Abstract
The geographical mosaic theory of coevolution predicts that species interactions vary between locales. Depending on who leads the coevolutionary arms race, the effectivity of parasite attack or host defence strategies will explain parasite prevalence. Here, we compare behaviour and brain transcriptomes of Temnothorax longispinosus ant workers when defending their nest against an invading social parasite, the slavemaking ant Temnothorax americanus. A full-factorial design allowed us to test whether behaviour and gene expression are linked to parasite pressure on host populations or to the ecological success of parasite populations. Albeit host defences had been shown before to covary with local parasite pressure, we found parasite success to be much more important. Our chemical and behavioural analyses revealed that parasites from high prevalence sites carry lower concentrations of recognition cues and are less often attacked by hosts. This link was further supported by gene expression analysis. Our study reveals that host–parasite interactions are strongly influenced by social parasite strategies, so that variation in parasite prevalence is determined by parasite traits rather than the efficacy of host defence. Gene functions associated with parasite success indicated strong neuronal responses in hosts, including long-term changes in gene regulation, indicating an enduring impact of parasites on host behaviour.
This article is part of the theme issue ‘The coevolutionary biology of brood parasitism: from mechanism to pattern’.
Keywords: coevolution, parasite prevalence, transcriptomics, social parasites, slavemaking ants, Temnothorax longispinosus
1. Introduction
The evolution of traits and species is largely driven by interactions with other species [1,2]. In particular, antagonistic host–parasite associations can escalate in Red Queen dynamics [3] or coevolutionary arms races [4–6], with parasites perfecting host exploitation and hosts mounting ever better defences. Depending on gene flow and differences in selection pressures between sites [7], hosts or parasites might locally adapt to the opponent [8] or develop more general resistance or offensive traits [9]. Escalation of the Red Queen dynamics can lead to population extinction and replacement, as recently shown for the pathogenic poplar rust fungus in France [10]. Often the species that is better adapted to its opponent is considered to be ahead in the evolutionary arms race. However, recent theoretical models indicate that this is not necessarily the case, as the magnitudes of local and global adaptation are often independent, and the association between local adaptation and coevolutionary advantage is less strong [11]. Antagonistic coevolution between parasites and hosts has been studied intensely from a theoretical perspective, in laboratory experimental evolution set-ups and in comparative field studies [12–14]. We are starting to gain insights into how hosts alter their gene expression in response to parasite contact [15–19], and into the transcriptomic and genomic basis of parasite evolution and host–parasite coevolution [20–24]. Yet, genes and pathways underlying traits important for coevolutionary interactions are often unknown. This is particularly true for some of the most fascinating coevolutionary field models, the brood parasites or social parasites, such as the cuckoos or slavemaking ants.
Brood parasites do not exploit the body of their hosts, but their social or care behaviours [25]. Well studied are the avian brood parasites, such as cuckoos or cowbirds, which take advantage of the care behaviour of other birds to circumvent the costs of parental care [26–28]. The social parasites of the ants and wasps parasitize entire animal societies [29,30]. As in other host–parasite systems, social parasites and their hosts engage in coevolutionary arms races [31–34]. According to Emery's rule, social parasites are often closely related to their hosts [35,36] (but see [37–39]), so that the population size, generation time and evolutionary potential of both opponents are largely similar [40], making the study of reciprocal adaptation particularly interesting.
Here, we focus on the coevolutionary interactions between the acorn ant, Temnothorax longispinosus, and its social parasite, Temnothorax americanus [41], a closely related slavemaking ant [21,42], hereafter referred to as ‘the parasite’. Via recurrent and destructive slave raids, this obligate social parasite exerts severe selection on its main host [43]. Local parasite pressure causes a reduction in host density and colony size and induces changes in the social structure, intra-colonial relatedness and allocation strategies of its host T. longispinosus [44,45]. Indeed, a geographical mosaic of coevolution [1] is evident in population differences in host defence portfolios, including behavioural, chemical and life-history traits of hosts, which can be chiefly explained by geographical variation in parasite pressure [34,46,47]. In low parasite pressure populations, Temnothorax hosts respond to the parasite with coordinated fights, while they move from a fight to a flight strategy in more highly parasitized locales. Temnothorax hosts also exhibit an inducible increase in aggression after parasite contact [48,49], suggesting that the permanent expression of high aggression in the absence of the enemy can be costly. Yet, host colonies responding highly aggressively towards non-nestmate conspecifics fare better during parasite attacks, possibly owing to interactions with T. longispinosus slaves that accompany slavemakers on raids [50,51]. Moreover, aggression increases with parasite pressure over the range of the two Temnothorax host species [52], pointing to convergent coadaptation.
To direct aggression towards invading social parasites, these enemies have to be recognized. In order to circumvent host counter-attacks, Temnothorax, Polyergus or Harpagoxenus slavemakers either mimic the chemical recognition profiles of their hosts [53–55], carry lower concentrations of recognition substances [56], or trace the chemical signature of their local host population [57]. Hosts can respond to these chemical adaptations by intercolonial diversification in cuticular hydrocarbon profiles, which has been shown in both Temnothorax and Formica ant hosts [47,58]. Social parasites also use the secretion of the Dufour's gland as a chemical weapon [46,59] to elicit fights among host defenders to deter host attacks. Parasites benefit from manipulating host aggression, because the likelihood that intruding parasites survive host encounters and the parasite prevalence in the field are linked to their ability to elicit skirmishes among hosts [57].
Many of our behavioural studies investigated geographical variation in host defences [34,46,52,60,61], while controlling for parasite population. The studies that did compare parasite populations [43,45,62] detected variation in behavioural or chemical strategies of parasites and at times even local adaptation. Investigating variation between parasite populations is more difficult as social parasites are less common than hosts and most T. americanus colonies are small, containing on average fewer than five slavemakers. Behavioural differences between host or parasite populations could be due to genetic differences in protein-coding sequences or due to variation in gene expression e.g. caused by changes in gene regulatory sequences. The latter is more likely, as rapid adaptation is often based on shifts in gene expression [63,64]. Indeed, if parasites manage to evade detection, they might not elicit any host attacks and this would leave a clear footprint in the transcriptomes of host defenders. A first gene expression study on the two focal species from a single locale identified interesting candidate genes, whose expression is associated with the raiding state in T. americanus and host nest defence in T. longispinosus [20].
Here, we combine behavioural experiments, chemical analyses of cuticular hydrocarbon profiles and transcriptomics aided by the newly sequenced genome of the ant T. longispinosus (which we publish alongside this study), to test whether host defences and brain gene expression of defending host workers are linked to parasite pressure on host populations or to the ecological success of parasite populations. We used a full-factorial design for our parasite intrusion experiment that allowed us to disentangle the influence of slavemaker origin from host origin on parasite–host interactions. Host populations differ in defence trait expression depending on local parasite pressure, with colonies under low parasite pressure fighting rather than fleeing [34,43,46,47,52,60]. Hence, we predicted that variation in host behaviour and gene expression should depend on parasite pressure, e.g. hosts from less parasitized sites that attack intruding slavemakers should upregulate the expression of aggression genes in their brains.
However, given the evidence of variation between parasite populations [43,45,62], it is also possible that parasite prevalence is more strongly linked to the efficacy of behavioural or chemical offensive strategies of parasite populations. If so, we predict that slavemakers from sites where they are common should manage to evade host detection and will be less frequently attacked. As hosts detect parasites by their chemical profile and recognition cues are known in T. longispinosus [47], we expected that a high parasite prevalence is linked to a lower expression of these recognition cues on the cuticle of slavemakers. Whatever will explain the behavioural responses of host colonies to slavemaker intruders, host or parasite origin, variation in host behaviour, especially attack versus no attack, should be reflected in the expression of behavioural defence genes in the brain.
2. Material and methods
(a). Collection sites and parasite pressure
Temnothorax longispinosus is a primary host of the obligate social parasite and slavemaking ant T. americanus and occurs in eastern North America. Colonies of T. longispinosus and its social parasite for the behaviour and transcriptome project were collected in May–June 2017 at eight sites across their range (electronic supplementary material-I, table T-1). The social parasite T. americanus was found at five sites with varying prevalence, which was estimated by the number of social parasite colonies per colony of the primary host in the community. The generation time of Temnothorax ants is about 10 years and population sizes are large, so that parasite prevalence should be relatively stable over time. However, as social parasites are patchily distributed [65], only intensive sampling will result in reliable estimates of parasite prevalence. We therefore combined our 2017 collection data with published long-term data [34,43,45,65], obtained previously at these locales. Some parasite colonies exploit slaves from two Temnothorax host species. In order to not overestimate parasite pressure on our focal host, we only included half of these mixed colonies when estimating parasite prevalence.
