Abstract
Periodontitis is a polymicrobial, biofilm-caused, inflammatory disease affecting the tooth-supporting tissues. It is not only the leading cause of tooth loss worldwide, but can also impact systemic health. The development of effective treatment strategies is hampered by the complicated disease pathogenesis which is best described by a polymicrobial synergy and dysbiosis model. This model classifies the Gram-negative anaerobe Tannerella forsythia as a periodontal pathogen, making it a prime candidate for interference with the disease. Tannerella forsythia employs a protein O-glycosylation system that enables high-density display of nonulosonic acids via the bacterium's two-dimensional crystalline cell surface layer. Nonulosonic acids are sialic acid-like sugars which are well known for their pivotal biological roles. This review summarizes the current knowledge of T. forsythia's unique cell envelope with a focus on composition, biosynthesis and functional implications of the cell surface O-glycan. We have obtained evidence that glycobiology affects the bacterium's immunogenicity and capability to establish itself in the polymicrobial oral biofilm. Analysis of the genomes of different T. forsythia isolates revealed that complex protein O-glycosylation involving nonulosonic acids is a hallmark of pathogenic T. forsythia strains and, thus, constitutes a valuable target for the design of novel anti-infective strategies to combat periodontitis.
Keywords: cell surface display, periodontitis, polymicrobial community, protein O-glycosylation, sialic acid-like sugar
1. Introduction
1.1. Periodontitis
To proliferate and persist in the oral cavity, bacteria tend to live in biofilms which are clinically described as oral plaque. As highly complex and dynamic polymicrobial communities these biofilms provide protection from shear forces and host immune responses. In a healthy individual, oral bacteria exist in a natural balance with the host. Different factors such as smoking, diabetes, genetic predisposition or poor dental hygiene can cause the community to become dysbiotic, enabling potentially pathogenic bacteria to increase in numbers and cause persistent infections, such as periodontitis. Periodontitis is a chronic inflammation of the gingiva and tooth-supporting tissues, including periodontal ligament and alveolar bone [1,2]. Up to 90% of the worldwide population is affected by periodontal disease, with 10–15% of patients suffering from severe forms, in which, if left untreated, inflammation causes irreversible tooth loss [3]. The dysbiotic oral microbial communities can mediate inflammatory pathology also at distant sites; periodontitis is associated with an increased risk to develop rheumatoid arthritis [4,5], atherosclerosis, cardiovascular diseases [6,7] and cancer [8]. Effective and targeted concepts for the treatment of the disease are currently unavailable. This might be due to the fact that the pathogenesis of periodontitis is still not fully understood. It involves a complex interplay of bacterial, genetic and environmental factors causing growth promotion of dysbiotic, predominantly Gram-negative, anaerobic bacteria that facilitate the progression of inflammation [3,9,10]. A number of potential pathogens promoting the onset of the disease have been identified through high-throughput sequencing, and metagenomic, metatranscriptomic as well as mechanistic studies [11–13]. These include the pathogens Tannerella forsythia, Porphyromonas gingivalis and Treponema denticola constituting the so-called ‘red complex’, a group of bacteria clearly associated with periodontal disease and classified as highly virulent [3,9,10,14].
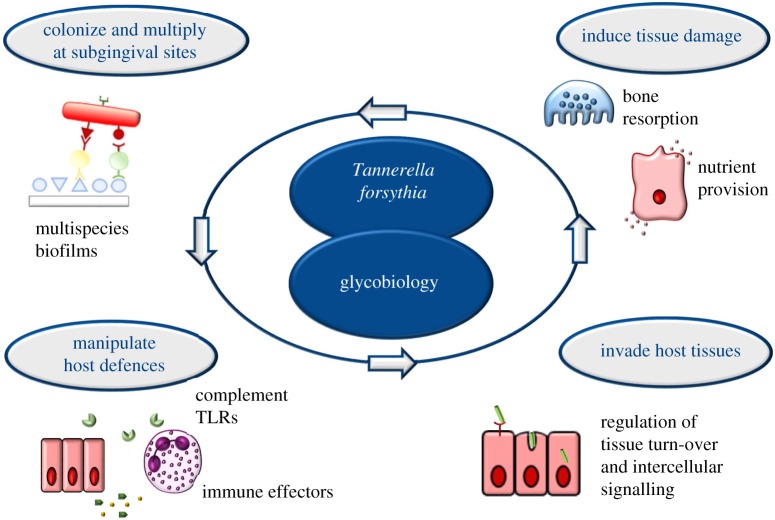
The dysbiotic periodontal community is faced with a survival conundrum (figure 1): on the one hand, these bacteria need to evade immune-mediated killing; on the other hand, they require inflammation to procure nutrients from tissue breakdown such as degraded collagen peptides and haem-containing compounds [10]. Hence, immunosuppression, though a common evasion strategy of many pathogens, is not a viable option for inflammophilic bacteria. Periodontal bacteria can manipulate the interaction with host immune responses—such as neutrophils and complement—to enhance bacterial fitness. Major immune-subversive organisms probably use additional strategies to protect bystander bacteria and elevate the virulence of the entire microbial community, although most of these putative mechanisms have not been confirmed in vivo. For instance, the capacity of P. gingivalis to degrade and inactivate antimicrobial peptides might confer in vivo protection to bystander bacteria [15]. In the interaction with the gingival tissues and underlying immune cells, such as monocytes and macrophages which are present in high numbers in periodontal lesions, the ‘red complex’ bacteria induce the secretion of cytokines such as IL-1β, IL-6, IL-8 and TNF-α, which contribute to the exacerbation of inflammation [16–18].
Figure 1.
Glycobiology underpins the pathogenicity of the oral bacterium Tannerella forsythia. The unique protein-linked O-glycan of the bacterium was found to be involved in (i) the bacterium's establishment in oral plaque, (ii) manipulation of host defence mechanisms, (iii) adhesion to and invasion of host tissues and, eventually, (iv) tissue damage. (Online version in colour.)
Protein glycosylation is employed by numerous pathogenic bacteria to modulate the host immune response, and especially cell surface glycoconjugates such as glycosylated appendages like pili or flagella, play a vital role in orchestrating invasion and infection [19]. We and others have obtained evidence that a complex, protein-bound O-glycan which is displayed at high density on the bacterial cell surface [20] underpins the pathogenicity of T. forsythia [21–24].
1.2. The cell envelope of Tannerella forsythia
Tannerella forsythia was first isolated from advanced periodontal lesions in human oral cavities by Tanner et al. at the Forsyth Dental Center in Boston, MA, USA [25]. Originally designated Bacteroides forsythus, the bacterium was classified as an anaerobic Gram-negative member of the Cytophaga–Bacteroides family. When taxonomic analyses based on 16S rRNA sequences revealed that the species was more closely related to Porphyromonas than to Bacteroides it was moved to the family Porphyromonadaceae and formally reclassified to Tannerella forsythia [26,27].
Tannerella forsythia is a non-motile bacterium with filamentous cell morphology when grown with external supply of the essential cell wall sugar N-acetylmuramic acid (MurNAc) [28]. Physiologically, human-derived T. forsythia strains can be identified based on the following criteria: positive activity for (i) α-glucosidase, (ii) β-glucosidase, (iii) sialidase, (iv) trypsin-like enzyme, (v) negative indole production, (vi) requirement for MurNAc, (vii) colonial morphology and (viii) Gram-stain morphology from blood agar medium deficient in MurNAc [29].
Currently the genome sequences of seven T. forsythia strains are available in public databases; these are the ATCC 43037 type strain (accession number: NZ_JUET00000000.1), the isolates FCD 92A2 (NC_016610.1), KS16 (NZ_AP013045.1), 3313 (NZ_AP013044.1), UB4 (FMMN01000000.1), UB20 (FMMM01000000.1) and UB22 (FMML01000000.1). Additionally, a periodontal health-associated Tannerella species was described recently—HOT-286 (clone BU063; NZ_CP017038.1)—which is the closest phylogenetic relative of T. forsythia [30–32].
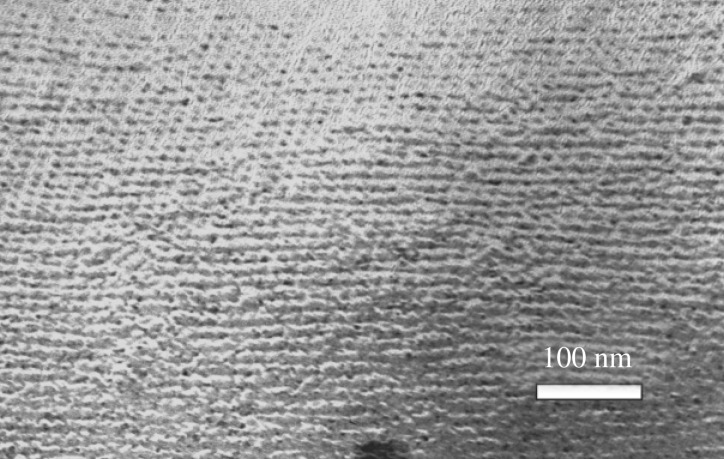
Tannerella forsythia shows a typical Gram-negative cell envelope consisting of a cytoplasmic membrane, a periplasm and an outer membrane that contains rough-type lipopolysaccharide [33]. In addition, the outer membrane of the bacterium is completely covered by a unique surface (S-) layer [34–36]. The S-layer represents a self-assembling, two-dimensional crystalline lattice formed of two different glycosylated proteins, TfsA and TfsB. The T. forsythia S-layer lattice has square symmetry with an overall lattice constant of 10.1 nm as revealed by a freeze-etched preparation of whole bacterial cells [35] (figure 2).
Figure 2.
Square S-layer lattice on a T. forsythia ATCC 43037 cell as visualized by freeze-etching and metal-shadowing.
Compared to other S-layer-carrying bacteria, T. forsythia's status is unique in two ways: (i) it is the only Gram-negative bacterium that is known to possess a glycosylated S-layer and (ii) its S-layer comprises two glycosylated proteins instead of just one [35,37]. Owing to their abundance and high molecular mass, TfsA (134.5 kDa) and TfsB (152.4 kDa) can be easily identified in SDS-polyacrylamide gels, where they migrate at apparent masses of roughly 230 and 270 kDa, respectively, owing to their glycosylation.
