Abstract
Combined with chemical synthesis, the use of glycoenzyme biocatalysts has shown great synthetic potential over recent decades owing to their remarkable versatility in terms of substrates and regio- and stereoselectivity that allow structurally controlled synthesis of carbohydrates and glycoconjugates. Nonetheless, the lack of appropriate enzymatic tools with requisite properties in the natural diversity has hampered extensive exploration of enzyme-based synthetic routes to access relevant bioactive oligosaccharides, such as cell-surface glycans or prebiotics. With the remarkable progress in enzyme engineering, it has become possible to improve catalytic efficiency and physico-chemical properties of enzymes but also considerably extend the repertoire of accessible catalytic reactions and tailor novel substrate specificities. In this review, we intend to give a brief overview of the advantageous use of engineered glycoenzymes, sometimes in combination with chemical steps, for the synthesis of natural bioactive oligosaccharides or their precursors. The focus will be on examples resulting from the three main classes of glycoenzymes specialized in carbohydrate synthesis: glycosyltransferases, glycoside hydrolases and glycoside phosphorylases.
Keywords: carbohydrate synthesis, enzyme engineering, chemo-enzymatic synthesis, glycans, bioactive oligosaccharides
1. Introduction
1.1. Carbohydrate chemical synthesis: limits and challenges
Carbohydrates are known to play major and complex roles in many biological systems and recognition processes [1]. As such, they have emerged as invaluable tools to probe carbohydrate–protein recognition and uncover novel therapeutic targets. Carbohydrates also have an increasing number of applications in the food or feed industries as prebiotic compounds able to stimulate the growth of beneficial bacteria in human and animal microflora [2–4]. Development of synthetic routes to access these carbohydrate structures is thus in high demand. However, in spite of many great advances in the field, conventional organic chemical synthesis of carbohydrates, especially oligosaccharides, remains highly challenging owing to their tremendous structural diversity and complexity in terms of constituting monosaccharides with numerous reactive hydroxyl groups and glycosidic linkages. Despite the development of novel methods and tremendous improvements in selectivities and yields (thoroughly reviewed in [5,6]), chemical methods still suffer from the challenge of protecting group manipulations of the precursors necessary to obtain a regioselective glycosylation, and the harsh methods, the use of heavy metal catalysts and often the requirement for intermediary purification steps to separate stereoisomers. To circumvent some of these limitations, chemical synthesis is being more and more frequently combined with (or sometimes replaced by) the use of highly selective enzymatic catalysts when advantageous for the desired synthetic transformation. Both chemical and enzymatic approaches used in combination should enable current synthetic bottlenecks to be overcome and convergent chemo-enzymatic synthetic routes to be provided to access carbohydrates more easily. In many cases though, chemical synthesis remains invaluable for either the synthesis of building blocks that are further used for enzymatic glycosylation or for the incorporation of rare monosaccharides for which no enzymatic transfer has been described.
1.2. Natural glycoenzyme catalysts for carbohydrate synthesis
Biosynthesis of carbohydrates is usually ensured by a combination of enzymes, mostly glycosyltransferases (GTs), glycoside hydrolases (GHs) or transglycosylases (TGs), and more rarely glycoside phosphorylases (GPs). These enzymes have been evolved by Nature to endow them with a wide array of substrate specificities and, in some cases, with improved stability in harsh environments [7,8]. Utilization of these enzyme catalysts has provided over recent decades an alternative to chemical synthesis in order to overcome some difficulties encountered in glycochemistry by taking advantage of their high stereo- and regioselectivities for non-protected substrates, and action in mild, aqueous conditions. However, utilization of natural enzymes is often hampered by a difficult recombinant production and the availability of substrates (in particular nucleotide-activated donors). To avoid these problems, in vivo production of carbohydrates, including the use of metabolically engineered whole cells, has been considered but has remained limited so far to the production of a few biologically relevant carbohydrate structures [9–11].
1.3. Contribution of enzyme engineering to the development of novel synthetic tools
In spite of the increasing number of glycoenzyme sequences identified from –omics technologies and the progress of automated annotation [12–15], the availability of enzymes with the requisite substrate specificity remains critical to performing highly specific glycosylations. With the remarkable progress of in vitro enzyme evolution, especially supported by in silico methods including computer-aided design [16,17], it has been possible to extend the repertoire of accessible reactions and improve catalytic efficiency and physico-chemical properties of enzymes.
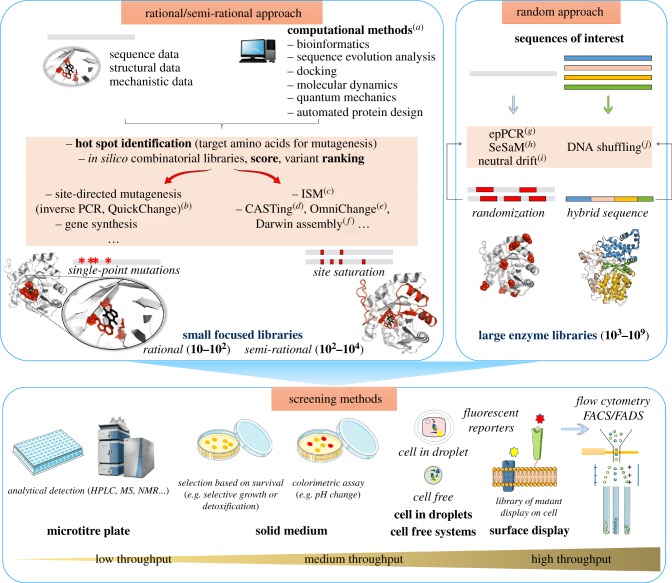
For a long time, engineering strategies to tune up enzyme properties have relied on site-directed mutagenesis [18] guided by various knowledge-based analyses derived from available sequences or three-dimensional structures to improve mostly catalytic activity or to alter substrate specificity. Directed evolution technologies (reviewed in [19]), mimicking in accelerated the natural evolution of enzymes by combining cycles of mutagenesis using, for example, error-prone polymerase chain reaction (epPCR) or gene shuffling with subsequent high-throughput screening (reviewed in [16]), have also been increasingly used over the years to enhance enzyme catalytic performances, solubility and stability to temperature or solvents. Controlled randomization at specific gene locations can also be achieved using degenerate oligonucleotides [20]. However, these methods (summarized in figure 1) require considerable screening efforts to sort out the requisite enzyme from very large mutant libraries (several thousand up to millions of mutants). With the development of more reliable predictive tools, rational and semi-rational strategies which combine several approaches have been developed. These methods allow the construction of a limited number of mutants or small-size libraries of controlled diversity focused on key regions for the desired property [31] (figure 1). Successful examples of enzyme engineering are abundant in the literature going up to the remarkable de novo computational design of biocatalysts able to catalyse new-to-Nature reactions [32]. So far, such computational strategies have been poorly explored with carbohydrate-active enzymes although they offer good new perspectives in chemo-enzymatic carbohydrate pathways, notably by tailoring enzymes to chemically modified substrates and allowing programing of most convenient chemical and enzymatic steps [33,34]. Furthermore, these enzymes acting on non-natural substrates could open the way to non-natural glycan structures of potential interest in different fields.
Figure 1.
Random and rational/semi-rational engineering approaches to generate genetic diversity combined with screening methods enabling access to enzyme biocatalysts with desired and/or improved properties. Semi-rational and rational approaches make use of sequence, structure and mechanism knowledge, often combined with (a) computational methods [17] in order to target regions or positions to insert mutations into the DNA template. Rational libraries (approx. 10–102 variants) can be constructed using simple techniques such as (b) inverse PCR or QuickChange [21] or directly by gene synthesis. Semi-rational libraries (approx. 102–104 variants) can be constructed using for example the NNK or NDT degenerate codons using subsequently different approaches such as (c) iterative saturation mutagenesis (ISM) [22], (d) combinatorial active site saturation testing (CASTing) [23], and using molecular biology methods such as: (e) OmniChange [24], or (f) the recently described Darwin assembly method [25]. The resulting smaller libraries can then be screened using low- or medium-throughput screening assays such as liquid assays in microtitre plates coupled with analytical detection or screening on a solid medium for the isolation of interesting mutants. Random approaches use either (g–i) random mutagenesis techniques that can insert errors randomly into a specific DNA template [19,26,27] or (j) shuffling in order to create hybrid sequences from different DNA templates [28]. The generated enzyme libraries (approx. 103–109 variants) require then a suitable high-throughput screening method such as cell-surface display or cell-free systems [29,30] coupled with flow cytometry–fluorescence activated cell/droplet sorting (FACS/FADS) to sort out interesting variants based on specific fluorescent reporters. (Online version in colour.)
In this review, we aim to provide a concise overview of the advantageous use of engineered glycoenzymes, sometimes in combination with chemical steps, for the synthesis of natural bioactive oligosaccharides or corresponding precursors, leaving aside glycoconjugates, glycopeptides and glycoproteins, which have been the subject of many recent studies using native and engineered glycosytransferases or oligosaccharyltransferases (OSTs) [35–38]. Here, the focus will be on examples resulting from the three main families (GTs, GHs and GPs) of carbohydrate-active enzymes specialized in carbohydrate synthesis [39].
2. Utilization of engineered Leloir-type glycosyltransferases
Leloir-type GTs are the most common enzymes involved in carbohydrate biosynthesis. GTs are responsible for the synthesis of most cell-surface glycoconjugates in mammalian systems and cell-wall carbohydrates in plants, fungi and bacteria [40]. These enzymes, which are mostly membrane associated, act on nucleotide-activated sugars as donor substrates to catalyse glycosylation reactions, in contrast to non-Leloir GTs, which use non-nucleotide sugars donors, such as lipid sugars. Although the in vitro utilization of these enzymes for synthetic purposes remains difficult owing to poor expression and the cost of sugar nucleotides, they remain of utmost interest because of the range of glycosyl donors and recognized acceptors used. The nine most common glycosyl moieties transferred by Leloir-type GTs are d-galactose (d-Gal), N-acetyl-d-galactosamine (d-GalNAc), d-glucose (d-Glc), N-acetyl-d-glucosamine (d-GlcNAc), d-glucuronic acid (d-GlcA), l-fucose (l-Fuc), d-mannose (d-Man), d-xylose (d-Xyl) and N-acetylneuraminic acid (Neu5Ac, the predominant form of sialic acid) [40]. The tremendous diversity of recognized acceptors (carbohydrates, lipids, polypeptides, DNA, antibiotics, etc.) is tightly related to the sequence variability and the structural topology of the acceptor binding site, which is itself strongly dependent on the overall fold of the enzyme [41]. Of great interest for synthetic purpose, GTs catalyse glycosyl transfer reactions with remarkable efficiency, regio-, stereo- and chemospecificities enabling the formation of the requisite glycosidic bonds. The Leloir-type GTs can proceed through either retention (double-displacement SN1- or dissociative SNi-like mechanisms) or inversion (SN2 mechanism) of the anomeric carbon configuration (figure 2) [40]. GTs isolated from natural organisms have been extensively explored in the synthesis of carbohydrates [53,54]. In addition, many instances of engineering of Leloir-type GTs, resulting either from rational mutagenesis or from directed evolution strategies, have been reported in recent years to synthesize efficiently various complex carbohydrates, glycoconjugates, glycolipids, glycoproteins and antibiotics, and further extend the repertoire of accessible reactions [42,55].
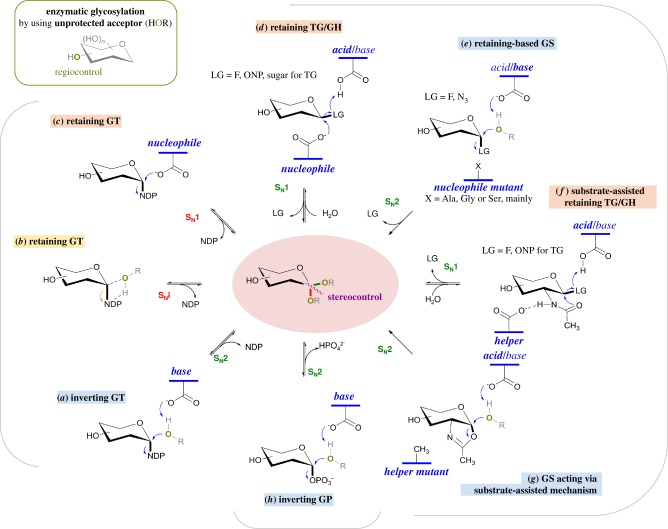
Figure 2.
