Abstract
Interspecific brood parasitism is common in many animal systems. Brood parasites enter the nests of other species and divert host resources for producing their own offspring, which can lead to strong antagonistic parasite–host coevolution. Here, we look at commonalities among social insect species that are victims of brood parasites, and use phylogenetic data and information on geographical range size to predict which species are most probably to fall victims to brood parasites in the future. In our analyses, we focus on three eusocial hymenopteran groups and their brood parasites: (i) bumblebees, (ii) Myrmica ants, and (iii) vespine and polistine wasps. In these groups, some, but not all, species are parasitized by obligate workerless inquilines that only produce reproductive-caste descendants. We find phylogenetic signals for geographical range size and the presence of parasites in bumblebees, but not in ants and wasps. Phylogenetic logistic regressions indicate that the probability of being attacked by one or more brood parasite species increases with the size of the geographical range in bumblebees, but the effect is statistically only marginally significant in ants. However, non-phylogenetic logistic regressions suggest that bumblebee species with the largest geographical range sizes may have a lower likelihood of harbouring social parasites than do hosts with medium-sized ranges. Our results provide new insights into the ecology and evolution of host–social parasite systems, and indicate that host phylogeny and geographical range size can be used to predict threats posed by social parasites, as well to design efficient conservation measures for both hosts and their parasites.
This article is part of the theme issue ‘The coevolutionary biology of brood parasitism: from mechanism to pattern’.
Keywords: Hymenoptera, social parasite, geographical range size, phylogeny, host–parasite interactions
1. Introduction
Owing to anthropogenically induced environmental changes, such as global warming, habitat destruction and pesticide use, species are increasingly becoming threatened [1–5]. This creates a need for intensified conservation efforts, but to effectively protect species, we need to know their habitat requirements and ecological features [1,2,6]. Protecting existing habitats is a logical first step, but because species may be able to shift their habitat requirements and, therefore, improve their chance of survival, alternative and perhaps less preferred habitats should also be protected [7]. However, identifying potential alternative habitats may be very difficult, calling for a viable method for assessing the usefulness of both existing and potential resources and habitats.
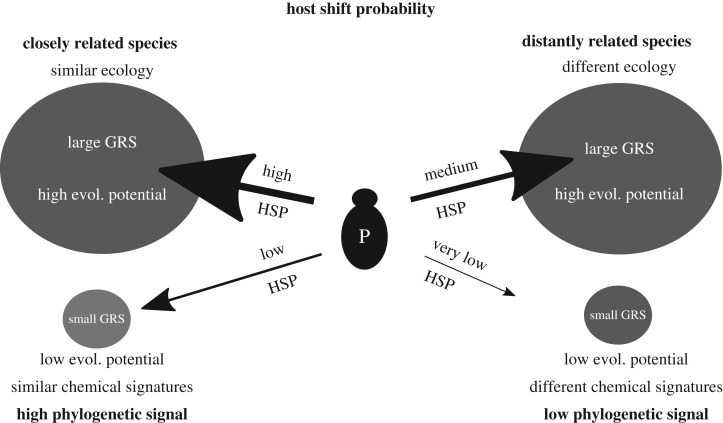
In the case of parasitic species, different host species constitute alternative habitats. Depending on the parasite species, the number of habitats (i.e. host species) varies from one (specialist) to many (generalist). When host numbers decline, e.g. owing to environmental deterioration, a parasite species may start using one or more additional hosts (host-range expansion), evolve a preference for a completely different host (host switch) (figure 1) [8] or disappear altogether. However, the causes and consequences of host switches and host-range expansions have received very little attention [9], and it is currently unknown whether certain traits of host species could increase the likelihood of host switching or host-range expansions. To this end, we decided here to examine whether host switching and the presence of a parasite species could be predicted based on the combination of host phylogeny and geographical range size (GRS) [9].
Figure 1.
Conceptual description of the effects of phylogenetic relatedness, geographical range size (GRS) (assumed to correlate positively with overall population size) and similarity of ecological traits and chemical signatures on the probability of shifts among current and potential host species by social brood parasites. The P denotes a parasite species facing a decision of whether or not to shift onto a new host, and the size of arrows indicates host shift probabilities (HSPs) onto different hosts. The consequences of realized shifts for phylogenetic signals are shown below the figures.
According to the resource distribution hypothesis [10,11], widespread species are able to support a higher parasite diversity than are species with more restricted geographical ranges. Large GRS and large population size of host species have often been shown to correlate with high parasite prevalence and diversity [12–16], so that rare hosts tend to have fewer [8] or no [15] parasite species. However, it is poorly known how GRS and phylogenetic relationships among actual and potential host species affect the probability of host switching by parasites [9].
There is a very tight coevolutionary relationship between hosts and their parasites, often described as an arms race or the Red Queen hypothesis [6,17–20]: hosts try to evolve stronger defensive capabilities, while parasites strive to increase their chances of successful infection and reproductive success. Host–parasite arms races function as accelerants for trait evolution [21–23] and genetic divergence [21,24], and may eventually lead to speciation, especially if the number of parasite species per host species is high [24]. Alternatively, a parasite species may simply switch to a new host species. The likelihood of a successful shift is influenced by two components: first, a shift is more probable if the new host species is closely related to the current host(s) [9,20,25], as close relatives tend to share ecological traits and probably have similar chemical signatures [19,26,27] (figure 1). Second, a large GRS and population size of the new host increase the probability of long-term survival of the parasite [9] (figure 1).
To investigate the likelihood of parasitism and the probability of host switching in relation to the GRS of host and non-host species, we selected three hymenopteran eusocial groups with species harbouring obligate interspecific workerless social parasites (inquilines): bumblebees (Apidae: Bombus), Myrmica ants (Formicidae) and wasps (Vespidae: Vespinae and Polistinae). In these focal groups, the parasite species commandeer the nests of their hosts and usurp their resources for reproduction of only their own sexual offspring, rendering the parasite species completely dependent on provisioning by their hosts. Owing to the tight relationship between the inquilines and their hosts, it has been suggested that parasites would tend to be close relatives of their host species. However, recent phylogenetic analyses have shown that this hypothesis, generalized as Emery's rule [28,29], does not hold in a wider perspective. In ants, inquilinism has evolved several times, which has resulted in a wide range of phylogenetic relationships between parasites and their hosts as well as between different parasite species [30,31]. In bumblebees, inquilinism is restricted to the subgenus Psithyrus, making them all close relatives of each other [32]. Inquiline wasps appear not to be the closest relatives of their hosts, although they are from the same clade as their host species [29].
By reanalysing previous studies on hosts and social parasites in bumblebees, Myrmica ants, and vespine and polistine wasps, we created a large dataset to answer the following questions: (i) does host GRS influence the presence and diversity of brood parasites, (ii) do obligate social parasite species have a tendency to parasitize the same host species, (iii) are closely related host species more similar in their parasitism than are hosts drawn at random, and (iv) can these results be used to predict the future parasitism of social insect species? Based on our previous results [15], we expected that host species with larger GRS are more often parasitized than are species with narrower ranges. We also expected a tendency for closely related species to be similarly parasitized or unparasitized [17] because previous studies [33] have suggested a phylogenetic signal in GRS.
2. Methods
(a). Host and parasite data
We compiled a dataset on host species and interspecific brood parasites in Bombus bumblebees, Myrmica ants, and vespine and polistine wasps based on a broad survey of relevant ecological and taxonomic literature. The focal host taxa and species were selected so that information was available on parasitism by social brood parasites, phylogenetic relationships (see below) and GRS of actual and potential hosts. Parasite species were considered if they are strict workerless inquilines, meaning that facultative inquilines were excluded. In all, our dataset includes information on parasitism in 230 bumblebee species [32–34] of which 182 have phylogenetic data [35], 63 ant species [31,36,37] and 37 wasp species [29,38,39] (electronic supplementary material, tables S1–S3).