We used a full-factorial design so that host colonies from each population were subjected to the intrusion by a slavemaker of each parasite population. This allowed us to disentangle the impact of host and parasite origin on the outcome of the behavioural interaction and associated gene expression. Therefore, we used information on parasite prevalence in the local ant community for both the host and the parasite population. We use the term ‘parasite prevalenceHOST’ to indicate parasite prevalence of the host colony's source population and ‘parasite prevalencePARASITE’ to specify parasite prevalence in the population of the slavemaker intruder. Whereas parasite prevalence in the host population provides information on the strength of parasite pressure on the host, as we discuss below, parasite prevalence could also be regarded as a measure of the ecological success of the social parasite population.
(b). Ant collection and maintenance
Ant colonies were detected in acorns, sticks and cracks of rocks. Each colony was separately transferred with some leaf litter into a Ziploc bag. Colonies were fed with cookie crumbs and moisture was maintained with water-soaked cotton. They were kept at 8°C until the transfer from the field to the laboratory, which was done within 16 days of collection. In the laboratory, each colony was given an artificial nest site to relocate to, made out of 4 mm Plexiglas, with a 50 × 10 mm cavity, sandwiched between two glass slides. Nests were kept in plastered three-chambered nest boxes to avoid desiccation. Ad libitum supply of water and honey was provided and ants were fed twice a week with crickets. Host colonies were kept at 18 to 22°C with a 16 L : 8 D cycle to slow down brood development. T. americanus colonies were kept at 25°C with a14 L : 10 D cycle. A week before the intrusion test, host colonies were transferred to the 25°C climate chamber for acclimation.
(c). Intrusion tests and sampling regime
Intrusion tests included a careful introduction of a live slavemaking worker (the parasite) into a host nest. We blocked the nest entrance for 1 h to ensure intense contact between parasite and hosts [34]. The intrusion test included 96 queenright Temnothorax longispinosus colonies from eight sites, each with at least 15 workers. We selected the experimental ant colonies so that there were no population differences in colony size (Kruskal–Wallis test: χ2 = 2.21, d.f. = 7, p = 0.947). We used 96 T. americanus workers from 31 colonies from four sites (on average, workers from 7.8 colonies per site) for the intrusion tests (see electronic supplementary material, Summary_tables_BEH_CHC_GCAvg n SD).
In the field, hosts encounter social parasites only during the raiding season from July to September. Parasite encounter alters host responses at least for a fortnight [51]. To ensure a similar non-induced state in host colonies, we (i) collected colonies a month before the onset of the raiding season, (ii) kept host and parasite colonies in separate boxes to avoid accidental encounters, and (iii) used each host colony only once. Host colonies had therefore no encounters with their slavemaker parasites for at least one year. We performed the intrusion tests during the raiding season over four consecutive days (18–21 July 2017). We controlled for light, temperature (25°C), humidity and time of the day by performing the experiments in a climate-controlled environmental chamber between 14.00 and 16.00. Each experiment was set up by the same experimenter and recorded on video. After 45 min, we recorded the number of workers involved in aggressive responses towards the intruder. An aggressive response included behaviours such as mandible opening (a threat behaviour), holding, biting and stinging. After 1 h we recorded whether the parasite was freely moving through the host nest or was under direct attack or killed by host workers. Behavioural analyses were conducted so that the experimenter was blind to the origin of the slavemaker.
Host colony aggression towards intruding parasite workers was measured as the proportion of colony members responding to the intruder. We ran a generalized linear model (GLM) with proportion of total responders as response variables and parasite prevalencePARASITE, colony size and parasite prevalenceHOST with two-way interactions as explanatory variables. We used the family parameter ‘quasi-binomial’ to control for overdispersion and checked a normal distribution of residuals graphically and statistically with the Shapiro–Wilk normality test. If one of the interaction terms was significant, we kept the full model. Host colony response was noted as under attack if the parasite was immobilized or killed by the host colony or as free if the parasite was alive and not currently attacked by host workers at the end of the intrusion test. To analyse host colony response, we used a generalized linear model (family binomial) and parasite prevalencePARASITE, colony size and parasite prevalenceHOST as explanatory variables. We started with a full model but owing to absence of any significant interactions, the model was reduced to a simple additive model. Statistical analysis were performed in R v. 3.4.1 [66].
(d). Cuticular hydrocarbon recognition cues and parasite prevalence
In the host T. longispinosus, nine hydrocarbons are particularly relevant for nestmate recognition and explain most of the variance in aggression between non-nestmates [47]. Slavemakers, including T. americanus, carry lower concentrations of these hydrocarbons on their cuticle compared with Temnothorax host species [56] and we analysed here whether slavemakers from high parasite prevalence sites carry less (or more) recognition substances on their cuticle. To this end, we analysed the cuticular hydrocarbon (CHC) profile of 7–12 parasite workers from our experiments (total N = 40, electronic supplementary material-I, table T-2) from each of the four parasite populations using gas chromatography and mass spectrometry (electronic supplementary material-I, §S-1). This was done blind, so that the observers did not know the samples' origin. We calculated the total proportion of these recognition cues of all cuticular hydrocarbons (electronic supplementary material-I, table T-3). Using a linear mixed-effects model (LMM), we tested whether this proportion is linked to parasite prevalence in their source population (parasite prevalencePARASITE) and/or the slavemaker population, and removed non-significant factors until the Akaike information criterion (AIC) was minimal, and included observer ID as random factor. Furthermore, we analysed the composition of the recognition cues depending on parasite prevalencePARASITE and slavemaker population using a permutational ANOVA (adonis, R package vegan; http://cc.oulu.fi/~jarioksa/softhelp/vegan/html/adonis.html).
(e). RNA samples, extraction and sequencing
For the transcriptome analysis we used 32 host colonies (electronic supplementary material-I, table T-2), four from each site, which did not differ in colony size (Kruskal–Wallis test: χ2 = 7.14, d.f. = 7, p = 0.41), which formed a subset of the intrusion trials. Experiments were conducted with host origin randomized during each day and tested against social parasites from a randomly selected population. Each of the four focal host colonies per population encountered a parasite from a different population. Directly thereafter host worker(s) in direct contact or closest to the social parasite were picked and transferred to a vial, which was immediately dipped in liquid nitrogen (−196°C) and later transferred to −80°C freezer. In some host colonies, none of the host workers responded with aggression to the parasitic intruder, in others many. Moreover, we found intra-colonial variation in that only some workers, the guards, focused on the intruder. We expect also variation in gene expression between workers of the same colony, which differed in their behavioural responses. However, here we focused on inter-population differences in gene expression in guards in response to a slavemaker intruder and to gain independent data we sampled only a single worker per colony for four colonies for each of the eight host populations.
To identify genes involved in host responses to social parasites, we focused on brain tissues for the transcriptome analysis. We prepared 32 RNA extractions from each of the 32 host brains (1 worker × 8 host populations × 4 parasite populations = 32 hosts). Ant brains were dissected on dry ice [16]. Brains were separately crushed in 50 µl of TRIZOL for storage at −20°C before RNA extraction. RNA was extracted from brain samples separately using the RNeasy Mini extraction kit (Qiagen), and libraries were constructed and sequenced on Illumina HiSeq 4000 by Beijing Genomics Institute (BGI), Shenzhen, PR China, resulting in about 46 Mio paired-end 100 bp long reads per library. Sequence data were already cleaned by BGI by removing adapter sequences and low-quality reads. The data obtained from BGI were checked using FASTQC v. 0.11.5 for read quality [67]. Raw read data can be obtained in the Sequence Read Archive (SRA, submission no. SUB4538765).
(f). Sequencing and annotation of the T. longispinosus genome
A single monogynous T. longispinosus colony was used to provide all samples for genome sequencing. DNA was isolated from male pupa using the DNeasy Mini kit (Qiagen). A single male pupa was used for the construction of the three paired-end libraries (300, 500 and 800 bp), and a pooled sample of eight male pupae for the mate pair libraries with 2, 5 and 8 kb insert sizes. Library construction and sequencing of 150 bp reads was conducted on an Illumina NextSeq 500 at StarSEQ, Mainz, Germany. Reads were assembled with AllPaths-LG [68] (v. r49967), yielding 3987 scaffolds and 26 795 contigs, with a scaffold N50 of 514 kb and a contig N50 of 30 kb. In total, 92.3% of the conserved arthropod orthologues (BUSCO [69]) were present in the assembly.
A T. longispinosus-specific repeat library was constructed (see electronic supplementary material-II) and used to identify repetitive elements (13.51% of the T. longispinosus assembly). In total, 13 028 protein-coding genes were annotated using a 3-pass iterative MAKER [70] workflow (v. 2.31.8). Functional annotations were obtained based on BLAST searches against the Uniprot-Arthropod database [71] (release 2018 04; e-value: 1e − 3) and Interproscan [72] (v. 5.24-63.0). For further details on genome assembly and annotation see electronic supplementary material-II. The Temnothorax longispinosus OGSv1.0 genome is accessible at GenBank (accession no. PRJNA449506).