Main enzymatic pathways for glycosidic bond formation by wild-type and engineered glycoenzymes mentioned in the manuscript (GT, TG/GH and GS, and GP). HOR, unprotected acceptor; LG, leaving group; GH, glycoside hydrolase; GP, glycoside phosphorylase; GS, glycosynthase; GT, glycosyltransferase; TG, transglycosylase; NDP, nucleotide diphosphate; ONP, O-nitrophenyl derivative. Only the initial stage of the glycoenzyme-catalysed reactions is summarized. Therefore, the involvement of the acceptor substrates (ROH) that occurs in the second step of the mechanism for retaining enzymes (c,d,f) is not depicted. (a) Inverting GTs use a single-displacement SN2 mechanism through a single oxocarbenium ion-like transition state formed under catalytic assistance from a general base. Retaining GTs follow either (b) an SNi-like mechanism, with the leaving phosphate group functioning as the catalytic base to deprotonate the ROH acceptor, or (c) a double-displacement mechanism involving the formation of a covalent glycosyl-enzyme intermediate [40,42]. (d) Classical Koshland two-step displacement mechanism of retaining TGs with enzyme nucleophile and acid/base residue proceeds via the formation of a covalent glycosyl-enzyme intermediate that opens access to transglycosylation even for innate hydrolytic GHs [43–45]. α-Retaining mechanism differs only from the depicted β-retaining one by the anomeric configurations of the α-substrate and α-product as well the more reactive β-covalent glycosyl-enzyme intermediate [46]. (e) Single-displacement mechanism of inverting GS combines the use of both an activated mimic of the covalent glycosyl-enzyme intermediate and a catalytic nucleophile mutant of the retaining GH [45,47]. (f) The two-step displacement mechanism with neighbouring group participation of retaining N-acetylhexosaminidases, also termed substrate-assisted catalysis, proceeds via an oxazoline or an oxazolinium ion intermediate. The 2-acetamido moiety of the donor substrate plays the role of an intramolecular nucleophile with the assistance of a helper residue to facilitate the formation of the intermediate [45]. (g) The mutation of the helper, which promotes the intermediate formation during the catalysis of retaining N-acetylhexosaminidases, coupled with the use of an activated oxazoline derivative leads to glycosynthase-like mutants [45,48]. (h) Inverting GPs promote synthesis via either axial-to-equatorial or equatorial-to-axial substitution on the anomeric carbon of the glycosyl-1-phosphate donor substrates through the base catalytic residue. Retaining GPs (not depicted here) are classified within both GH and GT families and, therefore, share common SN1- (c and d) or SNi-like (b) mechanisms [49–52]. (Online version in colour.)
In the following, we will exemplify the use of engineered Leloir-type GTs as synthetic tools, sometimes in combination with chemical synthesis, to access bioactive oligosaccharides, leaving aside reactions involving non-carbohydrate aglycones, which are reviewed elsewhere [42,56].
2.1. Synthesis of cell-surface glycans
Involved in a variety of biological functions such as structural and modulatory roles, complex carbohydrates, generally called glycans, are found in bacterial cell walls or attached to the lipids of the outer membrane of Gram-negative bacteria cells. These molecules are often targeted to treat inflammation, or to prevent bacterial infection. Glycans are also found at the surface of mammalian cells and constitute potential targets for the development of therapeutics against cancer [1]. The structure of these glycans is generally complex, involving a wide variety of glycosidic linkages, sugar types and arrangements of the glycosyl units. In vivo, these molecules are mostly assembled by the action of GTs.
As the d-galactosyl moiety is often found in glycan structures, linked through α- or β-linkages, some of the most explored GTs in oligosaccharide synthesis are galactosyltransferases (GalTs), which use uridine diphosphate galactose (UDP-Gal) as glycosyl donors to galactosylate a range of acceptor molecules linked via α-(1,3), α-(1,4), α-(1,6), β-(1,3) or β-(1,4) bonds. Most of these enzymes have been identified from natural biosynthetic pathways of O-antigens in bacteria (Escherichia coli, Helicobacter pylori, Neisseria meningitidis, Neisseria gonorrhoeae, etc.) [9,57] and they have been advantageously used, often in combination with a chemically prepared oligosaccharide precursor, for the synthesis of glycans [58–61].
Among them are the antigens of ABO blood-group systems that have shown remarkable differences in antigenicity related to changes in their structure [62]. In particular, oligosaccharide epitopes of antigens A and B composed of a disaccharide containing l-Fuc and d-Gal residues are differentiated by the presence of an additional d-GalNAc or a d-Gal residue, respectively, leading to α-d-GalNAc-(1,3)-[α-(l-Fuc-(1,2)]-β-d-GalOR and α-d-Gal-(1,3)-[α-l-Fuc-(1,2)]-β-d-GalOR antigens. GTs involved in A/B blood-group differentiation have been well studied, and were shown to be responsible for the formation of α-d-GalNAc-(1,3)-d-Gal and α-d-Gal-(1,3)-d-Gal linkages, respectively, on the α-l-Fuc-(1,2)-β-d-GalOR moieties of blood group O (H) antigens. The donor substrate specificity (UDP-Gal versus UDP-GalNAc) of human blood group A and B GTs was found to rely on only four amino acid differences located in the binding pocket of the donor substrate (R176 → G, G235 → S, L266 → M and G268 → A). Interchanges of these amino acids have led to remarkable changes in donor substrate recognition and catalytic efficiency of the GTs [63,64]. In particular, mutants, such as R176G/G235S/L266M, were found with dual specificity towards UDP-Gal and UDP-GalNAc donors. Others, like G235S/L266M, displayed broader promiscuity towards the donor substrate, and were found to accept UDP-Glc and UDP-GlcNAc as donor. These enzymes were used for the chemo-enzymatic synthesis of blood group A and B oligosaccharides and their corresponding analogues [65].
The α-(2,6)-sialosides found at the terminal or internal d-galactosyl unit of numerous glycan sequences are generally introduced in humans by two β-galactoside α-(2,6)-sialyltransferases (SiaTs) that have a narrow substrate specificity. Conversely, bacterial α-(2,6)-SiaTs such as β-galactoside α-(2,6)-SiaT from Photobacterium damselae (Pd2,6ST) have been widely used to produce sialosides as they have a more relaxed substrate specificity that could be further extended by enzyme engineering [66]. Combining a structure and sequence analysis, two non-conserved amino acid residues (A200 and S232) found at the bottom of the active site of Pd2,6ST, and potentially differentiating d-Gal/d-GalNAc moieties from the glycan acceptor, were targeted by site-directed mutagenesis to reshape the binding pocket. Out of 15 single and double mutants, a double-mutant A200Y/S232Y was found capable of regioselectively sialylating both the d-Gal and d-GalNAc at the non-reducing end of various glycans with higher efficiency than parental wild-type Pd2,6ST.
The active site of a GT80 α-(2,3)-SiaT, from Pasteurella dagmatis, was redesigned in order to switch its regioselectivity from (2,3) to (2,6). Sequence-based comparison with α-(2,6)-SiaT guided the identification of putative residues responsible for the (2,3)/(2,6) regioselectivity. The corresponding P7H/M117A mutant was constructed and used for the synthesis of sialyllactose (SL) from a CMP-Neu5Ac donor and lactose acceptor in a similar yield (72%) to that of the parental enzyme (75%). SL proved to be mainly 6′-sialyllactose and only a trace amount of 3′-sialyllactose was produced compared with the wild-type enzyme [67].
A random library of more than 106 variants of GT42 SiaT from Campylobacter jejuni, CStII, was generated by epPCR and expressed in modified E. coli strains which were further screened using a fluorescence-based high-throughput methodology [68]. Sialylated products were produced in vivo in the engineered cells, which were able to transport both donor (CMP-Neu5Ac) and fluorescent galactose-containing acceptor (e.g. bodipy-lactose). Cells accumulating the most transfer products were isolated by fluorescence activated cell sorting (FACS), leading to the isolation of a single mutant F91Y with improved transfer activity towards the labelled substrate [68].
An elegant strategy combining neutral genetic drift and site-directed mutagenesis was applied to engineer a polysialyltransferase from Neisseria meningitidis serogroup B and control polymer products [69]. This enzyme is responsible for transferring successively α-(2,8)-linked sialic acids onto the growing end of a polysaccharide chain, polySia. A single residue (K69) was found to control the size distribution of the polySia. In particular, mutant K69Q showed a distributive mechanism of chain elongation with a narrow-size product profile.
Several other examples of SiaT engineering have been reported for the production of sialylosides [70–72].
2.2. Production of human milk oligosaccharides
Human milk is composed of lipids, proteins, lactose and more complex oligosaccharides called HMOs, known to have a prebiotic effect, especially by providing an advantage for the growth of beneficial Bifidobacterium species [73], and also by having various immunomodulating effects [74]. The core structures of HMOs always display a lactose (Lac) moiety at the reducing end. This lactose can be branched with sialic acid (α-(2,3) or α-(2,6) linkages) or l-Fuc (α-(1,2) or α-(1,3) linkages), which corresponds to short-chain HMOs (trisaccharides). More complex HMOs are further extended in a linear or branched pattern with repetitions of lacto-N-biose (LNB; β-d-Gal-(1,3)-d-GlcNAc) or N-acetyl-lactosamine (LacNAc; β-d-Gal-β-(1,4)-d-GlcNAc) and can be decorated with sialic acid and/or l-Fuc residues [75–79].
In vivo, the pathway for the HMO biosynthesis involves several GTs [80]. Bacterial GTs, in native or engineered form, have been privileged to mammalian ones for the in vitro synthesis of HMOs (reviewed in [76]). Synthesis of HMOs has benefited from chemo-enzymatic strategies. Xiao et al. [81] produced a library of 31 simple and diverse HMOs from three chemically synthesized simple building blocks, comprising the core structure of more complex HMOs (lactose, d-GlcpNAc extension, and the presence or not of an additional d-Galp at the non-reducing end). Four native GTs were used sequentially or in one-pot multi-enzymatic synthesis in the presence of the chemical building blocks: a β-(1,4)-galactosyltransferase, an α-(2,6)-sialyltransferase, an α-(1,3)-fucosyltransferase and an α-(1,2)-fucosyltransferase. This work relied on the beneficial use of substrate promiscuity of these enzymes, but, undoubtedly, enzyme engineering technologies could provide access to more extended HMO structural diversity, improve enzyme activity, but also tackle recombinant expression challenges.
For instance, the α-(1,3)-fucosyltransferase from H. pylori that transfers the l-Fuc moiety from GDP-Fuc to LacNAc is of great interest to produce various antigen-like glycans (e.g. Lewis x, Lex, and Lewis y, Ley, epitopes) and HMO components, e.g. the trisaccharide block 3-fucosyllactose (3-FL). To improve soluble expression of this enzyme, Lin et al. [82] truncated the C-terminal region, which is constituted by 2–10 repeats (depending on the strain) of seven amino acids named heptad repeats, required for enzyme dimerization. By removing five out of the 10 heptad repeats together with 80 additional residues, the soluble expression of the enzyme was improved without any alteration of the structure or enzyme activity. A similar strategy was followed by Choi et al. [83] to improve the expression of an α-(1,3)-fucosyltransferase from another H. pylori strain. The C-terminal 52 amino acids truncated enzyme (Δ52 FutA) was left with only one heptad. The truncated enzyme gene was then synthesized after codon optimization for expression in E. coli. The solubility of the enzyme was found to be greatly improved, leading to better yields for the production of 3-FL (from 2% to 45%). This template was then used for further enzyme engineering using a two-step strategy of computer-aided analysis and iterative saturation mutagenesis (ISM) [22]. Among the several mutants tested, a quadruple mutant A128N/H129E/Y132I/S46F revealed a 15.5-fold increase in specific activity towards 3-FL with a 96% yield achieved (corresponding to a 40-fold improvement) in only 1 h of reaction.
2.3. Synthesis of GAG oligosaccharides
Glycosaminoglycans (GAGs, i.e. heparin, heparan sulfate, chondroitin, sulfate, dermatan sulfate, hyaluronans) are composed of repeating disaccharide building blocks formed by a hexosamine (d-GlcNAc or d-GalNAc) and a uronic acid residue (either d-glucuronic acid and/or l-iduronic acid) or d-Gal. Owing to the presence in their structure of negatively charged carboxylic groups and often sulfates, these polysaccharides exhibit a hydrophilic and anionic physico-chemical nature that confers on them key roles in biological interactions (cell adhesion, differentiation, signalling, etc.) and allows them to maintain the structural integrity of tissues [84]. Given the difficulty of producing large and complex GAG oligosaccharides by chemical synthesis, many chemo-enzymatic routes have been proposed using notably bifunctional GTs involved in GAG biosynthesis and sulfotransferases (O- and N-STases) that use high-cost 3′-phosphoadenosine-5′-phosphosulfate (PAPS) as donor as well as epimerases converting d-glucuronic acid (d-GlcA) into l-iduronic acid (l-IdoA) [85–89]. To diversify and better control the accessible structures of GAGs, limited studies report the use of engineered enzymes. While investigating the mechanism of the bifunctional chondroitin synthase K4CP that catalyses β-(1,3)-d-glucuronyl and β-(1,4)-N-acetyl-d-galactosaminyl transfer reactions to polymerize d-GlcA and d-GalNAc into a chondroitin chain, β-d-GlcA-(1,3)-β-d-GalNAc-(1,4), Sobhany et al. [90] identified amino acid residues in the N-terminal region that affect polymerase activity while retaining transfer activities. Targeting this region by site-directed mutagenesis, mutants were obtained that enable transfer of both d-GlcA and d-GalNAc moieties without any polymerization, or transfer only d-GalNAc with no d-GlcA transfer or polymerization, allowing a fine tuning of the reaction. Site-directed mutagenesis performed on the DXD amino acid motifs of processive bifunctional GTs involved in heparan sulfate (HS) synthesis resulted in mutants associated with a single action, processing one type of glycan unit only that could be used to better control the oligosaccharide synthesis [91].
