels. Several strains of Rhodococcus sp. (Khairy et al., 2015), Mycobacterium sp. (Kawaguchi et al., 2012), Brevibacterium sp. (van Afferden et al., 1990),Corynebacterium sp. (Maghsoudi et al., 2001), Paenibacillus sp. (Izumi and Ohshiro, 2001), Pseudomonas sp. (Van Keulen et al., 1998), Gordonia sp. (Chang et al., 2000), and Bacillus sp. (Kirimura et al., 2001) have been studied for their ability to metabolize various polyaromatic sulfur heterocycles (PASHs) including BT and DBT (Aggarwal et al., 2013). The process adopted by this organism is known as the 4S pathway and involves four enzymes: DszA, DszB, DszC, and DszD (Wang et al., 2013; Sousa et al., 2016). DszA and DszC are monooxygenases that insert oxygen into the sulfur compounds, while DszB is a desulphinase that removes sulfur in the form of sulfite. DszD supplies FMNH2 to the two monooxygenases and is responsible for reduction of FMN to FMNH2 through NADH oxidation to NAD+. The complete reaction as indicated in Figure 11 result in a phenolic product and SO32- (Gray et al., 2003; Kawaguchi et al., 2012).
FIGURE 11.
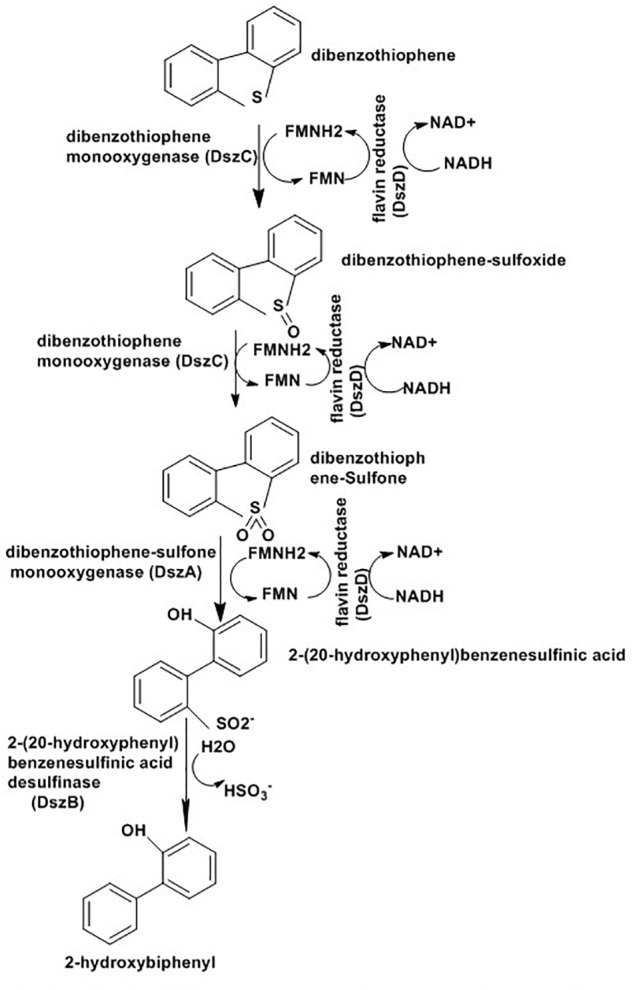
Desulfurization pathway of R. erythropolis strain IGTS8 (Gray et al., 2003).
Most desulfurization studies in the literature have used DBT as the model compound. While several rhodococci strains exhibit non-destructive desulfurization of DBT, R. erythropolis IGTS8 (Kilbane and Jackowski, 1992) was the first to be identified and has received the most attention. Even recent study highlighted their potential to desulfurization and denitrogenation of heavy gas oil by R. erythropolis ATCC 4277 (Maass et al., 2015). However, most rhodococci are unable to show high activity for the alkyl derivatives of DBT and show no activity for BT and other thiophenic compounds. various Gordonia species demonstrated greater desulfurization potential against broader range of PASHs compared to rhodococci (Alves et al., 2005). Of them, G. alkanivorans desulfurized DPT with a 4S enzyme system similar to R. erythropolis. Besides DBT, it can also specifically cleave the C–S bond in BT and other thiophenes with a reaction rate 2-10 times higher when compared to R. erythropolis (Mohebali et al., 2007; Aggarwal et al., 2013).
Microbial mechanisms also operate for degradation of N and O heterocycles. Carbazole is representative N heterocycles and bacteria such as pseudomonas sp. and Rhodococcus sp. (Maass et al., 2015) reported for mineralization of this compound. Metabolic pathways employed by most microorganisms for carbazole degradation are similar which involve ring cleavage of heterocycles to produce anthranilic acid as intermediates before their complete mineralization. In this pathway, carbazole is first degraded to 2′-aminobiphenyl-2, 3-diol by carbazole 1,9a-dioxygenase which breaks the first C–N bond. This process allow for the selective removal of refractory organonitrogen compounds from petroleum. Unlike other dioxygenases active against aromatic compound carbazole 1,9a-dioxygenase can catalyze cis-dihydroxylation, monooxygenation and angular dioxygenation on diverse aromatic compounds (Xu et al., 2006).
Dibenzofuran is representative of an O heterocyclic pollutant and microbes also developed mechanisms for biodegradation of these O heterocycles. The detail mechanisms were reviewed by Xu et al. (2006) and different monooxygenase and dioxygenases participate in angular dioxygenation, lateral dioxygenation and lateral oxygenation.
Conclusion
Unlike the traditional chemical based production of biologically active compounds, biocatalysts, particularly whole cell microbial biocatalysts, provide a cost effective and environmentally sound approach. Several candidate microorganisms displayed their ability to catalyze a range of substrates either to change them into usable compounds or to make them less toxic to the general community. Compared to other microorganisms, species in the phylum Actinobacteria are a priority for whole cell biocatalysts as this group of bacteria produce the highest percentage of biologically active compounds compared to any other and they are abundant in a range of environmental conditions. These inherent characteristics of actinobacteria encourage screening of various species in this phylum for their metabolic potential with the discovery of metabolic pathways applicable into different industries, including pharmaceuticals, food and bioenergy sectors. While most screening studies for whole cell actinobacteria as biocatalysts targeted only common species such as Rhodococcus, Streptomyces and Corynebacterium, future evaluation should consider other uncommon species such as Gordonia which has demonstrated promising bio-catalytic activities. Furthermore, most recent studies of whole cell actinobacteria as biocatalysts did not show the detailed mechanisms behind biotransformation such as what genes and enzymes are involved in the process. Future studies should focus on the investigation of the mechanisms behind the biotransformation of certain substrate to product, as this will help to manipulate each system to maximize the yield.