(g). Expression analysis and functional annotation
Clean reads were aligned against the T. longispinosus OGSv1.0 genome using HISAT2 2.1.0 with default settings [73]. Read counts of the mapped sequences were obtained using HTSeq-count v. 0.9.1 [74]. First, we controlled for parasite prevalencePARASITE and searched for genes differentially expressed based on parasite prevalenceHOST and then we reversed the procedure. We used DESeq2 v. 1.16.1 [75,76] to run these models, only taking genes as differentially expressed into account with an adjusted p-value of false discovery rate (FDR) < 0.05. Since both parasite prevalenceHOST and parasite prevalencePARASITE are continuous variables, the resulting lists of differentially expressed genes refer to genes that are e.g. overexpressed in hosts facing more parasites from populations with higher or lower parasite prevalence. The log fold change was measured in units of change of the continuous variable. To investigate modules of commonly expressed genes, we performed weighted gene co-expression network analysis (WGCNA) using the package WGCNA v. 1.63 provided by Bioconductor [77]. The soft-threshold power was set to 3 according to the scale free topology [78]. Fifty was chosen as the minimum number of genes in a module and the dissimilarity threshold was set to 0.25. We searched for functions of the different sets of genes associated with traits by performing a GO enrichment analysis. GO enrichment analysis was performed using TopGO v. 2.28.0, performing a Fisher's exact test on the different sets of genes compared with the whole genome using the weight01 algorithm [79]. To see which pathways were enriched in the set of genes that showed a positive association to a trait, we performed KEGG pathway analysis. Pathway analysis was performed using the online tool http://www.genome.jp/kegg/tool/map_pathway1.html; results can be obtained from the electronic supplementary material-I, table T-4.
3. Results
(a). Behaviour
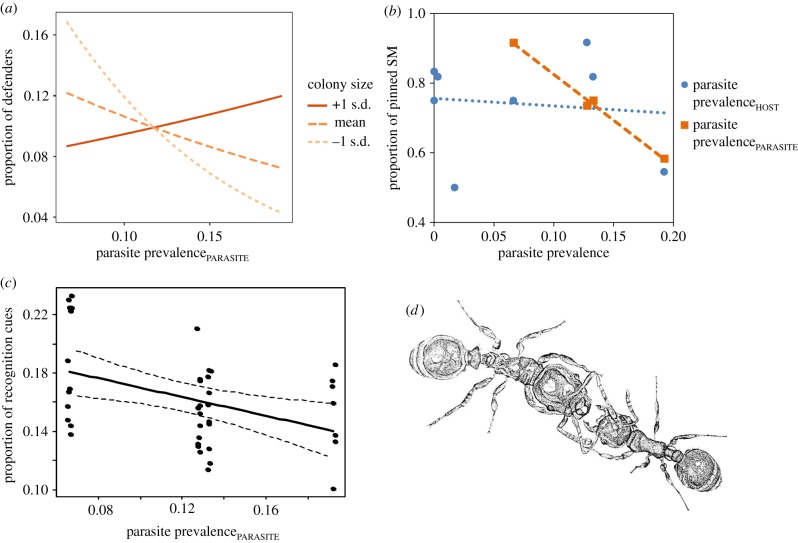
The proportion of workers responding to the intruder with aggression varied with the prevalence of the parasite population and colony size as well as their interaction (GLM: parasite prevalencePARASITE: t1,90 = −3.068, p = 0.003; colony size: t1,89 = −2.715, p = 0.008; parasite prevalencePARASITE × colony size: t1,87 = 2.991, p = 0.004). In small colonies, a larger proportion of workers defended their nest against parasites from low parasite prevalence sites compared with larger colonies. In contrast, when host colonies encountered a parasite from a high prevalence population, in small colonies often none or a few of the workers detected and attacked the intruder, while the proportion of attackers increased with colony size (figure 1a). Parasite prevalence on the host population and its interaction with the other variables did not affect the proportion of defenders in the host colony (GLM: parasite prevalenceHOST: t1,88 = −5.236, p = 0.269; parasite prevalenceHOST × colony size: t1,85 = −0.109, p = 0.335; parasite prevalenceHOST × parasite prevalencePARASITE: t1,86 = 2.746, p = 0.925). Moreover, the parasite's state i.e. whether it was ranging free or under attack by host defenders was also linked to the prevalence of the parasite population and colony size, but not to its interaction (GLM: parasite prevalencePARASITE: z1,89 = 2.54, p = 0.011; colony size: t1,88 = −2.27, p = 0.022; parasite prevalenceHOST: z1,87 = 0.557, p = 0.577). Parasites from high prevalence communities were less often pinned or killed by host defenders than parasites from populations with a lower parasite prevalence (figure 1b). For example, we found 42% of the parasites from the Ohio site moved freely through the host nest, but only 8% of the parasites from Vermont. Larger host colonies were more likely to detect and immobilize or kill the intruding parasite, compared with smaller ones.
Figure 1.
(a) Proportion of host workers involved in defence against the intruding slavemaker was affected by an interaction between the success of the slavemaking population (parasite prevalencePARASITE) and host colony size. Lines represent three different colony size classes (large = mean + 1 s.d.: solid line; midsized colonies = mean: long-dashed line; small colonies = mean − 1 s.d.: short-dashed line). (b) The percentage of slavemaker intruders that were immobilized or killed by host defenders (proportion of pinned SM) depended on the success of the slavemaking population (parasite prevalencePARASITE, brown squares, dashed line), but not on parasite pressure (blue circles, dotted line). (c) Total proportion of cuticular recognition cues in T. americanus in relation to its success. Each data point (n = 40) represents the proportion of recognition cues in the CHC profile of one T. americanus worker (i.e. nine CHCs identified as relevant for nestmate recognition in T. longispinosus). The graph shows a linear regression line with 95% confidence interval. (d) T. americanus slavemaker worker (left) interacting with a smaller T. longispinosus host worker (drawing by Inon Scharf). (Online version in colour.)
(b). Chemical recognition cues
Slavemakers from higher prevalence populations possessed fewer recognition cues compared with social parasites from populations with lower parasite prevalence (figure 1c). The proportion of recognition cues from all hydrocarbons decreased with increasing parasite prevalence (LMM: χ² = 8.053, d.f. = 1, p = 0.0045). When parasite prevalence was accounted for, slavemaker population had no further effect on the proportion of recognition cues and was not retained in the final model. In a multivariate analysis, both parasite prevalence and population of the slavemaker affected the composition (as opposed to the total proportion) of the recognition cues (ADONIS; parasite prevalencePARASITE: F = 4.16, d.f. = 1, p = 0.008; slavemaker population: F = 4.30, d.f. = 2, p = 0.002). We detected differences between all pairs of populations (all F > 2.7, FDR-adjusted p < 0.048).
(c). Gene expression analysis
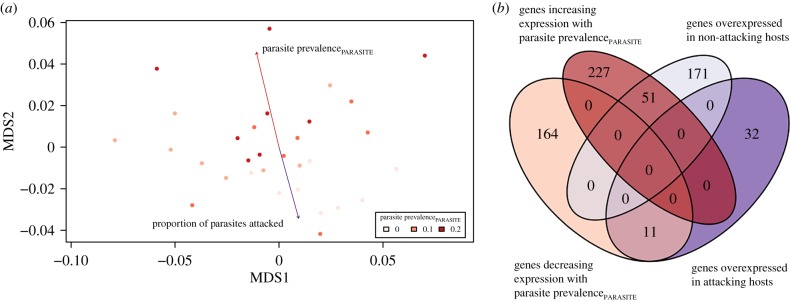
Gene expression in the brain of host defenders was mainly influenced by parasite prevalence in the population of the slavemaker intruder (figure 2a) and much less so by parasite prevalence in the host population. Indeed, differential gene expression analysis identified only a single gene that increased its expression with increasing parasite prevalenceHOST. This gene was annotated as armadillo repeat containing protein 4 in T. longispinosus, which is known to be involved the in Wnt signalling pathway in Drosophila [80] (table 1). Instead, 278 genes increased their expression with increasing parasite prevalence of the social parasite population and an additional 175 genes lowered their expression with parasite prevalencePARASITE (figure 2b).
Figure 2.
(a) Multi-dimensional scaling (MDS) plot showing sample distribution based on variance in gene expression including all contigs. Different shades of red denote variation in prevalence of the population of the parasite intruder (parasite prevalencePARASITE). As parasite prevalencePARASITE was negatively linked to whether or not an intruding slavemaker was immobilized or killed during host colony intrusion, the respective arrows point in the opposite direction. Distribution of samples did not vary with parasite prevalence in the host colony's source population (see Results for details). Plot created using the R package vegan v. 2.5-2. (b) Venn diagram showing the number of genes differentially expressed between hosts that do or do not attack the intruding slavemaker (right, bottom and top) and between hosts facing more or less successful slavemakers (left, top and bottom). Diagram created using the R package VennDiagram v. 1.6.20 (https://www.rdocumentation.org/packages/VennDiagram/versions/1.6.20).
Table 1.