3. Engineered glycoside hydrolases as synthetic tools
In Nature, GHs are responsible for the cleavage of glycosidic linkages between carbohydrate moieties or between a carbohydrate and a non-carbohydrate moiety. These retaining or inverting enzymes usually proceed to the hydrolysis along one of the two most common types of mechanism but with several additional variations, which lead to two different stereochemical outcomes, either the retention or the inversion of the anomeric configuration (figure 2) [43]. The catalytic reaction includes two key amino acid residues: a general acid/base (proton donor) and a catalytic nucleophile or a general acid and a general base. Both retaining and inverting glycosidases are also classified as either endo- or exo-glycosidases, depending on whether they cleave within the middle or at the end of an oligosaccharide chain, respectively. Most glycosidases used for synthetic purposes have an exo-mechanism, catalysing glycosyl transfer to the non-reducing terminal monosaccharide unit of acceptor substrates [92]. The use of endo-glycosidases is scarcer. These enzymes could be appealing for synthetic purposes because they are quite robust and tolerant to solvents [92,93]. However, their synthetic efficiency remains limited owing to their high activity towards water.
Despite their hydrolytic activity, some retaining GHs also display naturally efficient transferase activities and have thus been denominated ‘transglycosylases’. Out of the 153 GH families currently defined in the CAZy database [39], TGs can be mainly found in families GH2, 13, 16, 31, 70, 77, 23, 102, 103 and 104 [94]. They are known to function with diverse non-nucleotide sugar donors and preferentially transfer the glycosyl moiety in a regio- and stereoselective way onto a broad variety of acceptors distinct from water to synthesize a wide range of glycosides.
Both endo- and exo-GHs have also been engineered in order to limit the hydrolytic activity, meaning their ability to transfer the carbohydrate moiety onto water, and favour transglycosylation reactions [44]. Among them are the so-called ‘glycosynthases’ (GSs) that have been mutated on the catalytic nucleophile residue and are therefore devoid of their innate hydrolytic activity. The resulting enzymes are still able to catalyse the glycosyl transfer using a fluoride- or azide-activated glycosyl moiety, which mimics the covalent glycosyl-enzyme intermediate of the natural reaction, to perform the formation of glycosidic linkages (figure 2). GSs can be effective biocatalytic tools for carbohydrate synthesis, achieving high yields in spite of a limited donor substrate selectivity and the relatively poor stability and availability of glycosyl fluorides [95–97]. The GS approach has been implemented in many chemo-enzymatic pathways for the synthesis of oligosaccharides [47].
Overall, engineering of GHs has enabled not only improvements in catalytic activity, but also access to diversified oligosaccharides, often new to Nature, with altered chain length, polydispersity and glycosidic linkage specificities. It has also helped to remove unwanted side reactions, for example primary or secondary hydrolytic activities, or to improve enzyme expression, stability and solubility. This section will focus on the use of engineered GHs, mostly TGs and GSs, for the synthesis of a variety of relevant oligosaccharides.
3.1. Synthesis of cell-surface glycans
Endo-β-N-acetylglucosaminidase (Endo-M) is a retaining GH85 endo-glycosidase that cleaves the β-(1,4)-glycosidic linkage in the N,N′-diacetylchitobiose core of N-glycans. The enzyme can also transfer whole sugar chains onto a d-GlcNAc residue from a peptide or a protein. However, the secondary hydrolysis of the transglycosylation product obtained using Endo-M is a key limitation of the approach. To avoid hydrolysis of the product (secondary hydrolysis), the active site of the enzyme was engineered to yield GS-like enzymes. The putative helper residue, which acts to facilitate the formation of the oxazoline or oxazolinium ion intermediate, was substituted by Gln or His amino acids. The corresponding mutants N175H, N175Q and its evolved N175Q/W251N version showed diminished primary and secondary hydrolysis activity as well high transglycosylation activity to form the N,N′-diacetylchitobiose core in the presence of activated oligosaccharide oxazoline, which mimics the intermediate formed during the reaction [98,99]. Therefore, Endo-M and its related enzymes embody powerful tools for the glycoengineering and glycodiversification of glycoproteins [45].
The transglycosylating activity of GHs has also been engineered to produce bioactive oligosaccharides (such as HMOs, oligosaccharide antigens displayed on proteins or lipids and tumour-associated antigens) that often contain the core galacto-N-biose (GNB; β-d-Gal-(1,3)-d-GalNAc) and LNB moieties. As an example, the retaining GH35 β-galactosidase from Bacillus circulans (BgaC) suffering from low yields due to re-hydrolysis of the transfer products [100] was further engineered by Henze et al. [101] into a galactosynthase mutant by substituting the catalytic nucleophile glutamate into a glycine (E233G mutant), enabling transfer of the galactosyl residue from α-d-Gal fluoride to different β-linked N-acetyl-d-glucosamine acceptor substrates. The resulting β-(1,3)-linked d-galactosides, LNB conjugates, were stereo- and regioselectively synthesized in good yield (40–90%) without—as expected—any trace of secondary hydrolysis [101].
In order to recover its innate (1,3) regioselectivity during the β-d-Gal transfer for the synthesis of LNB, the substrate specificity of the TtbGly E338G GS derived from a GH1 retaining β-glycosidase from Thermus thermophilus was expanded. The use of the 2-amino-2-deoxy-β-d-glucoside instead of its bulky 2-acetamido derivative reoriented the regioselectivity of this GS-catalysed reaction towards the formation of (1,3) instead of (1,4) linkages; the use of the thiophenyl group at the anomeric position of the acceptor was already a prerequisite to ensure correct positioning of the acceptor in the active site and allowing the catalysis to proceed. Following the enzymatic coupling performed in 88% yield, additional sequential selective acylation of nitrogen followed by anomeric deprotection led to the production of pre-activated or free LNB disaccharides in 94% and then 97% yields [102].
Fucosynthases were obtained from the retaining GH29 α-(1,3/4)-fucosidase BbAfcB from Bifidobacterium bifidum. For instance, the D703S α-l-fucosynthase mutant regioselectively catalysed the synthesis of valuable biomimetic tri- and pentasaccharides, including the HMOs 3-FL and lacto-N-fucopentaose II (LNFP-II; β-d-Gal-(1,3)-[α-l-Fuc-(1,4)]-β-d-GlcNAc-(1,3)-β-d-Gal-(1,4)-d-Glc) in 13% and 41% yield, respectively) [103]. The D242S α-l-fucosynthase based on the GH29 retaining α-l-fucosidase from Sulfolobus solfataricus catalysed fucosylation of pNP-β-d-Gal to generate various regioisomers in an overall yield of 26% using the stable β-fucosyl azide as donor. Furthermore, pNP-β-d-GlcNAc was regioselectively fucosylated on its O-3 in 86% yield [104]. Likewise, β-d-Gal azide was converted with high regioselectivity to α-Gal-disaccharides in 33–51% yields, depending on the acceptor, using the D327G galactosynthase mutant based on the GH36 α-galactosidase from Thermotoga maritima [105].
A GH98 enzyme from Streptococcus pneumoniae SP3-BS71 was used to cleave respectively the A- and B-trisaccharides from type 2 core chains (LacNAc), responsible for determination of blood groups, with the objective of producing universal blood [106]. However, some linkages (e.g. LNB from type 1 core chains) were resistant to the activity of the wild-type enzyme. Following a structure-guided directed evolution strategy, involving several iterations of site-selected randomized mutagenesis combined with high-throughput screening, Kwan et al. [106] isolated a mutant displaying 170-fold higher activity towards the cleavage of the LNB linkage compared with parental wild-type enzyme, while preserving high activity towards type 2 core chains (LacNAc). This exemplifies the potential of building specific glycans or glycan building blocks with exo-enzymes by de-constructing more complex glycan structures.
3.2. Targeting bacterial polysaccharides
Pathogenic bacteria surface carbohydrates, either capsular polysaccharides (CPSs) when bacteria produce a capsule, cell wall-associated glycans in Gram-positive bacteria or lipopolysaccharides (LPSs) in Gram-negative bacteria are targets of choice for the development of glycoconjugate vaccines [107,108]. Various strategies have been proposed to access the relevant immunogenic oligosaccharides that could elicit an immune response and therefore be used in the composition of glycovaccines: extraction from natural sources, chemical synthesis (which remains the most used approach for the development of glycoconjugate vaccines in general), chemo-enzymatic synthesis and in vivo production by whole-cell biocatalysts.
Recently, programmed in vitro chemo-enzymatic routes have been reported by our group that take advantage of TG engineering to produce complex microbial cell-surface oligosaccharides and circumvent the synthetic boundaries of glycochemistry. The different studies targeted more specifically the synthesis of oligosaccharides, mimicking the O-antigen of several Shigella flexneri serotypes. Most known S. flexneri O-antigen repeats share a linear tri-l-rhamnosyl-N-acetyl-d-glucosamine tetrasaccharide backbone and the serotype specificity is partly defined by the α-d-glucosyl branched to the backbone. In the synthetic pathway foreseen for the production of the O-antigen repeats, regioselective glucosylation was considered using α-transglucosylases in order to overcome the poor α/β stereoselectivity of the chemical glucosylation process. Given that no native enzyme has been reported for this reaction, computer-aided engineering approaches were followed to redesign their active site in order to render them able to act on non-natural substrates, carrying protecting groups compatible with a subsequent chemical elongation. These chemically modified substrates were designed in order to integrate the enzymatic step at different stages of the synthesis, while taking into account the inherent constraints of chemical reactivity and enzyme-based catalysis. Different lightly chemically protected substrates derived from O-antigen repeats, and well suited for further chemical elongation leading to the S. flexneri haptens, were thus targeted to set up the programmed chemo-enzymatic routes of monosaccharide analogues such as methyl α-l-rhamnoside and allyl 2-acetamido-2-deoxy-α-d-glucoside [33,109], or disaccharide derivatives such as allyl 2-deoxy-2-trichloroacetamido-β-d-glucosyl-(1,2)-α-l-rhamnoside or allyl α-l-rhamnosyl-(1,2)-2-deoxy-2-trichloroacetamido-β-d-glucoside [34,110].
In this context, sucrose-using transglucosylases from GH13 and GH70 families, well known for their remarkable versatility regarding the acceptor substrate [46,111–113], were selected as starting scaffolds to adapt the active site to the target molecules. Recombinant amylosucrase from Neisseria polysaccharea (NpAS) was used as a scaffold to tailor enzymes for the glucosylation of the monosaccharides. By randomizing mutations at seven positions identified by molecular modelling in the acceptor binding site, a library of 133 single mutants followed by pairwise recombination of the mutations led to a library of approximately 20 000 mutants which was screened. Amylosucrase variants with either completely new specificity towards methyl α-l-rhamnoside or significantly enhanced (by up to a 400-fold compared with NpAS) toward allyl 2-N-acetyl-2-deoxy-α-d-glucoside were isolated. Although limited, the catalytic efficiency of the engineered enzymes towards the target acceptors was found to be considerably improved compared with the parental wild-type enzyme. The best variants were then used to synthesize glucosylated building blocks that were converted into acceptors and donors compatible with chemical elongation towards oligosaccharide fragments of the O-antigens of the targeted serotypes 1b and 3a [109].
A more ambitious design was then undertaken to re-engineer several binding subsites of NpAS and render it able to glucosylate a partially protected disaccharide acceptor (allyl (2-deoxy-2-trichloroacetamido-β-d-glucosyl)-(1,2)-α-l-rhamnoside) to generate a precursor of S. flexneri. An approach combining molecular docking, computational protein design and sequence coevolution-derived information was developed to design libraries containing a limited number of authorized mutations at 23 selected positions of the active site. From a designed library of 63 000 clones selected from a set of 2023 theoretical combinations, one NpAS mutant displaying seven mutations in the active site of the enzyme (R226L/I228V/F290Y/E300V/V331T/G396S/T398V) was able to glucosylate the disaccharide of interest using sucrose as a readily available donor substrate. A reaction for which there is no equivalent yet which has been reported in the literature was made possible, generating the S. flexneri type 1b glucosylation pattern [110].