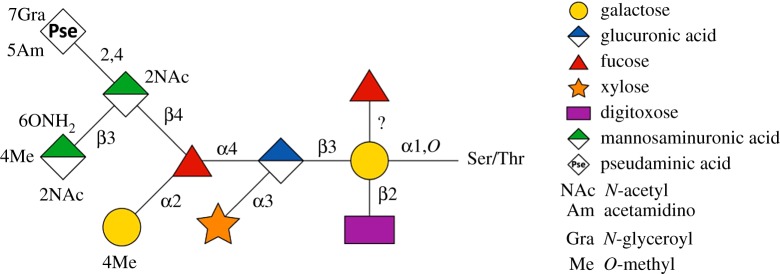
The S-layer glycoproteins are the most abundant glycoproteins of T. forsythia. These and other outer membrane glycoproteins are equipped with a unique, complex O-linked decasaccharide [20,21]. The T. forsythia O-glycan (figure 3) is bound to distinct Ser and Thr residues within the three-amino acid motif D(S/T)(A/I/L/M/T/V) [20]. Notably, the glycan terminates strain-specifically with a modified nonulosonic acid, which can be either a modified pseudaminic acid as shown for the ATCC 43037-type strain, where an N-acetimidoyl (Am) and N-glyceroyl (Gra) substitution is present (Pse5Am7Gra), or a modified legionaminic acid, exemplified by strain UB4 [39]. Besides these sialic acid mimics, other unique S-layer glycan sugars present in the T. forsythia O-glycan are α-l-fucose (Fuc), digitoxose (Dig), xylose (Xyl), N-acetyl mannosaminuronic acid (ManNAcA) and N-acetyl mannosaminuronamide (ManNAcCONH2). The T. forsythia O-glycan, and bacterial cell surface glycans in general, represent the immediate contact zone of bacteria with the environment/host and are, thus, prone to act as specific ligands for cell–cell or cell–bacterium interactions, or to serve as virulence factors.
Figure 3.
Structure of the unique T. forsythia O-glycan [21]. Monosaccharide symbols are shown according to the Symbol Nomenclature for Glycans [38]. (Online version in colour.)
1.3. Nonulosonic acids
Nonulosonic acids (NulOs) are a class of 9-carbon sugars that are α-keto acids. The most abundant naturally occurring NulOs are the sialic acids (Sias) and their derivatives [40–42]. It is established knowledge that Sias play crucial roles in the life cycles of various organisms from eukaryotes to prokaryotes [43]. The various functions of Sias are to some extent opposed, generating competition in which hosts needs to maintain Sias for critical endogenous functions, while constantly changing them to avoid rapidly evolving pathogens that are either binding to or mimicking them. Available data are consistent with this evolutionary scenario [44]. Among the interaction partners of Sias are a family of Sia-binding molecules, mostly expressed by cells of the immune system, which have the potential to interact with sialylated glycans expressed on host cells or certain pathogens [20,45–47].
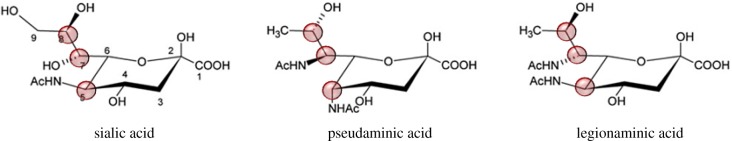
While Sias are commonly present as terminal residues of many eukaryotic glycoconjugates and have also been identified in several prokaryotic polysaccharides, a number of NulOs appear to be unique to bacterial species [48–52]. Owing to their structural and biosynthetic similarities to Sias, these NulOs are also often referred to as Sia-like sugars (figure 4), the most studied of which are pseudaminic acids (such as 5,7-diacetamido-3,5,7,9-tetradeoxy-l-glycero-l-manno-NulO, Pse5,7Ac2) and legionaminic acids (such as 5,7-diacetamido-3,5,7,9-tetradeoxy-d-glycero-d-galacto-NulO, Leg5,7Ac2). For both pseudaminic acid (Pse) and legionaminic acid (Leg), several naturally occurring derivatives have been identified. These most commonly include substitutions of the N-acyl groups at the C-5 and C-7 positions, e.g. with N-acetyl or acetamido (Ac), N-acetimidoyl or acetamidino (Am), N-formyl (Fo) and N-hydroxybutyryl (Hb) groups [50].
Figure 4.
Structures of Sia and bacterial Sia-like sugars (NulOs) with the C5, C7 and C8 highlighted for modifications and stereochemical differences. (Online version in colour.)
Apart from some evidence for involvement in bacterial fitness and pathogenicity of distinct organisms, the function of Sia-like sugars as part of bacterial cell surface components is still largely unknown. It was suggested that bacterial NulOs could play a role as molecular mimics of eukaryotic Sia, thereby contributing to the pathogen's ability to evade or modulate the host's immune responses [45,53,54]. A recent study, for instance, proposes that Pse derivatives on the flagella of Campylobacter jejuni interact with host Siglec-10 to increase IL-10 expression, thus promoting an anti-inflammatory response and host persistence [55].
In contrast to the scarcity of evidence for their function, an increasing number of studies suggest that Pse and Leg derivatives are widespread among prokaryotes, where more than 20% of 1000 microbial genomes examined were found to encode a predicted NulO biosynthesis pathway [45]. Despite the similarities of Pse and Leg to Sias [48], the potential roles of these sugars in host–pathogen interactions are still poorly defined [56], and their distribution among microbes has not yet been systematically investigated.
2. Virulence factors of Tannerella forsythia
Through the expression of virulence factors, periodontal pathogens are able to colonize and persist in the host and promote the destruction of gingival tissues [57].
Several virulence factors that likely contribute to the pathogenicity of T. forsythia have been identified [58,59], and among them is the bacterium's S-layer [34,60,61]. S-layers are present as the outermost cell envelope layer of many bacteria and archaea and have a characteristic ‘lattice-like’ appearance with nanometre-scale periodicity [62]. They cover bacterial cells during all stages of the growth cycle in the form of a closed monolayer. S-layers provide to bacteria a protective coat with molecular sieve properties and generally function in the maintenance of bacterial integrity, display of bacterial components and interaction with the host non-immune and immune cells [63–65].
Other virulence factors of T. forsythia include the outer membrane glycoprotein BspA [66–68], envelope lipoproteins [69], KLIKK-proteases [70], trypsin-like [71] and PrtH [72] proteases, sialidases [73–76] and NanH [77], fucosidase [78], glycosidases [79], methylglyoxal [80] and lipopolysaccharide [33,81,82].
Recently, evidence was obtained that T. forsythia might use a very potent means of ‘biological warfare’ supporting its pathogenicity, namely outer membrane vesicles (OMVs) carrying virulent cargo [83]. OMVs are nanoscopic, spherical particles that are ubiquitously produced by Gram-negative bacteria and have been shown to contribute to the pathogenicity of many bacteria by enriching virulence factors and delivering them over long distances, superseding direct contact with the host [84,85]. Tannerella forsythia produces OMVs with a mean diameter of approximately 100 nm. In a shotgun proteomics approach, these were found to contain a number of known virulence factors and glycoproteins, supportive of a role of T. forsythia's glycosylation in pathogenicity. Analysis of the expression and release of cytokines by macrophages and periodontal ligament cells challenged with T. forsythia OMVs showed a concentration-dependent inflammatory response that was significantly higher than that caused by whole cells, thus supporting the virulence potential of T. forsythia OMVs [83].
3. Nonulosonic acids and their potential association with periodontitis
3.1. Protein glycosylation in Tannerella forsythia
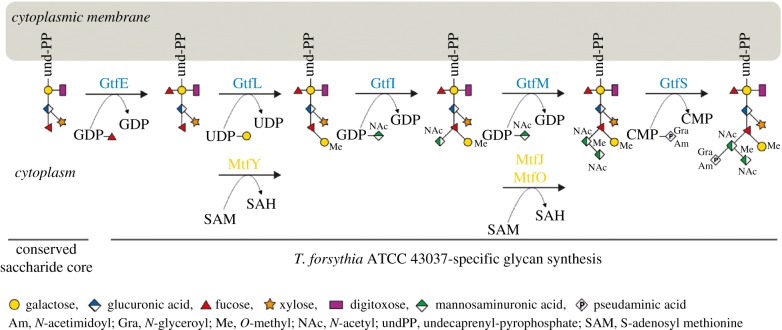
Protein O-glycosylation in T. forsythia is encoded in an approximate 27-kb, polycistronic glycosylation gene cluster working in concert with house-keeping functions of the bacterium [20,21]. Using a gene deletion approach targeted at predicted glycosyltransferases (Gtfs; named GtfSMILE) and methyltransferases (Mtfs; named MtfJOY) encoded in this gene cluster, in combination with mass spectrometry of the protein-released O-glycans, it was shown that the gene cluster encodes the species-specific part of the T. forsythia ATCC 43037 decasaccharide and that this is assembled on a pentasaccharide core from nucleotide-activated sugars in a step-wise fashion [21] (figure 5). The core was previously proposed to be conserved within the Bacteroidetes phylum, to which T. forsythia is affiliated [86], and its biosynthesis is encoded elsewhere on the bacterial genome. Upon synthesis of the pentasaccharide core on an undecaprenyl phosphate lipid carrier, the first carbohydrate residue of the species-specific glycan, a Fuc residue, is conferred by the glycosyltransferase GtfE to the reducing-end Gal of the glycan. The glycan is subsequently elongated with a Gal residue which is transferred by GtfL onto the core Fuc and methylated by MtfY. The assembly of the three sugar branch, consisting of a ManNAcA residue (transferred by GtfI), a ManNAcCONH2 residue (transferred by GtfM), which is methylated by either MtfJ or MtfO, and a Pse5Am7Gra residue (transferred by GtfS), completes the synthesis of the decasaccharide. A putative oligosaccharyltransferase (OTase) transferring the glycan onto the acceptor proteins could not be identified in the T. forsythia genome. This leaves the question open of whether this O-glycan is synthesized by an OTase-dependent or OTase-independent mechanism [87–90].