List of differentially expressed candidate genes. Details include explanatory variable, the BLAST annotation of each gene along with its log fold change and false discovery rate (FDR) values as well as functional annotation provided at uniprot.org along with the regarding species.
| upregulated with | BLAST annotation | logFC | FDR | UniProt annotation | UniProt species |
|---|---|---|---|---|---|
| increasing parasite pressure | armadillo repeat containing protein 4 | 14.3 | 0.048 | outer dynein arm assembly | Mus musculus |
| increasing parasite success | glutamate [NMDA] receptor subunit 1 | 1.5 | 0.032 | synaptic plasticity, memory acquisition, learning | Drosophila melanogaster |
| glutamate receptor | 4.0 | 0.032 | neurotransmitter receptor in central nervous system | Drosophila melanogaster | |
| huntingtin | 4.3 | 0.026 | synaptic vesicle transport | Drosophila melanogaster | |
| putative G-protein coupled receptor Mth-like 5 | 5.7 | 0.003 | biological ageing, stress response | Drosophila melanogaster | |
| putative G-protein coupled receptor no. 9 | 6.0 | 0.010 | G-protein coupled receptor activity | Ceratitis capitata | |
| neuronal acetylcholine receptor subunit alpha-5 | 4.4 | 0.005 | transmembrane signalling | Bactrocera latifrons | |
| putative tubulin polyglutamylase TTLL1 | 30.0 | 0.040 | protein polyglutamylation | Bactrocera latifrons | |
| decreasing parasite success | serine/threonine-protein kinase receptor | −1.8 | 0.044 | activin receptor signalling pathway | Drosophila melanogaster |
| fatty acyl-CoA reductase | −29.7 | 0.008 | reduction of fatty acyl-CoA to fatty alcohols | Drosophila melanogaster | |
| attacking the intruder | nuclease harbi1-like protein | 6.5 | 0.038 | potential nuclease activity | Danio rerio |
| not attacking the intruder | metabotropic glutamate receptor | −0.7 | 0.038 | learning, memory | Drosophila melanogaster |
| putative tyramine receptor 2 | −0.5 | 0.025 | signal transduction (olfactory system) | Drosophila melanogaster | |
| serine/threonine-protein kinase PLK | −0.6 | 0.014 | centriole duplication | Drosophila melanogaster | |
| G-protein coupled receptor | −0.7 | 0.010 | G-protein coupled receptor activity | Drosophila melanogaster |
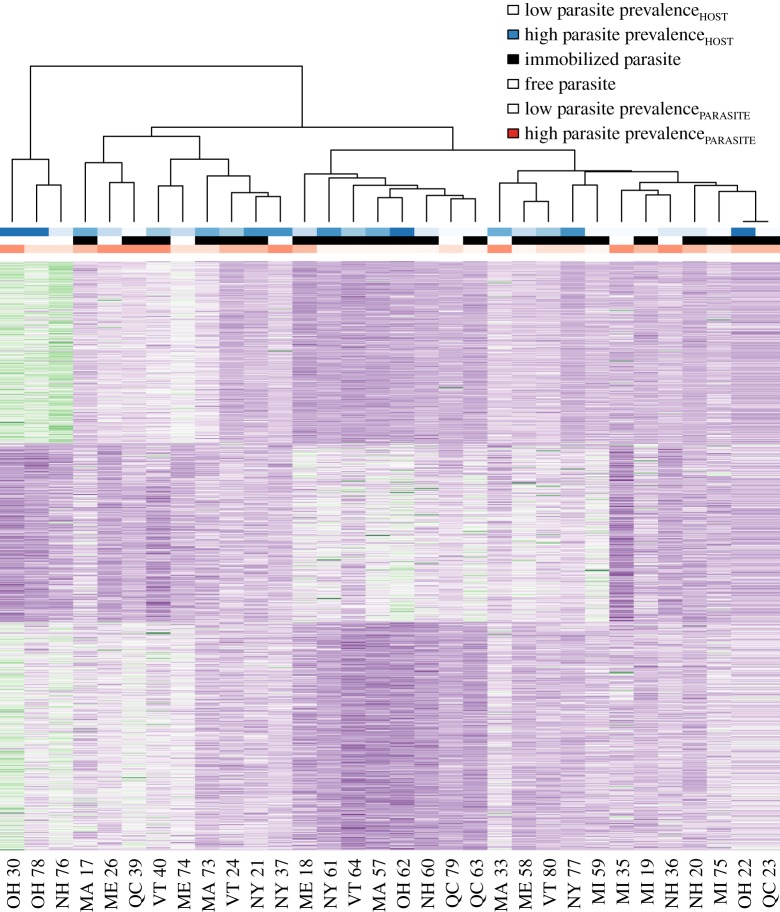
As some host colonies recognized, attacked or killed the intruding slavemaker and others ignored her, we additionally performed a gene expression analysis comparing brain gene expression of attacking versus non-attacking hosts. We found 43 genes to be overexpressed in hosts that attack the parasite, 29.8% of which overlapped with genes decreasing their expression with parasite prevalencePARASITE, which was more than expected by chance (hypergeometrical test; p = 1.6 × 10−11) (figure 2b, electronic supplementary material, Data-X1). Moreover, 222 genes were overexpressed in non-attacking hosts and 34.4% of those overlapped with genes increasing their expression with parasite prevalencePARASITE, which was more than expected by chance (hypergeometrical test; p = 4.2 × 10−37). A heatmap of all the differentially expressed genes confirms that variation is linked to parasite prevalencePARASITE and host attack, but not to parasite prevalenceHOST (figure 3). Furthermore, across all genes the log fold change depending on the prevalence of the parasite population was negatively correlated to the log fold change in respect to attack versus no attack (electronic supplementary material-I, figure F-1).
Figure 3.
Heatmap based on the expression patterns found in all differentially expressed genes. Dendrogram showing relationships between samples based on similarities in gene expression. Colour bars above dendrogram show the parasite prevalence in the host population (blue), whether the slavemaker was immobilized/killed by the hosts (black) and the parasite prevalence of the population of the intruding slavemaker (orange). Heatmap created using R v. 3.4.4.
We contrasted our findings to a gene expression study based on whole body transcriptomes [40] comparing New York T. longispinosus workers during a slave raiding attack by sympatric T. americanus with host workers not involved in a raid. Twenty-two genes overlapped between the differentially expressed gene lists of both studies, which amounts to 3.35% of all the genes found to be differentially expressed in the present study (electronic supplementary material-I, table T-5). In particular, we found five genes over-expressed in T. longispinosus defenders (leucine-rich repeat-containing protein 49, zinc finger MYND domain-containing protein 11, phospholipase, kinesin-like protein KIF14, putative RNA-binding protein EEED8.10) to increase their expression with decreasing parasite prevalenceHOST. Conversely, five genes overlapped between hosts before raiding season and hosts facing social parasites from sites with higher parasite prevalence (elongation factor 1-alpha, ring canal kelch-like protein, serine/threonine-protein kinases 32B, PLK and SIK2).
A repetition of the analysis including host and parasite populations, instead of parasite prevalenceHOST and parasite prevalencePARASITE, showed that indeed origin of the parasite population explains changes in gene expression in the host better than host colony origin (electronic supplementary material, Data-X2 and Data-X3).
(d). Weighted gene co-expression network analysis
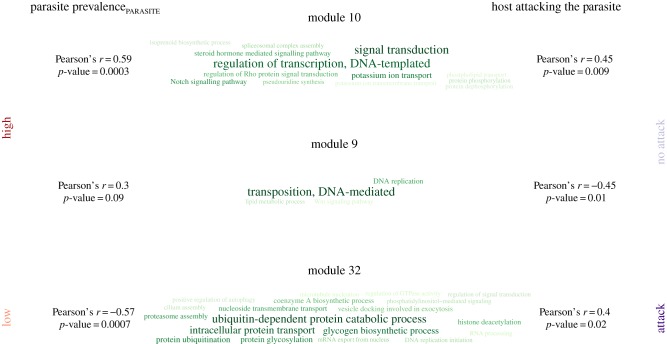
The WGCNA grouped genes with similar expression patterns into 37 modules. Parasite prevalenceHOST was unlinked to the eigengenes of any of these modules (Pearson correlation, n = 32, p < 0.05). In contrast, the eigengene values of two modules were significantly associated with parasite prevalencePARASITE of the intruder, with one showing a negative and one showing a positive correlation. The same two modules were also linked to whether host workers were attacking the parasite or not (Mann–Whitney U tests, n = 32, module 10: p = 0.009, module 32: p = 0.02). In addition, module 9 was linked to attack versus no attack and also tentatively associated with parasite prevalencePARASITE.
Since genes central in networks are of special importance, we assessed module membership of each gene in the co-expression network as a measure of network centrality. Module membership was calculated as the correlation between gene expression profile and module eigengene of each module [77]. For the three modules correlated to at least one trait (parasite prevalencePARASITE and/or attack), network centrality was highly correlated to gene significance (p < 0.0001 for all cases). We also assessed whether differentially expressed genes are more central in their respective modules compared with all other genes, which we found for genes both overexpressed with increasing parasite prevalencePARASITE and overexpressed in non-attacking hosts compared with attacking ones (electronic supplementary material-I, figures F-2a, F-2b, F-3a and F-3b).