Targeting another lightly protected disaccharide acceptor: allyl α-d-glucosyl-(1,4)-α-l-rhamnosyl-(1,3)-2-deoxy-2-trichloroacetamido-β-d-glucoside, which is well suited for further chemical elongation leading to the S. flexneri serotype 2a, an engineered dextransucrase (GBD-CD2 branching sucrase) was able to achieve a remarkably high yield of site-selective α-d-glucosylation (94%) of the targeted disaccharide. The product of enzymatic glucosylation was then chemically converted into the pentadecasaccharide hapten present in S. flexneri 2a-TT15, the first synthetic carbohydrate-based vaccine candidate against endemic shigellosis [34].
3.3. Production of human milk oligosaccharides
l-Fuc residues are present in 50% of the HMOs. Alternatively to the use of Leloir-type α-fucosyltransferases [78], fucosylation can also be achieved by α-fucosidases endowed with transfucosylating activity with yields between 5% and 40%, depending on the donor/acceptor ratio, although hydrolysis remains largely observed. To improve the transfucosylating activity, a directed evolution strategy was applied to introduce random mutations on the full-length gene corresponding to the enzyme TmαFuc from T. maritima by epPCR [114]. Five thousand clones of the α-fucosidase were first screened on a solid medium in the presence of the chromogenic X-α-l-Fuc susbtrate. From this first screening, 100 mutants were retained for further assessment of their transfucosylation activity using pNP-α-l-Fuc as the donor substrate and pNP-β-d-Gal or β-d-Gal-(1,3)-β-d-Glc-O-phenyl (phenyl laminaribioside) as the acceptor. From analysis of the mutations found in the best mutants, combined with structural analysis, a triple mutant T264A/Y267F/L322P was constructed that exhibited improved transfucosylation activity yielding 59% for the production of α-l-Fuc-(1,2)-β-d-Gal-OpNP compared with 7% for the parental wild-type enzyme, in agreement with a considerable decrease in the hydrolytic activity (around 100-fold) [114]. The same group later employed a semi-rational approach to design an α-l-transfucosylase starting from the retaining exo-GH α-l-fucosidase from Bifidobacterium longum ssp. infantis (BiAfcB). Structural and sequence comparison of the latter with TmαFuc guided the construction of several mutants containing the equivalent L321P substitution combined with neighbouring mutations at the donor binding site that aimed at improving the transglycosylation/hydrolysis ratio as devised by Teze et al. [115]. The double-mutant L321P/F34I yielded the best production of LNFP-II from the 3-FL donor and lacto-N-tetraose (LNT) acceptor going from 12% for the wild-type enzyme to 32%, and lowered secondary product hydrolysis. The mutant was also able to transfer the l-Fuc from the 3-FL donor to three other acceptors: lacto-N-neotetraose (LNnT), lacto-N-fucopentaose I (LNFP-I) and 2′-FL, opening the way for the synthesis of more complex fucosylated HMOs [116]. More recently, a loop engineering strategy was applied to enhance the transfucosylation/hydrolysis ratio of a GH29 α-(1,3/4)-fucosidase from B. bifidum (BbAfcB). A 23 amino acid long α-helical loop located near the active site was replaced by the corresponding 17 amino acid long loop from the α-(1,3/4)-fucosidase from Clostridium perfringens to confer better shielding from water molecules to BbAfcB. The hydrolytic activity on 3-FL was almost abolished (6000 times lower than for wild-type enzyme) and the engineered enzyme transferred l-Fuc with a 39% yield from LNT to LNFP-II using 3-FL as donor [117].
Synthetic routes to access sialylated HMOs have attracted increasing attention (see review [118]). A trans-sialidase from Trypanosoma cruzi (TcTS), able to transfer α-(2,3) sialic acid from sialoglyconjugate donors onto β-d-Gal glyconjugate acceptors, was used in various studies for the sialylation of different HMO precursors using a wide array of donors (casein glycomacropeptide, fetuin, α-Sia-(2,3′)-Lac, pNP-α-Sia) and acceptors (Lac, LNT, galacto-oligosaccharides (GOS), LacNAc, LNB, LNnT, lacto-N-fucopentaose V (LNFP-V), GNB, β-d-Gal-(1,6)-d-Gal, etc.) [118]. An issue encountered with the use of TcTS is related to the virulence factor of Trypanosoma cruzi, making it less interesting for food-related processes. A highly homologous enzyme, the sialidase from Trypanosoma rangeli (TrSA), was thus thought to be a good alternative to TcTS. To confer trans-sialidase activity to TrSA, various engineering approaches have been used over the years. Structural comparison of TrSA and TcTS revealed similar topology of the active site with the exception of three amino acid residues (S120, G249 and Q284 in TrSA, corresponding to Y119, Y248 and P283 in TcTS), assumed to be important for transferase activity [119–121]. The amino acid interchanges at these three positions were not sufficient to confer a trans-sialidase activity [121,122]. Additional mutations were thus introduced to recreate a favourable environment for binding the sialic acid moiety of the substrate. As a result, the variant TrSA5mut containing five mutations (G249Y/Q284P/S120Y/M96V/A98P) displayed 1% of trans-sialidase activity compared with TcTS, transferring sialic acid from α-sialyl-(2,3)-lactose to the lactose acceptor. Adding a sixth mutation in the active site led to mutant TrSA6mut having a trans-sialidase activity enhanced up to 11% [122]. In addition, Jers et al. [123] identified a positively charged motif in region 197–203 of TcTS, near the substrate binding cleft, that differed from TrSA. Hypothesizing its importance in trans-sialidase activity, a TrSA mutant incorporating this motif, Tr13mut, was constructed that contained a total of 13 mutations. This mutant showed a reduced hydrolytic activity compared with parental TrSA6mut enzyme (around 80% less hydrolysis of 3-SL), but still maintained trans-sialidase activity. It was thus used for the sialylation of various acceptors: GOS, isomalto-oligosaccharides (IMOs), lactulose, melibiose, maltose and l-Fuc. To further improve its trans-sialidation activity, a computer-aided design approach was applied to TrSA5mut, described earlier [124]. Five additional mutations were proposed (I37L, T39A, F59N, D285G, G342A) to help the stabilization of the transition state. Starting from Tr13mut, the best mutant described in the literature at the time, Nyffenegger et al. [125] added three of these mutations, T39A, F59N and D285G, resulting in mutant Tr16mut containing a total of 16 mutations and that showed a 13.6-fold improved overall transferase activity over hydrolysis ratio compared with parental TrSA5mut.
3.4. Production of other prebiotics
Food bioactive oligosaccharides have gained interest as potent health-benefitting ingredients over recent years. They are not digested by human hydrolytic enzymes and reach the lower gastrointestinal tract where they are metabolized by beneficial intestinal microbiota, which in turn produce short-chain fatty acids providing health benefits to the host, as well as preventing the growth of harmful bacteria [126]. Several studies have suggested that prebiotic intake plays a role in the immune system, may protect against colon cancer and may prevent cardiovascular diseases and metabolic syndromes. Their beneficial properties depend on the monosaccharide composition [127], and current applications mainly concern fructo-, galacto-, xylo-, gluco-oligosaccharides (FOS, GOS, XOS) and lactulose, especially for food and beverage processing, dietary supplements and animal feed [128]. Utilization of microbial GHs (including glycosidases and TGs) for a controlled regio- and stereoselective synthesis of prebiotic oligosaccharides has been extensively reviewed [129]. Nonetheless, the search for new TGs, the development of processes, as well as the improvement or redesign of catalysts by enzyme engineering is still relevant in order to develop new applications and/or to optimize the production yields.
For instance, an elegant engineering strategy of B. circulans β-galactosidase was proposed by Tanaka et al. [130] to alter its substrate specificity and enhance the production of short-chain GOS (DP2 and DP3 only) from lactose substrate. For that purpose, they designed and screened a library of synthetic binding proteins (monobodies) that specifically targeted the +3 acceptor subsite. This local constraint prevented the accommodation of oligosaccharides of DP3 in the active site and their further elongation into DP4. At the same time, enzyme properties for lactose recognition and production of short-chain GOS were not altered, allowing their accumulation in the medium [130].
Concerning XOS production, the X-ray structure resolution of an inactive mutant of the Bacillus pumilus IPO β-xylosidase in complex with xylobiose (DP2) enabled the identification of amino acid residues involved in xylobiose recognition. Notably, a tight binding of d-Xyl at subsite + 1 was reinforced by mutating a phenylalanine (F503) into tyrosine, leading to a 20% increase of xylobiose production from xylose, compared with the wild-type enzyme. Xylobiose presents the highest prebiotic properties among xylo-oligosaccharides, and this study opened the route for a similar engineering strategy within this family of enzymes [131].
Other interesting examples deal with microbial TGs that use readily available sucrose as a donor substrate to efficiently polymerize either the d-fructosyl or the d-glucosyl moiety of sucrose to produce β-d-fructan of α-d-glucan-type polysaccharides (i.e. fructansucrase or glucansucrase enzymes of GH families). By adding exogenous monosaccharides in the reaction medium with sucrose, these enzymes have the ability to transfer the d-fructosyl or d-glucosyl units onto these acceptor molecules to yield interesting dietary oligosaccharides. To lower the cost of such processes, enzyme engineering has been used to produce oligosaccharides from only sucrose. For instance, the production of levan-type FOS (FOS with β-(2,6) glycosidic linkages) from DP2 to DP10 was developed owing to the construction of a levansucrase–levanase fusion enzyme (levansucrase SacB from Bacillus subtilis and endolevanase LevB1 from Bacillus licheniformis). Interesting yields of about 40% were obtained from sucrose substrate, to access 6-kestose, levanbiose and blastose among other FOSs [132]. Concerning the production of gluco-oligosaccharides, determination of the three-dimensional structure of the dextransucrase DSR-M from Leuconostoc citreum NRRL B-1299, free or in complex with an isomalto-oligosaccharide (IMO) of DP4, allowed the design of variants producing short α-(1,6) chains of around 16 kg mol−1 (80% yield) by altering aromatic residues located in a glucan-binding domain shown to be involved in the anchoring of dextran chains [133]. Gluco-oligosaccharides rich in α-(1,6) linkages (IMO) are well-known prebiotic compounds that have been used in Asian countries for many years. However, their prebiotic properties would be enhanced by increasing the content of rare α-(1,2) glycosidic bonds. For that purpose, an engineered enzyme specialized in the α-d-(1,2) glucosylation of IMO acceptor molecules, called GBD-CD2 branching sucrase, was used for the controlled introduction of α-(1,2)-branched d-glucosyl moieties, from 1% to 37% following the reaction conditions used (sucrose/donor ratio) [134,135].
To finish, the amylosucrase from N. polysaccharea (NpAS) is another α-TG that uses sucrose as d-glucosyl donor to catalyse the formation of soluble malto-oligosaccharides and insoluble amylose-like polymer. Using a computer-aided approach, semi-rational libraries of 2.7 × 104 mutants targeting as many as 23 amino acid positions of the active site were constructed and screened in the presence of only sucrose [136]. This allowed identification of 17 mutants able to synthesize molecules which are not synthesized by the parental wild-type enzyme. Characterization of the three most promising mutants (namely 47A10, 37G4 and 39A8), containing, respectively, 7, 10 and 11 mutations in the active site, showed the production of a sucrose derivative, named erlose (α-d-glucosyl-(1,4)-α-d-glucosyl-(1,2)-β-d-fructose) and panose (α-d-glucosyl-(1,6)-α-d-glucosyl-(1→4)-α-d-glucose). These products, reported to have potential interest as sweeteners or prebiotic molecules, were never obtained using either natural amylosucrases previously characterized or engineered variants [137,138].
4. Exploitation of engineered glycoside phosphorylases in synthesis
Non-Leloir GTs called glycoside phosphorylases (GPs) cleave oligosaccharides in the presence of inorganic phosphate, producing a glycosyl phosphate and a shorter oligosaccharide (figure 2). This so-called phosphorolysis is reversible and can therefore be used to catalyse the stereospecific synthesis of glycosides using cheap phosphorylated glycosyl donors. When combined with a second appropriate phosphorylase, the said phosphorylated glycosyl donor can be produced in situ from the corresponding readily available glycosyl, making these enzymes suitable synthetic tools for the scaled-up production of carbohydrates [139]. GPs have scarcely been used for this purpose mainly because of a lack of diversity among the available enzymes. The known phosphorylases as well as their mechanism, structure and use in the synthesis of oligosaccharide have been extensively reviewed [49–52,140,141]. Around 30 phosphorylases have been reported and were classified in the CAZy database as either GT or GH members. Furthermore, GPs can adopt either retaining or inverting mechanisms. GPs have a broad acceptor specificity as they can act on various carbohydrates (such as trehalose, kojibiose, sucrose, nigerose, maltose, etc.) but their donor specificity is quite narrow [51,142]. Several recent studies report the characterization and the engineering of GPs that could be used for glycoside synthesis [143–146].