Author Contributions
CF provided the guidelines and wrote the outline and guided YA in the development of the review, sought a number of research papers on the topic, and did the Tables and the final proofreading. YA did most of the drafting of the manuscript and also sought relevant research papers.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
- Adams C. D., Scanlan P. A., Secrist N. D. (1994). Oxidation and biodegradability enhancement of 1,4-dioxane using hydrogen peroxide and ozone. Environ. Sci. Technol. 28 1812–1818. 10.1021/es00060a010 [DOI] [PubMed] [Google Scholar]
- Adegboye M., Babalola O. (2013). “Actinomycetes: a yet inexhaustive source of bioactive secondary metabolites,” in Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education, Microbiology Book Series, ed. Mendez-Vilas A. (Badajoz: Formatex; ),786–795. [Google Scholar]
- Aggarwal S., Karimi I., Ivan G. R. (2013). In silico modeling and evaluation of Gordonia alkanivorans for biodesulfurization. Mol. Biosyst. 9 2530–2540. 10.1039/c3mb70132h [DOI] [PubMed] [Google Scholar]
- Almeida B., Kjeldal H., Lolas I., Knudsen A. D., Carvalho G., Nielsen K. L., et al. (2013). Quantitative proteomic analysis of ibuprofen-degrading Patulibacter sp. strain I11. Biodegradation 24 615–630. 10.1007/s10532-012-9610-5 [DOI] [PubMed] [Google Scholar]
- Alvarez H., Kalscheuer R., Steinbüchel A. (2000). Accumulation and mobilization of storage lipids by Rhodococcus opacus PD630 and Rhodococcus ruber NCIMB 40126. Appl. Microbiol. Biotechnol. 54 218–223. 10.1007/s002530000395 [DOI] [PubMed] [Google Scholar]
- Alves L., Salgueiro R., Rodrigues C., Mesquita E., Matos J., Girio F. M. (2005). Desulfurization of dibenzothiophene, benzothiophene, and other thiophene analogs by a newly isolated bacterium, Gordonia alkanivorans strain 1B. Appl. Biochem. Biotechnol. 120 199–208. 10.1385/ABAB:120:3:199 [DOI] [PubMed] [Google Scholar]
- Arora P. K., Bae H. (2014). Bacterial degradation of chlorophenols and their derivatives. Microb Cell Fact. 13:31. 10.1186/1475-2859-13-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora P. K., Jain R. K. (2012). Metabolism of 2-chloro-4-nitrophenol in a Gram negative bacterium, Burkholderia sp. RKJ 800. PLoS One 7:e38676. 10.1371/journal.pone.0038676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atabani A. E., Silitonga A. S., Badruddin I. A., Mahlia T., Masjuki H., Mekhilef S. (2012). A comprehensive review on biodiesel as an alternative energy resource and its characteristics. Renew. Sustain. Energy Rev. 16 2070–2093. 10.1016/j.biortech.2014.10.030 [DOI] [PubMed] [Google Scholar]
- Atsumi S., Hanai T., Liao J. C. (2008). Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451 86–89. 10.1038/nature06450 [DOI] [PubMed] [Google Scholar]
- Babu V., Shilpi, Choudhury B. (2010). Nitrile-metabolizing potential of Amycolatopsis sp. IITR215. Process Biochem. 45 866–873. 10.1016/j.procbio.2010.02.008 [DOI] [Google Scholar]
- Banerjee A., Ghoshal A. K. (2010). Phenol degradation by Bacillus cereus: pathway and kinetic modeling. Bioresour. Technol. 101 5501–5507. 10.1016/j.biortech.2010.02.018 [DOI] [PubMed] [Google Scholar]
- Barka E. A., Vatsa P., Sanchez L., Gaveau-Vaillant N., Jacquard C., Meier-Kolthoff J. P., et al. (2015). Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 80 1–43. 10.1128/MMBR.00019-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter J., Cummings S. P. (2006). The current and future applications of microorganism in the bioremediation of cyanide contamination. Antonie Van Leeuwenhoek 90 1–17. 10.1007/s10482-006-9057-y [DOI] [PubMed] [Google Scholar]
- Beller H. R., Lee T. S., Katz L. (2015). Natural products as biofuels and bio-based chemicals: fatty acids and isoprenoids. Nat. Prod. Rep. 32 1508–1526. 10.1039/c5np00068h [DOI] [PubMed] [Google Scholar]
- Bérdy J. (2005). Bioactive microbial metabolites. J. Antibiot. 58 1–26. 10.1038/ja.2005.1 [DOI] [PubMed] [Google Scholar]
- Berlemont R., Martiny A. C. (2013). Phylogenetic distribution of potential cellulases in bacteria. Appl. Environ. Microbiol. 79 1545–1554. 10.1128/AEM.03305-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt D., Diekmann H. (1991). Degradation of dioxane, tetrahydrofuran and other cyclic ethers by an environmental Rhodococcus strain. Appl. Microbiol. Biotechnol. 36 120–123. 10.1007/BF00164711 [DOI] [PubMed] [Google Scholar]
- Bezborodov A., Zagustina N. (2016). Enzymatic biocatalysis in chemical synthesis of pharmaceuticals. Appl. Biochem. Microbiol. 52 237–249. 10.1134/S0003683816030030 [DOI] [PubMed] [Google Scholar]
- Bhalla T. C., Kumar H. (2005). Nocardia globerula NHB-2: a versatile nitrile-degrading organism. Can. J. Microbiol. 51 705–708. 10.1139/w05-046 [DOI] [PubMed] [Google Scholar]
- Birol F. (2017). Key World Energy Statistics. Paris: International Energy Agency. [Google Scholar]
- Blombach B., Riester T., Wieschalka S., Ziert C., Youn J. W., Wendisch V. F., et al. (2011). Corynebacterium glutamicum tailored for efficient isobutanol production. Appl. Environ. Microbiol. 77 3300–3310. 10.1128/aem.02972-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boadas-Vaello P., Jover E., Diez-Padrisa N., Bayona J. M., Llorens J. (2007). Differential role of CYP2E1-mediated metabolism in the lethal and vestibulotoxic effects of cis-crotononitrile in the mouse. Toxicol. Appl. Pharmacol. 225 310–317. 10.1016/j.taap.2007.07.014 [DOI] [PubMed] [Google Scholar]
- Boeck L. D., Fukuda D. S., Abbott B. J., Debono M. (1988). Deacylation of A21978C, an acidic lipopeptide antibiotic complex, by Actinoplanes utahensis. J. Antibiot. 41 1085–1092. 10.7164/antibiotics.41.1085 [DOI] [PubMed] [Google Scholar]
- Bommareddy R. R., Chen Z., Rappert S., Zeng A. P. (2014). A de novo NADPH generation pathway for improving lysine production of Corynebacterium glutamicum by rational design of the coenzyme specificity of glyceraldehyde 3-phosphate dehydrogenase. Metab. Eng. 25 30–37. 10.1016/j.ymben.2014.06.005 [DOI] [PubMed] [Google Scholar]
- Brinkmann M., Blenkle H., Salowsky H., Bluhm K., Schiwy S., Tiehm A., et al. (2014). Genotoxicity of heterocyclic PAHs in the micronucleus assay with the fish liver cell line RTL-W1. PLoS One 9:e85692. 10.1371/journal.pone.0085692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun G. L., MacDonald R. M., Verge J., Aubé J. (2008). Long-term atmospheric deposition of current-use and banned pesticides in Atlantic Canada; 1980–2000. Chemosphere 71 314–327. 10.1016/j.chemosphere.2007.09.003 [DOI] [PubMed] [Google Scholar]
- Bull A. T., Stach J. E. (2007). Marine Actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol. 15 491–499. 10.1016/j.tim.2007.10.004 [DOI] [PubMed] [Google Scholar]
- Burkovski A. (2008). Corynebacteria: Genomics and Molecular Biology. Poole: Horizon Scientific Press. [Google Scholar]