(e). GO enrichment analysis
Enriched functions of genes both overexpressed with both high and low parasite prevalencePARASITE were linked to signal transduction and signalling (electronic supplementary material-I, figure F-4). In contrast, genes overexpressed with higher parasite prevalencePARASITE were enriched for protein phosphorylation, genes overexpressed with lower parasite prevalencePARASITE were enriched for protein dephosphorylation. Functions linked to immune response like ‘activation of innate immune response’ and ‘positive regulation of type I interferon production’ were enriched in genes overexpressed in hosts confronted with parasites from sites with higher parasite prevalence.
We only found functions to be enriched in the WGCNA module of genes positively correlated with parasite prevalencePARASITE and negatively with host attacks, which are summarized as DNA-mediated transposition, including the Wnt-pathway (figure 4). The module negatively associated with parasite prevalencePARASITE (and positively with host attacks) showed functions linked to ubiquitin-dependent protein catabolism to be enriched, like protein ubiquitination, and epigenetic processes, such as histone deacetylation. This module also contained an enrichment of genes linked to intracellular protein transport and positive regulation of autophagy. These enrichments indicated that hosts that detect and attack intruding parasites undergo a major turnover in protein composition and change their gene expression strongly and for the longer term.
Figure 4.
Enriched GO terms in three WGCNA modules which were significantly associated with prevalence in the parasite population and whether or not a slavemaker was attacked/killed by host defenders. Genes in modules were either positively or negatively linked to parasite prevalencePARASITE or under attack/no attack state of slavemakers, visualized using the R package tagcloud v. 0.6 (https://CRAN.R-project.org/package=tagcloud). The type size denotes the number of genes with this annotation (in green) found in the respective module. (Online version in colour.)
4. Discussion
The behavioural responses of a host colony during the attack of an intruding parasite are critical for host fitness. If a parasite scout is allowed to return and recruit nestmates for a slave raid, most host brood will be lost, many workers will die and often the queen will be killed [81]. However, if hosts are able to detect and overwhelm the intruder, the host colony will remain unharmed. This behavioural interaction therefore drives the coevolutionary arms race between parasite and host, with parasites evolving ever better invasion strategies, while hosts improve detection mechanisms. We demonstrate here that the outcome of the parasite–host interaction depends much more on the expression of traits in the parasite than in the host. Parasite success of slavemaking ants can be measured directly as intrusion or raiding success, but in the long run a successful social parasite population will increase in density and a higher fraction of hosts will be parasitized, that is, parasite prevalence should go up. Therefore, a high parasite prevalence can be regarded as evidence for the ecological success of a parasite population [46,82] as well as a sign of high parasite pressure on the host population.
In this study we show that the likelihood that the attacker is subdued is linked to the success rate of the parasite population in the field and not to parasite pressure on host populations. This parasite success in turn is associated with a low expression of recognition substances on the cuticle of the parasite, allowing slavemakers to circumvent detection by the host. Moreover, this dependency on parasite success is clearly reflected in brain gene expression patterns of the host. That is, how genes are expressed in host defenders' brains does not depend on their own origin, but on the origin of the opponent they are facing. This finding fits our studies revealing differences between parasite populations in behaviour, cuticular chemistry and ecological success [43,45,62]. Yet the absence of an effect of parasite pressure on the host is surprising as previous work showed that host defence portfolios vary with parasite pressure [34,46]. However, these studies did not compare multiple parasite populations, but rather standardized parasite population to reveal differences among host populations. Our data here indicate that parasite origin overrides any behavioural or transcriptome differences between host populations.
(a). Parasite success influences host behavioural response
Intruding social parasites have to be recognized by hosts as enemies and T. longispinosus colonies from all populations studied so far respond to T. americanus intruders with high levels of aggression [49,52]. However, the number of host defenders was found to decline with parasite pressure on host populations, with hosts from highly parasitized sites shifting to flight rather than fight [34]. Albeit our current study included many, but not all, the host populations studied before, we found that the proportion of host workers involved in nest defence did not depend on the geographical origin of the hosts or local parasite pressure. Rather, the fraction of workers involved in parasite defence depended on an interaction between colony size and the ecological success of the social parasite population. When parasites originated from low prevalence sites, the proportion of workers involved in defence decreased with colony size. This makes sense as social parasite cues might spread only locally in the nest and there is only limited space around the parasite for host workers to attack. Interestingly, this association with colony size was reversed against highly successful parasites. Here, we find a lower proportion of defending host workers in small colonies and a higher one in larger colonies. This is probably because successful parasites are rather able to evade detection by individual host workers and thus perform better against small colonies. Differences in host response to slavemakers of various T. americanus populations were quite strong, with parasites from the least successful Vermont population (parasite prevalence 6%) being nearly always attacked or killed (92%), while fewer than 60% of highly successful Ohio parasites (parasite prevalence 20%) were attacked in our experiments.
(b). Parasite success is linked to chemical recognition cues
Avoiding detection by the host is essential for the ecological success of slavemakers. Former studies have shown that Temnothorax slavemakers, including our focal species, generally exhibit a cuticular hydrocarbon profile with lower proportions of hydrocarbons relevant for recognition in the host T. longispinosus [56]. Here we demonstrate that interpopulation variance in this trait is further linked to parasite success. Slavemakers from high prevalence sites exhibit a cuticular hydrocarbon profile deprived of recognition cues, which should help to avoid host detection and aggressive attacks and therefore could explain their ecological success in the field. Instead, parasites from less successful populations carry more recognition substances and might therefore more easily induce host aggression. In addition, slavemakers from high prevalence sites might effectively manipulate a small number of hosts with their Dufour's gland secretion. Indeed, hosts from highly parasitized populations are more easily tricked to attack their nestmates by chemical manipulation by the Dufour's gland secretion than those from populations where social parasites are rare or absent [46]. In contrast, here we find that the behavioural responses of the host depend on the ecological success of the parasite population and not on parasite pressure of the host populations, indicating that differences in offensive traits, be they behavioural or chemical, mask any differences between host populations. Similar inter-population variation in the efficacy of the Dufour's gland secretion to manipulate hosts into intra-colonial fights was found in the European social parasite Harpagoxenus sublaevis [62].
(c). Parasite success influences gene expression in the host
Gene expression analysis based on brain transcriptomes of defending host workers confirmed our behavioural findings as it showed that more genes altered their expression depending on the ecological success of the intruding parasite than with parasite pressure on the host. Indeed, the expression of only a single gene covaried with parasite pressure, when controlling for parasite success. In contrast, we found several hundred differently expressed genes linked to the ecological success of the parasite population, which indicates that host brain gene expression depends more on the origin of the opponent than on the geographical origin of the sampled worker. As our behavioural analysis showed that these effects were driven by whether or not hosts recognize intruding parasites, we analysed gene expression in host brains depending on whether these host workers attacked the intruder or not. Again, we identified hundreds of differentially expressed genes with host attack, which strongly overlapped with the analysis based on parasite success. Finally, some of our overexpressed genes were already detected to play a role in regulating host defences in T. longispinosus [40] against T. americanus slave raids.
During the coevolutionary arms race between social parasites and hosts [33], the parasites have perfected their raiding strategies, while hosts are under strong selection pressure to develop effective defence strategies [34,45,47,61,81]. As we had ample evidence especially for the latter process, our initial expectation was that gene expression would also shift with parasite pressure. Our cross-fostering experiment allowed us to disentangle parasite pressure on the host from parasite success, and revealed that parasite success affects gene expression in the host much more than parasite pressure. The results of the WGCNA support the gene expression analysis: here, we were able to find two modules positively and negatively correlated with parasite success respectively, but none was linked to parasite pressure. Overall, our analysis suggests that host responses and their gene expression in the brain depend on the type of intruder and especially on its traits, which are likely linked to its ecological success. An important social parasite trait is its cuticular hydrocarbon profile [72], in particular the expression of recognition substances, which we show here to be negatively linked to parasite success. In social insects, and in ants in particular, the cuticular hydrocarbon profile is the main trait used to recognize, and fend off, non-nestmates and parasites, which is vital to maintain colony integrity [83].