4.1. Production of oligosaccharides
De Groeve et al. [147] engineered both the donor and acceptor specificities of a cellobiose phosphorylase (CP), first discovered in extracts from the bacterium Clostridium thermocellum and belonging to the GH94 family. This enzyme catalyses the reversible phosphorolysis of cellobiose into α-d-glucosyl 1-phosphate (α-d-Glc-1P). To diversify the products obtained by GPs, the donor specificity of CP from Cellulomonas uda was changed from cellobiose to lactose using a two-step strategy of directed evolution. First, random mutagenesis was applied by epPCR on part of the gene, targeting more specifically amino acid residues contained between T216 and V757 (out of the 822 amino acid enzyme), which includes all residues located within 15 Å of the catalytic site. Altogether, 10 000 mutants were generated and subsequently selected for their activity towards lactose using a growth-selective assay on minimal solid medium containing lactose as the sole carbon source. Three active mutants were retained and further sequenced, showing the presence of a set of six amino acid substitutions in all mutants. All mutations except N667T occurred at more than 15 Å from the donor subsite. The variant LP1 (A397V/T508A/A512T/D557N/N667T/G681S) showed a threefold improved activity towards lactose and a sixfold decreased activity towards cellobiose compared with parental wild-type enzyme. Mutant LP1 was then subjected to additional rounds of site-directed mutagenesis in order to decipher the role of each mutation by partial deconvolution. Mutations T508A present near the entrance to the active site and N667T located close to the donor site were the only mutations that conferred an activity towards lactose. The corresponding double-mutant T508A/N667T named LP2 was further engineered on these two positions by site-directed saturation mutagenesis. This yielded the double-mutant T508I/N667A, named LP3, which had the best activity towards lactose with a 50% increase compared with LP2, leading to a 7.5-fold increase compared with that of the parental enzyme [148]. Mutant LP3 was ultimately used for the production of lactose in a completely new way compared with the usual GalT enzymes that use expensive UDP-Gal as a donor.
CPs have a broad acceptor promiscuity and they are able to recognize several mono- and disaccharides such as d-Glc, d-Man, d-GlcNAc, d-Xyl, l-Fuc, d-Ara, melibiose, gentiobiose, isomaltose, … which is particularly valuable for glycoside synthesis [147]. To further expand the acceptor specificity, De Groeve et al. [149] identified a residue (E649) in the structure of CP from C. uda, whose side chain has hydrogen bonding with the d-Glc acceptor. Upon saturation mutagenesis of this residue, the mutant E649C was identified as being able to recognize new glucoside acceptors. Mutation E649C was then combined with mutations present in mutant LP3. Three additional amino acid positions located at the entrance to the active site were then identified for saturation mutagenesis on the basis of structure analysis. The resulting mutant (N156D, N163D, T508I, E649G and N667A) revealed a broad acceptor specificity towards α- and β-glycosides [149].
Using the ISM technology, Chen et al. [150] engineered the specificity of a trehalose phosphorylase for the production of β-d-Gal-1P instead of β-d-Glc-1P produced by the parental enzyme. Computational modelling of the lactotrehalose binding in the enzyme active site enabled identification of three amino acid residues that were further targeted by mutagenesis. Altogether, 600 variants were screened and the triple mutant L649G/A693Q/W371Y was identified with a 2196-fold increase in activity for the release of β-d-Gal-1P. The reverse reaction allowed lactotrehalose to be produced from β-d-Gal-1P and d-Glc.
5. Outlook and future directions
Significant advances have been made over recent years regarding the enzyme-based in vitro synthesis of complex carbohydrates with the development of enzyme cascades, one-pot synthesis, and the combined use of chemical and enzymatic approaches. Progress has been noticeably boosted by the advent of enzyme engineering technologies that enabled a whole new diversity of enzymes to be accessed. This has strongly benefitted in recent years from the tremendous developments in bioinformatics [156,157] and computer-aided design of enzymes [17] that nowadays allows reprogramming of the substrate specificity towards the desired glycosylation reaction, with sometimes the tailoring on purpose of enzymes to be able to act on non-natural chemically modified carbohydrate precursors. Despite the impressive advances in rational engineering, semi-rational approaches to medium to large libraries of variants still require the development of high-throughput screening methods. In the field of glycoenzymes, those methods cannot always be easily implemented when looking for very specific sugar structures. They often necessitate the adjunction of analytical techniques, which are highly accurate but remain time consuming. Progress in nuclear magnetic resonance and mass spectrometry should help to tackle this question more efficiently in the near future. The field is rapidly evolving with the democratization of these technologies, which could also be combined with post-synthesis and specific chemical labelling of the reaction products to facilitate product analysis at high throughput. In parallel, cell development for specific carbohydrate production inspired from existing biosynthetic pathways is also rapidly progressing [9,158]. The incorporation of engineered enzymes gives total freedom to even more creative pathways to produce in vivo bioactive oligosaccharides and glycoconjugates at large scale and offers exciting perspectives.
When targeting a specific carbohydrate structure, several bio- and/or chemo-synthetic approaches can thus be employed. Of course, there is no easy way to anticipate the most appropriate one as the choice will depend on many factors, including the structural complexity of the target molecules, the chemical difficulties, the enzyme and substrate availability, the environmental impact, regulation and ethical considerations, and most importantly the economic advantage of the process knowing that production of oligosaccharides is expensive. All these constraints and issues will have to be considered. To develop innovative processes with the highest possible discernment, there is no doubt that strong interactions and interfacing between all these disciplines will be essential, with hybrid and composite approaches constituting the future of synthetic glycobiology.
Data accessibility
This article has no additional data.
Authors' contributions
M.B., R.F., M.R.S., C.M. and I.A. planned and drafted the manuscript. All authors gave final approval for publication.
Competing interests
We declare no competing interests.
Funding
This work was supported by the French National Research Agency (ANR Project CARBUNIVAX and ANR-15-CE07-0019-01).
References
- 1.Varki A. 2017. Biological roles of glycans. Glycobiology 27, 3–49. ( 10.1093/glycob/cww086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monsan P, Paul F. 1995. Enzymatic synthesis of oligosaccharides. FEMS Microbiol. Rev. 16, 187–192. ( 10.1111/j.1574-6976.1995.tb00165.x) [DOI] [Google Scholar]
- 3.Zhu Y, Romain C, Williams CK. 2016. Sustainable polymers from renewable resources. Nature 540, 354–362. ( 10.1038/nature21001) [DOI] [PubMed] [Google Scholar]
- 4.Schmid J. 2018. Recent insights in microbial exopolysaccharide biosynthesis and engineering strategies. Curr. Opin. Biotechnol. 53, 130–136. ( 10.1016/j.copbio.2018.01.005) [DOI] [PubMed] [Google Scholar]
- 5.Nielsen MM, Pedersen CM. 2018. Catalytic glycosylations in oligosaccharide synthesis. Chem. Rev. 118, 8285–8358. ( 10.1021/acs.chemrev.8b00144) [DOI] [PubMed] [Google Scholar]
- 6.Hsu CH, Hung SC, Wu CY, Wong CH. 2011. Toward automated oligosaccharide synthesis. Angew. Chemie Int. Ed. 50, 11 872–11 923. ( 10.1002/anie.201100125) [DOI] [PubMed] [Google Scholar]
- 7.Brown SD, Babbitt PC. 2014. New insights about enzyme evolution from large scale studies of sequence and structure relationships. J. Biol. Chem. 289, 30 221–30 228. ( 10.1074/jbc.R114.569350) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antranikian G, Egorova K. 2003. Extremophiles, a unique resource of biocatalysts for industrial biotechnology. In Physiology and biochemistry of extremophiles (eds Gerday C, Glansdorff N), pp. 361–406. Washington, DC: American Society of Microbiology. [Google Scholar]
- 9.Chen R. 2018. Enzyme and microbial technology for synthesis of bioactive oligosaccharides: an update. Appl. Microbiol. Biotechnol. 102, 3017–3026. ( 10.1007/s00253-018-8839-2) [DOI] [PubMed] [Google Scholar]
- 10.Geremia RA, Samain E. 2000. Production of heterologous oligosaccharides by recombinant bacteria (recombinant oligosaccharides). Carbohydrates Chem. Biol. 2–4, 845–860. ( 10.1002/9783527618255.ch30) [DOI] [Google Scholar]
- 11.Priem B, Gilbert M, Wakarchuk WW, Heyraud A, Samain E. 2002. A new fermentation process allows large-scale production of human milk oligosaccharides by metabolically engineered bacteria. Glycobiology 12, 235–240. ( 10.1093/glycob/12.4.235) [DOI] [PubMed] [Google Scholar]
- 12.Uchiyama T, Miyazaki K. 2009. Functional metagenomics for enzyme discovery: challenges to efficient screening. Curr. Opin. Biotechnol. 20, 616–622. ( 10.1016/j.copbio.2009.09.010) [DOI] [PubMed] [Google Scholar]
- 13.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238. ( 10.1093/nar/gkn663) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. 2012. DbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40, 445–451. ( 10.1093/nar/gks479) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park BH, Karpinets TV, Syed MH, Leuze MR, Uberbacher EC. 2010. CAZymes Analysis Toolkit (CAT): Web service for searching and analyzing carbohydrate-active enzymes in a newly sequenced organism using CAZy database. Glycobiology 20, 1574–1584. ( 10.1093/glycob/cwq106) [DOI] [PubMed] [Google Scholar]
- 16.André I, Potocki-Véronèse G, Barbe S, Moulis C, Remaud-Siméon M. 2014. CAZyme discovery and design for sweet dreams. Curr. Opin. Chem. Biol. 19, 17–24. ( 10.1016/j.cbpa.2013.11.014) [DOI] [PubMed] [Google Scholar]
- 17.Khan FI, Wei DQ, Gu KR, Hassan MI, Tabrez S. 2016. Current updates on computer aided protein modeling and designing. Int. J. Biol. Macromol. 85, 48–62. ( 10.1016/j.ijbiomac.2015.12.072) [DOI] [PubMed] [Google Scholar]
- 18.Hsieh P-C, Vaisvila R. 2013. Protein engineering: single or multiple site-directed mutagenesis. Methods Mol. Biol. 978, 173–186. ( 10.1007/978-1-62703-293-3_13) [DOI] [PubMed] [Google Scholar]
- 19.Packer MS, Liu DR. 2015. Methods for the directed evolution of proteins. Nat. Rev. Genet. 16, 379–394. ( 10.1038/nrg3927) [DOI] [PubMed] [Google Scholar]
- 20.Chusacultanachai S, Yuthavong Y. 2004. Random mutagenesis strategies for construction of large and diverse clone libraries of mutated DNA fragments. In Parasite genomics protocols, pp. 319–334. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- 21.Hogrefe HH, Cline J, Youngblood GL, Allen RM. 2002. Creating randomized amino acid libraries with the QuikChange® multi site-directed mutagenesis kit. Biotechniques 33, 1158–1165. ( 10.2144/02335pf01) [DOI] [PubMed] [Google Scholar]