For bumblebees, we considered cuckoo bumblebee species (Bombus subgenus Psithyrus spp.) which produce only sexual castes (queens and males), but not workers [32]. A total of 24 Psithyrus species have been found in the nests of 34 of the focal 230 bumblebee species. However, because direct observations of breeding inquiline queens are not available for all presumed hosts [32], we performed statistical analyses using also a dataset that considered as hosts only those 24 bumblebee species for which nests with breeding Psithyrus queens had been found [32].
Geographical range sizes were obtained by counting the number of 611 000 km2 squares that each species occupies, based on data from [34] for bumblebees and AntMaps [40,41] for ants. We added Bombus glacialis to one of our datasets owing to the fact that it occurs only on two arctic islands [32]. Reliable data on GRS were not available for the focal wasp species.
(b). Phylogenetic trees
To control for the effect of relatedness among species, which can confound comparative analyses [42,43], we estimated phylogenetic signal in GRS and parasitism and performed phylogenetic logistic regression based on ultrametric phylogenetic trees of actual and potential hosts. A time-calibrated phylogeny for Bombus from [35] was pruned down in Mesquite v. 3.40 [44] to 182 species for which data on parasitism and GRS were available. For Myrmica ants, we estimated an ultrametric tree based on the 3886 bp sequence data matrix and tree topology of Jansen et al. [31], both of which are available in TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S10277). The original topology was ultrametricized based on penalized likelihood with the chronos command (correlated rate model with λ = 1) in the ape v. 4.0 [45] package in R v. 3.3.2 [46]. The resultant tree was then used as a starting tree and topological constraint in an analysis in BEAST v. 2.5.1 [47], in which branch lengths were estimated by implementing a GTR + I + G substitution model and assuming an uncorrelated lognormal molecular clock. The analysis was run for 10 million steps while sampling trees and parameters every 1000 steps. After confirming in Tracer v. 1.7.1 [48] that the run had converged and that ESS values were greater than 200, the final maximum clade credibility tree with median node heights was calculated in TreeAnnotator (part of the BEAST package) based on the 9001 trees sampled after a 10% burnin. Outgroups and species with missing data were then pruned in Mesquite, leaving a tree with 63 terminals. For vespine and polistine wasps, we supplemented the 6568 bp sequence matrix of Lopez-Osorio et al. [29] with 658 bp of COI barcode data downloaded from GenBank for four species (Vespula rufa KU496858, Dolichovespula alpicola KM566926, Dolichovespula norwegica HM860327 and Vespa velutina JQ780455). A maximum-likelihood phylogeny including the additional species was then calculated using RAxML BlackBox [49] on the CIPRES server [50] based on a GTR + G substitution model. The resulting topology was then ultrametricized and used as a constraint when re-estimating branch lengths in BEAST in the same way as described above for ants, and, finally, parasite taxa were removed, leaving a tree with 37 actual and potential wasp host species.
(c). Statistical analyses
Pagel's λ [28,51] was used to measure phylogenetic signal in geographical range size and in the number of social brood parasites per host species. A λ of 0 indicates that trait values vary randomly across a phylogeny (i.e. absence of phylogenetic signal), while a value of 1 indicates Brownian-motion evolution (i.e. the presence of phylogenetic signal) [51]. To measure phylogenetic conservatism in the binary variable of whether a species is parasitized or not, we used the D-statistic [52]. In this case, a D close to 0 indicates a phylogenetically clustered pattern expected under a Brownian threshold model, while a value of 1 indicates a phylogenetically random pattern [52]. These two metrics of phylogenetic signal perform well in statistical tests for evolutionary trait conservatism [53]. We estimated index values and tested for deviations from 0 and 1 in R, using the phytools package [54] for Pagel's λ and the caper package [55] for the D-statistic.
For bumblebee and ant species that were included in the phylogenetic trees, we used phylogenetic logistic regression models to assess whether species-specific GRS influences the presence of social brood parasites. The presence or absence of parasites was explained using GRS and GRS2 using the pgls function of the phylolm v. 2.6 package [56] in R.