- Burkovski A. (ed.) (2015). “Trends in Corynebacterium glutamicum research and application,” in Corynebacterium glutamicum: From Systems Biology to Biotechnological Applications, (Poole: Caister Academic Press; ), 1–9. 10.21775/9781910190050.01 [DOI] [Google Scholar]
- Buschke N., Schroder H., Wittmann C. (2011). Metabolic engineering of Corynebacterium glutamicum for production of 1,5-diaminopentane from hemicellulose. Biotechnol. J. 6 306–317. 10.1002/biot.201000304 [DOI] [PubMed] [Google Scholar]
- Buser H.-R., Poiger T., Müller M. D. (1999). Occurrence and environmental behavior of the chiral pharmaceutical drug ibuprofen in surface waters and in wastewater. Environ. Sci. Technol. 33 2529–2535. 10.1021/es981014w [DOI] [PubMed] [Google Scholar]
- Caspeta L., Nielsen J. (2013). Economic and environmental impacts of microbial biodiesel. Nat. Biotechnol. 31 789–793. 10.1038/nbt.2683 [DOI] [PubMed] [Google Scholar]
- Chang J. H., Chang Y. K., Ryu H. W., Chang H. N. (2000). Desulfurization of light gas oil in immobilized-cell systems of Gordona sp. CYKS1 and Nocardia sp. CYKS2. FEMS Microbiol. Lett. 182 309–312. 10.1111/j.1574-6968.2000.tb08913.x [DOI] [PubMed] [Google Scholar]
- Chater K. F. (2016). Recent advances in understanding Streptomyces. F1000Res. 5:2795. 10.12688/f1000research.9534.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Rosazza J. P. (1994). Microbial transformation of ibuprofen by a Nocardia species. Appl. Environ. Microbiol. 60 1292–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-H., Chen L.-L., Shang N.-C. (2009). Photocatalytic degradation of dimethyl phthalate in an aqueous solution with Pt-doped TiO2-coated magnetic PMMA microspheres. J. Hazard. Mater. 172 20–29. 10.1016/j.jhazmat.2009.06.122 [DOI] [PubMed] [Google Scholar]
- Chen Z., Huang J., Wu Y., Liu D. (2016). Metabolic engineering of Corynebacterium glutamicum for the de novo production of ethylene glycol from glucose. Metab. Eng. 33 12–18. 10.1016/j.ymben.2015.10.013 [DOI] [PubMed] [Google Scholar]
- Christen P., Davidson S., Combet-Blanc Y., Auria R. (2011). Phenol biodegradation by the thermoacidophilic archaeon Sulfolobus solfataricus 98/2 in a fed-batch bioreactor. Biodegradation 22 475–484. 10.1007/s10532-010-9420-6 [DOI] [PubMed] [Google Scholar]
- Cy H., Lh F., Mh S., Cf H., Jp W., Hw K. (2018). Ibuprofen biodegradation by hospital, municipal, and distillery activated sludges. Environ. Technol. 10.1080/09593330.2018.1493146 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Dargnat C., Teil M. J., Chevreuil M., Blanchard M. (2009). Phthalate removal throughout wastewater treatment plant: case study of Marne Aval station (France). Sci. Total Environ. 407 1235–1244. 10.1016/j.scitotenv.2008.10.027 [DOI] [PubMed] [Google Scholar]
- de Carvalho C. C. (2017). Whole cell biocatalysts: essential workers from Nature to the industry. Microb. Biotechnol. 10 250–263. 10.1111/1751-7915.12363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeb R. A., Alvarez-Cohen L. (1999). Temperature effects and substrate interactions during the aerobic biotransformation of BTEX mixtures by toluene-enriched consortia and Rhodococcus rhodochrous. Biotechnol. Bioeng. 62 526–536. [DOI] [PubMed] [Google Scholar]
- Demain A. L., Sanchez S. (2009). Microbial drug discovery: 80 years of progress. J. Antibiot. 62 5–16. 10.1038/ja.2008.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Fong S. S. (2011). Metabolic engineering of Thermobifida fusca for direct aerobic bioconversion of untreated lignocellulosic biomass to 1-propanol. Metab. Eng. 13 570–577. 10.1016/j.ymben.2011.06.007 [DOI] [PubMed] [Google Scholar]
- Donova M. V., Egorova O. V. (2012). Microbial steroid transformations: current state and prospects. Appl. Microbiol. Biotechnol. 94 1423–1447. 10.1007/s00253-012-4078-0 [DOI] [PubMed] [Google Scholar]
- Dourson M., Reichard J., Nance P., Burleigh-Flayer H., Parker A., Vincent M., et al. (2014). Mode of action analysis for liver tumors from oral 1, 4-dioxane exposures and evidence-based dose response assessment. Regul. Toxicol. Pharmacol. 68 387–401. 10.1016/j.yrtph.2014.01.011 [DOI] [PubMed] [Google Scholar]
- Elander R. T., Dale B. E., Holtzapple M., Ladisch M. R., Lee Y., Mitchinson C., et al. (2009). Summary of findings from the biomass refining consortium for applied fundamentals and innovation (CAFI): corn stover pretreatment. Cellulose 16 649–659. 10.1007/s10570-009-9308-y [DOI] [Google Scholar]
- Ettireddy S., Chandupatla V., Veeresham C. (2017). Enantioselective resolution of (R,S)-Carvedilol to (S)-(-)-Carvedilol by biocatalysts. Nat. Prod. Bioprospect. 7 171–179. 10.1007/s13659-016-0118-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang C.-R., Yao J., Zheng Y.-G., Jiang C.-J., Hu L.-F., Wu Y.-Y., et al. (2010). Dibutyl phthalate degradation by Enterobacter sp. T5 isolated from municipal solid waste in landfill bioreactor. Int. Biodeterior. Biodegradation 64 442–446. 10.1016/j.ibiod.2010.04.010 [DOI] [Google Scholar]
- Fang S., An X., Liu H., Cheng Y., Hou N., Feng L., et al. (2015). Enzymatic degradation of aliphatic nitriles by Rhodococcus rhodochrous BX2, a versatile nitrile-degrading bacterium. Bioresour. Technol. 185 28–34. 10.1016/j.biortech.2015.02.078 [DOI] [PubMed] [Google Scholar]
- Feng Y. S., Lee C. M. (2009). The potential of the acetonitrile biodegradation by Mesorhizobium sp. F28. J. Hazard. Mater. 164 646–650. 10.1016/j.jhazmat.2008.08.039 [DOI] [PubMed] [Google Scholar]
- Fernandes P., Cruz A., Angelova B., Pinheiro H., Cabral J. (2003). Microbial conversion of steroid compounds: recent developments. Enzyme Microb. Technol. 32 688–705. 10.1016/S0141-0229(03)00029-2 [DOI] [Google Scholar]
- Foti P., Erba D., Riso P., Spadafranca A., Criscuoli F., Testolin G. (2005). Comparison between daidzein and genistein antioxidant activity in primary and cancer lymphocytes. Arch. Biochem. Biophys. 433 421–427. 10.1016/j.abb.2004.10.008 [DOI] [PubMed] [Google Scholar]
- Fulton L. M., Lynd L. R., Körner A., Greene N., Tonachel L. R. (2015). The need for biofuels as part of a low carbon energy future. Biofuel. Bioprod. Biorefin. 9 476–483. 10.1021/ic3008848 [DOI] [PubMed] [Google Scholar]
- Gadler P., Faber K. (2007). New enzymes for biotransformations: microbial alkyl sulfatases displaying stereo- and enantioselectivity. Trends Biotechnol. 25 83–88. 10.1016/j.tibtech.2006.11.006 [DOI] [PubMed] [Google Scholar]
- Gagyi L., Gyeresi A., Kilar F. (2008). Role of chemical structure in stereoselective recognition of beta-blockers by cyclodextrins in capillary zone electrophoresis. J. Biochem. Biophys. Methods 70 1268–1275. 10.1016/j.jbbm.2007.10.004 [DOI] [PubMed] [Google Scholar]