- 22.Reetz MT, Carballeira JD. 2007. Iterative saturation mutagenesis (ISM) for rapid directed evolution of functional enzymes. Nat. Protoc. 2, 891–903. ( 10.1038/nprot.2007.72) [DOI] [PubMed] [Google Scholar]
- 23.Reetz MT, Bocola M, Carballeira JD, Zha D, Vogel A. 2005. Expanding the range of substrate acceptance of enzymes: combinatorial active-site saturation test. Angew. Chemie Int. Ed. 44, 4192–4196. ( 10.1002/anie.200500767) [DOI] [PubMed] [Google Scholar]
- 24.Dennig A, Shivange AV, Marienhagen J, Schwaneberg U. 2011. Omnichange: the sequence independent method for simultaneous site-saturation of five codons. PLoS ONE 6, e0026222 ( 10.1371/journal.pone.0026222) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cozens C, Pinheiro VB. 2018. Darwin assembly: fast, efficient, multi-site bespoke mutagenesis. Nucleic Acids Res. 46, 67 ( 10.1093/nar/gky067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mundhada H, Marienhagen J, Scacioc A, Schenk A, Roccatano D, Schwaneberg U. 2011. SeSaM-Tv-II generates a protein sequence space that is unobtainable by epPCR. Chembiochem 12, 1595–1601. ( 10.1002/cbic.201100010) [DOI] [PubMed] [Google Scholar]
- 27.Aharoni A, Gaidukov L, Khersonsky O, Gould SM, Roodveldt C, Tawfik DS. 2005. The ‘evolvability’ of promiscuous protein functions. Nat. Genet. 37, 73–76. ( 10.1038/ng1482) [DOI] [PubMed] [Google Scholar]
- 28.Stemmer WP. 1994. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc. Natl. Acad. Sci. 91, 10 747–10 751. ( 10.1111/j.1399-6576.2007.01374.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Becker S, Schmoldt HU, Adams TM, Wilhelm S, Kolmar H. 2004. Ultra-high-throughput screening based on cell-surface display and fluorescence-activated cell sorting for the identification of novel biocatalysts. Curr. Opin. Biotechnol. 15, 323–329. ( 10.1016/j.copbio.2004.06.001) [DOI] [PubMed] [Google Scholar]
- 30.Smith MR, Khera E, Wen F. 2015. Engineering novel and improved biocatalysts by cell surface display. Ind. Eng. Chem. Res. 54, 4021–4032. ( 10.1021/ie504071f) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jimenez-Rosales A, Flores-Merino MV. 2018. Tailoring proteins to re-evolve Nature: a short review. Mol. Biotechnol. 60, 946–974. ( 10.1007/s12033-018-0122-3) [DOI] [PubMed] [Google Scholar]
- 32.Huang PS, Boyken SE, Baker D. 2016. The coming of age of de novo protein design. Nature 537, 320–327. ( 10.1038/nature19946) [DOI] [PubMed] [Google Scholar]
- 33.Champion E, et al. 2009. Design of α-transglucosidases of controlled specificity for programmed chemoenzymatic synthesis of antigenic oligosaccharides. J. Am. Chem. Soc. 131, 7379–7389. ( 10.1021/ja900183h) [DOI] [PubMed] [Google Scholar]
- 34.Salamone S, Guerreiro C, Cambon E, André I, Remaud-Siméon M, Mulard LA. 2015. Programmed chemo-enzymatic synthesis of the oligosaccharide component of a carbohydrate-based antibacterial vaccine candidate. Chem. Commun. 51, 2581–2584. ( 10.1039/c4cc08805k) [DOI] [PubMed] [Google Scholar]
- 35.Oldrini D, et al. 2018. Combined chemical synthesis and tailored enzymatic elongation provide fully synthetic and conjugation-ready Neisseria meningitidis serogroup X vaccine antigens. ACS Chem. Biol. 13, 984–994. ( 10.1021/acschembio.7b01057) [DOI] [PubMed] [Google Scholar]
- 36.Ihssen J, Haas J, Kowarik M, Wiesli L, Wacker M, Schwede T, Thony-Meyer L. 2015. Increased efficiency of Campylobacter jejuni N-oligosaccharyltransferase PglB by structure-guided engineering. Open Biol. 5, 140227 ( 10.1098/rsob.140227) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ollis AA, Zhang S, Fisher AC, DeLisa MP. 2014. Engineered oligosaccharyltransferases with greatly relaxed acceptor-site specificity. Nat. Chem. Biol. 10, 816–822. ( 10.1038/nchembio.1609) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hunter CD, Guo T, Daskhan G, Richards MR, Cairo CW. 2018. Synthetic strategies for modified glycosphingolipids and their design as probes. Chem. Rev. 118, 8188–8241. ( 10.1021/acs.chemrev.8b00070) [DOI] [PubMed] [Google Scholar]
- 39.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, 490–495. ( 10.1093/nar/gkt1178) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lairson LL, Henrissat B, Davies GJ, Withers SG. 2008. Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555. ( 10.1146/annurev.biochem.76.061005.092322) [DOI] [PubMed] [Google Scholar]
- 41.Chang A, Singh S, Phillips GN, Thorson JS. 2011. Glycosyltransferase structural biology and its role in the design of catalysts for glycosylation. Curr. Opin. Biotechnol. 22, 800–808. ( 10.1016/j.copbio.2011.04.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nidetzky B, Gutmann A, Zhong C. 2018. Leloir glycosyltransferases as biocatalysts for chemical production. ACS Catal. 8, 6283–6300. ( 10.1021/acscatal.8b00710) [DOI] [Google Scholar]
- 43.Koshland DE. 1953. Stereochemistry and the mechanism of enzymatic reactions. Biol. Rev. 28, 416–436. ( 10.1111/j.1469-185X.1953.tb01386.x) [DOI] [Google Scholar]
- 44.Bissaro B, Monsan P, Fauré R, O'Donohue MJ. 2015. Glycosynthesis in a waterworld: new insight into the molecular basis of transglycosylation in retaining glycoside hydrolases. Biochem. J. 467, 17–35. ( 10.1042/BJ20141412) [DOI] [PubMed] [Google Scholar]
- 45.Li C, Wang LX. 2018. Chemoenzymatic methods for the synthesis of glycoproteins. Chem. Rev. 118, 8359–8413. ( 10.1021/acs.chemrev.8b00238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.André I, Potocki-Véronèse G, Morel S, Monsan P, Remaud-Siméon M. 2010. Sucrose-utilizing transglucosidases for biocatalysis. Top. Curr. Chem. 294, 25–48. ( 10.1007/128_2010_52) [DOI] [PubMed] [Google Scholar]
- 47.Cobucci-Ponzano B, Strazzulli A, Rossi M, Moracci M. 2011. Glycosynthases in biocatalysis. Adv. Synth. Catal. 353, 2284–2300. ( 10.1002/adsc.201100461) [DOI] [Google Scholar]
- 48.Umekawa M, Huang W, Li B, Fujita K, Ashida H, Wang LX, Yamamoto K. 2008. Mutants of Mucor hiemalis endo-β-N-acetylglucosaminidase show enhanced transglycosylation and glycosynthase-like activities. J. Biol. Chem. 283, 4469–4479. ( 10.1074/jbc.M707137200) [DOI] [PubMed] [Google Scholar]
- 49.Luley-Goedl C, Nidetzky B. 2010. Carbohydrate synthesis by disaccharide phosphorylases: reactions, catalytic mechanisms and application in the glycosciences. Biotechnol. J. 5, 1324–1338. ( 10.1002/biot.201000217) [DOI] [PubMed] [Google Scholar]
- 50.Nakai H, Kitaoka M, Svensson B, Ohtsubo K. 2013. Recent development of phosphorylases possessing large potential for oligosaccharide synthesis. Curr. Opin. Chem. Biol. 17, 301–309. ( 10.1016/j.cbpa.2013.01.006) [DOI] [PubMed] [Google Scholar]
- 51.O'Neill EC, Field RA. 2015. Enzymatic synthesis using glycoside phosphorylases. Carbohydr. Res. 403, 23–37. ( 10.1016/j.carres.2014.06.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Puchart V. 2015. Glycoside phosphorylases: structure, catalytic properties and biotechnological potential. Biotechnol. Adv. 33, 261–276. ( 10.1016/j.biotechadv.2015.02.002) [DOI] [PubMed] [Google Scholar]
- 53.Schmaltz RM, Hanson SR, Wong CH. 2011. Enzymes in the synthesis of glycoconjugates. Chem. Rev. 111, 4259–4307. ( 10.1021/cr200113w) [DOI] [PubMed] [Google Scholar]
- 54.Wen L, et al. 2018. Toward automated enzymatic synthesis of oligosaccharides. Chem. Rev. 118, 8151–8187. ( 10.1021/acs.chemrev.8b00066) [DOI] [PubMed] [Google Scholar]
- 55.Gantt RW, Peltier-Pain P, Thorson JS. 2011. Enzymatic methods for glyco(diversification/randomization) of drugs and small molecules. Nat. Prod. Rep. 28, 1811–1853. ( 10.1039/c1np00045d) [DOI] [PubMed] [Google Scholar]
- 56.Desmet T, Soetaert W, Bojarová P, Křen V, Dijkhuizen L, Eastwick-Field V, Schiller A. 2012. Enzymatic glycosylation of small molecules: challenging substrates require tailored catalysts. Chem. Eur. J. 18, 10 786–10 801. ( 10.1002/chem.201103069) [DOI] [PubMed] [Google Scholar]
- 57.Weijers CAGM, Franssen MCR, Visser GM. 2008. Glycosyltransferase-catalyzed synthesis of bioactive oligosaccharides. Biotechnol. Adv. 26, 436–456. ( 10.1016/j.biotechadv.2008.05.001) [DOI] [PubMed] [Google Scholar]
- 58.Koeller KM, Wong C. 2000. Synthesis of complex carbohydrates and glycoconjugates: enzyme-based and programmable one-pot strategies. Chem. Rev. 100, 4465–4494. ( 10.1021/cr990297n) [DOI] [PubMed] [Google Scholar]
- 59.Palcic MM. 1999. Biocatalytic synthesis of oligosaccharides. Curr. Opin. Biotechnol. 10, 616–624. ( 10.1016/S0958-1669(99)00044-0) [DOI] [PubMed] [Google Scholar]
- 60.Niggemann J, Kamerling JP, Vliegenthart JFG. 1998. β-1,4-galactosyltransferase-catalyzed synthesis of the branched tetrasaccharide repeating unit of Streptococcus pneumoniae type 14. Bioorg. Med. Chem. 6, 1605–1612. ( 10.1016/S0968-0896(98)00095-9) [DOI] [PubMed] [Google Scholar]
- 61.Fang J, et al. 1998. Highly efficient chemoenzymatic synthesis of α-galactosyl epitopes with a recombinant α(1→3)-galactosyltransferase. J. Am. Chem. Soc. 120, 6635–6638. ( 10.1021/ja9808898) [DOI] [Google Scholar]
- 62.Stanley P, Cummings DR. 2017. Structures common to different glycans. In Essentials of glycobiology [Internet], 3rd edn (eds Varki A, Cummings RD, Esko J, Al E). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 63.Seto NOL, Palcic MM, Compston CA, Li H, Bundle DR, Narang SA. 1997. Sequential interchange of four amino acids from blood group B to blood group A glycosyltransferase boosts catalytic activity and progressively modifies substrate recognition in human recombinant enzymes. J. Biol. Chem. 272, 14 133–14 138. ( 10.1074/jbc.272.22.14133) [DOI] [PubMed] [Google Scholar]
- 64.Marcus SL, Polakowski R, Seto NOL, Leinala E, Borisova S, Blancher A, Roubinet F, Evans SV, Palcic MM. 2003. A single point mutation reverses the donor specificity of human blood group B-synthesizing galactosyltransferase. J. Biol. Chem. 278, 12 403–12 405. ( 10.1074/jbc.M212002200) [DOI] [PubMed] [Google Scholar]
- 65.Seto NO, Compston CA, Szpacenko A, Palcic MM. 2000. Enzymatic synthesis of blood group A and B trisaccharide analogues. Carbohydr. Res. 324, 161–169. ( 10.1016/S0008-6215(99)00297-9) [DOI] [PubMed] [Google Scholar]
- 66.Xu Y, Fan Y, Ye J, Wang F, Nie Q, Wang L, Wang PG, Cao H, Cheng J. 2018. Successfully engineering a bacterial sialyltransferase for regioselective α2,6-sialylation. ACS Catal. 8, 7222–7227. ( 10.1021/acscatal.8b01993) [DOI] [Google Scholar]
- 67.Schmolzer K, Czabany T, Luley-Goedl C, Pavkov-Keller T, Ribitsch D, Schwab H, Gruber K, Weber H, Nidetzky B. 2015. Complete switch from α-2,3- to α-2,6-regioselectivity in Pasteurella dagmatis β-D-galactoside sialyltransferase by active-site redesign. Chem. Commun. 51, 3083–3086. ( 10.1039/c4cc09772f) [DOI] [PubMed] [Google Scholar]
- 68.Aharoni A, Thieme K, Chiu CPC, Buchini S, Lairson LL, Chen H, Strynadka NCJ, Wakarchuk WW, Withers SG. 2006. High-throughput screening methodology for the directed evolution of glycosyltransferases. Nat. Methods 3, 609–614. ( 10.1038/nmeth899) [DOI] [PubMed] [Google Scholar]
- 69.Keys TG, Fuchs HLS, Ehrit J, Alves J, Freiberger F, Gerardy-Schahn R. 2014. Engineering the product profile of a polysialyltransferase. Nat. Chem. Biol. 10, 437–442. ( 10.1038/nchembio.1501) [DOI] [PubMed] [Google Scholar]