Figure 5.
Biosynthesis pathway of the T. forsythia ATCC 43037 O-glycan. Modified after [21]. (Online version in colour.)
Since, in addition to the S-layer proteins TfsA and TfsB, also other proteins of T. forsythia are targeted by this protein O-glycosylation system [20], it is referred to as ‘general protein O-glycosylation system’. The species-wide conservation of the respective general protein O-glycosylation gene cluster was assessed by sequence comparison of the ATCC 43037 type strain with six other publicly available T. forsythia genomes (i.e. T. forsythia UB20, FDC 92A2, UB4, KS16, UB22, 3313). In all of the analysed T. forsythia genomes, general protein O-glycosylation gene clusters of comparable size, content and organization were identified [21]. Notably, these gene clusters start at the 5'-end with a Wzx-like flippase encoding gene for translocation of the glycan moiety to the periplasmic space prior to protein transfer, and terminate at the 3'-end with the gtfE gene encoding a fucosyltransferase initiating the biosynthesis of the T. forsythia-specific part of the O-linked decasaccharide. Further, two genes encoding enzyme proteins for the synthesis of UDP-linked ManNAcA (WecB, WecC) and one putatively involved in the Am-modification of the Pse residue were identified in the gene clusters. The major difference found between the analysed protein glycosylation gene clusters was in the alternate presence of six genes encoding the biosynthesis pathway for either CMP-Pse (in strains ATCC 43037 and UB20) or CMP-Leg (in strains FDC 92A2, UB4, KS16 and UB22), respectively.
The corresponding region in the genome of the periodontal health-associated Tannerella species isolate HOT-286 [31] showed a different gene composition lacking CMP-NulO pathway genes and most of the other genes commonly found in the pathogenic strains. Ongoing research on this difficult-to-grow Tannerella sp. [30] is assessing its glycosylation potential [32] as a basis to dissect a potential role in periodontal health and disease.
3.2. Biosynthesis and occurrence of nonulosonic acids in Tannerella forsythia isolates
For the incorporation of Pse or Leg at terminal position of an otherwise mostly identical O-glycan (except for one methyl group) [39], their CMP-activated forms are necessary. These are encoded by six pathway genes, each, named pseBCHGIF and legBCHGIF, respectively. The elucidation of the NulO biosynthetic pathways of T. forsythia relied primarily on knowledge of the Pse and Leg biosynthesis pathways in Helicobacter pylori [91] and Campylobacter jejuni [92], respectively. In a study by Fiedrich et al., CMP-NulO pathway enzymes from T. forsythia ATCC 43037 (CMP-Pse) and UB4 (CMP-Leg) were produced as recombinant proteins and functionally characterized [39]. In both cases, CMP-NulO is formed by the subsequent action of six enzymes starting from nucleotide-activated N-acetylglucosamine (GlcNAc). While the pathway to Pse uses UDP-GlcNAc as a precursor, Leg biosynthesis involves GDP-GlcNAc. Apart from using different sugar nucleotides, the biosynthetic routes for Pse and Leg differ in the reactions performed at the C-2, C-4 and C-5 positions of the hexose intermediates, which results in stereochemical differences at C-5, C-7 and C-8 positions in the final NulO [39]. In the first enzymatic step of the Leg pathway, the dehydratase LegB catalyses a C-4,6 dehydration, while the corresponding enzyme of the Pse pathway, PseB, additionally performs a 5-epimerization reaction. In the subsequent amino transfer at C-4 a stereochemical distinction as an axial (PseC) versus an equatorial (LegC) addition of the amino group is performed. After N-4 acetylation (PseH/LegH), NDP removal catalysed by LegG results in C-2 epimerization, whereas the corresponding enzyme of the Pse biosynthesis pathway, PseG, only possesses NDP-sugar hydrolase activity. In both NulO routes, the formation of the 9-carbon-backbone sugar is catalysed by homologous enzymes (PseI/LegI) via the condensation of a 6-carbon hexose intermediate with the 3-carbon molecule pyruvate. In the final enzymatic step, accomplished by PseF/LegF, CMP activates the free NulO residue using cytidine triphosphate. Considering that in T. forsythia strains unique NulO derivatives are found, i.e. Pse5Am7Gra in strain ATCC 43037 or modified Leg in strain UB4, but no modifying enzyme candidate could be reliably identified in the respective T. forsythia genomes, deviations from the outlined scenario would be anticipated to occur either within the NulO biosynthetic pathway or post CMP-NulO biosynthesis [39]. Since knowledge about the identity and function of NulO-modifying enzymes is scarce, the elucidation of the biochemical mechanism and genetic basis for the Am- and Gra-substitution observed in the ATCC 43037 type strain remains a major challenge.
In the so far analysed, limited number of T. forsythia genomes, the genetic potential for CMP-Leg biosynthesis seems to be more prevalent than that for CMP-Pse biosynthesis [21]. However, this finding needs to be validated by bioinformatic inspection of further genomes and accompanied with glycan analysis of the respective T. forsythia glycans. It should be noted that the genes of either CMP-NulO pathway have been found to be transcribed at a high level in different oral plaque samples containing T. forsythia (S. Bloch, S. Eick, C. Schäffer, unpublished data); this might underline the biological importance of NulOs in the physiology and/or pathogenicity of T. forsythia.
3.3. Protein O-glycosylation in Tannerella forsythia implicates stringent nonulosonic acid transferases
Based on the identification of CMP-NulO biosynthesis genes in all analysed genomes of pathogenic T. forsythia strains and the absence of those in the genome of the periodontal health-associated Tannerella isolate [21], we sought to characterize the NulO transferase from strains ATCC 43037 and UB4, the glycans of which terminate in a Pse5Am7Gra and modified Leg residue, respectively [39]. The enzyme would catalyse the transfer of either NulO on a proximal ManNAcA residue within the O-glycan structure (cf. figure 3).
Despite the predictably wide occurrence of NulOs in bacteria, no experimental evidence of a corresponding NulO transferase has been reported in the literature until research was carried out with the T. forsythia enzymes. The motility-associated factor Maf1 predicted to be involved in the transfer of Pse onto the flagellin of Aeromonas caviae was considered as a candidate Pse transferase [93,94]. With regard to Leg transferases, no predictions are presently available, neither in the literature nor in databases. Interestingly, selected sialyltransferases, i.e. porcine ST3Gal1, Pasteurella multocida sialyltransferase, Photobacterium α2,6-sialyltransferase and Neisseria meningitidis MC58 α2,3-sialyltransferase, were shown to accept CMP-Leg5Ac7Ac as a donor substrate to replace Sia as terminal sugar [95,96].
Bioinformatic analyses of the general protein O-glycosylation gene clusters provided the candidate genes gtfS (Tanf_01245; strain ATCC 43037) and TFUB4_00887 (strain UB4), encoding a putative Pse and a Leg derivative transferase, respectively. The predicted transferases GtfS (445 amino acids; calculated molecular weight, 51.9 kDa) and TFUB4_00887 (442 amino acids; calculated molecular weight, 51.3 kDa) share an identical 241-amino acid long C-terminal domain and have an overall amino acid sequence identity of 81% [97]. A DXD motif starting at position D205 (ATCC 43037) and D202 (UB4), respectively, is present at the beginning of the conserved C-terminal domain. This short motif is found in many glycosyltransferase families, which add a range of different sugars to other sugars, phosphates and proteins. All DXD-containing glycosyltransferases use nucleoside diphosphate sugars as donors and require divalent cations [98]. However, DXD-motifs are usually absent in sialyltransferases [99], which do not require divalent metal ions for enzymatic activity. Instead, two recently identified functional motifs (D/E-D/E-G and HP), found in Neisseria meningitidis (NmB-polyST) and Pasteurella multocida (PmST1), are highly conserved in bacterial sialyltransferases and important to enzyme catalysis and CMP-Neu5Ac binding [100]; these are also present in the putative NulO transferases of the analysed T. forsythia strains. Thus, motif-wise the T. forsythia enzymes seem to be hybrids of glycosyltransferases and sialyltransferases.
Single-gene knock-out mutants targeted at either NulO transferase were analysed for S-layer O-glycan composition by ESI-MS, revealing the loss of the Pse5Am7Gra residue (ATCC 43037) and the Leg derivative (UB4), respectively, and thereby confirming the predicted activity of the enzymes in the glycan biosynthesis pathway. The substrate of either transferase is the fully modified CMP-NulO as was confirmed by the analysis of the cellular pool of nucleotide-activated sugars, where in T. forsythia ATCC 43037, CMP-Pse5Am7Gra with m/z = 683.2 and in T. forsythia UB4, modified CMP-Led with m/z = 654.3 were identified [39].
To learn about the substrate specificity of the T. forsythia Pse5Am7Gra and Leg derivative transferases cross-complementation experiments were performed. Complementation of T. forsythia ATCC 43037 ΔgtfS and T. forsythia UB4 ΔTFUB4_00887 with the non-native enzyme could not restore the native O-glycan phenotype, despite identity of the underlying O-glycan structure. Thus, it is conceivable to assume that GtfS from T. forsythia ATCC 43037 and TFUB4_00887 from T. forsythia UB4 possess high stringency for the CMP-activated NulO substrate. Whether stringency relates to the stereoisomery or the modifications of the NulOs or both remains to be investigated.