(d). Candidate genes and enriched functions linked to parasite success
Among the genes overexpressed in hosts encountering a successful parasite was glutamate [NMDA] receptor subunit 1, which was found to be overexpressed in the aggressive Africanized honeybee compared with the European honeybee, pointing to a role in regulating aggressive behaviours [84]. Additionally, a gene encoding an ionotropic glutamate receptor was also found to be upregulated across three species of T. americanus host workers defending their nest during slave raids [20]. In honeybees the expression of the huntingtin gene, which is one of our candidate genes for high parasite success, was linked to highly defensive bees [85]. That we find overexpression of genes linked to aggression in hosts encountering successful parasites, which are less often attacked, is at first surprising. However, it could be explained by successful parasites exhibiting lower concentrations of recognition substances on their cuticle and possibly also manipulating hosts via the Dufour's gland secretion into attacking each other. We did not note intra-colonial attacks here, but an earlier study found that the degree to which host workers are manipulated by this chemical weapon to attack each other explains survival of parasite intruders and their success in the field [46]. The GO enrichment of these genes showed many functions linked to signal transduction, indicating a high neuronal activity in hosts encountering parasites from highly successful populations. A comparison of our results with a former study investigating raiding-specific expression differences in our two focal species [20] shows overlap in differentially expressed genes, suggesting that hosts facing a successful parasite share genes overexpressed in hosts not currently under attack by social parasites, while genes overexpressed during raids mostly overlapped with hosts facing less successful parasites. By expressing fewer recognition cues, successful parasites manage to evade host recognition and avoid aggressive responses. This is why their hosts fail to express genes important for nest defence. In turn, less successful parasites induce host aggression and cause the expression of defence genes. A few genes of the attack/no attack analysis overlapped with the earlier raiding analysis, but no clear trend was visible here. Expression of genes linked to aggression does not necessarily imply that this aggression is addressed to the parasite, and this could be why we do not find a strong overlap between genes overexpressed during raiding defence and attacking hosts. The WGCNA results demonstrate that genes linked to histone modifications like histone deacetylation are enriched in a module correlated negatively with parasite success and positively with hosts attacking the intruder. Histone modifications, such as acetylation, methylation or phosphorylation regulate gene expression by facilitating or impeding the access of transcription factors to the DNA [86]. The consequential changes in gene expression can alter the phenotype, especially behaviour. For example, histone acetylation was found to control the expression of genes linked to foraging in ants [87]. Also in our focal species, T. longispinosus, histone acetylation appears to play a role for behavioural flexibility: after removal of old workers, inhibition of histone acetyltransferase facilitated the shift of young workers to foraging, but impeded the reverse shift (Kohlmeier et al. 2018, personal communication). Similarly, under histone acetyltransferase inhibition T. longispinosus colonies were unable to adapt their circadian rhythms to changes in the day/night cycle (Libbrecht et al. 2018, personal communication). Therefore, the upregulation of histone deacetylation genes following the encounter with slavemakers from less successful populations, which are most likely recognized and attacked, indicates long-term changes in gene regulation and expression, supporting earlier work showing that aggression behaviour of T. longispinosus workers is altered for over two weeks following a parasite encounter [51].
Interestingly, while our study revealed the importance of variation in gene expression for the interactions between parasites and hosts, differences in gene regulation were not found between host populations or depending on local parasite pressure on the host. Thus, there is no evidence for selection on regulatory regions in the host during host–parasite coevolution. Rather, expression differences in the brain of host workers depended on the traits of the slavemaker they were facing and on the latter's population of origin. If anything, gene expression in host worker brains thus could be interpreted as the extended phenotype of the parasite [88].
5. Conclusion
Parasite–host interactions are models for studying the dynamics of adaptation during coevolutionary arms races. Parasite prevalence can be shaped by many factors, including local adaptation of the host [34,47], climatic conditions and parasite virulence [89]. Variation in parasite traits important for the interaction was found lately to be quite important in avian brood parasites, especially when both host resistance and tolerance are taken into account [90–92]. Despite previous studies revealing that host defence portfolios covary with parasite pressure in the T. longispinosus-T. americanus system [34], we show here that the origin of the parasite determines the outcome of its interactions with the host. Indeed, the expression of genes in the brain of host defenders did not depend on parasite pressure or geographical origin of the host, but on the ecological success of the parasite they are facing. This indicates that variation in parasite virulence is more important for the species interaction than reported variation in host defence traits.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Jürgen Heinze, Claudia Gstöttl, Matteo Negroni, Heike Stypa and Marion Kever for help in the field and laboratory, and Inon Scharf for the drawing of our ants.
Ethics
Ant collection permits were obtained from parks/preserves or we asked private land owners for permission to collect ant colonies. Import and export licences are not required for the transport of our study species. We followed the guidelines of the Study of Animal Behaviour and the legal and institutional rules.
Data accessibility
All raw data presented here are provided in the electronic supplementary material and available in the BioSample database, accession nos SAMN10250252, SAMN10250253, SAMN10250254, SAMN10250255, SAMN10250256, SAMN10250257, SAMN10250258, SAMN10250259, SAMN10250260, SAMN10250261.
Authors' contributions
R.K. and S.F. designed the experimental set-up and collected the ant colonies. R.K. conducted the behavioural experiments, chemical analyses and statistics. R.K. and M.S. analysed the gene expression data. Genome assembly and annotation was conducted by E.J. with support from B.F. and E.B.-B. F.M. contributed to the chemical analyses. R.K., M.S. and S.F. wrote a first draft of the paper and all authors revised it.
Competing interests
We have no competing interests.
Funding
R.K. was funded by the Alexander von Humboldt Foundation, Bonn, Germany. Funding for transcriptome and genome analysis came from a JGU grant, Mainz, Germany, the Alexander von Humboldt Foundation and two DFG grants nos FO 298/17-1 and BO 2544/12-1. The field collection trip was partly funded by the E. N. Huyck Preserve, Albany, NY, USA and the Equal Opportunity Commission, JGU, Mainz, Germany.
References
- 1.Thompson J. 1999. Specific hypotheses on the geographic mosaic of coevolution. Am. Nat. 153, S1–S14. ( 10.1086/303208) [DOI] [Google Scholar]
- 2.Betts A, Gray C, Zelek M, MacLean R, King K. 2018. High parasite diversity accelerates host adaptation and diversification. Science 360, 907–911. ( 10.1126/science.aam9974) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Valen L. 1973. A new evolutionary law. Evol. Theory 1, 1–30. [Google Scholar]
- 4.Dawkins R, Krebs J. 1979. Arms races between and within species. Proc. R. Soc. Lond. B 205, 489–511. ( 10.1098/rspb.1979.0081) [DOI] [PubMed] [Google Scholar]
- 5.Tellier A, Moreno-Gámez S, Stephan W. 2014. Speed of adaptation and genomic footprints of host-parasite coevolution under arms race and trench warfare dynamics. Evolution 68, 2211–2224. ( 10.1111/evo.12427) [DOI] [PubMed] [Google Scholar]
- 6.Nash D, Als T, Maile R, Jones G, Boomsma J. 2008. A mosaic of chemical coevolution in a large blue butterfly. Science 319, 88–90. ( 10.1126/science.1149180) [DOI] [PubMed] [Google Scholar]
- 7.Thompson J. 2005. The geographic mosaic of coevolution. Chicago, IL: University of Chicago Press. [Google Scholar]
- 8.Gurney J, Aldakak L, Betts A, Gougat-Barbera C, Poisot T, Kaltz O, Hochberg ME. 2017. Network structure and local adaptation in co-evolving bacteria–phage interactions. Mol. Ecol. 26, 1764–1777. ( 10.1111/mec.14008) [DOI] [PubMed] [Google Scholar]