Analogous non-phylogenetic logistic regression analyses (for all bumblebee and ant species with data on parasitism, GRS and GRS2) were performed using IBM SPSS for Windows v. 20.0. For the larger bumblebee dataset, we also investigated whether the GRS of a species can be used to estimate whether it is likely to be infected by brood parasites in the future, using the parasitism probabilities of different bumblebee species predicted by the non-phylogenetic logistic regression. We matched current host species with their closest unparasitized relatives (electronic supplementary material, tables S2 (ants) and S4 (bumblebees)), and then compared GRS and estimated probability of parasitism across the two groups using paired-samples t-tests.
3. Results
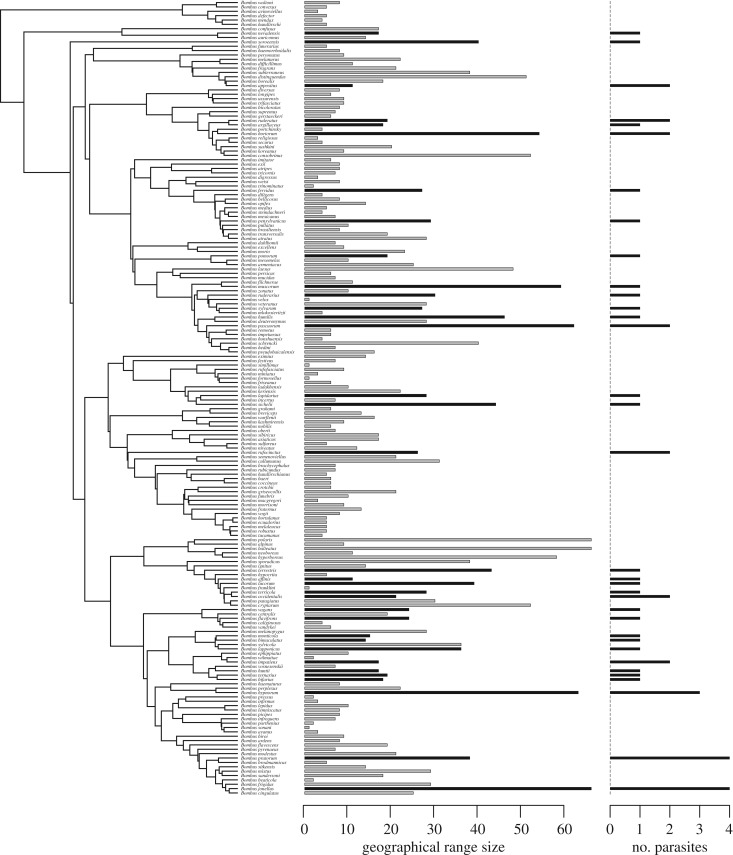
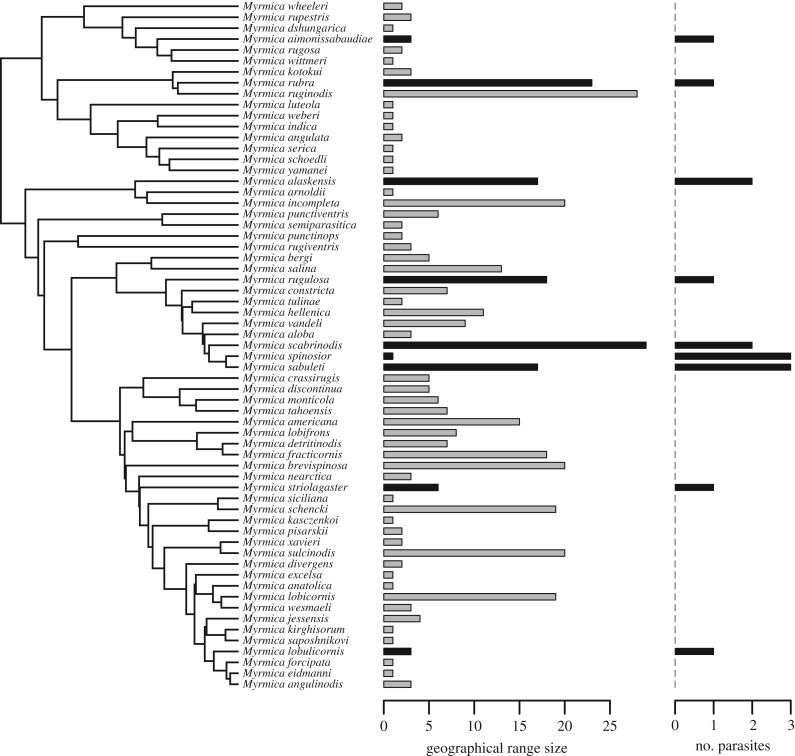
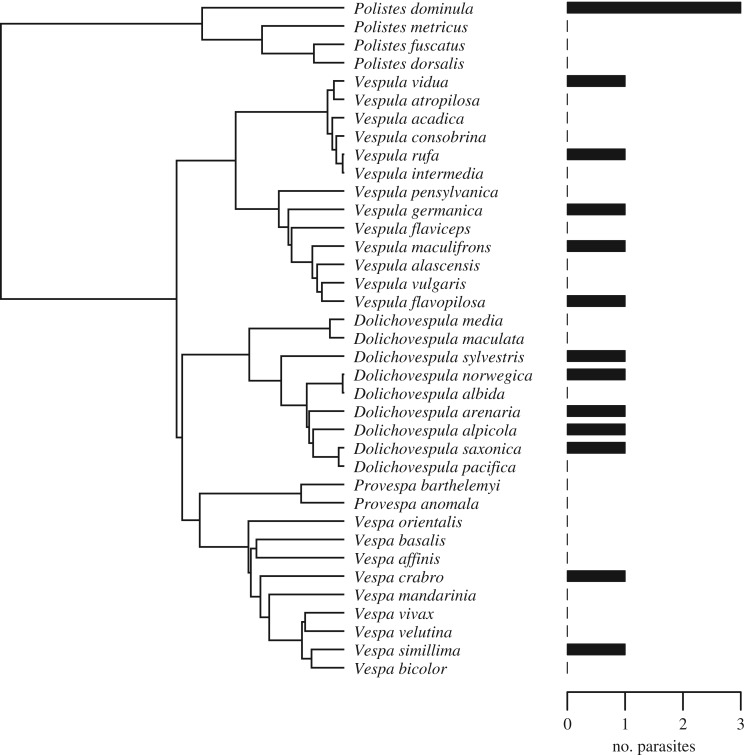
Brood parasitism existed in all three hymenopteran groups to a varying degree. The number of cuckoo bumblebee species per bumblebee host varied from 0 to 4 (figure 2), with a mean of 0.23 (s.d. = 0.62). The number of inquiline Myrmica ants per host species ranged from 0 to 3 (figure 3), with an average of 0.24 (s.d. = 0.67). In the studied vespine and polistine wasps, each host species harboured a single parasite species, except for Polistes dominula, which is attacked by three inquiline species (figure 4); the mean number of parasites per host species was 0.43 (s.d. = 0.64).
Figure 2.
Phylogenetic tree of 182 eusocial bumblebee species, with bar plots showing species-specific geographical range sizes (measured as the number of 611 000 km2 squares occupied) and numbers of associated social brood parasite species. In both plots, species attacked by parasites are indicated by black bars.
Figure 3.
Phylogenetic tree of 63 eusocial Myrmica ant species, with bar plots showing species-specific geographical range sizes (measured as the number of 611 000 km2 squares occupied) and numbers of associated social brood parasite species. Parasitized species are indicated by black bars.
Figure 4.
Phylogenetic tree of 37 eusocial vespine and polistine wasp species, with bar plot showing numbers of associated social brood parasite species.
In bumblebees, statistically significant phylogenetic signal was present for GRS and the presence of Psithyrus inquilines when considering all observations of parasitism, but not when considering only the smaller number of cases in which a breeding inquiline queen had been observed; phylogeny had no effect on the number of parasite species (table 1). In Myrmica ants, GRS varied randomly across the phylogeny, and phylogenetic signals for the existence of parasitism and the number of associated social parasite species were statistically only marginally significant (table 1). In the case of vespine and polistine wasps, both the existence of parasitism and the number of parasite species were random with respect to phylogeny (table 1).
Table 1.
Estimated phylogenetic signals for the presence or absence of social brood parasitism (measured using the D-statistic) and numbers of associated parasite species and geographical range sizes (GRS) (measured using Pagel's λ) in the three focal hymenopteran groups. N denotes the number of species in each host phylogeny, p-values are given for deviations from the absence of phylogenetic signal (D = 1, λ = 0) and from pure Brownian-model evolution (D = 0). In the case of bumblebees, phylogenetic signal is estimated for a full dataset of parasitized hosts (n = 34 parasitized bumblebee species) and for hosts with observations of breeding cuckoo bumblebee queens (breeding queen) (n = 24 parasitized bumblebee species).
| taxon and trait | N | D | p (D = 0) | p (D = 1) | λ | p (λ = 0) |
|---|---|---|---|---|---|---|
| bumblebees | ||||||
| parasitism | 182 | 0.747 | <0.001 | 0.028 | ||
| breeding queen | 182 | 0.976 | <0.001 | 0.456 | ||
| number of parasite species | 182 | 0.015 | 0.910 | |||
| GRS | 182 | 0.292 | 0.004 | |||
| ants | ||||||
| parasitism | 63 | 0.491 | 0.189 | 0.052 | ||
| number of parasite species | 63 | 0.266 | 0.064 | |||
| GRS | 63 | 0.243 | 0.227 | |||
| wasps | ||||||
| parasitism | 37 | 1.16 | <0.001 | 0.715 | ||
| number of parasite species | 37 | 0.001 | 0.999 | |||
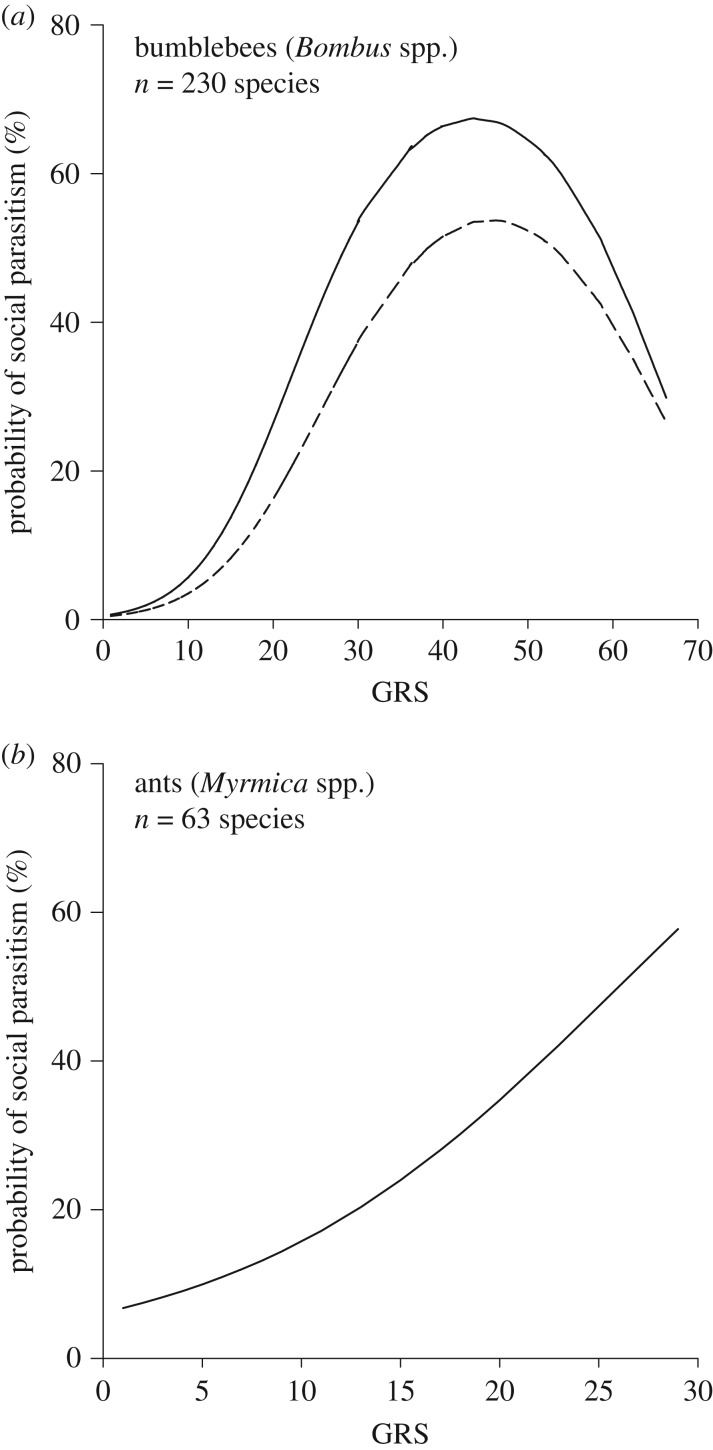
According to our logistic and phylogenetic logistic regressions, the risk of being attacked by at least one social parasite species increases statistically significantly with GRS in both bumblebees and Myrmica ants, although the effect is statistically only marginally significant for ants when taking phylogeny into account (table 2). However, the non-phylogenetic analyses based on all parasitized species as well as all species with direct observations of breeding inquiline queens suggest that bumblebee species with the largest GRS are less often parasitized than are species with a medium GRS (figure 5a and table 2).