4. Nonulosonic acids in the biofilm lifestyle of Tannerella forsythia
4.1. Monospecies biofilm
As part of the multispecies biofilms that constitute the dental plaque [3,9,10], the ability of T. forsythia to form biofilms is a vital prerequisite for the bacterium's survival in its natural environment. The presence of the T. forsythia O-glycan and its charged sugar residues (cf. figure 3) on the S-layer influences the physicochemical properties and hydrophobicity of the cellular surface and, thereby, has implications for the interaction of the bacterium with its environment. Nonulosonic acids in particular have long been implicated in facilitating bacterial virulence and survival within the host and impacting biofilm formation [101,102]. For T. forsythia, there has been obtained evidence that the glycosylated S-layer as an entity plays a role in the bacterium's biofilm lifestyle. In a proteomic analysis, the two S-layer proteins TfsA and TfsB were found to be upregulated in biofilms in comparison with planktonic cells [103], while in a different study, deletion of the T. forsythia S-layer led to an increase in monospecies biofilm formation attributing an inhibitory effect of the S-layer proteins on biofilm growth [104]. While these studies did not take into consideration a potential role of the individual sugars present in the attached O-glycan, first insight into the role of the O-glycan in this process was gained by studies on glycan mutants showing that a three-sugar truncation of the O-glycan including the Pse, ManNAcA and ManNAcCONH2 of T. forsythia ATTC 43037 (T. forsythia ΔwecC; UDP-N-acetyl-d-mannosaminuronic dehydrogenase deletion mutant) resulted in an increased biofilm formation when the bacteria were cultivated in an untreated polystyrene culture dish [105]. Friedrich et al., on the other hand, reported on a promoting role of the terminal NulO derivatives (both Pse and Leg derivative in the different T. forsythia strains) in monospecies biofilm formation on a mucin-coated surface [39]. Not only the deletion of the terminal NulO, as well as the three-sugar truncation in T. forsythia ATTC 43037 ΔwecC, but also the complete deletion of the S-layer and attached glycan in T. forsythia ATTC 43037 ΔtfsAB resulted in a decreased capability of these mutants to form monospecies biofilms [106]. These contradictory results of monospecies biofilm experiments, with an increase in biofilm formation on untreated polystyrene plates upon glycan truncation [105] and a reduction of it on a mucin-coated plate [39,106], showed that biofilm behaviour is influenced by the properties of the surface provided for attachment. The coating with mucin renders the otherwise hydrophobic polystyrene surface highly hydrophilic [107], and influences how T. forsythia strains lacking one or more charged sugar residues can adhere to this surface. Through hydrophobic interactions, steric forces and charge effects, adhesion and bacterial interaction are influenced [108] and this effect becomes especially apparent when the bacterial cell surface composition is altered. The contribution of the cell surface, in particular the terminal Pse and Leg derivatives on interbacterial interaction was reflected in an enhanced autoaggregation of T. forsythia strains lacking the terminal NulO as well as the terminal three-sugar branch [106].
Functional analyses characterizing the growth and biofilm behaviour of T. forsythia strains differing in the display of either Pse5Am7Gra (ATCC 43037) or Leg derivative (UB4) on their surface as well as select cell surface mutants thereof, not only showed a contribution of the cell surface composition to the growth behaviour of T. forsythia, but also discovered differences in the growth characteristics of the two wild-type strains [106]. In a planktonic and monospecies biofilm setting as well as in a multispecies biofilm model, the two strains differed in their growth characteristics, with the clinical isolate T. forsythia UB4 generally growing to higher cell numbers than the T. forsythia ATCC 43037-type strain, suggesting strain-specific differences in the adaptation to varying environments [106].
4.2. Multispecies subgingival biofilm model
In the oral cavity, T. forsythia has to be able to survive in the environment of the multispecies biofilms. Using a subgingival biofilm model incorporating 10 different species of oral bacteria and thereby mimicking the native situation in the oral cavity [109], the extent to which T. forsythia uses its surface glycosylation to persist in its natural habitat was analysed [106].
As already observed in monospecies biofilm experiments, this study showed that the two T. forsythia strains—ATCC 43037 and UB4—differed considerably in their behaviour in the 10-species biofilms [106]. Comparison between the two strains showed that the latter was detected in much higher cell numbers in the multispecies consortium and both strains exhibited a different localization within the biofilm structure, as determined by confocal laser scanning microscopy (CLSM) [106].
Contrary to expectations raised by findings of monospecies biofilm studies, truncation of the O-glycan or ablation of the S-layer as a whole did not influence the bacterium's capability to grow in the multispecies biofilms but modulated interbacterial interaction in the microcolonies within the biofilms, again highlighting the importance of the bacterial cell surface composition in this process [106]. The presence of the S-layer deficient T. forsythia ATCC 43037 ΔtfsAB in the multispecies biofilms led to an decrease in Campylobacter rectus cell numbers suggesting a growth impeding effect of the S-layer on this bacterium; however CLSM analysis and autoaggregation assays revealed this to be independent of a direct contact between the two species [106]. In CLSM, however, a strong co-localization of T. forsythia ATCC 43037 ΔwecC with P. gingivalis was noticeable [106]. Interestingly, a synergistic interaction between the two species had also been described using the same in vitro biofilm model showing reduced growth of T. forsythia in multispecies biofilms containing a P. gingivalis Lys-gingipain-deficient strain [110]. Outside of the multispecies consortium however, increased coaggregation between P. gingivalis and this mutant compared to the other T. forsythia strains analysed could not be identified [106]. These multispecies biofilm experiments suggest that the glycosylated S-layer is not vital for the establishment of the bacterium within the polymicrobial consortium but can influence the interaction with other species such as C. rectus and P. gingivalis. Contrary to a monospecies setting, the NulOs were not found to influence biofilm formation in the in vitro multispecies model [106] (figure 6).
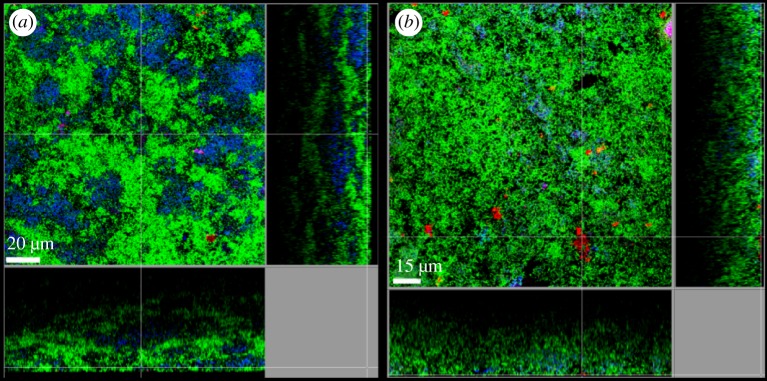
Figure 6.
Fluorescence-in-situ-hybridization staining of fixed multispecies biofilms showing the localization of T. forsythia ATCC 43073 WT (a) and the NulO deficient mutant T. forsythia ATCC 43037 ΔpseC (b). Red: T. forsythia; cyan: P. gingivalis (a)/ T. denticola (b); green: non-hybridized cells (DNA staining YoPro-1 + Sytox). A representative area for one disc each is shown with a top view in the middle panel and side views with the biofilm–disc interface directed towards the top view. Scale bars, (a) 20 µm and (b) 15 µm. Both T. forsythia strains can be clearly detected in the form of microcolonies at the surface of the biofilm. Alteration of the surface glycosylation does not influence the bacteria's capability to grow in the multispecies consortium.
Notably, decoupled from its cell surface composition, in the absence of early colonizing streptococci from the multispecies oral biofilm, the structural arrangement of T. forsythia within the biofilm is affected, changing from the formation of tight bacterial clusters within the biofilm to a more dispersed distribution [111].
5. Tannerella forsythia cell surface composition and immune response
5.1. Immune response to the Tannerella forsythia S-layer
The immunogenic potential of the T. forsythia S-layer was recognized in early studies investigating the IgG responses of human sera to purified TfsA and TfsB [112]. Their role as virulence factor was proposed following experiments showing that the S-layer proteins possess haemagglutination activity, mediate adherence and facilitate the invasion of epithelial KB cells [113].
By using T. forsythia ATCC 43037 wild-type cells in comparison to the S-layer-deficient ΔtfsAB mutant, Sakakibara et al. supported the evidence that the glycosylated S-layer of T. forsythia ATCC 43037 is involved in the adherence to and invasion of human epithelial cell-like gingival carcinoma cells (Ca9–22) and KB cells; this suggested that the T. forsythia S-layer plays an important role in the initiation stage of periodontal disease [60].
Another study comparing the T. forsythia ATCC 43037 wild-type with the ΔtfsAB mutant found that the S-layer conferred increased resistance to both calf and human non-heat-inactivated serum and inhibited deposition of complement factor C3b, a potent agent in the opsonization of pathogens, on the surface of the bacteria [114].
Further, the glycosylated S-layer of T. forsythia ATCC 43037 was shown to suppress the production of proinflammatory mediators IL-1β, TNF-α and IL-8 in U937 macrophages and human gingival fibroblasts [61], at least at the early stage of infection. These cytokines are released by macrophages during the early phase of host cell stimulation and are associated with the acute phase of host response [115]. Both IL-1β and TNF-α may directly stimulate bone resorption in vitro and in vivo [116] or stimulate production of prostaglandin E2 [117,118], which, in turn, is a potent stimulator of bone resorption [119]. IL-8 attracts neutrophils into the inflamed tissue, promoting the development of acute inflammation [120].
A recent study revealed the macrophage-inducible C-type lectin receptor (Mincle) as an important receptor for macrophage sensing of T. forsythia ATCC 43037 and modulation of the immune response to the bacterium in terms of induction of both pro- and anti-inflammatory cytokine secretion [121]. Mincle is an FcRγ-coupled pathogen recognition receptor that recognizes in a Ca2+-dependent manner proinflammatory stimuli from fungal and bacterial pathogens, with suggested mannose/fucose/N-acetylglucosamine/glucose specificity [122]. This might be indicative of the T. forsythia ATCC 43037 S-layer O-glycan with branching fucose residues serving as a ligand for the receptor. However, in-depth research is needed to support this assumption.
5.2. Immunogenicity of the Tannerella forsythia O-glycan
The presence of a sialic acid-like sugar residue as a terminal decoration of the T. forsythia O-glycan [123] suggests a role of the glycan in the pathogenicity of the bacterium, especially in the context of immune evasion and molecular mimicry.
As discussed above, the presence of the T. forsythia S-layer as an entity is a prerequisite for the bacterium to adhere to and invade gingival epithelial cells [60] and delays the immune response by macrophages and gingival fibroblasts [61]. A possible contribution to this process by specifically the S-layer O-glycosylation was first presented in a study using a T. forsythia ATCC 43037 ΔwecC mutant [124]. The terminal branch of the O-glycan was found to regulate dendritic cell effector function, suppress Th17 responses and neutrophil infiltration into the gingival tissues, thereby facilitating the persistence of the bacterium in the host [23,124]. The availability of deletion mutants of T. forsythia ATCC 43037 and T. forsythia UB4 lacking only the terminal NulO derivative further allowed a dissection of the function of Pse and Leg derivative in the interaction with the host [22].