- Garzón-Posse F., Becerra-Figueroa L., Hernández-Arias J., Gamba-Sánchez D. (2018). Whole cells as biocatalysts in organic transformations. Molecules 23:1265. 10.3390/molecules23061265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring C., Wessel M., Schaffer S., Thum O. (2016). The power of biocatalysis: a one-pot total synthesis of rhamnolipids from butane as the sole carbon and energy source. ChemistryOpen 5 513–516. 10.1002/open.201600127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getenga Z., Dorfler U., Iwobi A., Schmid M., Schroll R. (2009). Atrazine and terbuthylazine mineralization by an Arthrobacter sp. isolated from a sugarcane-cultivated soil in Kenya. Chemosphere 77 534–539. 10.1016/j.chemosphere.2009.07.031 [DOI] [PubMed] [Google Scholar]
- Ghosh P. K., Philip L. (2004). Atrazine degradation in anaerobic environment by a mixed microbial consortium. Water Res. 38 2276–2283. 10.1016/j.watres.2003.10.059 [DOI] [PubMed] [Google Scholar]
- Gilliom R. J., Barbash J. E., Crawford C. G., Hamilton P. A., Martin J. D., Nakagaki N., et al. (2006). Pesticides in the Nation’s Streams and Ground Water, 1992-2001. Reston, VA: US Geological Survey. [Google Scholar]
- Gopinath V., Murali A., Dhar K. S., Nampoothiri K. M. (2012). Corynebacterium glutamicum as a potent biocatalyst for the bioconversion of pentose sugars to value-added products. Appl. Microbiol. Biotechnol. 93 95–106. 10.1007/s00253-011-3686-4 [DOI] [PubMed] [Google Scholar]
- Gray K. A., Mrachko G. T., Squires C. H. (2003). Biodesulfurization of fossil fuels. Curr. Opin. Microbiol. 6 229–235. 10.1016/S1369-5274(03)00065-1 [DOI] [PubMed] [Google Scholar]
- Gupta N., Roychoudhury P., Deb J. (2005). Biotechnology of desulfurization of diesel: prospects and challenges. Appl. Microbiol. Biotechnol. 66 356–366. 10.1007/s00253-004-1755-7 [DOI] [PubMed] [Google Scholar]
- Hao J., Kim C.-H. (2010). Comparing the effects of carvedilol enantiomers on regression of established cardiac hypertrophy induced by pressure overload. Lab. Anim. Res. 26 75–82. 10.5625/lar.2010.26.1.75 [DOI] [Google Scholar]
- He Y. C., Ma C. L., Zhang X., Li L., Xu J. H., Wu M. X. (2013). Highly enantioselective oxidation of racemic phenyl-1,2-ethanediol to optically pure (R)-(-)-mandelic acid by a newly isolated Brevibacterium lutescens CCZU12-1. Appl. Microbiol. Biotechnol. 97 7185–7194. 10.1007/s00253-013-4989-4 [DOI] [PubMed] [Google Scholar]
- He Y. C., Tao Z. C., Zhang D. P., Yang Z. X., Gao S., Ma C. L. (2015). Biotransformation of 1,3-propanediol cyclic sulfate and its derivatives to diols by Rhodococcus sp. Biotechnol. Lett. 37 183–188. 10.1007/s10529-014-1670-7 [DOI] [PubMed] [Google Scholar]
- He Y. C., Tao Z. C., Zhang X., Yang Z. X., Xu J. H. (2014). Highly efficient synthesis of ethyl (S)-4-chloro-3-hydroxybutanoate and its derivatives by a robust NADH-dependent reductase from E. coli CCZU-K14. Bioresour. Technol. 161 461–464. 10.1016/j.biortech.2014.03.133 [DOI] [PubMed] [Google Scholar]
- He Z., Niu C., Lu Z. (2014). Individual or synchronous biodegradation of di-n-butyl phthalate and phenol by Rhodococcus ruber strain DP-2. J. Hazard. Mater. 273 104–109. 10.1016/j.jhazmat.2014.03.033 [DOI] [PubMed] [Google Scholar]
- Hecker M., Park J. W., Murphy M. B., Jones P. D., Solomon K. R., Van Der Kraak G., et al. (2005). Effects of atrazine on CYP19 gene expression and aromatase activity in testes and on plasma sex steroid concentrations of male African clawed frogs (Xenopus laevis). Toxicol. Sci. 86 273–280. 10.1093/toxsci/kfi203 [DOI] [PubMed] [Google Scholar]
- Hollender J., Hopp J., Dott W. (1997). Degradation of 4-Chlorophenol via the meta cleavage pathway by Comamonas testosteroni JH5. Appl. Environ. Microbiol. 63 4567–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W.-K., Rairakhwada D., Seo P.-S., Park S.-Y., Hur B.-K., Kim C. H., et al. (2011). Production of lipids containing high levels of docosahexaenoic acid by a newly isolated microalga, Aurantiochytrium sp. KRS101. Appl. Biochem. Biotechnol. 164 1468–1480. 10.1007/s12010-011-9227-x [DOI] [PubMed] [Google Scholar]
- Hou J., Liu F., Wu N., Ju J., Yu B. (2016). Efficient biodegradation of chlorophenols in aqueous phase by magnetically immobilized aniline-degrading Rhodococcus rhodochrous strain. J. Nanobiotechnology 14:5. 10.1186/s12951-016-0158-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hronska H., Tokosova S., Pilnikova A., Kristofikova L., Rosenberg M. (2015). Bioconversion of fumaric acid to L-malic acid by the bacteria of the genus Nocardia. Appl. Biochem. Biotechnol. 175 266–273. 10.1007/s12010-014-1251-1 [DOI] [PubMed] [Google Scholar]
- Huang G., Li Q., Zhang X. (2003). Adsorption and desorption of atrazine by three soils. Bull. Environ. Contam. Toxicol. 71 655– Adsorption and desorption of atrazine by three soils 661. 10.1007/s00128-003-0183-1 [DOI] [PubMed] [Google Scholar]
- Igbinosa E. O., Odjadjare E. E., Chigor V. N., Igbinosa I. H., Emoghene A. O., Ekhaise F. O., et al. (2013). Toxicological profile of chlorophenols and their derivatives in the environment: the public health perspective. Sci. World J. 2013:460215. 10.1155/2013/460215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M., Takeno S. (2013). “Amino acid production by Corynebacterium glutamicum,” in Corynebacterium glutamicum. Microbiology Monographs Vol. 23 eds Yukawa H., Inui M. (Berlin: Springer; ), 107–147. [Google Scholar]
- Inoue D., Tsunoda T., Sawada K., Yamamoto N., Saito Y., Sei K., et al. (2016). 1,4-Dioxane degradation potential of members of the genera Pseudonocardia and Rhodococcus. Biodegradation 27 277–286. 10.1007/s10532-016-9772-7 [DOI] [PubMed] [Google Scholar]
- Inui M., Murakami S., Okino S., Kawaguchi H., Vertès A. A., Yukawa H. (2004). Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J. Mol. Microbiol. Biotechnol. 7 182–196. 10.1159/000079827 [DOI] [PubMed] [Google Scholar]
- Ismailsab M., Reddy P. V., Nayak A. S., Karegoudar T. B. (2017). Biotransformation of aromatic and heterocyclic amides by amidase of whole cells of Rhodococcus sp. MTB5: biocatalytic characterization and substrate specificity. Biocatal. Biotransformation 35 74–85. 10.1080/10242422.2017.1282467 [DOI] [Google Scholar]
- Izumi Y., Ohshiro T. (2001). Purification and characterization of enzymes involved in desulfurization of dibezothiophene in fossil fuels. J. Mol. Catal. B Enzym. 11 1061–1064. 10.1016/S1381-1177(00)00063-1 [DOI] [Google Scholar]
- Jablonowski N. D., Schaffer A., Burauel P. (2011). Still present after all these years: persistence plus potential toxicity raise questions about the use of atrazine. Environ. Sci. Pollut. Res. Int. 18 328–331. 10.1007/s11356-010-0431-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemli S., Ayadi-Zouari D., Hlima H. B., Bejar S. (2016). Biocatalysts: application and engineering for industrial purposes. Crit. Rev. Biotechnol. 36 246–258. 10.3109/07388551.2014.950550 [DOI] [PubMed] [Google Scholar]