Table 2.
Results from analyses in which the presence of social parasites in bumblebees and Myrmica ants is modelled using logistic regression (all species as independent observations, 229 bumblebee species and 63 Myrmica ant species) and phylogenetic logistic regressions (182 bumblebee species and 63 Myrmica ant species). Two different models were used for bumblebees: one with every species known to host parasites (‘all parasitism’, 34 parasitized bumblebee species) and one including only host species where breeding cuckoo bumblebee queens have been observed (‘breeding queen’, 24 parasitized bumblebee species). Explanatory variables were geographical range size (GRS, number of 611 000 km2 grids) and GRS2. If GRS2 was not statistically significant, only the model with GRS is presented.
| variable | logistic regression |
phylogenetic logistic regression |
||||||
|---|---|---|---|---|---|---|---|---|
| estimate | s.e. | Wald | p-value | estimate | s.e. | t | p-value | |
| bumblebees | ||||||||
| all parasitism | ||||||||
| intercept | −5.213 | 0.739 | 49.78 | <0.001 | 0.008 | 0.446 | 0.02 | 0.985 |
| GRS | 0.272 | 0.052 | 27.60 | <0.001 | 0.012 | 0.002 | 6.57 | <0.001 |
| GRS2 | −0.003 | 0.001 | 17.16 | <0.001 | ||||
| breeding queen | ||||||||
| intercept | −5.525 | 0.873 | 40.08 | <0.001 | 0.014 | 0.438 | 0.03 | 0.974 |
| GRS | 0.249 | 0.058 | 18.49 | <0.001 | 0.010 | 0.053 | 4.72 | <0.001 |
| GRS2 | −0.003 | 0.001 | 11.02 | 0.001 | ||||
| ants | ||||||||
| intercept | −2.727 | 0.607 | 20.199 | <0.001 | 0.072 | 0.242 | 0.30 | 0.766 |
| GRS | 0.105 | 0.043 | 5.960 | 0.015 | 0.009 | 0.005 | 1.85 | 0.069 |
Figure 5.
Probability (%) of the presence of social brood parasitism in (a) bumblebee species and (b) ant species in relation to species-specific geographical range size (number of 611 000 km2 grids). In (a), the continuous line is based on all host species for which social brood parasites have been observed, whereas the dashed line is based only on cases in which a breeding queen of a social brood parasite has been observed [32].
Reflecting the results of the logistic regressions, average GRS of unparasitized bumblebee species (10.8 grids, s.d. = 11.9, n = 196) was statistically significantly smaller than that of parasitized species (30.9, s.d. = 15.8, n = 34; t-test with unequal variances, t = −7.06, d.f. = 39.8, p < 0.001). Because the probability of social parasitism in bumblebee species was not linear (figure 5a), we also compared the estimated probabilities of parasitism between unparasitized and parasitized bumblebee species. This comparison revealed that the average estimated probability of parasitism in unparasitized species (10.5%, s.d. = 15.8%, n = 196) was smaller than in parasitized species (30.9%, s.d. = 19.6, n = 34; t-test with unequal variances, t = −8.07, d.f. = 41.8, p < 0.001). Reanalysing the data using only hosts for which nests with breeding Psithyrus queens had been found did not change these results (results not shown).
In the case of Myrmica ants, the difference in average GRS of unparasitized (5.7 grids, s.d = 6.7, n = 54) and parasitized species (13.0, s.d. = 10.0, n = 9) was only marginally significant (t-test with unequal variances, t = −2.11, d.f. = 9.2, p = 0.064).
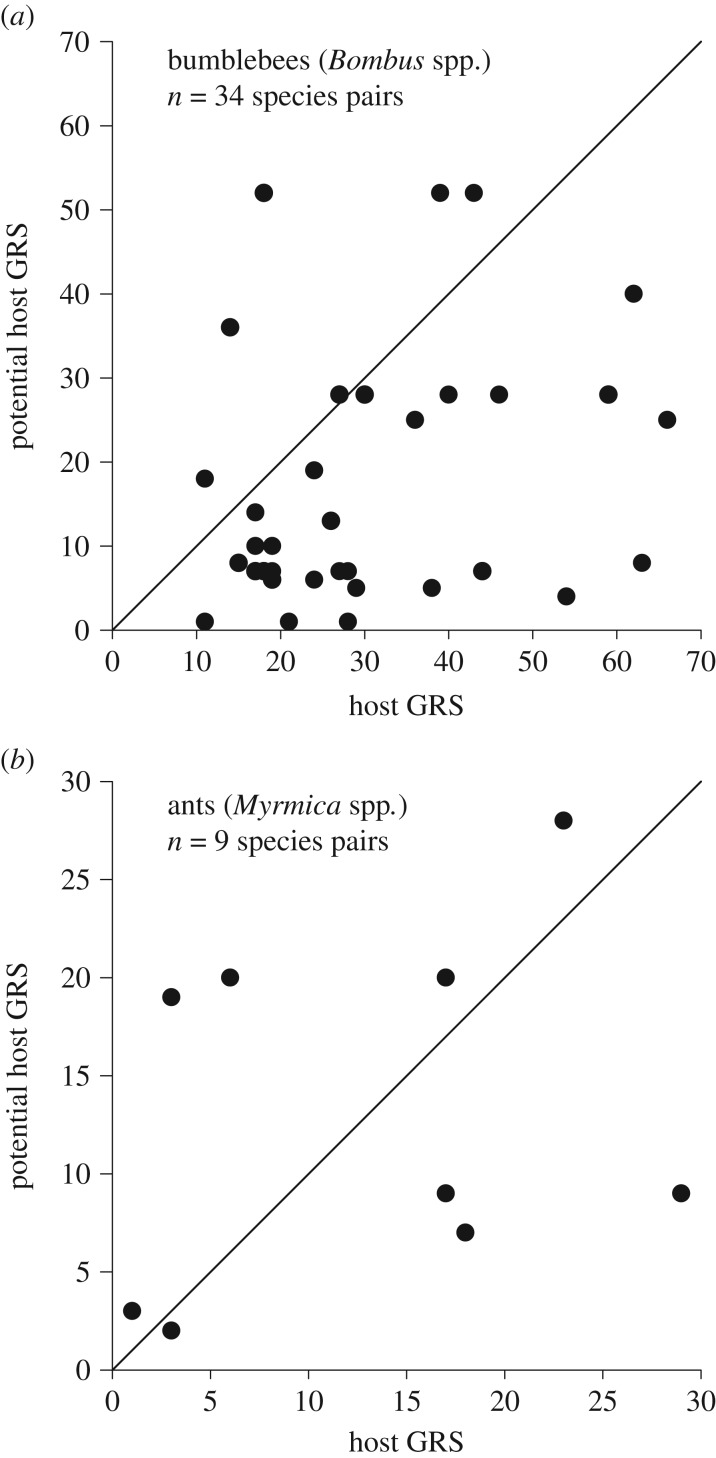
In 34 bumblebee species pairs consisting of closely related parasitized and unparasitized species (electronic supplementary table S4), parasite-free species had on average a smaller GRS (17.4, s.d. = 15.2) than did their closest parasitized relative (30.9, s.d. = 15.8) (paired-samples t-test, t = 4.21, d.f. = 33, p < 0.001; figure 6a). By contrast, in nine corresponding pairs of Myrmica ants (table S2), the average GRS of non-parasitized species (13.0, s.d. = 10.0) did not differ statistically significantly from parasitized species (13.0, s.d. = 9.0) (paired-samples t-test, t = 0.00, d.f. = 8, p = 0.999; figure 6b).