- 9.Koskella B, Parr N. 2015. The evolution of bacterial resistance against bacteriophages in the horse chestnut phyllosphere is general across both space and time. Phil. Trans. R. Soc. B 370, 20140297 ( 10.1098/rstb.2014.0297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Persoons A, Hayden K, Fabre B, Frey P, Mita S, Tellier A, Halkett F. 2017. The escalatory Red Queen: population extinction and replacement following arms race dynamics in poplar rust. Mol. Ecol. 26, 1902–1918. ( 10.1111/mec.13980) [DOI] [PubMed] [Google Scholar]
- 11.Nuismer S. 2017. Introduction to coevolutionary theory. New York, NY: W. H. Freeman. [Google Scholar]
- 12.Lion S, Gandon S. 2015. Evolution of spatially structured host–parasite interactions. J. Evol. Biol. 28, 10–28. ( 10.1111/jeb.12551) [DOI] [PubMed] [Google Scholar]
- 13.Kerstes NAG, Martin OY. 2014. Insect host–parasite coevolution in the light of experimental evolution. Insect Sci. 21, 401–414. ( 10.1111/1744-7917.12064) [DOI] [PubMed] [Google Scholar]
- 14.Feeney WE, Welbergen JA, Langmore NE. 2014. Advances in the study of coevolution between avian brood parasites and their hosts. Annu. Rev. Ecol. Evol. Syst. 45, 227–246. ( 10.1146/annurev-ecolsys-120213-091603) [DOI] [Google Scholar]
- 15.Dennis A, Patel V, Oliver K, Vorburger C. 2017. Parasitoid gene expression changes after adaptation to symbiont-protected hosts. Evolution 17, 2599–2617. ( 10.1111/evo.13333) [DOI] [PubMed] [Google Scholar]
- 16.Feldmeyer B, Mazur J, Beros S, Lerp H, Binder H, Foitzik S. 2016. Gene expression patterns underlying parasite-induced alterations in host behaviour and life history. Mol. Ecol. 25, 648–660. ( 10.1111/mec.13498) [DOI] [PubMed] [Google Scholar]
- 17.Martinson EO, Wheeler D, Wright J, Siebert AL, Werren JH. 2014. Nasonia vitripennis venom causes targeted gene expression changes in its fly host. Mol. Ecol. 23, 5918–5930. ( 10.1111/mec.12967) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barribeau S, Sadd B, du Plessis L, Schmid-Hempel P. 2014. Gene expression differences underlying genotype-by-genotype specificity in a host–parasite system. Proc. Natl Acad. Sci. USA 111, 3496–3501. ( 10.1073/pnas.1318628111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Libersat F, Kaiser M, Emanuel S. 2018. Mind control: how parasites manipulate cognitive functions in their insect hosts. Front. Psychol. 9, 572 ( 10.3389/fpsyg.2018.00572) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alleman A, Feldmeyer B, Foitzik S. 2018. Comparative analyses of co-evolving host-parasite associations reveal unique gene expression patterns underlying slavemaker raiding and host defensive phenotypes. Sci. Rep. 8, 1951 ( 10.1038/s41598-018-20262-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feldmeyer B, Elsner D, Alleman A, Foitzik S. 2017. Species-specific genes under selection characterize the co-evolution of slavemaker and host lifestyles. BMC Evol. Biol. 17, 1–11. ( 10.1186/s12862-017-1078-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feis ME, John U, Lokmer A, Luttikhuizen PC, Wegner KM. 2018. Dual transcriptomics reveals co-evolutionary mechanisms of intestinal parasite infections in blue mussels Mytilus edulis. Mol. Ecol. 27, 1505–1519. ( 10.1111/mec.14541) [DOI] [PubMed] [Google Scholar]
- 23.Cini A, Patalano S, Segonds-Pichon A, Busby GBJ, Cervo R, Sumner S. 2015. Social parasitism and the molecular basis of phenotypic evolution. Front. Genet. 6, 32 ( 10.3389/fgene.2015.00032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith CR, et al. 2015. How do genomes create novel phenotypes? Insights from the loss of the worker caste in ant social parasites. Mol. Biol. Evol. 32, 2919–2931. ( 10.1093/molbev/msv165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies NB, Bourke AF, Brooke MD. 1989. Cuckoos and parasitic ants: interspecific brood parasitism as an evolutionary arms race. Trends Ecol. Evol. 4, 274–278. ( 10.1016/0169-5347(89)90202-4) [DOI] [PubMed] [Google Scholar]
- 26.Brooke M, Davies N. 1988. Egg mimicry by cuckoos Cuculus canorus in relation to discrimination by hosts. Nature 335, 630–632. ( 10.1038/335630a0) [DOI] [Google Scholar]
- 27.Davies N, Brooke M, Kacelnik A. 1996. Recognition errors and probability of parasitism determine whether reed warblers should accept or reject mimetic cuckoo eggs. Proc. R. Soc. Lond. B 263, 925–931. ( 10.1098/rspb.1996.0137) [DOI] [Google Scholar]
- 28.Stoddard MMC, Hauber MME. 2017. Colour, vision and coevolution in avian brood parasitism. Phil. Trans. R. Soc. B 372, 20160339 ( 10.1098/rstb.2016.0339) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hölldobler B, Wilson EO. 1990. The ants. Cambridge, MA: Harvard University Press. [Google Scholar]
- 30.Buschinger A. 2009. Social parasitism among ants: a review (Hymenoptera: Formicidae). Myrmecol. News 12, 219–235. [Google Scholar]
- 31.Soler M, Møller A. 1990. Duration of sympatry and coevolution between the great spotted cuckoo and its magpie host. Nature 343, 748–750. ( 10.1038/343748a0) [DOI] [Google Scholar]
- 32.Davies N. 2000. Cuckoos, cowbirds and other cheats. London, UK: T & AD Poyser. [Google Scholar]
- 33.Brandt M, Foitzik S, Fischer-Blass B, Heinze J. 2005. The coevolutionary dynamics of obligate ant social parasite systems—between prudence and antagonism. Biol. Rev. 80, 251–267. ( 10.1017/S1464793104006669) [DOI] [PubMed] [Google Scholar]
- 34.Jongepier E, Kleeberg I, Job S, Foitzik S. 2014. Collective defence portfolios of ant hosts shift with social parasite pressure. Proc. R. Soc. B 281, 20140225 ( 10.1098/rspb.2014.0225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emery C.1909. [On the origin of dulotic, parasitic and myrmecophylic ants]. Biol. Zent. Bl.29, 352–362. (In German.)
- 36.Smith JA, Chenoweth LB, Tierney SM, Schwarz MP. 2013. Repeated origins of social parasitism in allodapine bees indicate that the weak form of Emery's rule is widespread, yet sympatric speciation remains highly problematic. Biol. J. Linn. Soc. 109, 320–331. ( 10.1111/bij.12043) [DOI] [Google Scholar]
- 37.Carpenter JM, Perera EP. 2006. Phylogenetic relationships among yellowjackets and the evolution of social parasitism (Hymenoptera: Vespidae, Vespinae). Am. Mus. Novit. 3507, 1 ( 10.1206/0003-0082(2006)3507[1:PRAYAT]2.0.CO;2) [DOI] [Google Scholar]
- 38.Sumner S, Aanen DK, Delabie J, Boomsma JJ. 2004. The evolution of social parasitism in Acromyrmex leaf-cutting ants: a test of Emery's rule. Insectes Sociaux 51, 37–42. ( 10.1007/s00040-003-0723-z) [DOI] [Google Scholar]
- 39.Huang MH, Dornhaus A. 2008. A meta-analysis of ant social parasitism: host characteristics of different parasitism types and a test of Emery's rule. Ecol. Entomol. 33, 589–596. ( 10.1111/j.1365-2311.2008.01005.x) [DOI] [Google Scholar]
- 40.Pennings PS, Achenbach A, Foitzik S. 2011. Similar evolutionary potentials in an obligate ant parasite and its two host species. J. Evol. Biol. 24, 871–886. ( 10.1111/j.1420-9101.2010.02223.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward PS, Brady SG, Fisher BL, Schultz TR. 2015. The evolution of myrmicine ants: phylogeny and biogeography of a hyperdiverse ant clade (Hymenoptera: Formicidae). Syst. Entomol. 40, 61–81. ( 10.1111/syen.12090) [DOI] [Google Scholar]
- 42.Beibl J, Stuart RJ, Heinze J, Foitzik S. 2005. Six origins of slavery in formicoxenine ants. Insectes Sociaux 52, 291–297. ( 10.1007/s00040-005-0808-y) [DOI] [Google Scholar]
- 43.Brandt M, Foitzik S. 2004. Community context and specialization influence coevolution between a slavemaking ant and its hosts. Ecology 85, 2997–3009. ( 10.1890/03-0778) [DOI] [Google Scholar]
- 44.Foitzik S, Herbers JM. 2001. Colony structure of a slavemaking ant. II. Frequency of slave raids and impact on the host population. Evolution 55, 316–323. ( 10.2307/2640753) [DOI] [PubMed] [Google Scholar]
- 45.Foitzik S, Achenbach A, Brandt M. 2009. Locally adapted social parasite affects density, social structure, and life history of its ant hosts. Ecology 90, 1195–1206. ( 10.1890/08-0520.1) [DOI] [PubMed] [Google Scholar]
- 46.Jongepier E, Kleeberg I, Foitzik S. 2015. The ecological success of a social parasite increases with manipulation of collective host behaviour. J. Evol. Biol. 28, 2152–2162. ( 10.1111/jeb.12738) [DOI] [PubMed] [Google Scholar]
- 47.Jongepier E, Foitzik S. 2016. Ant recognition cue diversity is higher in the presence of slavemaker ants. Behav. Ecol. 27, 304–311. ( 10.1093/beheco/arv153) [DOI] [Google Scholar]
- 48.Pamminger T, Scharf I, Pennings PS, Foitzik S. 2011. Increased host aggression as an induced defense against slave-making ants. Behav. Ecol. 22, 255–260. ( 10.1093/beheco/arq191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scharf I, Pamminger T, Foitzik S. 2011. Differential response of ant colonies to intruders: attack strategies correlate with potential threat. Ethology 117, 731–739. ( 10.1111/j.1439-0310.2011.01926.x) [DOI] [Google Scholar]