- Jensen P. R., Mincer T. J., Williams P. G., Fenical W. (2005). Marine actinomycete diversity and natural product discovery. Antonie Van Leeuwenhoek 87 43–48. 10.1007/s10482-004-6540-1 [DOI] [PubMed] [Google Scholar]
- Jianlong W., Xiangchun Q., Liping H., Yi Q., Hegemann W. (2002). Microbial degradation of quinoline by immobilized cells of Burkholderia pickettii. Water Res. 36 2288–2296. 10.1016/S0043-1354(01)00457-2 [DOI] [PubMed] [Google Scholar]
- Jin D. C., Liang R. X., Dai Q. Y., Zhang R. Y., Wu X. L., Chao W. L. (2010). Biodegradation of di-n-butyl phthalate by Rhodococcus sp. JDC-11 and molecular detection of 3, 4-phthalate dioxygenase gene. J. Microbiol. Biotechnol. 20 1440–1445. 10.4014/jmb.1004.04034 [DOI] [PubMed] [Google Scholar]
- Jojima T., Inui M., Yukawa H. (2013). “Biorefinery applications of Corynebacterium glutamicum,” in Corynebacterium glutamicum. Microbiology Monographs Vol. 23 eds Yukawa H., Inui M. (Berlin: Springer; ),149–172. [Google Scholar]
- Jonsson S., Vavilin V., Svensson B. (2006). Phthalate hydrolysis under landfill conditions. Water Sci. Technol. 53 119–127. 10.2166/wst.2006.242 [DOI] [PubMed] [Google Scholar]
- Kamjam M., Sivalingam P., Deng Z., Hong K. (2017). Deep sea actinomycetes and their secondary metabolites. Front. Microbiol. 8:760 10.3389/fmicb.2017.00760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao C. M., Chen K. F., Liu J. K., Chou S. M., Chen S. C. (2006). Enzymatic degradation of nitriles by Klebsiella oxytoca. Appl. Microbiol. Biotechnol. 71 228–233. 10.1007/s00253-005-0129-0 [DOI] [PubMed] [Google Scholar]
- Kato D.-I., Mitsuda S., Ohta H. (2003). Microbial deracemization of α-substituted carboxylic acids: substrate specificity and mechanistic investigation. J. Org. Chem. 68 7234–7242. 10.1021/jo034253x [DOI] [PubMed] [Google Scholar]
- Kawaguchi H., Kobayashi H., Sato K. (2012). Metabolic engineering of hydrophobic Rhodococcus opacus for biodesulfurization in oil-water biphasic reaction mixtures. J. Biosci. Bioeng. 113 360–366. 10.1016/j.jbiosc.2011.10.017 [DOI] [PubMed] [Google Scholar]
- Khairy H., Wubbeler J. H., Steinbuchel A. (2015). Biodegradation of the organic disulfide 4,4′-dithiodibutyric acid by Rhodococcus spp. Appl. Environ. Microbiol. 81 8294–8306. 10.1128/aem.02059-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khedkar S., Shanker R. (2015). Isolation and classification of a soil actinomycete capable of sulphur-specific biotransformation of dibenzothiophene, benzothiophene and thianthrene. J. Appl. Microbiol. 118 62–74. 10.1111/jam.12665 [DOI] [PubMed] [Google Scholar]
- Kilbane J. J., Jackowski K. (1992). Biodesulfurization of water-soluble coal-derived material by Rhodococcus rhodochrous IGTS8. Biotechnol. Bioeng. 40 1107–1114. 10.1002/bit.260400915 [DOI] [PubMed] [Google Scholar]
- Kim S.-K., Toldrá F., Prihanto A., Wakayama M., Trivedi N., Reddy C., et al. (2016). Marine Enzymes Biotechnology: Production and Industrial Applications, Part II-Marine Organisms Producing Enzymes. New York, NY: Elsevier. [Google Scholar]
- Kirimura K., Furuya T., Nishii Y., Ishii Y., Kino K., Usami S. (2001). Biodesulfurization of dibenzothiophene and its derivatives through the selective cleavage of carbon-sulfur bonds by a moderately thermophilic bacterium Bacillus subtilis WU-S2B. J. Biosci. Bioeng. 91 262–266. 10.1016/S1389-1723(01)80131-6 [DOI] [PubMed] [Google Scholar]
- Kishimoto N., Nakagawa T., Asano M., Abe M., Yamada M., Ono Y. (2008). Ozonation combined with electrolysis of 1,4-dioxane using a two-compartment electrolytic flow cell with solid electrolyte. Water Res. 42 379–385. 10.1016/j.watres.2007.07.029 [DOI] [PubMed] [Google Scholar]
- Kolekar P. D., Phugare S. S., Jadhav J. P. (2014). Biodegradation of atrazine by Rhodococcus sp. BCH2 to N-isopropylammelide with subsequent assessment of toxicity of biodegraded metabolites. Environ. Sci. Pollut. Res. Int. 21 2334–2345. 10.1007/s11356-013-2151-6 [DOI] [PubMed] [Google Scholar]
- Kumaresan C. (2010). Ibuprofen (Dexibuprofen): the superior non-steroidal anti-inflammatory agents for development of pharmaceuticals. Int. J. Curr. Pharm. Res. 2 1–3. [Google Scholar]
- Ladkau N., Schmid A., Bühler B. (2014). The microbial cell—functional unit for energy dependent multistep biocatalysis. Curr. Opin. Biotechnol. 30 178–189. 10.1016/j.copbio.2014.06.003 [DOI] [PubMed] [Google Scholar]
- Lam K. S. (2007). New aspects of natural products in drug discovery. Trends Microbiol. 15 279–289. 10.1016/j.tim.2007.04.001 [DOI] [PubMed] [Google Scholar]
- Lau T., Chu W., Graham N. (2005). The degradation of endocrine disruptor di-n-butyl phthalate by UV irradiation: a photolysis and product study. Chemosphere 60 1045–1053. 10.1016/j.chemosphere.2005.01.022 [DOI] [PubMed] [Google Scholar]
- Leuchtenberger W., Huthmacher K., Drauz K. (2005). Biotechnological production of amino acids and derivatives: current status and prospects. Appl. Microbiol. Biotechnol. 69 1–8. 10.1007/s00253-005-0155-y [DOI] [PubMed] [Google Scholar]
- Lewin G. R., Carlos C., Chevrette M. G., Horn H. A., McDonald B. R., Stankey R. J., et al. (2016). Evolution and ecology of actinobacteria and their bioenergy applications. Annu. Rev. Microbiol. 70 235–254. 10.1146/annurev-micro-102215-095748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Hu Y., Shen X., Dai X., Han X. (2010). Combined effects of two environmental endocrine disruptors nonyl phenol and di-n-butyl phthalate on rat Sertoli cells in vitro. Reprod. Toxicol. 30 438–445. 10.1016/j.reprotox.2010.06.003 [DOI] [PubMed] [Google Scholar]
- Lievano R., Perez H. I., Manjarrez N., Solis A., Solis-Oba M. (2012). Hydrolysis of ibuprofen nitrile and ibuprofen amide and deracemisation of ibuprofen using Nocardia corallina B-276. Molecules 17 3148–3154. 10.3390/molecules17033148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B., Tao Y. (2017). Whole-cell biocatalysts by design. Microb Cell Fact. 16 106–106. 10.1186/s12934-017-0724-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B.-X., Zhang Z.-J., Liu W.-F., Dong Z.-Y., Tao Y. (2013). Enhanced production of N-acetyl-D-neuraminic acid by multi-approach whole-cell biocatalyst. Appl. Microbiol. Biotechnol. 97 4775–4784. 10.1007/s00253-013-4754-8 [DOI] [PubMed] [Google Scholar]