Figure 6.
The geographical range size (number of 611 000 km2 grids) of parasitized (a) bumblebee species and (b) Myrmica ant species plotted against the GRS of their most closely related unparasitized species. The continuous line indicates an equal GRS in host and non-host species. Below the line are species pairs in which the GRS of the parasitized species is larger than that of its closest unparasitized relative.
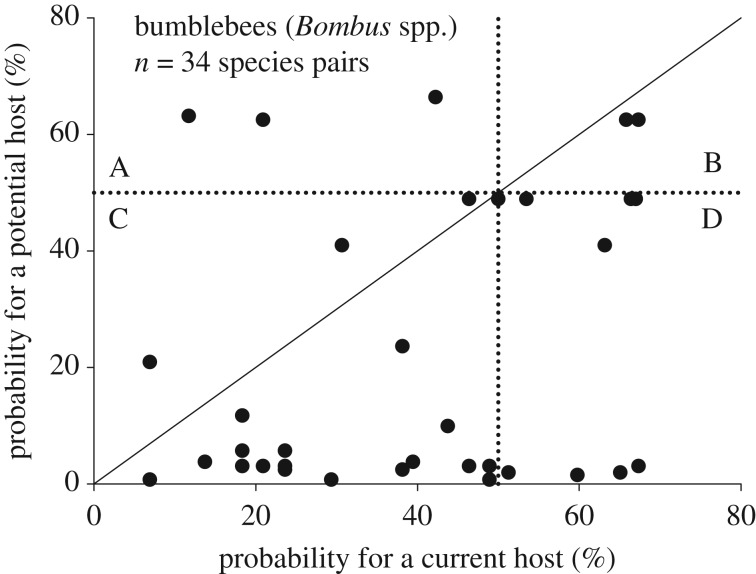
In pairwise analyses of the probability of parasitism in bumblebees estimated based on the non-phylogenetic logistic regression model, the estimated probability was on average lower in unparasitized (mean = 22.3%, s.d. = 24.6%, n = 34) than in parasitized species (mean = 39.3%, s.d. = 19.6%) (n = 34, paired-samples t-test, t = 3.69, d.f. = 33, p = 0.001; figure 7). Based on the pairwise plot, three of the studied parasitized bumblebee species (Bombus argillaceus, Bombus bimaculatus and Bombus pascuorum) have a low probability of parasitism (figure 7, sector A), while their currently non-parasitized relatives have high probability of serving as new potential hosts in the future (see also electronic supplementary material, table S4). Two of the current host species (Bombus lucorum and Bombus terrestris) are not likely to lose their social parasites (figure 7, sector B). The situation is worst for cuckoo bumblebee species located in sector C of figure 7: their host species have a high probability of losing their social brood parasites, but alternative host species either are not available or have a low estimated probability of parasitism (less than 50% and in most of cases less than 20%; figure 7, sector C). A slightly better situation for cuckoo bumblebees is present within sector D of figure 7, as the current hosts are unlikely to lose their social brood parasite species.
Figure 7.
Probability (%) of social parasitism of potential bumblebee host species in relation to parasitized bumblebee host species. The continuous line indicates an equal probability between host and non-host species. The symbols below the continuous line represent a situation where the most closely related unparasitized species has a smaller probability than the parasitized bumblebee host species. Dotted lines indicate a 50% probability of infection. Species in sector A are likely to have lost their social parasite but have an alternative closely related potential host, in B have a social parasite and have also a closely related potential host, in C may lose their social parasite and do not have a closely related potential host, and in D have a social parasite, but do not have a closely related potential host.
We found that 13 bumblebee species assumed to be unparasitized may in fact act as hosts for cuckoo bumblebees, as their model-based probability of parasitism is over 50% (table 3). Three of these potential host species occur in the Holarctic region, seven in the Palaearctic region and three in the Nearctic region. Furthermore, we found that 23 currently parasitized bumblebee species have a high probability of losing their social brood parasites in the future, as their estimated probability of parasitism is below 50% (table 3). More than half (14) of these species occur in the Nearctic region, eight in the Palaearctic region and only one in the Holarctic region (table 3). The lowest estimated probability (7%) of parasitism in an actually parasitized species was found for Bombus affinis, meaning that it may lose its inquiline Bombus ashtoni. Also, Bombus appositus is likely to lose its social brood parasites Bombus insularis and Bombus flavidus (table 3).
Table 3.
Bumblebee (Bombus) species that are likely to be parasitized or to lose their social parasite, and the geographical area of a specific social parasite species that is a potential parasite for a given host species. The probability was calculated using non-phylogenetic logistic regression that was based on the geographical range size of current and potential hosts (figure 5a and table 1). ‘GR’ indicates the geographical region of each species.
| Bombus species | probability (%) | GR | |
|---|---|---|---|
| likely parasitized | potential parasite species | ||
| B. consobrinus | 63 | Palaearctic | B. barbutellus |
| B. cryptarum | 80 | Holarctic | B. bohemicus, B. vestalisi |
| B. cullumanus | 56 | Palaearctic | western Palaearctic species |
| B. distinguendus | 64 | Holarctic | B. barbutellus |
| B. frigidus | 51 | Nearctic | Nearctic species |
| B. hyperboreus | 52 | Holarctic | western Palaearctic species |
| B. laesus | 66 | Palaearctic | western Palaearctic species |
| B. mixtus | 51 | Nearctic | B. insularis, Neartic species |
| B. patagiatus | 53 | Palaearctic | western Palaearctic species |
| B. schrencki | 66 | Palaearctic | B. rubestris, B. campestris |
| B. sporadicus | 65 | Palaearctic | western Palaearctic species |
| B. subterraneus | 65 | Palaearctic | B. barbutellus |
| B. sylvicola | 63 | Nearctic | B. citrus, Neartic species |
| likely to lose parasite | parasite species | ||
| B. affinis | 7 | Nearctic | B. ashtoni |
| B. appositus | 7 | Nearctic | B. insularis, B. flavidus |
| B. argillaceus | 21 | Palaearctic | B. barbutellus |
| B. bifarius | 21 | Nearctic | B. insularis |
| B. bimaculatus | 12 | Nearctic | B. citrinus |
| B. fervidus | 46 | Nearctic | B. insularis |
| B. flavifrons | 38 | Nearctic | B. insularis |
| B. huntii | 18 | Nearctic | B. insularis |
| B. hypnorum | 39 | Palaearctic | B. barbatellus, B. sylvestris |
| B. impatiens | 18 | Nearctic | B. citrinus, B. insularis |
| B. jonellus | 31 | Holarctic | B. flavidus, B. quadricolor |
| B. lapidarius | 49 | Palaearctic | B. ruperstris |
| B. monticola | 14 | Palaearctic | B. sylvestris |
| B. nevadensis | 18 | Nearctic | B. insularis |
| B. occidentalis | 29 | Nearctic | B. suckleyi, B. flavidus |
| B. pascuorum | 42 | Palaearctic | B. campestris, B. rupestris |
| B. pomorum | 24 | Palaearctic | B. campestris |
| B. ruderatus | 24 | Palaearctic | B. inexspectatus, B. barbutellus |
| B. rufocinctus | 44 | Nearctic | B. insularis, B. flavidus |
| B. sylvarum | 46 | Palaearctic | B. campestris |
| B. ternarius | 24 | Nearctic | B. insularis |
| B. terricola | 49 | Nearctic | B. ashtoni |
| B. vagans | 38 | Nearctic | B. citrinus |
4. Discussion
Our objective was to evaluate how host phylogeny and species-specific GRS influence the occurrence of brood parasitism in eusocial insects. We found five main results: first, the number of social brood parasite species varied widely among actual and potential hosts, so that most species are parasite-free, whereas others may be attacked by up to four different inquiline species. Second, geographical range size and the presence of one or more parasites were phylogenetically conserved in bumblebees, but phylogenetic signal was less clear or absent in Myrmica ants and vespine and polistine wasps. Third, phylogenetic logistic regressions showed that the probability of attack by one or more parasite species increased with GRS in bumblebees, but the effect was only marginally significant in ants; comparisons of range sizes across pairs of related parasitized and unparasitized species supported these conclusions. Fourth, non-phylogenetic logistic regressions suggested that parasitism increases with increasing range size in bumblebees and in Myrmica ants, but also that bumblebee species with the broadest distributions have a lower probability of being parasitized than species with a medium-sized range. Finally, we used our logistic regression model to estimate the likelihood of parasitism or loss of parasites for specific bumblebee species. Our study provides new insights into brood parasitism of eusocial insects, how host GRS and phylogeny affect parasitism, how likely parasites are to switch among hosts, and how we can use this information for evaluating threats of host and parasite species in conservation biology.