In human oral keratinocytes (HOK) and human monocytes, both representing cells of the first line of defence against invading microorganisms, infection with the two T. forsythia wild-type strains displaying either a Pse or Leg derivative yields preferential immune responses. Infection with T. forsythia UB4 (Leg) dampens the IL-1β response in monocytes when compared to the T. forsythia ATCC 43037-type strain (Pse), but contrary to the latter strongly induces IL-7, which had not been observed in response to the species before; in HOK challenged with either wild-type, T. forsythia UB4 elicits a higher IL-8 release [22]. The cell surface composition of the bacterium seems to be especially relevant in the interaction with HOK where the ablation of the NulO, truncation of the O-glycan by three sugars and the deletion of the S-layer in T. forsythia ATCC 43037 significantly increase IL-8 release by HOK when compared with the parental wild-type strain [22]. These findings are in accordance with previous experiments attesting to the S-layer having a role in delaying the immune response during T. forsythia infection [61], however for the first time demonstrating a contribution of the terminal Pse derivative in this process [22]. The data suggest that the surface glycosylation of T. forsythia plays a prominent role in the initial phase of infection. By the suppression of proinflammatory immune responses on the one hand it might not only enable T. forsythia itself to persist in the host, but also other biofilm bacteria could benefit from it. Tomek et al. further analysed the role of the T. forsythia O-glycan at the immune interface using glycosyltransferase mutants of the T. forsythia ATCC 43037-type strain displaying truncated glycans on their surface [21]. While these mutants, lacking either the terminal Pse5Am7Gra residue, the terminal trisaccharide branch or the complete species-specific part of the glycan exposing a conserved pentasaccharide core with a terminal Fuc residue, did not affect dendritic cell maturation, the cell surface composition did have an effect on T-cell maturation [21]. Truncation of the glycan down to the pentasaccharide core resulted in an enhanced differentiation of Th17 cells [21] confirming the suppressive role of the T. forsythia-specific glycan portion and possibly the terminal Pse5Am7Gra residue in this process [23,124]. While the display of NulOs at the cell surface seems to be especially vital for the bacterium's interaction with the gingival tissues, the complete O-glycan might promote T. forsythia's persistence within the host through dampening the initial response to bacterial infection and maintaining a favourable T-cell environment, thereby orchestrating T. forsythia's virulence potential and pathogenicity.
6. Critical discussion
The periodontal pathogen T. forsythia synthesizes an elaborate O-linked decasaccharide as decoration of its abundant S-layer and other cell surface proteins. This glycan terminates strain-specifically in a modified NulO, which can either be a Pse or a Leg derivative. The O-glycan can be dissected into a T. forsythia-specific portion containing the NulO, and a core region which [21] is proposed to be conserved over the Bacteroidetes phylum [86], to which T. forsythia is affiliated. Considering the increasing documentation of the occurrence of NulOs in pathogenic bacteria [45,49], the functional proof the NulO derivative transferases in T. forsythia strains is an important and necessary step to fully understand the biosynthesis of NulO-containing glycan structures [97] and to exploit these enzymes for engineering of novel sialoglycoconjugates.
For NulOs, major roles have been attributed in biology and disease, including involvement in bacterial biofilm formation and motility. The structural similarity of Pse and Leg to eukaryotic Sias indicates molecular mimicry as a basic strategy these pathogens may employ to evade the host immune response [48,125]. However, it should be noted here that Pse and Leg have different stereochemistry, making Leg a potentially better mimic of host Sias than Pse. Interestingly, T. forsythia strain specifically varies the NulO type on an otherwise overall identical glycan. A comparable situation has so far only been described for C. jejuni, Campylobacter concisus [126], Acinetobacter spp. [127] and Photobacterium profundum [45]. The evolutionary purpose for such structural variation remains a matter of speculation. It is conceivable that different NulO types play distinct roles in the molecular mimicry of host tissues, thus enabling the pathogen to avoid immune detection, modulate the host immune response and enter into host cells to escape immune surveillance [128]. Whether for T. forsythia this correlates with the status of oral plaque or the stage of periodontal disease remains to be investigated.
It has been doubtlessly shown that T. forsythia employs its unique, NulO-containing cell surface to colonize its niche within the polymicrobial oral biofilm and to orchestrate the immune response of resident host tissue and the immune system [22,106]. Studies with human macrophages and gingival fibroblasts demonstrated that the S-layer attenuates the host immune response by evading recognition by the innate immune system, at least at the early stage of infection [61]. There are indications that specifically the S-layer O-glycan is crucial for the modulation of host immunity through Th17 suppression [124]. With regard to the modulation of dendritic cell effector functions, it was specifically shown that the T. forsythia-specific glycan portion suppresses and the pentasaccharide core activates a Th17 response. A very recent study suggests a role specifically of the modified Pse residue (Pse5Am7Gra) present at terminal position on the T. forsythia ATCC 43037 O-glycan in facilitating immune evasion by dampening the response of epithelial tissues to initial infection [22]. Changes in the S-layer and surface glycosylation of T. forsythia might actually contribute to the bacterium's virulence potential by promoting structural arrangements within the biofilm. Whether this contributes to the immune evasion of the biofilm-associated species needs to be tested in functional interaction assays with host cells [106].
From bioinformatic analysis of available genomes of seven pathogenic T. forsythia strains and one periodontal health-associated Tannerella isolate for the presence of genes encoding the biosynthesis of CMP-activated NulOs as required for the incorporation of NulOs in the S-layer O-glycan, it can be concluded that complex, NulO-containing protein O-glycosylation is a hallmark of pathogenic T. forsythia strains; thus, we propose it as a valuable target for the design of novel antimicrobials against periodontitis.
Data accessibility
This article has no additional data.
Authors' contributions
C.S. planned the manuscript. S.B., V.F., M.B.T. contributed data. S.B., P.M. and C.S. wrote the manuscript. All authors gave final approval for publication.
Competing interests
The authors have no competing interests.
Funding
This work was supported by the Austrian Science Fund FWF, projects P20605-B20, P24317-B22, P26836-B11, I2875-B22 (to C.S.), and the Doctoral Programme ‘Biomolecular Technology of Proteins’ W1224.
References
- 1.Socransky S, Haffajee A, Cugini M, Smith C, Kent R. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25, 134–144. ( 10.1111/j.1600-051X.1998.tb02419.x) [DOI] [PubMed] [Google Scholar]
- 2.Darveau RP. 2010. Periodontitis: a polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 8, 481–490. ( 10.1038/nrmicro2337) [DOI] [PubMed] [Google Scholar]
- 3.Hajishengallis G. 2015. Periodontitis: from microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 15, 30–44. ( 10.1038/nri3785) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scher JU, Bretz WA, Abramson SB. 2014. Periodontal disease and subgingival microbiota as contributors for rheumatoid arthritis pathogenesis: modifiable risk factors? Curr. Opin Rheumatol. 26, 424–429. ( 10.1097/BOR.0000000000000076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joseph R, Rajappan S, Nath SG, Paul BJ. 2013. Association between chronic periodontitis and rheumatoid arthritis: a hospital-based case-control study. Rheumatol. Int. 33, 103–109. ( 10.1007/s00296-011-2284-1) [DOI] [PubMed] [Google Scholar]
- 6.Friedewald VE, et al. 2009. The American Journal of Cardiology and Journal of Periodontology Editors' Consensus: periodontitis and atherosclerotic cardiovascular disease. Am. J. Cardiol. 104, 59–68. ( 10.1016/j.amjcard.2009.05.002) [DOI] [PubMed] [Google Scholar]
- 7.Bahekar AA, Singh S, Saha S, Molnar J, Arora R. 2007. The prevalence and incidence of coronary heart disease is significantly increased in periodontitis: a meta-analysis. Am. Heart J. 154, 830–837. ( 10.1016/j.ahj.2007.06.037) [DOI] [PubMed] [Google Scholar]