- Litsanov B., Brocker M., Bott M. (2012). Toward homosuccinate fermentation: metabolic engineering of Corynebacterium glutamicum for anaerobic production of succinate from glucose and formate. Appl. Environ. Microbiol. 78 3325–3337. 10.1128/AEM.07790-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Parales R. E. (2009). Bacterial chemotaxis to atrazine and related s-triazines. Appl. Environ. Microbiol. 75 5481–5488. 10.1128/aem.01030-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wang F., Tan T. (2009). Cyclic resolution of racemic ibuprofen via coupled efficient lipase and acid-base catalysis. Chirality 21 349–353. 10.1002/chir.20578 [DOI] [PubMed] [Google Scholar]
- Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., Henrissat B. (2013). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42 D490–D495. 10.1093/nar/gkt1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi P. (2002). Exemestane, a new steroidal aromatase inhibitor of clinical relevance. Biochim. Biophys. Acta 1587 326–337. 10.1016/S0925-4439(02)00096-0 [DOI] [PubMed] [Google Scholar]
- Lottrup G., Andersson A. M., Leffers H., Mortensen G. K., Toppari J., Skakkebaek N., et al. (2006). Possible impact of phthalates on infant reproductive health. Int. J. Androl. 29 172–180. 10.1111/j.1365-2605.2005.00642.x [DOI] [PubMed] [Google Scholar]
- Lu Y., Tang F., Wang Y., Zhao J., Zeng X., Luo Q., et al. (2009). Biodegradation of dimethyl phthalate, diethyl phthalate and di-n-butyl phthalate by Rhodococcus sp. L4 isolated from activated sludge. J. Hazard. Mater. 168 938–943. 10.1016/j.jhazmat.2009.02.126 [DOI] [PubMed] [Google Scholar]
- Lu Y., Wang J., Deng Z., Wu H., Deng Q., Tan H., et al. (2013). Isolation and characterization of fatty acid methyl ester (FAME)-producing Streptomyces sp. S 161 from sheep (Ovis aries) faeces. Lett. Appl. Microbiol. 57 200–205. 10.1111/lam.12096 [DOI] [PubMed] [Google Scholar]
- Ma T., Li G., Li J., Liang F., Liu R. (2006). Desulfurization of dibenzothiophene by Bacillus subtilis recombinants carrying dszABC and dszD genes. Biotechnol. Lett. 28 1095–1100. 10.1007/s10529-006-9056-0 [DOI] [PubMed] [Google Scholar]
- Maass D., Todescato D., Moritz D. E., Oliveira J. V., Oliveira D., Ulson de Souza A. A., et al. (2015). Desulfurization and denitrogenation of heavy gas oil by Rhodococcus erythropolis ATCC 4277. Bioprocess Biosyst. Eng. 38 1447–1453. 10.1007/s00449-015-1386-7 [DOI] [PubMed] [Google Scholar]
- Maghsoudi S., Vossoughi M., Kheirolomoom A., Tanaka E., Katoh S. (2001). Biodesulfurization of hydrocarbons and diesel fuels by Rhodococcus sp. strain P32C1. Biochem. Eng. J. 8 151–156. 10.1016/S1369-703X(01)00097-3 [DOI] [Google Scholar]
- Mandelbaum R. T., Allan D. L., Wackett L. P. (1995). Isolation and characterization of a Pseudomonas sp. that mineralizes the s-triazine herbicide atrazine. Appl. Environ. Microbiol. 61 1451–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui R., Takagi K., Sakakibara F., Abe T., Shiiba K. (2016). Identification and characterization of 1,4-dioxane-degrading microbe separated from surface seawater by the seawater-charcoal perfusion apparatus. Biodegradation 27 155–163. 10.1007/s10532-016-9763-8 [DOI] [PubMed] [Google Scholar]
- Matsumura E., Sakai M., Hayashi K., Murakami S., Takenaka S., Aoki K. (2006). Constitutive expression of catABC genes in the aniline-assimilating bacterium Rhodococcus species AN-22: production, purification, characterization and gene analysis of CatA, CatB and CatC. Biochem. J. 393(Pt 1), 219–226. 10.1042/bj20050740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattam A. J., Clomburg J. M., Gonzalez R., Yazdani S. S. (2013). Fermentation of glycerol and production of valuable chemical and biofuel molecules. Biotechnol. Lett. 35 831–842. 10.1007/s10529-013-1240-4 [DOI] [PubMed] [Google Scholar]
- Meyer H., Ruesing M. (2008). “Lonza-Examples of bioprocesses for the production of nutraceuticals,” in Paper Presented at the World Congress on Industrial Biotechnology & Bioprocessing, Chicago, IL. [Google Scholar]
- Mitsukura K., Kondo Y., Yoshida T., Nagasawa T. (2006). Regioselective hydroxylation of adamantane by Streptomyces griseoplanus cells. Appl. Microbiol. Biotechnol. 71 502–504. 10.1007/s00253-005-0167-7 [DOI] [PubMed] [Google Scholar]
- Mitsukura K., Suzuki M., Tada K., Yoshida T., Nagasawa T. (2010). Asymmetric synthesis of chiral cyclic amine from cyclic imine by bacterial whole-cell catalyst of enantioselective imine reductase. Org. Biomol. Chem. 8 4533–4535. 10.1039/c0ob00353k [DOI] [PubMed] [Google Scholar]
- Mitsukura K., Yoshida T., Nagasawa T. (2002). Synthesis of (R)-2-phenylpropanoic acid from its racemate through an isomerase-involving reaction by Nocardia diaphanozonaria. Biotechnol. Lett. 24 1615–1621. 10.1023/A:1020353631566 [DOI] [Google Scholar]
- Mohamed Mel S., Al-Yacoub Z. H., Vedakumar J. V. (2015). Biocatalytic desulfurization of thiophenic compounds and crude oil by newly isolated bacteria. Front. Microbiol. 6:112. 10.3389/fmicb.2015.00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohebali G., Ball A. S., Rasekh B., Kaytash A. (2007). Biodesulfurization potential of a newly isolated bacterium, RIPI90A. Enzyme Microb. Technol. 40 578–584. 10.1016/j.enzmictec.2006.05.012 [DOI] [Google Scholar]
- Mukhtar S., Zaheer A., Aiysha D., Malik K., Mehnaz S. (2017). Actinomycetes: a source of industrially important enzymes. J. Proteomics Bioinform. 10 316–319. 10.4172/jpb.1000456 [DOI] [Google Scholar]
- Müller A. L. H., Picoloto R. S., de Azevedo Mello P., Ferrão M. F., dos Santos M. D. F. P., Guimarães R. C. L., et al. (2012). Total sulfur determination in residues of crude oil distillation using FT-IR/ATR and variable selection methods. Spectrochim. Acta A Mol. Biomol. Spectrosc. 89 82–87. 10.1016/j.saa.2011.12.001 [DOI] [PubMed] [Google Scholar]
- Murohisa T., Iida M. (1993). Some new intermediates in microbial side chain degradation of β-sitosterol. J. Ferment. Bioeng. 76 174–177. 10.1016/0922-338X(93)90003-Q [DOI] [Google Scholar]
- Mycroft Z., Gomis M., Mines P., Law P., Bugg T. D. (2015). Biocatalytic conversion of lignin to aromatic dicarboxylic acids in Rhodococcus jostii RHA1 by re-routing aromatic degradation pathways. Green Chem. 17 4974–4979. 10.1039/C5GC01347J [DOI] [Google Scholar]
- Neumann G., Teras R., Monson L., Kivisaar M., Schauer F., Heipieper H. J. (2004). Simultaneous degradation of atrazine and phenol by Pseudomonas sp. strain ADP: effects of toxicity and adaptation. Appl. Environ. Microbiol. 70 1907–1912. 10.1128/AEM.70.4.1907-1912.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigam V. K., Khandelwal A. K., Gothwal R. K., Mohan M. K., Choudhury B., Vidyarthi A. S., et al. (2009). Nitrilase-catalysed conversion of acrylonitrile by free and immobilized cells of Streptomyces sp. J. Biosci. 34 21–26. 10.1007/s12038-009-0005-7 [DOI] [PubMed] [Google Scholar]