- 70.Ding L, Zhao C, Qu J, Li Y, Sugiarto G, Yu H, Wang J, Chen X. 2015. A Photobacterium sp. α2-6-sialyltransferase (Psp2,6ST) mutant with an increased expression level and improved activities in sialylating Tn antigens. Carbohydr. Res. 408, 127–133. ( 10.1016/j.carres.2014.12.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Choi YH, Kim JH, Park JH, Lee N, Kim DH, Jang KS, Park IH, Kim BG. 2014. Protein engineering of α2,3/2,6-sialyltransferase to improve the yield and productivity of in vitro sialyllactose synthesis. Glycobiology 24, 159–169. ( 10.1093/glycob/cwt092) [DOI] [PubMed] [Google Scholar]
- 72.Guo Y, Jers C, Meyer AS, Li H, Kirpekar F, Mikkelsen JD. 2015. Modulating the regioselectivity of a Pasteurella multocida sialyltransferase for biocatalytic production of 3′- and 6'-sialyllactose. Enzyme Microb. Technol. 78, 54–62. ( 10.1016/j.enzmictec.2015.06.012) [DOI] [PubMed] [Google Scholar]
- 73.Zivkovic AM, German JB, Lebrilla CB, Mills DA. 2011. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc. Natl Acad. Sci. USA 108, 4653–4658. ( 10.1073/pnas.1000083107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Newburg DS, Ruiz-Palacios GM, Morrow AL. 2005. Human milk glycans protect infants against enteric pathogens. Annu. Rev. Nutr. 25, 37–58. ( 10.1146/annurev.nutr.25.050304.092553) [DOI] [PubMed] [Google Scholar]
- 75.Kobata A. 2010. Structures and application of oligosaccharides in human milk. Proc. Japan Acad. Ser. B 86, 731–747. ( 10.2183/pjab.86.731) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sprenger GA, Baumgärtner F, Albermann C. 2017. Production of human milk oligosaccharides by enzymatic and whole-cell microbial biotransformations. J. Biotechnol. 258, 79–91. ( 10.1016/j.jbiotec.2017.07.030) [DOI] [PubMed] [Google Scholar]
- 77.Han NS, Kim TJ, Park YC, Kim J, Seo JH. 2012. Biotechnological production of human milk oligosaccharides. Biotechnol. Adv. 30, 1268–1278. ( 10.1016/j.biotechadv.2011.11.003) [DOI] [PubMed] [Google Scholar]
- 78.Petschacher B, Nidetzky B. 2016. Biotechnological production of fucosylated human milk oligosaccharides: prokaryotic fucosyltransferases and their use in biocatalytic cascades or whole cell conversion systems. J. Biotechnol. 235, 61–83. ( 10.1016/j.jbiotec.2016.03.052) [DOI] [PubMed] [Google Scholar]
- 79.Urashima T, Hirabayashi J, Sato S, Kobata A. 2018. Human milk oligosaccharides as essential tools for basic and application studies on galectins. Trends Glycosci. Glycotechnol. 30, SE51–SE65. ( 10.4052/tigg.1734.1SJ) [DOI] [Google Scholar]
- 80.Bode L. 2012. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology 22, 1147–1162. ( 10.1093/glycob/cws074) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xiao Z, et al. 2016. Chemoenzymatic synthesis of a library of human milk oligosaccharides. J. Org. Chem. 81, 5851–5865. ( 10.1021/acs.joc.6b00478) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lin S-W, Yuan T, Li J, Lin C. 2006. Carboxyl terminus of Helicobacter pylori α1,3-fucosyltransferase determines the structure and stability. Biochemistry 45, 8108–8116. ( 10.1021/bi0601297) [DOI] [PubMed] [Google Scholar]
- 83.Choi YH, Kim JH, Park BS, Kim BG. 2016. Solubilization and iterative saturation mutagenesis of α1,3-fucosyltransferase from Helicobacter pylori to enhance its catalytic efficiency. Biotechnol. Bioeng. 113, 1666–1675. ( 10.1002/bit.25944) [DOI] [PubMed] [Google Scholar]
- 84.Zhang F, Zhang Z, Linhardt RJ. 2010. Glycosaminoglycans. In Handbook of glycomics (eds Cummings R, Pierce J), pp. 59–80. London, UK: Academic Press. [Google Scholar]
- 85.Lu W, Zong C, Chopra P, Pepi LE, Xu Y, Amster IJ, Liu J, Boons GJ. 2018. Controlled chemoenzymatic synthesis of heparan sulfate oligosaccharides. Angew. Chemie Int. Ed. 57, 5340–5344. ( 10.1002/anie.201800387) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li J, Su G, Liu J. 2017. Enzymatic synthesis of homogeneous chondroitin sulfate oligosaccharides. Angew. Chemie Int. Ed. 56, 11 784–11 787. ( 10.1002/anie.201705638) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang X, Pagadala V, Jester HM, Lim AM, Pham TQ, Goulas AMP, Liu J, Linhardt RJ. 2017. Chemoenzymatic synthesis of heparan sulfate and heparin oligosaccharides and NMR analysis: paving the way to a diverse library for glycobiologists. Chem. Sci. 8, 7932–7940. ( 10.1039/c7sc03541a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Suflita M, Fu L, He W, Koffas M, Linhardt RJ. 2015. Heparin and related polysaccharides: synthesis using recombinant enzymes and metabolic engineering. Appl. Microbiol. Biotechnol. 99, 7465–7479. ( 10.1007/s00253-015-6821-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Deangelis PL, Liu J, Linhardt RJ. 2013. Chemoenzymatic synthesis of glycosaminoglycans: re-creating, re-modeling and re-designing nature's longest or most complex carbohydrate chains. Glycobiology 23, 764–777. ( 10.1093/glycob/cwt016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sobhany M, Kakuta Y, Sugiura N, Kimata K, Negishi M. 2012. The structural basis for a coordinated reaction catalyzed by a bifunctional glycosyltransferase in chondroitin biosynthesis. J. Biol. Chem. 287, 36 022–36 028. ( 10.1074/jbc.M112.375873) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chavaroche AAE, van den Broek LAM, Springer J, Boeriu C, Eggink G. 2011. Analysis of the polymerization initiation and activity of Pasteurella multocida heparosan synthase PmHS2, an enzyme with glycosyltransferase and UDP-sugar hydrolase activity. J. Biol. Chem. 286, 1777–1785. ( 10.1074/jbc.M110.136754) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kobata A. 2013. Exo- and endoglycosidases revisited. Proc. Jpn. Acad. Ser. B. Phys. Biol. Sci. 89, 97–117. ( 10.2183/pjab.89.97) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li C, Wang L-X. 2016. Endoglycosidases for the synthesis of polysaccharides and glycoconjugates. Adv. Carbohydr. Chem. Biochem. 73, 73–116. ( 10.1016/bs.accb.2016.07.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Williams S. 2015. Transglycosylases. See http://www.cazypedia.org/index.php/Transglycosylases.
- 95.Mackenzie LF, Wang Q, Warren RAJ, Withers SG. 1998. Glycosynthases: mutant glycosidases for oligosaccharide synthesis. J. Am. Chem. Soc. 120, 5583–5584. ( 10.1021/ja980833d) [DOI] [Google Scholar]
- 96.Malet C, Planas A. 1998. From β-glucanase to β-glucansynthase: glycosyl transfer to α-glycosyl fluorides catalyzed by a mutant endoglucanase lacking its catalytic nucleophile. FEBS Lett. 440, 208–212. ( 10.1016/S0014-5793(98)01448-3) [DOI] [PubMed] [Google Scholar]
- 97.Moracci M, Trincone A, Perugino G, Ciaramella M, Rossi M. 1998. Restoration of the activity of active-site mutants of the hyperthermophilic β-glycosidase from Sulfolobus solfataricus: dependence of the mechanism on the action of external nucleophiles. Biochemistry 37, 17 262–17 270. ( 10.1021/bi981855f) [DOI] [PubMed] [Google Scholar]
- 98.Umekawa M, Li C, Higashiyama T, Huang W, Ashida H, Yamamoto K, Wang LX. 2010. Efficient glycosynthase mutant derived from Mucor hiemalis endo-β-N-acetylglucosaminidase capable of transferring oligosaccharide from both sugar oxazoline and natural N-glycan. J. Biol. Chem. 285, 511–521. ( 10.1074/jbc.M109.059832) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sakaguchi K, Katoh T, Yamamoto K. 2016. Transglycosidase-like activity of Mucor hiemalis endoglycosidase mutants enabling the synthesis of glycoconjugates using a natural glycan donor. Biotechnol. Appl. Biochem. 63, 812–819. ( 10.1002/bab.1433) [DOI] [PubMed] [Google Scholar]
- 100.Warmerdam A, Paudel E, Jia W, Boom RM, Janssen AEM. 2013. Characterization of β-galactosidase isoforms from Bacillus circulans and their contribution to GOS production. Appl. Biochem. Biotechnol. 170, 340–358. ( 10.1007/s12010-013-0181-7) [DOI] [PubMed] [Google Scholar]
- 101.Henze M, You D-J, Kamerke C, Hoffmann N, Angkawidjaja C, Ernst S, Pietruszka J, Kanaya S, Elling L. 2014. Rational design of a glycosynthase by the crystal structure of β-galactosidase from Bacillus circulans (BgaC) and its use for the synthesis of N-acetyllactosamine type 1 glycan structures. J. Biotechnol. 191, 78–85. ( 10.1016/j.jbiotec.2014.07.003) [DOI] [PubMed] [Google Scholar]
- 102.D'Almeida A, Ionata M, Tran V, Tellier C, Dion M, Rabiller C. 2009. An expeditious and efficient synthesis of β-D-galactopyranosyl-(1→3)-D-N-acetylglucosamine (lacto-N-biose) using a glycosynthase from Thermus thermophilus as a catalyst. Tetrahedron: Asymmetry 20, 1243–1246. ( 10.1016/j.tetasy.2009.05.007) [DOI] [Google Scholar]
- 103.Sakurama H, et al. 2012. 1,3-1,4-α-L-Fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains. J. Biol. Chem. 287, 16 709–16 719. ( 10.1074/jbc.M111.333781) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cobucci-Ponzano B, et al. 2009. β-glycosyl azides as substrates for α-glycosynthases: preparation of efficient α-L-fucosynthases. Chem. Biol. 16, 1097–1108. ( 10.1016/j.chembiol.2009.09.013) [DOI] [PubMed] [Google Scholar]
- 105.Cobucci-Ponzano B, et al. 2011. A novel α-D-galactosynthase from Thermotoga maritima converts β-D-galactopyranosyl azide to α-galacto-oligosaccharides. Glycobiology 21, 448–456. ( 10.1093/glycob/cwq177) [DOI] [PubMed] [Google Scholar]
- 106.Kwan DH, Constantinescu I, Chapanian R, Higgins MA, Kötzler MP, Samain E, Boraston AB, Kizhakkedathu JN, Withers SG. 2015. Toward efficient enzymes for the generation of universal blood through structure-guided directed evolution. J. Am. Chem. Soc. 137, 5695–5705. ( 10.1021/ja5116088) [DOI] [PubMed] [Google Scholar]
- 107.Micoli F, Costantino P, Adamo R. 2018. Potential targets for next generation antimicrobial glycoconjugate vaccines. FEMS Microbiol. Rev. 42, 388–423. ( 10.1093/femsre/fuy011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Phalipon A, Tanguy M, Grandjean C, Guerreiro C, Belot F, Cohen D, Sansonetti PJ, Mulard LA. 2009. A synthetic carbohydrate-protein conjugate vaccine candidate against Shigella flexneri 2a infection. J. Immunol. 182, 2241–2247. ( 10.4049/jimmunol.0803141) [DOI] [PubMed] [Google Scholar]
- 109.Champion E, et al. 2012. Applying pairwise combinations of amino acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides. J. Am. Chem. Soc. 134, 18 677–18 688. ( 10.1021/ja306845b) [DOI] [PubMed] [Google Scholar]
- 110.Vergès A, Cambon E, Barbe S, Salamone S, Le Guen Y, Moulis C, Mulard LA, Remaud-Siméon M, André I.. 2015. Computer-aided engineering of a transglycosylase for the glucosylation of an unnatural disaccharide of relevance for bacterial antigen synthesis. ACS Catal. 5, 1186–1198. ( 10.1021/cs501288r) [DOI] [Google Scholar]
- 111.Daudé D, Champion E, Morel S, Guieysse D, Remaud-Siméon M, André I. 2013. Probing substrate promiscuity of amylosucrase from Neisseria polysaccharea. ChemCatChem 5, 2288–2295. ( 10.1002/cctc.201300012) [DOI] [Google Scholar]
- 112.Daudé D, André I, Monsan P. 2014. Successes in engineering glucansucrases to enhance glycodiversification. Carbohydr. Chem. 40, 624–645. ( 10.1039/9781849739986) [DOI] [Google Scholar]
- 113.Moulis C, André I, Remaud-Simeon M. 2016. GH13 amylosucrases and GH70 branching sucrases, atypical enzymes in their respective families. Cell. Mol. Life Sci. 73, 2661–2679. ( 10.1007/s00018-016-2244-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Osanjo G, Dion M, Drone J, Solleux C, Tran V, Rabiller C, Tellier C. 2007. Directed evolution of the α-L-fucosidase from Thermotoga maritima into an α-L-transfucosidase. Biochemistry 46, 1022–1033. ( 10.1021/bi061444w) [DOI] [PubMed] [Google Scholar]