- 8.Whitmore SE, Lamont RJ. 2014. Oral bacteria and cancer. PLoS Pathog. 10, e1003933 ( 10.1371/journal.ppat.1003933) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hajishengallis G. 2014. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol. 35, 3–11. ( 10.1016/j.it.2013.09.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamont RJ, Hajishengallis G. 2015. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol. Med. 21, 172–183. ( 10.1016/j.molmed.2014.11.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, Gamonal J, Diaz PI. 2013. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. Multidisc. J. Microb. Ecol. 7, 1016–1025. ( 10.1038/ismej.2012.174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ. 2012. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 6, 1176–1185. ( 10.1038/ismej.2011.191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jorth P, Turner KH, Gumus P, Nizam N, Buduneli N, Whiteley M. 2014. Metatranscriptomics of the human oral microbiome during health and disease. MBio 5, e01012–e01014. ( 10.1128/mBio.01012-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trindade F, Oppenheim FG, Helmerhorst EJ, Amado F, Gomes PS, Vitorino R. 2014. Uncovering the molecular networks in periodontitis. Proteomics Clin. Appl. 8, 748–761. ( 10.1002/prca.201400028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hajishengallis G, Lamont RJ. 2014. Breaking bad: manipulation of the host response by Porphyromonas gingivalis. Eur. J. Immunol. 44, 328–338. ( 10.1002/eji.201344202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bostanci N, Belibasakis GN. 2012. Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen. FEMS Microbiol. Lett. 333, 1–9. ( 10.1111/j.1574-6968.2012.02579.x) [DOI] [PubMed] [Google Scholar]
- 17.Birkedal-Hansen H. 1993. Role of cytokines and inflammatory mediators in tissue destruction. J. Periodontal Res. 28, 500–510. ( 10.1111/j.1600-0765.1993.tb02113.x) [DOI] [PubMed] [Google Scholar]
- 18.Okada H, Murakami S. 1998. Cytokine expression in periodontal health and disease. Crit. Rev. Oral Biol. Med. 9, 248–266. ( 10.1177/10454411980090030101) [DOI] [PubMed] [Google Scholar]
- 19.Bernardi A, et al. 2013. Multivalent glycoconjugates as anti-pathogenic agents. Chem. Soc. Rev. 42, 4709–4727. ( 10.1039/c2cs35408j) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Posch G, Pabst M, Brecker L, Altmann F, Messner P, Schäffer C. 2011. Characterization and scope of S-layer protein O-glycosylation in Tannerella forsythia. J. Biol. Chem. 286, 38 714–38 724. ( 10.1074/jbc.M111.284893) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomek MB, et al. 2018. A general protein O-glycosylation gene cluster encodes the species-specific glycan of the oral pathogen Tannerella forsythia: O-glycan biosynthesis and immunological implications. Front. Microbiol. 9, 2008 ( 10.3389/fmicb.2018.02008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bloch S, Zwicker S, Bostanci N, Sjöling A, Bostrom EA, Belibasakis GN, Schäffer C. 2018. Immune response profiling of primary monocytes and oral keratinocytes to different Tannerella forsythia strains and their cell surface mutants. Mol. Oral Microbiol. 33, 155–167. ( 10.1111/omi.12208) [DOI] [PubMed] [Google Scholar]
- 23.Settem RP, Honma K, Sharma A. 2014. Neutrophil mobilization by surface-glycan altered th17-skewing bacteria mitigates periodontal pathogen persistence and associated alveolar bone loss. PLoS ONE 9, e108030 ( 10.1371/journal.pone.0108030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Settem RP, Honma K, Stafford GP, Sharma A. 2013. Protein-linked glycans in periodontal bacteria: prevalence and role at the immune interface. Front. Microbiol. 4, 310 ( 10.3389/fmicb.2013.00310) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanner ACR, Listgarten MA, Ebersole JL, Strezempko MN. 1986. Bacteroides forsythus sp. nov., a slow-growing, fusiform Bacteroides sp. from the human oral cavity. Int. J. Syst. Bacteriol. 36, 213–221. ( 10.1099/00207713-36-2-213) [DOI] [Google Scholar]
- 26.Maiden MF, Cohee P, Tanner AC. 2003. Proposal to conserve the adjectival form of the specific epithet in the reclassification of Bacteroides forsythus Tanner et al. 1986 to the genus Tannerella Sakamoto et al. 2002 as Tannerella forsythia corrig., gen. nov., comb. nov. Request for an opinion. Int. J. Syst. Evol. Microbiol. 53, 2111–2112. ( 10.1099/ijs.0.02641-0) [DOI] [PubMed] [Google Scholar]
- 27.Sakamoto M, Suzuki M, Umeda M, Ishikawa I, Benno Y. 2002. Reclassification of Bacteroides forsythus (Tanner et al. 1986) as Tannerella forsythensis corrig., gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 52, 841–849. ( 10.1099/00207713-52-3-841) [DOI] [PubMed] [Google Scholar]
- 28.Wyss C. 1989. Dependence of proliferation of Bacteroides forsythus on exogenous N-acetylmuramic acid. Infect. Immun. 57, 1757–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braham PH, Moncla BJ. 1992. Rapid presumptive identification and further characterization of Bacteroides forsythus. J. Clin. Microbiol. 30, 649–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vartoukian SR. 2016. Cultivation strategies for growth of uncultivated bacteria. J. Oral Biosci. 58, 142–149. ( 10.1016/j.job.2016.08.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beall CJ, Campbell AG, Dayeh DM, Griffen AL, Podar M, Leys EJ. 2014. Single cell genomics of uncultured, health-associated Tannerella BU063 (oral taxon 286) and comparison to the closely related pathogen Tannerella forsythia. PLoS ONE 9, e89398 ( 10.1371/journal.pone.0089398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frey AM, Ansbro K, Kamble NS, Pham TK, Stafford GP. 2018. Characterisation and pure culture of putative health-associated oral bacterium BU063 (Tannerella sp. HOT-286) reveals presence of a potentially novel glycosylated S-layer. FEMS Microbiol. Lett. 365, 180 ( 10.1093/femsle/fny180) [DOI] [PubMed] [Google Scholar]
- 33.Posch G, Andrukhov O, Vinogradov E, Lindner B, Messner P, Holst O, Schäffer C. 2013. Structure and immunogenicity of the rough-type lipopolysaccharide from the periodontal pathogen Tannerella forsythia. Clin. Vaccine Immunol. 20, 945–953. ( 10.1128/CVI.00139-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee SW, Sabet M, Um HS, Yang J, Kim HC, Zhu W. 2006. Identification and characterization of the genes encoding a unique surface (S-) layer of Tannerella forsythia. Gene 371, 102–111. ( 10.1016/j.gene.2005.11.027) [DOI] [PubMed] [Google Scholar]
- 35.Sekot G, Posch G, Oh YJ, Zayni S, Mayer HF, Pum D, Messner P, Hinterdorfer P, Schäffer C. 2012. Analysis of the cell surface layer ultrastructure of the oral pathogen Tannerella forsythia. Arch. Microbiol. 194, 525–539. ( 10.1007/s00203-012-0792-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kerosuo E. 1988. Ultrastructure of the cell envelope of Bacteroides forsythus strain ATCC 43037T. Oral Microbiol. Immunol. 3, 134–137. ( 10.1111/j.1399-302X.1988.tb00098.x) [DOI] [PubMed] [Google Scholar]
- 37.Sekot G, Schuster D, Messner P, Pum D, Peterlik H, Schäffer C. 2013. Small-angle X-ray scattering for imaging of surface layers on intact bacteria in the native environment. J. Bacteriol. 195, 2408–2414. ( 10.1128/jb.02164-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varki A, et al. 2015. Symbol nomenclature for graphical representations of glycans. Glycobiology 25, 1323–1324. ( 10.1093/glycob/cwv091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedrich V, et al. 2017. Tannerella forsythia strains display different cell-surface nonulosonic acids: biosynthetic pathway characterization and first insight into biological implications. Glycobiology 27, 342–357. ( 10.1093/glycob/cww129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varki A. 1992. Diversity in the sialic acids. Glycobiology 2, 25–40. ( 10.1093/glycob/2.1.25) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varki A. 2008. Sialic acids in human health and disease. Trends Mol. Med. 14, 351–360. ( 10.1016/j.molmed.2008.06.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Traving C, Schauer R. 1998. Structure, function and metabolism of sialic acids. Cell. Mol. Life Sci. 54, 1330–1349. ( 10.1007/s000180050258) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varki A, Cummings RD, Esko JD, Hudson FH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. 2009. Essentials of glycobiology, 2nd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- 44.Angata T, Varki A. 2002. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem. Rev. 102, 439–469. ( 10.1021/cr000407m) [DOI] [PubMed] [Google Scholar]
- 45.Lewis AL, Desa N, Hansen EE, Knirel YA, Gordon JI, Gagneux P, Nizet V, Varki A. 2009. Innovations in host and microbial sialic acid biosynthesis revealed by phylogenomic prediction of nonulosonic acid structure. Proc. Natl Acad. Sci. USA 106, 13 552–13 557. ( 10.1073/pnas.0902431106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ram S, et al. 2018. A novel sialylation site on Neisseria gonorrhoeae lipooligosaccharide links heptose II lactose expression with pathogenicity. Infect. Immun. 86, e00285-18 ( 10.1128/IAI.00285-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bensing BA, et al. 2016. Novel aspects of sialoglycan recognition by the Siglec-like domains of streptococcal SRR glycoproteins. Glycobiology 26, 1222–1234. ( 10.1093/glycob/cww042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knirel YA, Shashkov AS, Tsvetkov YE, Jansson PE, Zähringer U. 2003. 5,7-Diamino-3,5,7,9-tetradeoxynon-2-ulosonic acids in bacterial glycopolymers: chemistry and biochemistry. Adv. Carbohydr. Chem. Biochem. 58, 371–417. ( 10.1016/S0065-2318(03)58007-6) [DOI] [PubMed] [Google Scholar]
- 49.Morrison MJ, Imperiali B. 2014. The renaissance of bacillosamine and its derivatives: pathway characterization and implications in pathogenicity. Biochemistry 53, 624–638. ( 10.1021/bi401546r) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zunk M, Kiefel MJ. 2014. The occurrence and biological significance of the alpha-keto-sugars pseudaminic acid and legionaminic acid within pathogenic bacteria. RSC Adv. 4, 3413–3421. ( 10.1039/c3ra44924f) [DOI] [Google Scholar]
- 51.Kenyon JJ, Marzaioli AM, De Castro C, Hall RM.. 2015. 5,7-Di-N-acetyl-acinetaminic acid: a novel non-2-ulosonic acid found in the capsule of an Acinetobacter baumannii isolate. Glycobiology 25, 644–654. ( 10.1093/glycob/cwv007) [DOI] [PubMed] [Google Scholar]