- Nordin K., Unell M., Jansson J. K. (2005). Novel 4-chlorophenol degradation gene cluster and degradation route via hydroxyquinol in Arthrobacter chlorophenolicus A6. Appl. Environ. Microbiol. 71 6538–6544. 10.1128/AEM.71.11.6538-6544.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaniran A. O., Igbinosa E. O. (2011). Chlorophenols and other related derivatives of environmental concern: properties, distribution and microbial degradation processes. Chemosphere 83 1297–1306. 10.1016/j.chemosphere.2011.04.009 [DOI] [PubMed] [Google Scholar]
- Oyama M., Kakinoki T., Inoue D., Michihiko I. (2013). Isolation and characterization of tetrahydrofuran-degrading bacteria for 1, 4-dioxane-containing wastewater treatment by co-metabolic degradation. J. Water Environ. Technol. 11 11–19. 10.2965/jwet.2013.11 [DOI] [Google Scholar]
- Pandey J., Heipieper H. J., Chauhan A., Arora P. K., Prakash D., Takeo M., et al. (2011). Reductive dehalogenation mediated initiation of aerobic degradation of 2-chloro-4-nitrophenol (2C4NP) by Burkholderia sp. strain SJ98. Appl. Microbiol. Biotechnol. 92 597–607. 10.1007/s00253-011-3254-y [DOI] [PubMed] [Google Scholar]
- Parales R. E., Adamus J. E., White N., May H. D. (1994). Degradation of 1,4-dioxane by an actinomycete in pure culture. Appl. Environ. Microbiol. 60 4527–4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta-Yahya P. P., Zhang F., Del Cardayre S. B., Keasling J. D. (2012). Microbial engineering for the production of advanced biofuels. Nature 488 320–328. 10.1038/nature11478 [DOI] [PubMed] [Google Scholar]
- Peters-Wendisch P., Stolz M., Etterich H., Kennerknecht N., Sahm H., Eggeling L. (2005). Metabolic engineering of Corynebacterium glutamicum for L-serine production. Appl. Environ. Microbiol. 71 7139–7144. 10.1128/AEM.71.11.7139-7144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan R. M., Sekurova O. N., Keasling J. D., Zotchev S. B. (2014). Engineering terpene biosynthesis in Streptomyces for production of the advanced biofuel precursor bisabolene. ACS Synth. Biol. 4 393–399. 10.1021/sb5002517 [DOI] [PubMed] [Google Scholar]
- Pignatello R., Panto V., Salmaso S., Bersani S., Pistara V., Kepe V., et al. (2008). Flurbiprofen derivatives in Alzheimer’s disease: synthesis, pharmacokinetic and biological assessment of lipoamino acid prodrugs. Bioconjug. Chem. 19 349–357. 10.1021/bc700312y [DOI] [PubMed] [Google Scholar]
- Pimentel-Elardo S. M., Kozytska S., Bugni T. S., Ireland C. M., Moll H., Hentschel U. (2010). Anti-parasitic compounds from Streptomyces sp. strains isolated from Mediterranean sponges. Mar. Drugs 8 373–380. 10.3390/md8020373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash D., Nawani N., Prakash M., Bodas M., Mandal A., Khetmalas M., et al. (2013). Actinomycetes: a repertory of green catalysts with a potential revenue resource. Biomed Res. Int. 2013:264020. 10.1155/2013/264020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadharsini P., Dhanasekaran D. (2015). Diversity of soil allelopathic Actinobacteria in Tiruchirappalli district, Tamilnadu, India. J. Saudi Soc. Agric. Sci. 14 54–60. 10.1016/j.jssas.2013.07.001 [DOI] [Google Scholar]
- Raj J., Seth A., Prasad S., Bhalla T. C. (2007). Bioconversion of butyronitrile to butyramide using whole cells of Rhodococcus rhodochrous PA-34. Appl. Microbiol. Biotechnol. 74 535–539. 10.1007/s00253-006-0693-y [DOI] [PubMed] [Google Scholar]
- Ralebits T. K., Senior E., van Verseveld H. W. (2002). Microbial aspects of atrazine degradation in natural environments. Biodegradation 13 11–19. 10.1023/A:1016329628618 [DOI] [PubMed] [Google Scholar]
- Ramteke P. W., Maurice N. G., Joseph B., Wadher B. J. (2013). Nitrile-converting enzymes: an eco-friendly tool for industrial biocatalysis. Biotechnol. Appl. Biochem. 60 459–481. 10.1002/bab.1139 [DOI] [PubMed] [Google Scholar]
- Roh C., Seo S. H., Choi K. Y., Cha M., Pandey B. P., Kim J. H., et al. (2009). Regioselective hydroxylation of isoflavones by Streptomyces avermitilis MA-4680. J. Biosci. Bioeng. 108 41–46. 10.1016/j.jbiosc.2009.02.021 [DOI] [PubMed] [Google Scholar]
- Rohr J. R., McCoy K. A. (2010). A qualitative meta-analysis reveals consistent effects of atrazine on freshwater fish and amphibians. Environmen. Health Perspect. 118 20–32. 10.1289/ehp.0901164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseaux S., Hartmann A., Soulas G. (2001). Isolation and characterisation of new Gram-negative and Gram-positive atrazine degrading bacteria from different French soils. FEMS Microbiol. Ecol. 36 211–222. 10.1111/j.1574-6941.2001.tb00842.x [DOI] [PubMed] [Google Scholar]
- Saa L., Jaureguibeitia A., Largo E., Llama M. J., Serra J. L. (2010). Cloning, purification and characterization of two components of phenol hydroxylase from Rhodococcus erythropolis UPV-1. Appl. Microbiol. Biotechnol. 86 201–211. 10.1007/s00253-009-2251-x [DOI] [PubMed] [Google Scholar]
- Santhi V. A., Mustafa A. M. (2013). Assessment of organochlorine pesticides and plasticisers in the Selangor River basin and possible pollution sources. Environ. Monit. Assess. 185 1541–1554. 10.1007/s10661-012-2649-2 [DOI] [PubMed] [Google Scholar]
- Santoshkumar M., Veeranagouda Y., Lee K., Karegoudar T. B. (2011). Utilization of aliphatic nitrile by Paracoccus sp SKG isolated from chemical waste samples. Int. Biodeterior. Biodegradation 65 153–159. 10.1016/j.ibiod.2010.10.008 [DOI] [Google Scholar]
- Sen S. E., Anliker K. S. (1996). 1H NMR analysis of R/S ibuprofen by the formation of diasteriomeric pairs: microscale stereochemistry experiment for the undergraduate organic laboratory. J. Chem. Educ. 73:569 10.1021/ed073p569 [DOI] [Google Scholar]
- Sgobba E., Blöbaum L., Wendisch V. F. (2018). Production of food and feed additives from non-food-competing feedstocks: valorizing N-acetylmuramic acid for amino acid and carotenoid fermentation with Corynebacterium glutamicum. Front. Microbiol. 9:2046. 10.3389/fmicb.2018.02046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafi R., Hutchings G. J. (2000). Hydrodesulfurization of hindered dibenzothiophenes: an overview. Catal. Today 59 423–442. 10.1016/S0920-5861(00)00308-4 [DOI] [Google Scholar]
- Smith K. M., Cho K.-M., Liao J. C. (2010). Engineering Corynebacterium glutamicum for isobutanol production. Appl. Microbiol. Biotechnol. 87 1045–1055. 10.1007/s00253-010-2522-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell D., Colby J. (1999). Enantioselective hydrolysis of racemic ibuprofen amide to s-(+)-ibuprofen by Rhodococcus AJ270. Enzyme Microb. Technol. 24 160–163. 10.1016/S0141-0229(98)00097-0 [DOI] [Google Scholar]
- Solecka J., Zajko J., Postek M., Rajnisz A. (2012). Biologically active secondary metabolites from Actinomycetes. Open Life Sci. 7 373–390. 10.2478/s11535-012-0036-1 [DOI] [Google Scholar]
- Solomon K. R., Carr J. A., Du Preez L. H., Giesy J. P., Kendall R. J., Smith E. E., et al. (2008). Effects of atrazine on fish, amphibians, and aquatic reptiles: a critical review. Crit. Rev. Toxicol. 38 721–772. 10.1080/10408440802116496 [DOI] [PubMed] [Google Scholar]