- 115.Teze D, Hendrickx J, Czjzek M, Ropartz D, Sanejouand Y-H, Tran V, Tellier C, Dion M. 2014. Semi-rational approach for converting a GH1 beta-glycosidase into a beta-transglycosidase. Protein Eng. Des. Sel. 27, 13–19. ( 10.1093/protein/gzt057) [DOI] [PubMed] [Google Scholar]
- 116.Saumonneau A, Champion E, Peltier-Pain P, Molnar-Gabor D, Hendrickx J, Tran V, Hederos M, Dekany G, Tellier C. 2015. Design of an α-L-transfucosidase for the synthesis of fucosylated HMOs. Glycobiology 26, 261–269. ( 10.1093/glycob/cwv099) [DOI] [PubMed] [Google Scholar]
- 117.Zeuner B, Vuillemin M, Holck J, Muschiol J, Meyer AS. 2018. Loop engineering of an α-1,3/4-L-fucosidase for improved synthesis of human milk oligosaccharides. Enzyme Microb. Technol. 115, 37–44. ( 10.1016/j.enzmictec.2018.04.008) [DOI] [PubMed] [Google Scholar]
- 118.Zeuner B, Jers C, Mikkelsen JD, Meyer AS. 2014. Methods for improving enzymatic trans-glycosylation for synthesis of human milk oligosaccharide biomimetics. J. Agric. Food Chem. 62, 9615–9631. ( 10.1021/jf502619p) [DOI] [PubMed] [Google Scholar]
- 119.Buschiazzo A. 2000. Structural basis of sialyltransferase activity in trypanosomal sialidases. EMBO J. 19, 16–24. ( 10.1093/emboj/19.1.16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Amaya MF, et al. 2004. Structural insights into the catalytic mechanism of Trypanosoma cruzi trans-sialidase. Structure 12, 775–784. ( 10.1016/j.str.2004.02.036) [DOI] [PubMed] [Google Scholar]
- 121.Paris G, Cremona ML, Amaya MF, Buschiazzo A, Giambiagi S, Frasch ACC, Alzari PM. 2001. Probing molecular function of trypanosomal sialidases: single point mutations can change substrate specificity and increase hydrolytic activity. Glycobiology 11, 305–311. ( 10.1093/glycob/11.4.305) [DOI] [PubMed] [Google Scholar]
- 122.Paris G, Ratier L, Amaya MF, Nguyen T, Alzari PM, Frasch ACC. 2005. A sialidase mutant displaying trans-sialidase activity. J. Mol. Biol. 345, 923–934. ( 10.1016/j.jmb.2004.09.031) [DOI] [PubMed] [Google Scholar]
- 123.Jers C, Michalak M, Larsen DM, Kepp KP, Li H, Guo Y, Kirpekar F, Meyer AS, Mikkelsen JD. 2014. Rational design of a new Trypanosoma rangeli trans-sialidase for efficient sialylation of glycans. PLoS ONE 9, e0083902 ( 10.1371/journal.pone.0083902) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pierdominici-Sottile G, Palma J, Roitberg AE. 2014. Free-energy computations identify the mutations required to confer trans-sialidase activity into Trypanosoma rangeli sialidase. Proteins Struct. Funct. Bioinforma. 82, 424–435. ( 10.1002/prot.24408) [DOI] [PubMed] [Google Scholar]
- 125.Nyffenegger C, Nordvang RT, Jers C, Meyer AS, Mikkelsen JD. 2017. Design of Trypanosoma rangeli sialidase mutants with improved trans-sialidase activity. PLoS One 12, 1–11. ( 10.1371/journal.pone.0171585) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gibson GR, Roberfroid MB. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125, 1401–1412. ( 10.1093/jn/125.6.1401) [DOI] [PubMed] [Google Scholar]
- 127.Sanz ML, Polemis N, Morales V, Corzo N, Drakoularakou A, Gibson GR, Rastall RA. 2005. In vitro investigation into the potential prebiotic activity of honey oligosaccharides. J. Agric. Food Chem. 53, 2914–2921. ( 10.1021/jf0500684) [DOI] [PubMed] [Google Scholar]
- 128.Mano MCR, Neri-Numa IA, da Silva JB, Paulino BN, Pessoa MG, Pastore GM. 2018. Oligosaccharide biotechnology: an approach of prebiotic revolution on the industry. Appl. Microbiol. Biotechnol. 102, 17–37. ( 10.1007/s00253-017-8564-2) [DOI] [PubMed] [Google Scholar]
- 129.Díez-Municio M, Herrero M, Olano A, Moreno FJ. 2014. Synthesis of novel bioactive lactose-derived oligosaccharides by microbial glycoside hydrolases. Microb. Biotechnol. 7, 315–331. ( 10.1111/1751-7915.12124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tanaka SI, Takahashi T, Koide A, Ishihara S, Koikeda S, Koide S. 2015. Monobody-mediated alteration of enzyme specificity. Nat. Chem. Biol. 11, 762–764. ( 10.1038/nchembio.1896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hong S, Kyung M, Jo I, Kim YR, Ha NC. 2018. Structure-based protein engineering of bacterial β-xylosidase to increase the production yield of xylobiose from xylose. Biochem. Biophys. Res. Commun. 501, 703–710. ( 10.1016/j.bbrc.2018.05.051) [DOI] [PubMed] [Google Scholar]
- 132.Porras-Domínguez JR, Rodríguez-Alegría ME, Ávila-Fernández Á, Montiel-Salgado S, López-Munguía A. 2017. Levan-type fructooligosaccharides synthesis by a levansucrase-endolevanase fusion enzyme (LevB1SacB). Carbohydr. Polym. 177, 40–48. ( 10.1016/j.carbpol.2017.08.040) [DOI] [PubMed] [Google Scholar]
- 133.Claverie M, Cioci G, Vuillemin M, Monties N, Roblin P, Lippens G, Remaud-Simeon M, Moulis C. 2017. Investigations on the determinants responsible for low molar mass dextran formation by DSR-M dextransucrase. ACS Catal. 7, 7106–7119. ( 10.1021/acscatal.7b02182) [DOI] [Google Scholar]
- 134.Fabre E, Bozonnet S, Arcache A, Willemot R-M, Vignon M, Monsan P, Remaud-Simeon M. 2005. Role of the two catalytic domains of DSR-E dextransucrase and their involvement in the formation of highly α-1,2 branched dextran. J. Bacteriol. 187, 296–303. ( 10.1128/JB.187.1.296-303.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Brison Y, Fabre E, Moulis C, Portais JC, Monsan P, Remaud-Siméon M. 2010. Synthesis of dextrans with controlled amounts of α-1,2 linkages using the transglucosidase GBD-CD2. Appl. Microbiol. Biotechnol. 86, 545–554. ( 10.1007/s00253-009-2241-z) [DOI] [PubMed] [Google Scholar]
- 136.Vergès A, Cambon E, Barbe S, Moulis C, Remaud-Siméon M, André I. 2017. Novel product specificity toward erlose and panose exhibited by multisite engineered mutants of amylosucrase. Protein Sci. 26, 566–577. ( 10.1002/pro.3106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Prapulla SG, Subhaprada V, Karanth NG. 2000. Microbial production of oligosaccharides: a review. Adv. Appl. Microbiol. 47, 299–343. ( 10.1016/S0065-2164(00)47008-5) [DOI] [PubMed] [Google Scholar]
- 138.Mäkeläinen H, Hasselwander O, Rautonen N, Ouwehand AC. 2009. Panose, a new prebiotic candidate. Lett. Appl. Microbiol. 49, 666–672. ( 10.1111/j.1472-765X.2009.02698.x) [DOI] [PubMed] [Google Scholar]
- 139.Nishimoto M, Kitaoka M. 2007. Practical preparation of lacto-N-biose I, a candidate for the bifidus factor in human milk. Biosci. Biotechnol. Biochem. 71, 2101–2104. ( 10.1271/bbb.70320) [DOI] [PubMed] [Google Scholar]
- 140.Kitaoka M. 2015. Diversity of phosphorylases in glycoside hydrolase families. Appl. Microbiol. Biotechnol. 99, 8377–8390. ( 10.1007/s00253-015-6927-0) [DOI] [PubMed] [Google Scholar]
- 141.Pergolizzi G, Kuhaudomlarp S, Kalita E, Field RA. 2017. Glycan phosphorylases in multi-enzyme synthetic processes. Protein Pept. Lett. 24, 696–709. ( 10.2174/0929866524666170811125109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Desmet T, Soetaert W. 2012. Broadening the synthetic potential of disaccharide phosphorylases through enzyme engineering. Process Biochem. 47, 11–17. ( 10.1016/j.procbio.2011.10.039) [DOI] [Google Scholar]
- 143.Kuhaudomlarp S, Patron NJ, Henrissat B, Rejzek M, Saalbach G, Field RA. 2018. Identification of Euglena gracilis β-1,3-glucan phosphorylase and establishment of a new glycoside hydrolase (GH) family GH149. J. Biol. Chem. 293, 2865–2876. ( 10.1074/jbc.RA117.000936) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Macdonald SS, Patel A, Larmour VLC, Morgan-Lang C, Hallam SJ, Mark BL, Withers SG. 2018. Structural and mechanistic analysis of a β-glycoside phosphorylase identified by screening a metagenomic library. J. Biol. Chem. 293, 3451–3467. ( 10.1074/jbc.RA117.000948) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mukherjee K, Narindoshvili T, Raushel FM. 2018. Discovery of a kojibiose phosphorylase in Escherichia coli K-12. Biochemistry 57, 2857–2867. ( 10.1021/acs.biochem.8b00392) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Awad FN, Laborda P, Wang M, Lu AM, Li Q, Cai ZP, Liu L, Voglmeir J. 2017. Discovery and biochemical characterization of a mannose phosphorylase catalyzing the synthesis of novel β-1,3-mannosides. Biochim. Biophys. Acta Gen. Subj. 1861, 3231–3237. ( 10.1016/j.bbagen.2017.09.013) [DOI] [PubMed] [Google Scholar]
- 147.De Groeve MRM, Desmet T, Soetaert W.. 2011. Engineering of cellobiose phosphorylase for glycoside synthesis. J. Biotechnol. 156, 253–260. ( 10.1016/j.jbiotec.2011.07.006) [DOI] [PubMed] [Google Scholar]
- 148.De Groeve MRM, De Baere M, Hoflack L, Desmet T, Vandamme EJ, Soetaert W.. 2009. Creating lactose phosphorylase enzymes by directed evolution of cellobiose phosphorylase. Protein Eng. Des. Sel. 22, 393–399. ( 10.1093/protein/gzp017) [DOI] [PubMed] [Google Scholar]
- 149.De Groeve MRM, Remmery L, Van Hoorebeke A, Stout J, Desmet T, Savvides SN, Soetaert W.. 2010. Construction of cellobiose phosphorylase variants with broadened acceptor specificity towards anomerically substituted glucosides. Biotechnol. Bioeng. 107, 413–420. ( 10.1002/bit.22818) [DOI] [PubMed] [Google Scholar]
- 150.Chen C, Van Der Borght J, De Vreese R, D'Hooghe M, Soetaert W, Desmet T. 2014. Engineering the specificity of trehalose phosphorylase as a general strategy for the production of glycosyl phosphates. Chem. Commun. 50, 7834–7836. ( 10.1039/c4cc02202e) [DOI] [PubMed] [Google Scholar]
- 151.Yamamoto T, Yamashita H, Mukai K, Watanabe H, Kubota M, Chaen H, Fukuda S. 2006. Construction and characterization of chimeric enzymes of kojibiose phosphorylase and trehalose phosphorylase from Thermoanaerobacter brockii. Carbohydr. Res. 341, 2350–2359. ( 10.1016/j.carres.2006.06.024) [DOI] [PubMed] [Google Scholar]
- 152.Nakai H, Petersen BO, Westphal Y, Dilokpimol A, Abou Hachem M, Duus JO, Schols HA, Svensson B. 2010. Rational engineering of Lactobacillus acidophilus NCFM maltose phosphorylase into either trehalose or kojibiose dual specificity phosphorylase. Protein Eng. Des. Sel. 23, 781–787. ( 10.1093/protein/gzq055) [DOI] [PubMed] [Google Scholar]
- 153.Ye X, Zhang C, Zhang YHP. 2012. Engineering a large protein by combined rational and random approaches: stabilizing the Clostridium thermocellum cellobiose phosphorylase. Mol. Biosyst. 8, 1815–1823. ( 10.1039/c2mb05492b) [DOI] [PubMed] [Google Scholar]
- 154.Van der Borght J, Soetaert W, Desmet T.. 2012. Engineering the acceptor specificity of trehalose phosphorylase for the production of trehalose analogs. Biotechnol. Prog. 28, 1257–1262. ( 10.1002/btpr.1609) [DOI] [PubMed] [Google Scholar]
- 155.Hamura K, Saburi W, Matsui H, Mori H. 2013. Modulation of acceptor specificity of Ruminococcus albus cellobiose phosphorylase through site-directed mutagenesis. Carbohydr. Res. 379, 21–25. ( 10.1016/j.carres.2013.06.010) [DOI] [PubMed] [Google Scholar]
- 156.Liu G, Neelamegham S. 2014. A computational framework for the automated construction of glycosylation reaction networks. PLoS ONE 9, e0100939 ( 10.1371/journal.pone.0100939) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sánchez-Rodríguez A, Tytgat HLP, Winderickx J, Vanderleyden J, Lebeer S, Marchal K. 2014. A network-based approach to identify substrate classes of bacterial glycosyltransferases. BMC Genomics 15, 349 ( 10.1186/1471-2164-15-349) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ruffing A, Chen RR. 2006. Metabolic engineering of microbes for oligosaccharide and polysaccharide synthesis. Microb. Cell Fact. 5, 1–9. ( 10.1186/1475-2859-5-25) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.