- 52.Schäffer C, Messner P. 2017. Emerging facets of prokaryotic glycosylation. FEMS Microbiol. Rev. 41, 49–91. ( 10.1093/femsre/fuw036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carlin AF, Uchiyama S, Chang YC, Lewis AL, Nizet V, Varki A. 2009. Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood 113, 3333–3336. ( 10.1182/blood-2008-11-187302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Avril T, Wagner ER, Willison HJ, Crocker PR. 2006. Sialic acid-binding immunoglobulin-like lectin 7 mediates selective recognition of sialylated glycans expressed on Campylobacter jejuni lipooligosaccharides. Infect. Immun. 74, 4133–4141. ( 10.1128/IAI.02094-05) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stephenson HN, et al. 2014. Pseudaminic acid on Campylobacter jejuni flagella modulates dendritic cell IL-10 expression via Siglec-10 receptor: a novel flagellin-host interaction. J. Infect. Dis. 210, 148 714–148 798. ( 10.1093/infdis/jiu287) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poole J, Day CJ, von Itzstein M, Paton JC, Jennings MP.. 2018. Glycointeractions in bacterial pathogenesis. Nat. Rev. Microbiol. 16, 440–452. ( 10.1038/s41579-018-0007-2) [DOI] [PubMed] [Google Scholar]
- 57.O'Brien-Simpson NM, Veith PD, Dashper SG, Reynolds EC. 2004. Antigens of bacteria associated with periodontitis. Periodontology 35, 101–134. ( 10.1111/j.0906-6713.2004.003559.x) [DOI] [PubMed] [Google Scholar]
- 58.Sharma A. 2010. Virulence mechanisms of Tannerella forsythia. Periodontology 54, 106–116. ( 10.1111/j.1600-0757.2009.00332.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Veith PD, O'Brien-Simpson NM, Tan Y, Djatmiko DC, Dashper SG, Reynolds EC. 2009. Outer membrane proteome and antigens of Tannerella forsythia. J. Proteome Res. 8, 4279–4292. ( 10.1021/pr900372c) [DOI] [PubMed] [Google Scholar]
- 60.Sakakibara J, Nagano K, Murakami Y, Higuchi N, Nakamura H, Shimozato K, Yoshimura F. 2007. Loss of adherence ability to human gingival epithelial cells in S-layer protein-deficient mutants of Tannerella forsythensis. Microbiology 153, 866–876. ( 10.1099/mic.0.29275-0) [DOI] [PubMed] [Google Scholar]
- 61.Sekot G, Posch G, Messner P, Matejka M, Rausch-Fan X, Andrukhov O, Schäffer C. 2011. Potential of the Tannerella forsythia S-layer to delay the immune response. J. Dent. Res. 90, 109–114. ( 10.1177/0022034510384622) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Messner P, Schäffer C, Egelseer EM, Sleytr UB. 2010. Occurrence, structure, chemistry, genetics, morphogenesis, and functions of S-layers. In Prokaryotic cell wall compounds: structure and biochemistry (eds König H, Claus H, Varma A), pp. 53–109. Berlin, Germany: Springer. [Google Scholar]
- 63.Sára M, Sleytr UB. 2000. S-Layer proteins. J. Bacteriol. 182, 859–868. ( 10.1128/JB.182.4.859-868.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sleytr UB, Schuster B, Egelseer EM, Pum D. 2014. S-layers: principles and applications. FEMS Microbiol. Rev. 38, 823–864. ( 10.1111/1574-6976.12063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fagan RP, Fairweather NF. 2014. Biogenesis and functions of bacterial S-layers. Nat. Rev. Microbiol. 12, 211–222. ( 10.1038/nrmicro3213) [DOI] [PubMed] [Google Scholar]
- 66.Onishi S, Honma K, Liang S, Stathopoulou P, Kinane D, Hajishengallis G, Sharma A. 2008. Toll-like receptor 2-mediated interleukin-8 expression in gingival epithelial cells by the Tannerella forsythia leucine-rich repeat protein BspA. Infect. Immun. 76, 198–205. ( 10.1128/iai.01139-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sharma A, Sojar HT, Glurich I, Honma K, Kuramitsu HK, Genco RJ. 1998. Cloning, expression, and sequencing of a cell surface antigen containing a leucine-rich repeat motif from Bacteroides forsythus ATCC 43037. Infect. Immun. 66, 5703–5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee HR, Jun HK, Choi BK. 2014. Tannerella forsythia BspA increases the risk factors for atherosclerosis in ApoE(−/−) mice. Oral Dis. 20, 803–808. ( 10.1111/odi.12214) [DOI] [PubMed] [Google Scholar]
- 69.Hasebe A, et al. 2004. Biological activities of Bacteroides forsythus lipoproteins and their possible pathological roles in periodontal disease. Infect. Immun. 72, 1318–1325. ( 10.1128/IAI.72.3.1318-1325.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ksiazek M, Mizgalska D, Eick S, Thøgersen IB, Enghild JJ, Potempa J. 2015. KLIKK proteases of Tannerella forsythia: putative virulence factors with a unique domain structure. Front. Microbiol. 6, 312 ( 10.3389/fmicb.2015.00312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Song Q, Zhang X, Li N, Shen J, Cheng J. 2017. A propeptide-independent protease from Tannerella sp.6_1_58FAA_CT1 displays trypsin-like specificity. J. Basic Microbiol. 57, 50–56. ( 10.1002/jobm.201600486) [DOI] [PubMed] [Google Scholar]
- 72.Hamlet SM, Ganashan N, Cullinan MP, Westerman B, Palmer JE, Seymour GJ. 2008. A 5-year longitudinal study of Tannerella forsythia prtH genotype: association with loss of attachment. J. Periodontol. 79, 144–149. ( 10.1902/jop.2008.070228) [DOI] [PubMed] [Google Scholar]
- 73.Moncla BJ, Braham P, Hillier SL. 1990. Sialidase (neuraminidase) activity among Gram-negative anaerobic and capnophilic bacteria. J. Clin. Microbiol. 28, 422–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saito T, Ishihara K, Kato T, Okuda K. 1997. Cloning, expression, and sequencing of a protease gene from Bacteroides forsythus ATCC 43037 in Escherichia coli. Infect. Immun. 65, 4888–4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roy S, Honma K, Douglas CW, Sharma A, Stafford GP. 2011. Role of sialidase in glycoprotein utilization by Tannerella forsythia. Microbiology 157, 3195–3202. ( 10.1099/mic.0.052498-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ishikura H, Arakawa S, Nakajima T, Tsuchida N, Ishikawa I. 2003. Cloning of the Tannerella forsythensis (Bacteroides forsythus) siaHI gene and purification of the sialidase enzyme. J. Med. Microbiol. 52, 1101–1107. ( 10.1099/jmm.0.05349-0) [DOI] [PubMed] [Google Scholar]
- 77.Thompson H, Homer KA, Rao S, Booth V, Hosie AH. 2009. An orthologue of Bacteroides fragilis NanH is the principal sialidase in Tannerella forsythia. J. Bacteriol. 191, 3623–3628. ( 10.1128/JB.01618-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Megson ZA, et al. 2015. Characterization of an a-L-fucosidase from the periodontal pathogen Tannerella forsythia. Virulence 6, 282–292. ( 10.1080/21505594.2015.1010982) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hughes CV, Malki G, Loo CY, Tanner AC, Ganeshkumar N. 2003. Cloning and expression of a-D-glucosidase and N-acetyl-b-glucosaminidase from the periodontal pathogen Tannerella forsythensis (Bacteroides forsythus). Oral Microbiol. Immunol. 18, 309–312. ( 10.1034/j.1399-302X.2003.00091.x) [DOI] [PubMed] [Google Scholar]
- 80.Maiden MF, Pham C, Kashket S. 2004. Glucose toxicity effect and accumulation of methylglyoxal by the periodontal anaerobe Bacteroides forsythus. Anaerobe 10, 27–32. ( 10.1016/j.anaerobe.2003.12.001) [DOI] [PubMed] [Google Scholar]
- 81.Bodet C, Chandad F, Grenier D. 2006. Inflammatory responses of a macrophage/epithelial cell co-culture model to mono and mixed infections with Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia. Microbes Infect. 8, 27–35. ( 10.1016/j.micinf.2005.05.015) [DOI] [PubMed] [Google Scholar]
- 82.Bodet C, Grenier D. 2010. Synergistic effects of lipopolysaccharides from periodontopathic bacteria on pro-inflammatory cytokine production in an ex vivo whole blood model. Mol. Oral Microbiol. 25, 102–111. ( 10.1111/j.2041-1014.2010.00566.x) [DOI] [PubMed] [Google Scholar]
- 83.Friedrich V, et al. 2015. Outer membrane vesicles of Tannerella forsythia: biogenesis, composition, and virulence. Mol. Oral Microbiol. 30, 451–473. ( 10.1111/omi.12104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Amano A, Takeuchi H, Furuta N. 2010. Outer membrane vesicles function as offensive weapons in host-parasite interactions. Microbes Infect. 12, 791–798. ( 10.1016/j.micinf.2010.05.008) [DOI] [PubMed] [Google Scholar]
- 85.Berleman J, Auer M. 2013. The role of bacterial outer membrane vesicles for intra- and interspecies delivery. Environ. Microbiol. 15, 347–354. ( 10.1111/1462-2920.12048) [DOI] [PubMed] [Google Scholar]
- 86.Coyne MJ, Fletcher CM, Chatzidaki-Livanis M, Posch G, Schäffer C, Comstock LE. 2013. Phylum-wide general protein O-glycosylation system of the Bacteroidetes. Mol. Microbiol. 88, 772–783. ( 10.1111/mmi.12220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Choi KJ, Grass S, Paek S, St Geme JW 3rd, Yeo HJ. 2010. The Actinobacillus pleuropneumoniae HMW1C-like glycosyltransferase mediates N-linked glycosylation of the Haemophilus influenzae HMW1 adhesin. PLoS ONE 5, e15888 ( 10.1371/journal.pone.0015888) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grass S, Lichti CF, Townsend RR, Gross J, St Geme JW 3rd. 2010. The Haemophilus influenzae HMW1C protein is a glycosyltransferase that transfers hexose residues to asparagine sites in the HMW1 adhesin. PLoS Pathog. 6, e1000919 ( 10.1371/journal.ppat.1000919) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schwarz F, Fan Y-Y, Schubert M, Aebi M. 2011. Cytoplasmic N-glycosyltransferase of Actinobacillus pleuropneumoniae is an inverting enzyme and recognizes the NX(S/T) consensus sequence. J. Biol. Chem. 286, 35 267–35 274. ( 10.1074/jbc.M111.277160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Iwashkiw JA. et al. 2012. Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PLoS Pathog. 8, e1002758 ( 10.1371/journal.ppat.1002758) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schoenhofen IC, McNally DJ, Brisson JR, Logan SM. 2006. Elucidation of the CMP-pseudaminic acid pathway in Helicobacter pylori: synthesis from UDP-N-acetylglucosamine by a single enzymatic reaction. Glycobiology 16, 8C–14C. ( 10.1093/glycob/cwl010) [DOI] [PubMed] [Google Scholar]