While most parasitized species are attacked by a single social parasite species, others harbour as many as four species. The most severely parasitized species are apparently very poor at defending against their parasites, because their parasites either are good at mimicking the chemical signatures of the host, or are otherwise able to evade the host's defences [19,26]. Currently, there is very little research on this topic, and it needs to be studied thoroughly in the future. In host species that are used by more than one brood parasite species, interspecific competition is likely to occur between the different parasite species. This will most probably reduce the reproductive success of all brood parasites and thereby lower their population sizes and the overall prevalence. In the long run, interspecific competition among parasites may even lead to extinction of weaker competitors, unless they are able to shift onto a new species. For the host species, multi-parasite attack is especially demanding because they need to defend themselves against two, three or even four parasite species. It seems unlikely that a host species could develop a defensive strategy that would be simultaneously effective against all parasite species. Therefore, it is likely that the main coevolutionary processes in multispecies host–parasite systems involve interspecific competition between parasite species rather than escalating defences and counter-defences of hosts and parasites.
The phylogenetic signal present in bumblebee parasitism and its tendency in Myrmica ants suggest that brood parasite species have been able to shift among closely related hosts and non-hosts. At least some social brood parasites, such as Myrmica karavajevi, are generalists capable of using multiple host species. Such generalist parasite species may occasionally be forced to shift from a previously exploited host to a new one. In our previous studies, we have found phylogenetic signals in water mite and gregarine parasitism in damselfly and dragonfly hosts [57]. This begs for further phylogenetic studies on other host–parasite systems, in order to find possible general patterns. That vespine and polistine wasps did not exhibit phylogenetic signal with regard to social brood parasitism suggests that their parasites have not been restricted to shifting among closely related hosts in the past, at least not to the extent that we expected. It is possible that wasp parasites are specialized to their host species, to the extent that interspecific competition has not forced parasites to shift from one host to another. Wasps are somewhat problematic for our study owing to the lack of reliable information on species-specific geographical ranges and to the fact that some widely distributed species, especially Vespula germanica and Vespula vulgaris, are invasive alien species throughout most part of their range [58]. A larger dataset and more data on the distributions of wasp species are required before we can rule out the possibility that the patterns we observed in bumblebees and Myrmica ants do not occur in wasps.
As expected, our different analyses consistently revealed that the GRS of bumblebee species explains the occurrence of brood parasite species. Reflecting this general pattern, unparasitized bumblebee species tended to have a smaller GRS than closely related parasitized species. Shifts onto new hosts that have a small GRS [15] may be rare because the chance of encounter may be low, but also because local abundance of species tends to drop with decreasing GRS [59], making the potential host harder to locate and parasitize.
However, our logistic regressions suggest that bumblebee species with a large geographical range may be less often parasitized than species with a medium-sized distribution. This indicates that some species with large GRS are not attacked by brood parasites. There are several mutually non-exclusive explanations for why hosts with larger range sizes would not have brood parasites. It is possible that certain species with large GRS have had parasites, but for some reason have lost them, possibly because a widely distributed host is a superior competitor against potential brood parasite species and is often better adapted to fight against parasites. This may be supported by the fact that reproduction by inquiline queens has been observed in the nests of only 24 out of 34 of host bumblebee species, while successful reproduction remains to be confirmed in 10 host species [32]. Analogous examples have also been found in birds and their brood parasites [60]. However, it is also possible that, in some cases, the existing parasitism has not been confirmed owing to low parasite prevalence, which might be the case with the 10 bumblebee species with observations of non-reproducing Psithyrus queens in their nests. Evidently, more studies are needed to confirm the status of such inquiline–possible host species pairs.
Conservation biology of parasites is challenging, because observing parasite species is always more difficult than observing their hosts. In general, parasitism depends on host GRS [61], and seems to be lower near the edges of host geographical ranges [61]. Therefore, the GRS of parasites tends to be smaller than those of their hosts [15]. Under this assumption, we developed a method to evaluate whether a given parasite species is likely to be lost from its current host species, based on the GRS of both current and potential host species. We found that several bumblebee species will probably lose their parasite species in the future. In this respect, our results support observations of a general decline of bumblebee species and their social brood parasites in North America [3,33,62] and Europe [8,15,33]. We also found that relatively few parasite species have the potential to find a new suitable host that is closely related to their current host (sectors A and B in figure 7). On the other hand, most host species seem to have a high risk of losing their parasite species (sector C in figure 7). Those host species should be a high priority for conservation efforts, as otherwise both the bumblebees and their social parasite species will be lost [8]. Finally, we found that there are probably several bumblebee species that have cuckoo bumblebee parasites that have not yet been discovered (table 3). To date, the hosts of less than half of the known cuckoo bumblebee species have been confirmed [32]. Evidently, identifying the host species of cuckoo bumblebees is vitally important for evaluating their conservation priorities; in table 3, we have listed bumblebee species that probably act as hosts of cuckoo bumblebees with still-unknown hosts.
5. Conclusion
Parasites are more vulnerable to extinction than are their hosts [8,63], and this seems to be true also for social brood parasites. Human-induced environmental changes and global warming will cause declines in the population sizes of hosts as well as in ranges of brood parasites, because the probability of host shifts decreases. Our results highlight the fact that the risk of extinction is highest in brood parasites that rely on rare or threatened host species [8]. Thus, to lower the risk of extinction of brood parasite species, it is important to conserve their current hosts, especially in cases in which suitable closely related hosts do not exist or have restricted geographical ranges. Our results show that GRS of host species can be used to estimate the extinction risk of brood parasites, especially if information on phylogenetic relationships among actual and potential hosts is available. However, long-term datasets and more up-to-date information from larger geographical areas are required to provide more insightful data in order to understand the proximate and ultimate reasons for the risks of extinction of hosts and their social brood parasites.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank H. Hines for kindly providing the Bombus chronogram used in our analyses. We thank also Jenni Kauppi and Sylvia Suhonen, who helped us in datamining and estimating species-specific geographical range sizes from distribution maps.
Data accessibility
All relevant data are within the paper. Ecological data and information on phylogenetic relationships in this study were compiled based on a literature survey; see cited publications.
Authors' contributions
J.Su. and J.So. collected data and designed the study. J.S.u coordinated the study and compiled the database. J.Su., T.N. and J.J.I. conducted statistical tests and phylogenetic analyses. All the authors contributed to manuscript preparation. All the authors read and approved the final manuscript.