- Solyanikova I., Golovleva L. (2011). Biochemical features of the degradation of pollutants by Rhodococcus as a basis for contaminated wastewater and soil cleanup. Microbiology 80 579–594. 10.1134/S0026261711050158 [DOI] [PubMed] [Google Scholar]
- Sousa S. F., Sousa J. F., Barbosa A. C., Ferreira C. E., Neves R. P., Ribeiro A. J., et al. (2016). Improving the biodesulfurization of crude oil and derivatives: a qm/mm investigation of the catalytic mechanism of NADH-FMN oxidoreductase (DszD). J. Phys. Chem. A 120 5300–5306. 10.1021/acs.jpca.6b01536 [DOI] [PubMed] [Google Scholar]
- Sripalakit P., Wichai U., Saraphanchotiwitthaya A. (2006). Biotransformation of various natural sterols to androstenones by Mycobacterium sp. and some steroid-converting microbial strains. J. Mol. Catal. B Enzym. 41 49–54. 10.1016/j.molcatb.2006.04.007 [DOI] [Google Scholar]
- Steinmann J. G., Phillips J. H., Sanders W. J., Kiessling L. L. (2001). Synthesis of cyclic sulfates by halocyclization. Org. Lett. 3 3557–3559. 10.1021/ol016674b [DOI] [PubMed] [Google Scholar]
- Stepien D. K., Diehl P., Helm J., Thoms A., Puttmann W. (2014). Fate of 1,4-dioxane in the aquatic environment: from sewage to drinking water. Water Res. 48 406–419. 10.1016/j.watres.2013.09.057 [DOI] [PubMed] [Google Scholar]
- Szentirmai A. (1990). Microbial physiology of sidechain degradation of sterols. J. Ind. Microbiol. 6 101–115. 10.1007/BF01576429 [DOI] [Google Scholar]
- Tappe W., Groeneweg J., Jantsch B. (2002). Diffuse atrazine pollution in German aquifers. Biodegradation 13 3–10. 10.1023/A:1016325527709 [DOI] [PubMed] [Google Scholar]
- Topp E., Mulbry W. M., Zhu H., Nour S. M., Cuppels D. (2000). Characterization of s-triazine herbicide metabolism by a Nocardioides sp. isolated from agricultural soils. Appl. Environ. Microbiol. 66 3134–3141. 10.1128/AEM.66.8.3134-3141.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trung T. Q., Kim J. M., Kim K. H. (2006). Preparative method of R-(-)-lbuprofen by diastereomer crystallization. Arch. Pharm. Res. 29 108–111. 10.1007/BF02977477 [DOI] [PubMed] [Google Scholar]
- Udaka S. (1960). Screening method for microorganisms accumulating metabolites and its use in the isolation of Micrococcus glutamicus. J. Bacteriol. 79 754–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich E. M., Morrison C. N., Goldsmith M. R., Foreman W. T. (2012). Chiral pesticides: identification, description, and environmental implications. Rev. Environ. Contam. Toxicol. 217 1–74. 10.1007/978-1-4614-2329-4_1 [DOI] [PubMed] [Google Scholar]
- van Afferden M., Schacht S., Klein J., Trüper H. G. (1990). Degradation of dibenzothiophene by Brevibacterium sp. DO. Arch. Microbiol. 153 324–328. 10.1007/BF00249000 [DOI] [Google Scholar]
- Van Hamme J. D., Singh A., Ward O. P. (2003). Recent advances in petroleum microbiology. Microbiol. Mol. Biol. Rev. 67 503–549. 10.1128/MMBR.67.4.503-549.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Keulen F., Correia C., Da Fonseca M. (1998). Solvent selection for the biotransformation of terpenes by Pseudomonas putida. J. Mol. Catal. B Enzym. 5 295–299. 10.1016/S1381-1177(98)00054-X [DOI] [Google Scholar]
- Vesela A. B., Pelantova H., Sulc M., Mackova M., Lovecka P., Thimova M., et al. (2012). Biotransformation of benzonitrile herbicides via the nitrile hydratase-amidase pathway in rhodococci. J. Ind. Microbiol. Biotechnol. 39 1811–1819. 10.1007/s10295-012-1184-z [DOI] [PubMed] [Google Scholar]
- Wadhwa L., Smith K. E. (2000). Progesterone side-chain cleavage by Bacillus sphaericus. FEMS Microbiol. Lett. 192 179–183. 10.1111/j.1574-6968.2000.tb09379.x [DOI] [PubMed] [Google Scholar]
- Wang W., Ma T., Lian K., Zhang Y., Tian H., Ji K., et al. (2013). Genetic analysis of benzothiophene biodesulfurization pathway of Gordonia terrae strain C-6. PLoS One 8:e84386. 10.1371/journal.pone.0084386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward T. R., Köhler V. (2015). Editorial overview: biocatalysis and biotransformation: bio-inspired, bio-based and bio-linked catalysis. Curr. Opin. Chem. Biol. 25 v–vi. 10.1016/j.cbpa.2015.02.022 [DOI] [PubMed] [Google Scholar]
- Webster N. A., Ramsden D. K., Hughes J. (2001). Comparative characterisation of two Rhodococcus species as potential biocatalysts for ammonium acrylate production. Biotechnol. Lett. 23 95–101. 10.1023/A:1010389115549 [DOI] [Google Scholar]
- Whited G. M., Feher F. J., Benko D. A., Cervin M. A., Chotani G. K., McAuliffe J. C., et al. (2010). Technology update: development of a gas-phase bioprocess for isoprene-monomer production using metabolic pathway engineering. Ind. Biotechnol. 6 152–163. 10.1089/ind.2010.6.152 [DOI] [Google Scholar]
- Wu X., Wang Y., Liang R., Dai Q., Jin D., Chao W. (2011). Biodegradation of an endocrine-disrupting chemical di-n-butyl phthalate by newly isolated Agrobacterium sp. and the biochemical pathway. Process Biochem. 46 1090–1094. 10.1016/j.procbio.2011.01.031 [DOI] [Google Scholar]
- Xu P., Yu B., Li F. L., Cai X. F., Ma C. Q. (2006). Microbial degradation of sulfur, nitrogen and oxygen heterocycles. Trends Microbiol. 14 398–405. 10.1016/j.tim.2006.07.002 [DOI] [PubMed] [Google Scholar]
- Xun L., Topp E., Orser C. (1992). Purification and characterization of a tetrachloro-p-hydroquinone reductive dehalogenase from a Flavobacterium sp. J. Bacteriol. 174 8003–8007. 10.1128/jb.174.24.8003-8007.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Suda M., Niimi S., Inui M., Yukawa H. (2013). Strain optimization for efficient isobutanol production using Corynebacterium glutamicum under oxygen deprivation. Biotechnol. Bioeng. 110 2938–2948. 10.1002/bit.24961 [DOI] [PubMed] [Google Scholar]
- Yu L. L., Li Z. Y., Peng C. S., Li Z. Y., Guo Y. W. (2009). Neobacillamide A, a novel thiazole-containing alkaloid from the marine bacterium Bacillus vallismortis C89, associated with South China Sea sponge Dysidea avara. Helv. Chim. Acta 92 607–612. 10.1002/hlca.200800349 [DOI] [Google Scholar]
- Zahoor A., Lindner S. N., Wendisch V. F. (2012). Metabolic engineering of Corynebacterium glutamicum aimed at alternative carbon sources and new products. Comput. Struct. Biotechnol. J. 3:e201210004. 10.5936/csbj.201210004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Yu Y. (2013). Dehydration of ethanol to ethylene. Ind. Eng. Chem. Res. 52 9505–9514. 10.1021/ie401157c [DOI] [Google Scholar]
- Zheng Y. G., Chen J., Liu Z. Q., Wu M. H., Xing L. Y., Shen Y. C. (2008). Isolation, identification and characterization of Bacillus subtilis ZJB-063, a versatile nitrile-converting bacterium. Appl. Microbiol. Biotechnol. 77 985–993. 10.1007/s00253-007-1236-x [DOI] [PubMed] [Google Scholar]