- 50.Pamminger T, Modlmeier AP, Suette S, Pennings PS, Foitzik S. 2012. Raiders from the sky: slavemaker founding queens select for aggressive host colonies. Biol. Lett. 8, 748–750. ( 10.1098/rsbl.2012.0499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kleeberg I, Pamminger T, Jongepier E, Papenhagen M, Foitzik S. 2014. Forewarned is forearmed: aggression and information use determine fitness costs of slave raids. Behav. Ecol. 25, 1058–1063. ( 10.1093/beheco/aru084) [DOI] [Google Scholar]
- 52.Kleeberg I, Jongepier E, Job S, Foitzik S. 2015. Geographic variation in social parasite pressure predicts intraspecific but not interspecific aggressive responses in hosts of a slavemaking ant. Ethology 121, 694–702. ( 10.1111/eth.12384) [DOI] [Google Scholar]
- 53.D'Ettorre P, Mondy N, Lenoir A, Errard C. 2002. Blending in with the crowd: social parasites integrate into their host colonies using a flexible chemical signature. Proc. R. Soc. Lond. B 269, 1911–1918. ( 10.1098/rspb.2002.2110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brandt M, Heinze J, Schmitt T, Foitzik S. 2005. A chemical level in the coevolutionary arms race between an ant social parasite and its hosts. J. Evol. Biol. 18, 576–586. ( 10.1111/j.1420-9101.2004.00867.x) [DOI] [PubMed] [Google Scholar]
- 55.Bauer S, Witte V, Böhm M, Foitzik S. 2009. Fight or flight? A geographic mosaic in host reaction and potency of a chemical weapon in the social parasite Harpagoxenus sublaevis. Behav. Ecol. Sociobiol. 64, 45–56. ( 10.1007/s00265-009-0817-3) [DOI] [Google Scholar]
- 56.Kleeberg I, Menzel F, Foitzik S. 2017. The influence of slavemaking lifestyle, caste and sex on chemical profiles in Temnothorax ants: insights into the evolution of cuticular hydrocarbons. Proc. R. Soc. B 284, 20162249 ( 10.1098/rspb.2016.2249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Achenbach A, Witte V, Foitzik S. 2010. Brood exchange experiments and chemical analyses shed light on slave rebellion in ants. Behav. Ecol. 21, 948–956. ( 10.1093/beheco/arq008) [DOI] [Google Scholar]
- 58.Martin SJ, Helanterä H, Drijfhout FP. 2011. Is parasite pressure a driver of chemical cue diversity in ants? Proc. R. Soc. B 278, 496–503. ( 10.1098/rspb.2010.1047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Allies A, Bourke AFG, Franks NR. 1986. Propaganda substances in the cuckoo ant Leptothorax kutteri and the slave-maker Harpagoxenus sublaevis. J. Chem. Ecol. 12, 1285–1293. ( 10.1007/BF01012348) [DOI] [PubMed] [Google Scholar]
- 60.Pamminger T, Leingärtner A, Achenbach A, Kleeberg I, Pennings PS, Foitzik S. 2013. Geographic distribution of the anti-parasite trait ‘slave rebellion’. Evol. Ecol. 27, 39–49. ( 10.1007/s10682-012-9584-0) [DOI] [Google Scholar]
- 61.Jongepier E, Foitzik S. 2016. Fitness costs of worker specialization for ant societies. Proc. R. Soc. B 283, 20152572 ( 10.1098/rspb.2015.2572) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Foitzik S, Fischer B, Heinze J. 2003. Arms races between social parasites and their hosts: geographic patterns of manipulation and resistance. Behav. Ecol. 14, 80–88. ( 10.1093/beheco/14.1.80) [DOI] [Google Scholar]
- 63.Campbell-Staton S, Cheviron Z, Rochette N, Catchen J, Losos J, Edwards S. 2017. Winter storms drive rapid phenotypic, regulatory, and genomic shifts in the green anole lizard. Science 357, 495–498. ( 10.1126/science.aam5512) [DOI] [PubMed] [Google Scholar]
- 64.Ghalambor C, Hoke K, Ruell E, Fischer E, Reznick D, Highes K. 2015. Non-adaptive plasticity potentiates rapid adaptive evolution of gene expression in nature. Nature 525, 372–375. ( 10.1038/nature15256) [DOI] [PubMed] [Google Scholar]
- 65.Herbers JM, Foitzik S. 2002. The ecology of slavemaking ants and their hosts in north temperate forests. Ecology 83, 148–163. ( 10.1890/0012-9658(2002)083[0148:TEOSAA]2.0.CO;2) [DOI] [Google Scholar]
- 66.R Development Core Team. 2011. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical computing. See http://www.R-project.org/. (doi:10.1007/978-3-540-74686-7)
- 67.Andrews S. 2010. FastQC: A quality control tool for high throughput sequence data. See https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (doi:citeulike-article-id:11583827)
- 68.Butler J, MacCallum I, Kleber M, Shlyakhter I, Belmonte M, Lander E, Nusbaum C, Jaffe D. 2008. ALLPATHS: de novo assembly of whole-genome shotgun microreads. Genome Res. 18, 810–820. ( 10.1101/gr.7337908) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simão F, Waterhouse R, Ioannidis P, Kriventseva E, Zdobnov E. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. ( 10.1093/bioinformatics/btv351) [DOI] [PubMed] [Google Scholar]
- 70.Holt C, Yandell M. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12, 491 ( 10.1186/1471-2105-12-491) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.UniProt Consortium. 2017. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169. ( 10.1093/nar/gkw1099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Finn R, et al. 2017. InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res. 45, D190–D199. ( 10.1093/nar/gkw1107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. ( 10.1038/nmeth.3317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. ( 10.1093/bioinformatics/btu638) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Love MI, Anders S, Huber W. 2016. Differential analysis of count data—the DESeq2 package. Genome Biol. 15, 550 ( 10.1186/s13059-014-0550-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gentleman R, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 ( 10.1186/gb-2004-5-10-r80) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 ( 10.1186/1471-2105-9-559) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang B, Horvath S. 2005. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4, 17 ( 10.2202/1544-6115.1128) [DOI] [PubMed] [Google Scholar]
- 79.Alexa A, Rahnenführer J.2009. topGO: Enrichment analysis for gene ontology. R package version 2.28.0.
- 80.Bejsovec A. 2013. Wingless/Wnt signaling in Drosophila: the pattern and the pathway. Mol. Reprod. Dev. 80, 882–894. ( 10.1002/mrd.22228) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Foitzik S, DeHeer CJ, Hunjan DN, Herbers JM. 2001. Coevolution in host–parasite systems: behavioural strategies of slave-making ants and their hosts. Proc. R. Soc. Lond. B 268, 1139–1146. ( 10.1098/rspb.2001.1627) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Novak CW, Goater TM. 2013. Introduced bullfrogs and their parasites: Haematoloechus longiplexus (Trematoda) exploits diverse damselfly intermediate hosts on Vancouver Island. J. Parasitol. 99, 59–63. ( 10.1645/GE-3145.1) [DOI] [PubMed] [Google Scholar]
- 83.Guerrieri FJ, Nehring V, Jørgensen CG, Nielsen J, Galizia CG, D'Ettorre P. 2009. Ants recognize foes and not friends. Proc. R. Soc. B 276, 2461–2468. ( 10.1098/rspb.2008.1860) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alaux C, Sinha S, Hasadsri L, Hunt G. 2009. Honey bee aggression supports a link between gene regulation and behavioral evolution. Proc. Natl Acad. Sci. USA 106, 15 400–15 405. ( 10.1073/pnas.0907043106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hunt GJ, et al. 2007. Behavioral genomics of honeybee foraging and nest defense. Naturwissenschaften 94, 247–267. ( 10.1007/s00114-006-0183-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bell O, Schwaiger M, Oakeley EJ, Lienert F, Beisel C, Stadler MB, Schübeler D. 2010. Accessibility of the Drosophila genome discriminates PcG repression, H4K16 acetylation and replication timing. Nat. Struct. Mol. Biol. 17, 894–900. ( 10.1038/nsmb.1825) [DOI] [PubMed] [Google Scholar]
- 87.Simola DF, et al. 2016. Epigenetic (re)programming of caste-specific behavior in the ant Camponotus floridanus. Science 351, aac6633 ( 10.1126/science.aac6633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dawkins R. 1982. The extended phenotype. Oxford, UK: Oxford University Press. [Google Scholar]
- 89.Szöllosi E, et al. 2011. Determinants of distribution and prevalence of avian malaria in blue tit populations across Europe: separating host and parasite effects. J. Evol. Biol. 24, 2014–2024. ( 10.1111/j.1420-9101.2011.02339.x) [DOI] [PubMed] [Google Scholar]
- 90.Medina I, Langmore NE. 2016. The evolution of acceptance and tolerance in hosts of avian brood parasites. Biol. Rev. 91, 569–577. ( 10.1111/brv.12181) [DOI] [PubMed] [Google Scholar]
- 91.Svensson E, Råberg L. 2010. Resistance and tolerance in animal enemy–victim coevolution. Trends Ecol. Evol. 25, 267–274. ( 10.1016/j.tree.2009.12.005) [DOI] [PubMed] [Google Scholar]
- 92.Soler J, Martín-Gálvez D, Martinez J, Soler M, Canestrani D, Abad-Gomez J, Moller A. 2011. Evolution of tolerance by magpies to brood parasitism by great spotted cuckoos. Proc. R. Soc. B 278, 2047–2052. ( 10.1098/rspb.2010.2218) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data presented here are provided in the electronic supplementary material and available in the BioSample database, accession nos SAMN10250252, SAMN10250253, SAMN10250254, SAMN10250255, SAMN10250256, SAMN10250257, SAMN10250258, SAMN10250259, SAMN10250260, SAMN10250261.