Competing interests
We have no competing interest.
Funding
No financial support.
References
- 1.Urban MC. 2015. Accelerating extinction risk from climate change . Science 348, 571–573. ( 10.1126/science.aaa4984) [DOI] [PubMed] [Google Scholar]
- 2.Thomas C, et al. 2004. Extinction risk from climate change . Nature 427, 145–148. ( 10.1038/nature02121) [DOI] [PubMed] [Google Scholar]
- 3.Szabo ND, Colla SR, Wagner DL, Gall LF, Kerr JT. 2012. Do pathogen spillover, pesticide use, or habitat loss explain recent North American bumblebee declines? Conserv. Lett. 5, 232–239. ( 10.1111/j.1755-263X.2012.00234.x) [DOI] [Google Scholar]
- 4.Woodcock BA, et al. 2017. Country-specific effects of neonicotinoid pesticides on honey bees and wild bees . Science 356, 1393 ( 10.1126/science.aaa1190) [DOI] [PubMed] [Google Scholar]
- 5.Kerr JT, et al. 2015. Climate change impacts on bumblebees converge across continents . Science 349, 177–180. ( 10.1126/science.aaa7031) [DOI] [PubMed] [Google Scholar]
- 6.Pimm SL, Jenkins CN, Abell R, Brooks TM, Gittleman JL, Joppa LN, Raven PH, Roberts CM, Sexton JO. 2014. The biodiversity of species and their rates of extinction, distribution, and protection . Science 344, 987 ( 10.1126/science.1246752) [DOI] [PubMed] [Google Scholar]
- 7.Suhonen J, Hilli-Lukkarinen M, Korkeamäki E, Kuitunen M, Kullas J, Penttinen J, Salmela J. 2010. Local extinction of dragonfly and damselfly populations in low- and high-quality habitat patches . Conserv. Biol. 24, 1148–1153. ( 10.1111/j.1523-1739.2010.01504.x) [DOI] [PubMed] [Google Scholar]
- 8.Suhonen J, Rannikko J, Sorvari J. 2015. The rarity of host species affects the co-extinction risk in socially parasitic bumblebee Bombus (Psithyrus) species . Ann. Zool. Fenn. 52, 236–242. ( 10.5735/086.052.0402) [DOI] [Google Scholar]
- 9.Hoberg EP, Brooks DR. 2008. A macroevolutionary mosaic: episodic host-switching, geographical colonization and diversification in complex host–parasite systems . J. Biogeogr. 35, 1533–1550. ( 10.1111/j.1365-2699.2008.01951.x) [DOI] [Google Scholar]
- 10.Marques E, Price P, Cobb N. 2000. Resource abundance and insect herbivore diversity on woody fabaceous desert plants . Environ. Entomol. 29, 696–703. ( 10.1603/0046-225X-29.4.696) [DOI] [Google Scholar]
- 11.Christman M, Culver D. 2001. The relationship between cave biodiversity and available habitat . J. Biogeogr. 28, 367–380. ( 10.1046/j.1365-2699.2001.00549.x) [DOI] [Google Scholar]
- 12.Gregory R. 1990. Parasites and host geographic range as illustrated by waterfowl . Funct. Ecol. 4, 645–654. ( 10.2307/2389732) [DOI] [Google Scholar]
- 13.Krasnov B, Shenbrot G, Khokhlova I, Degen A. 2004. Flea species richness and parameters of host body, host geography and host ‘milieu’ . J. Anim. Ecol. 73, 1121–1128. ( 10.1111/j.0021-8790.2004.00883.x) [DOI] [Google Scholar]
- 14.Lindenfors P, Nunn CL, Jones KE, Cunningham AA, Sechrest W, Gittleman JL. 2007. Parasite species richness in carnivores: effects of host body mass, latitude, geographical range and population density . Global Ecol. Biogeogr. 16, 496–509. ( 10.1111/j.1466-8238.2006.00301.x) [DOI] [Google Scholar]
- 15.Suhonen J, Rannikko J, Sorvari J. 2016. Species richness of cuckoo bumblebees is determined by the geographical range area of the host bumblebee . Insect Conserv. Divers. 9, 529–535. ( 10.1111/icad.12196) [DOI] [Google Scholar]
- 16.Papkou A, Gokhale CS, Traulsen A, Schulenburg H. 2016. Host–parasite coevolution: why changing population size matters . Zoology 119, 330–338. ( 10.1016/j.zool.2016.02.001) [DOI] [PubMed] [Google Scholar]
- 17.Kilner RM, Langmore NE. 2011. Cuckoos versus hosts in insects and birds: adaptations, counter-adaptations and outcomes . Biol. Rev. 86, 836–852. ( 10.1111/j.1469-185X.2010.00173.x) [DOI] [PubMed] [Google Scholar]
- 18.King KC, Delph LF, Jokela J, Lively CM. 2009. The geographic mosaic of sex and the Red Queen . Curr. Biol. 19, 1438–1441. ( 10.1016/j.cub.2009.06.062) [DOI] [PubMed] [Google Scholar]
- 19.Grüter C, Jongepier E, Foitzik S. 2018. Insect societies fight back: the evolution of defensive traits against social parasites . Phil. Trans. R. Soc. B 373, 20170200 ( 10.1098/rstb.2017.0200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bourke A, Franks N. 1991. Alternative adaptations, sympatric speciation and the evolution of parasitic, inquiline ants . Biol. J. Linn. Soc. 43, 157–178. ( 10.1111/j.1095-8312.1991.tb00591.x) [DOI] [Google Scholar]
- 21.Schulte RD, Makus C, Hasert B, Michiels NK, Schulenburg H. 2010. Multiple reciprocal adaptations and rapid genetic change upon experimental coevolution of an animal host and its microbial parasite . Proc. Natl Acad. Sci. USA 107, 7359–7364. ( 10.1073/pnas.1003113107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marston MF, Pierciey FJ Jr, Shepard A, Gearin G, Qi J, Yandava C, Schuster SC, Henn MR, Martiny JBH. 2012. Rapid diversification of coevolving marine Synechococcus and a virus . Proc. Natl Acad. Sci. USA 109, 4544–4549. ( 10.1073/pnas.1120310109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurze C, Le Conte Y, Dussaubat C, Erler S, Kryger P, Lewkowski O, Müller T, Widder M, Moritz RFA. 2015. Nosema tolerant honeybees (Apis mellifera) escape parasitic manipulation of apoptosis . PLoS One 10, e0140174 ( 10.1371/journal.pone.0140174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Betts A, Gray C, Zelek M, MacLean RC, King KC. 2018. High parasite diversity accelerates host adaptation and diversification . Science 360, 907 ( 10.1126/science.aam9974) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krasnov B, Shenbrot G, Khokhlova I, Poulin R. 2004. Relationships between parasite abundance and the taxonomic distance among a parasite's host species: an example with fleas parasitic on small mammals . Int. J. Parasitol. 34, 1289–1297. ( 10.1016/j.ijpara.2004.08.003) [DOI] [PubMed] [Google Scholar]
- 26.Martin SJ, Carruthers JM, Williams PH, Drijfhout FP. 2010. Host specific social parasites (Psithyrus) indicate chemical recognition system in bumblebees . J. Chem. Ecol. 36, 855–863. ( 10.1007/s10886-010-9805-3) [DOI] [PubMed] [Google Scholar]
- 27.Ayasse M, Jarau S. 2014. Chemical ecology of bumble bees . Annu. Rev. Entomol. 59, 299–319. ( 10.1146/annurev-ento-011613-161949) [DOI] [PubMed] [Google Scholar]
- 28.Pagel M. 1999. Inferring the historical patterns of biological evolution . Nature 401, 877–884. ( 10.1038/44766) [DOI] [PubMed] [Google Scholar]
- 29.Lopez-Osorio F, Perrard A, Pickett KM, Carpenter JM, Agnarsson I. 2015. Phylogenetic tests reject Emery's rule in the evolution of social parasitism in yellowjackets and hornets (Hymenoptera: Vespidae, Vespinae). R. Soc. open sci. 2, 150159 ( 10.1098/rsos.150159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savolainen R, Vepsäläinen K. 2003. Sympatric speciation through intraspecific social parasitism . Proc. Natl Acad. Sci. USA 100, 7169–7174. ( 10.1073/pnas.1036825100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jansen G, Savolainen R, Vepsäläinen K. 2010. Phylogeny, divergence-time estimation, biogeography and social parasite–host relationships of the Holarctic ant genus Myrmica (Hymenoptera: Formicidae) . Mol. Phylogenet. Evol. 56, 294–304. ( 10.1016/j.ympev.2010.01.029) [DOI] [PubMed] [Google Scholar]
- 32.Lhomme P, Hines HM. In press. Ecology and evolution of cuckoo bumble bees . Ann. Soc. Am., say031 ( 10.1093/aesa/say031) [DOI] [Google Scholar]
- 33.Arbetman MP, Gleiser G, Morales CL, Williams P, Aizen MA. 2017. Global decline of bumblebees is phylogenetically structured and inversely related to species range size and pathogen incidence . Proc. R. Soc. B 284, 20170204 ( 10.1098/rspb.2017.0204) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arbetman MP, Gleiser G, Morales CL, Williams P, Aizen MA. 2017. Data from: Global decline of bumblebees is phylogenetically structured and inversely related to species range size and pathogen incidence Dryad Digital Repository. ( 10.5061/dryad.71q32) [DOI] [PMC free article] [PubMed]
- 35.Cameron SA, Hines HM, Williams PH. 2007. A comprehensive phylogeny of the bumble bees (Bombus) . Biol. J. Linn. Soc. 91, 161–188. ( 10.1111/j.1095-8312.2007.00784.x) [DOI] [Google Scholar]
- 36.Anonymous. 2018. AntWiki. See http://www.antwiki.org/wiki/Welcome_to_AntWiki.