- 92.Schoenhofen IC, Vinogradov E, Whitfield DM, Brisson JR, Logan SM. 2009. The CMP-legionaminic acid pathway in Campylobacter: biosynthesis involving novel GDP-linked precursors. Glycobiology 19, 715–725. ( 10.1093/glycob/cwp039) [DOI] [PubMed] [Google Scholar]
- 93.Parker JL, Day-Williams MJ, Tomás JM, Stafford GP, Shaw JG. 2012. Identification of a putative glycosyltransferase responsible for the transfer of pseudaminic acid onto the polar flagellin of Aeromonas caviae Sch3N. Microbiologyopen 1, 149–160. ( 10.1002/mbo3.19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Parker JL, Lowry RC, Couto NAS, Wright PC, Stafford GP, Shaw JG. 2014. Maf-dependent bacterial flagellin glycosylation occurs before chaperone binding and flagellar T3SS export. Mol. Microbiol. 92, 258–272. ( 10.1111/mmi.12549) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Watson DC, Leclerc S, Wakarchuk WW, Young NM. 2011. Enzymatic synthesis and properties of glycoconjugates with legionaminic acid as a replacement for neuraminic acid. Glycobiology 21, 99–108. ( 10.1093/glycob/cwq135) [DOI] [PubMed] [Google Scholar]
- 96.Watson DC, Wakarchuk WW, Leclerc S, Schur MJ, Schoenhofen IC, Young NM, Gilbert M. 2015. Sialyltransferases with enhanced legionaminic acid transferase activity for the preparation of analogs of sialoglycoconjugates. Glycobiology 25, 767–773. ( 10.1093/glycob/cwv017) [DOI] [PubMed] [Google Scholar]
- 97.Tomek MB, Janesch B, Maresch D, Windwarder M, Altmann F, Messner P, Schäffer C. 2017. A pseudaminic acid or a legionaminic acid derivative transferase is strain-specifically implicated in the general protein O-glycosylation system of the periodontal pathogen Tannerella forsythia. Glycobiology 27, 555–567. ( 10.1093/glycob/cwx019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Breton C, Šnajdrová L, Jeanneau C, Koča J, Imberty A. 2006. Structures and mechanisms of glycosyltransferases. Glycobiology 16, 29R–37R. ( 10.1093/glycob/cwj016) [DOI] [PubMed] [Google Scholar]
- 99.Brockhausen I. 2014. Crossroads between bacterial and mammalian glycosyltransferases. Front. Immunol. 5, 492 ( 10.3389/fimmu.2014.00492) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Freiberger F, et al. 2007. Biochemical characterization of a Neisseria meningitidis polysialyltransferase reveals novel functional motifs in bacterial sialyltransferases. Mol. Microbiol. 65, 1258–1275. ( 10.1111/j.1365-2958.2007.05862.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Guerry P, Ewing CP, Schirm M, Lorenzo M, Kelly J, Pattarini D, Majam G, Thibault P, Logan S. 2006. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol. Microbiol. 60, 299–311. ( 10.1111/j.1365-2958.2006.05100.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Guerry P. 2007. Campylobacter flagella: not just for motility. Trends Microbiol. 15, 456–461. ( 10.1016/j.tim.2007.09.006) [DOI] [PubMed] [Google Scholar]
- 103.Pham TK, Roy S, Noirel J, Douglas I, Wright PC, Stafford GP. 2010. A quantitative proteomic analysis of biofilm adaptation by the periodontal pathogen Tannerella forsythia. Proteomics 10, 3130–3141. ( 10.1002/pmic.200900448) [DOI] [PubMed] [Google Scholar]
- 104.Narita Y, Sato K, Yukitake H, Shoji M, Nakane D, Nagano K, Yoshimura F, Naito M, Nakayama K. 2014. Lack of a surface layer in Tannerella forsythia mutants deficient in the type IX secretion system. Microbiology 160, 2295–2303. ( 10.1099/mic.0.080192-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Honma K, Inagaki S, Okuda K, Kuramitsu HK, Sharma A. 2007. Role of a Tannerella forsythia exopolysaccharide synthesis operon in biofilm development. Microb. Pathog. 42, 156–166. ( 10.1016/j.micpath.2007.01.003) [DOI] [PubMed] [Google Scholar]
- 106.Bloch S, Thurnheer T, Murakami Y, Belibasakis GN, Schäffer C. 2017. Behavior of two Tannerella forsythia strains and their cell surface mutants in multispecies oral biofilms. Mol. Oral Microbiol. 32, 404–418. ( 10.1111/omi.12182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Crouzier T, Jang H, Ahn J, Stocker R, Ribbeck K. 2013. Cell patterning with mucin biopolymers. Biomacromolecules 14, 3010–3016. ( 10.1021/bm400447z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Renner LD, Weibel DB. 2011. Physicochemical regulation of biofilm formation. MRS Bull. 36, 347–355. ( 10.1557/mrs.2011.65) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guggenheim B, Gmür R, Galicia JC, Stathopoulou PG, Benakanakere MR, Meier A, Thurnheer T, Kinane DF. 2009. In vitro modeling of host-parasite interactions: the ‘subgingival’ biofilm challenge of primary human epithelial cells. BMC Microbiol. 9, 280 ( 10.1186/1471-2180-9-280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bao K, Belibasakis GN, Thurnheer T, Aduse-Opoku J, Curtis MA, Bostanci N. 2014. Role of Porphyromonas gingivalis gingipains in multi-species biofilm formation. BMC Microbiol. 14, 258 ( 10.1186/s12866-014-0258-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ammann TW, Belibasakis GN, Thurnheer T. 2013. Impact of early colonizers on in vitro subgingival biofilm formation. PLoS ONE 8, e83090 ( 10.1371/journal.pone.0083090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yoneda M, et al. 2003. Humoral immune responses to S-layer-like proteins of Bacteroides forsythus. Clin. Diagn. Lab. Immunol. 10, 383–387. ( 10.1128/CDLI.10.3.383-387.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sabet M, Lee S-W, Nauman RK, Sims T, Um H-S. 2003. The surface (S-) layer is a virulence factor of Bacteroides forsythus. Microbiology 149, 3617–3627. ( 10.1099/mic.0.26535-0) [DOI] [PubMed] [Google Scholar]
- 114.Shimotahira N, Oogai Y, Kawada-Matsuo M, Yamada S, Fukutsuji K, Nagano K, Yoshimura F, Noguchi K, Komatsuzawa H. 2013. The surface layer of Tannerella forsythia contributes to serum resistance and oral bacterial coaggregation. Infect. Immun. 81, 1198–1206. ( 10.1128/IAI.00983-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Le J, Vilcek J.. 1987. Tumor necrosis factor and interleukin 1: cytokines with multiple overlapping biological activities. Lab. Invest. 56, 234–248. [PubMed] [Google Scholar]
- 116.Mundy GR. 1993. Role of cytokines in bone resorption. J. Cell. Biochem. 53, 296–300. ( 10.1002/jcb.240530405) [DOI] [PubMed] [Google Scholar]
- 117.Nakao S, Ogtata Y, Shimizu E, Yamazaki M, Furuyama S, Sugiya H. 2002. Tumor necrosis factor alpha (TNF-a)-induced prostaglandin E2 release is mediated by the activation of cyclooxygenase-2 (COX-2) transcription via NFkB in human gingival fibroblasts. Mol. Cell. Biochem. 238, 11–18. ( 10.1023/A:1019927616000) [DOI] [PubMed] [Google Scholar]
- 118.Rausch-Fan X, Ulm C, Jensen-Jarolim E, Schedle A, Boltz-Nitulescu G, Rausch WD, Matejka M. 2005. Interleukin-1b-induced prostaglandin E2 production by human gingival fibroblasts is upregulated by glycine. J. Periodontol. 76, 1182–1188. ( 10.1902/jop.2005.76.7.1182) [DOI] [PubMed] [Google Scholar]
- 119.Offenbacher S, Heasman PA, Collins JG. 1993. Modulation of host PGE2 secretion as a determinant of periodontal disease expression. J. Periodontol. 64, 432–444. ( 10.1902/jop.1993.64.5s.432) [DOI] [PubMed] [Google Scholar]
- 120.Baggiolini M, Dewald B, Moser B. 1993. Interleukin-8 and related chemotactic cytokines—CXC and CC chemokines. Adv. Immunol. 55, 97–179. ( 10.1016/S0065-2776(08)60509-X) [DOI] [PubMed] [Google Scholar]
- 121.Chinthamani S, Settem RP, Honma K, Kay JG, Sharma A. 2017. Macrophage inducible C-type lectin (Mincle) recognizes glycosylated surface (S)-layer of the periodontal pathogen Tannerella forsythia. PLoS ONE 12, e0173394 ( 10.1371/journal.pone.0173394) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Richardson MB, Williams SJ. 2014. MCL and Mincle: C-type lectin receptors that sense damaged self and pathogen-associated molecular patterns. Front. Immunol. 5, 288 ( 10.3389/fimmu.2014.00288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Messner P, Schäffer C, Kosma P. 2013. Bacterial cell-envelope glycoconjugates. Adv. Carbohydr. Chem. Biochem. 69, 209–272. ( 10.1016/B978-0-12-408093-5.00006-X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Settem RP, Honma K, Nakajima T, Phansopa C, Roy S, Stafford GP, Sharma A. 2013. A bacterial glycan core linked to surface (S)-layer proteins modulates host immunity through Th17 suppression. Mucosal Immunol. 6, 415–426. ( 10.1038/mi.2012.85) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. 2004. Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 68, 132–153. ( 10.1128/MMBR.68.1.132-153.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kaakoush NO, Deshpande NP, Wilkins MR, Raftery MJ, Janitz K, Mitchell H. 2011. Comparative analyses of Campylobacter concisus strains reveal the genome of the reference strain BAA-1457 is not representative of the species. Gut Pathog. 3, 15 ( 10.1186/1757-4749-3-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Scott NE, Kinsella RL, Edwards AV, Larsen MR, Dutta S, Saba J, Foster LJ, Feldman MF. 2014. Diversity within the O-linked protein glycosylation systems of acinetobacter species. Mol. Cell. Proteomics 13, 2354–2370. ( 10.1074/mcp.M114.038315) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Amano A, Chen C, Honma K, Li C, Settem RP, Sharma A. 2014. Genetic characteristics and pathogenic mechanisms of periodontal pathogens. Adv. Dent. Res. 26, 15–22. ( 10.1177/0022034514526237) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.