- 37.Radchenko AG, Elmes GW. 2010. Myrmica ants (Hymenoptera: Formicidae) of the Old World. Fauna mundi 3. Warsaw, Poland: Natura Optima Dux Foundation. [Google Scholar]
- 38.Persson S. 2015. Phylogeny and taxonomy of the subfamily Vespinae (Hymenoptera: Vespidae), based on five molecular markers. MSc thesis, Department of Biological and Environmental Sciences, University of Gothenburg. [Google Scholar]
- 39.Cervo R. 2006. Polistes wasps and their social parasites: an overview . Ann. Zool. Fenn. 43, 531–549. [Google Scholar]
- 40.Guenard B, Weiser MD, Gomez K, Narula N, Economo EP. 2017. The Global Ant Biodiversity Informatics (GABI) database: synthesizing data on the geographic distribution of ant species (Hymenoptera: Formicidae) . Myrmecol. News 24, 83–89. [Google Scholar]
- 41.Janicki J, Narula N, Ziegler M, Guenard B, Economo EP. 2016. Visualizing and interacting with large-volume biodiversity data using client-server web-mapping applications: the design and implementation of antmaps.org . Ecol. Inform. 32, 185–193. ( 10.1016/j.ecoinf.2016.02.006) [DOI] [Google Scholar]
- 42.Harvey PH, Pagel MD. 1991. The comparative method in evolutionary biology. Oxford, UK: Oxford University Press. [Google Scholar]
- 43.Freckleton R. 2000. Phylogenetic tests of ecological and evolutionary hypotheses: checking for phylogenetic independence . Funct. Ecol. 14, 129–134. ( 10.1046/j.1365-2435.2000.00400.x) [DOI] [Google Scholar]
- 44.Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 3.51. See http://www.mesquiteproject.org .
- 45.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language . Bioinformatics 20, 289–290. ( 10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- 46.R Core Team. 2018. R, a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org/. [Google Scholar]
- 47.Bouckaert R, Heled J, Kuehnert D, Vaughan T, Wu C, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis . PloS Comput. Biol. 10, e1003537 ( 10.1371/journal.pcbi.1003537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7 . Syst. Biol. 67, 901–904. ( 10.1093/sysbio/syy032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies . Bioinformatics 30, 1312–1313. ( 10.1093/bioinformatics/btu033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Gateway Computing Environments Workshop (GCE), New Orleans, LA, 14 November 2010, pp. 1–8. Piscataway, NJ: IEEE. ( 10.1109/GCE.2010.5676129) [DOI]
- 51.Freckleton R, Harvey P, Pagel M. 2002. Phylogenetic analysis and comparative data: a test and review of evidence . Am. Nat. 160, 712–726. ( 10.1086/343873) [DOI] [PubMed] [Google Scholar]
- 52.Fritz SA, Purvis A. 2010. Selectivity in mammalian extinction risk and threat types: a new measure of phylogenetic signal strength in binary traits . Conserv. Biol. 24, 1042–1051. ( 10.1111/j.1523-1739.2010.01455.x) [DOI] [PubMed] [Google Scholar]
- 53.Muenkemueller T, Lavergne S, Bzeznik B, Dray S, Jombart T, Schiffers K, Thuiller W. 2012. How to measure and test phylogenetic signal . Methods Ecol. Evol. 3, 743–756. ( 10.1111/j.2041-210X.2012.00196.x) [DOI] [Google Scholar]
- 54.Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things) . Methods Ecol. Evol. 3, 217–223. ( 10.1111/j.2041-210X.2011.00169.x) [DOI] [Google Scholar]
- 55.Orme D.2018. The caper package: comparative analyses of phylogenetics and evolution in R.
- 56.Ho LST, Ane C. 2014. A linear-time algorithm for Gaussian and non-Gaussian trait evolution models . Syst. Biol. 63, 397–408. ( 10.1093/sysbio/syu005) [DOI] [PubMed] [Google Scholar]
- 57.Ilvonen JJ, Suhonen J. 2016. Phylogeny affects host's weight, immune response and parasitism in damselflies and dragonflies . R. Soc. open sci. 3, 160421 ( 10.1098/rsos.160421) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beggs JR, Brockerhoff EG, Corley JC, Kenis M, Masciocchi M, Muller F, Rome Q, Villemant C. 2011. Ecological effects and management of invasive alien Vespidae . Biocontrol 56, 505–526. ( 10.1007/s10526-011-9389-z) [DOI] [Google Scholar]
- 59.Soler J, Møller A, Soler M. 1999. A comparative study of host selection in the European cuckoo Cuculus canorus . Oecologia 118, 265–276. ( 10.1007/s004420050727) [DOI] [PubMed] [Google Scholar]
- 60.Nakamura H, Kubota S, Suzuki R. 1998. Coevolution between the common cuckoo and its major hosts in Japan. In Parasitic birds and their hosts (eds Rothstein SI, Robinson SK), pp. 94–112. New York, NY: Oxford University Press. [Google Scholar]
- 61.Kaunisto KM, Kaunisto P, Vahtera V, Suhonen J. 2015. Populations of the damselfly Coenagrion hastulatum at the edge of the species range have fewer gregarine and water mite parasites. Freshwat. Biol. 60, 794–801. ( 10.1111/fwb.12534) [DOI] [Google Scholar]
- 62.Cameron SA, Lozier JD, Strange JP, Koch JB, Cordes N, Solter LF, Griswold TL. 2011. Patterns of widespread decline in North American bumble bees . Proc. Natl Acad. Sci. USA 108, 662–667. ( 10.1073/pnas.1014743108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koh L, Dunn R, Sodhi N, Colwell R, Proctor H, Smith V. 2004. Species coextinctions and the biodiversity crisis . Science 305, 1632–1634. ( 10.1126/science.1101101) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Arbetman MP, Gleiser G, Morales CL, Williams P, Aizen MA. 2017. Data from: Global decline of bumblebees is phylogenetically structured and inversely related to species range size and pathogen incidence Dryad Digital Repository. ( 10.5061/dryad.71q32) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
All relevant data are within the paper. Ecological data and information on phylogenetic relationships in this study were compiled based on a literature survey; see cited publications.