Abstract
Adolescent idiopathic scoliosis (AIS) is the most common spinal deformity disease in adolescents but its etiology and pathogenesis are still unclear. The current study aims to identify the relationship between single nucleotide polymorphisms (SNPs) of G protein-coupled receptor 126 (GPR126) gene and AIS predisposition. GPR126 contains 26 exons and alternative splicing of exon 6 and exon 25 produces 4 protein-coding transcripts. We genotyped SNPs of GPR126 gene around exon 6 and exon 25 in 131 Chinese AIS patients and 132 healthy controls and provided evidence that SNP rs41289839 G>A is strongly associated with AIS susceptibility. Linkage disequilibrium analysis suggests that rs41289839 and other AIS-related SNPs were in strong LD. Next, we demonstrated that rs41289839 G>A inhibits the inclusion of exon 6 during alternative splicing, resulting in a decreased expression level of exon 6-included transcript (GPR126-exon6in) relative to the exon 6 excluded transcript (GPR126-exon6ex) by minigene assay. Chondrogenic differentiation experiment showed that GPR126-exon6in has a high expression level relative to GPR126-exon6ex during chondrogenic differentiation of hMSCs. Our findings indicate that newly discovered SNP is related to cartilage development and may provide valuable insights into the etiology and pathogenesis of adolescent idiopathic scoliosis.
1. Introduction
Adolescent idiopathic scoliosis (AIS) is a structural, tridimensional spinal deformity with a Cobb angle greater than 10° [1, 2]. It occurs in 1–3% of the adolescent populations [3]. AIS has limited treatment options and is currently mainly used for brace or surgery. The etiology of AIS remains unknown, but genetic factors are considered to be the most important cause in many studies [4–6]. GWAS study in 1819 Japanese AIS cases and 25,939 controls by Kou et al. demonstrated that SNP rs6570507 of GPR126 is a new susceptibility locus for AIS [7]. Subsequent follow-up studies further confirmed the relationship between GPR126 and AIS [8–10]. The GPR126 protein is a member of the adhesion-G protein-coupled receptor (GPCR) family [11]. It is widely hypothesized that scoliosis is caused by abnormal skeletal growth [12]. A recent study reported that GPR126-null mice have limb posture abnormalities and growth failure [13]. GWAS studies suggest that human height is associated with SNPs in GPR126 in both children and adults [14–17]. In addition, an AIS related SNP rs6570507 was also associated with trunk length in European populations [15]. However, there is currently no research to explore the specific functions of the GPR126-SNP.
We further analyzed how GPR126-SNP regulates the gene function and even spinal development. The results of this research can be used for early diagnosis and development of new treatment methods for AIS.
2. Materials and Methods
2.1. Study Population
A total of 131 adolescent idiopathic scoliosis patients and 132 healthy controls in Shanghai Changzheng Hospital were enrolled in this study between October 2014 and February 2018. The standing posteroanterior radiographs were taken for each AIS patient. Cobb angle of the curves was measured, and the most severe curve was selected if more than one curve was discovered in one patient (Table 1). All patients in scoliosis group have at least one curve with a Cobb angle greater than 10 degrees. Blood samples were collected after obtaining informed consent from all participants or their parents. The study has been approved by the Ethical Committee of Shanghai Changzheng Hospital and conformed to the tenets of the Declaration of Helsinki.
Table 1.
General data of patients and controls.
| Variables | AIS cases | Controls |
|---|---|---|
| Ethnic group | Chinese Han | Chinese Han |
| Female/Male | 124/7 | 62/70 |
| Mean age ± SD (years) | 13.94 ± 1.93 | 36.72 ± 16.84 |
| Age range (years) | 11-18 | 20-75 |
| Age at diagnosis (years) | 12.91 ± 1.31 | NA |
| Mean Cobb angle ± SD (°) | 37.40 ± 13.90 | NA |
| Cobb angle range (°) | 20-105 | NA |
2.2. PCR and Sequencing
GPR126 contains 26 exons and alternative splicing of exon 6 and exon 25 produces 4 protein-coding transcripts. We suspect that some SNPs in GPR126 may regulate the alternative splicing and finally alter the protein function of GPR126. Genomic DNA was extracted from peripheral blood of patients using the TIANamp Genomic DNA Kit (TIANGEN, China). The SNPs around exon 6 and exon 25 associated with splicing were analyzed. Sequencing primers: exon 6-F: 5′-TCTTTTGACAGACTCAGGAAACCA-3′; exon 6-R: 5′- AACTTGTTTCCTGCAGCAAATAAT-3′; exon 25-F: 5′- CTCAAACTCCTGGGCTCAAG -3′; exon 25-R: 5′- TCCTAGAAGGAGCCGCTTGC-3′. We also identified the genotype of rs6570507 by sequencing: primer: rs6570507-F: 5′-GAAAGATTTTCTGTGACATTCTC-3′; rs6570507-R: 5′-TGGTCAGGCTGGTCTCAA-3′. PCR conditions were set as follows: 1 min initial denaturation at 94°C, 30 cycles of 15 s denaturation at 94°C, 10 s annealing at 58°C, 1min extension at 72°C, and 10 min final extension at 72°C. The PCR products were analyzed using the sequencing system ABI3100 (Applied Biosystems).
2.3. Minigene Constructs
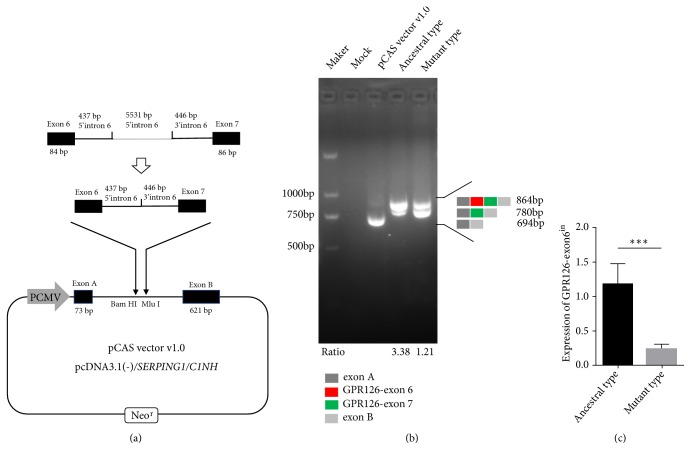
The exon trapping vector pCAS vector v1.0 (pcDNA3.1(-)/C1NH/SERPING1) was constructed as described by Gaildrat et al. [18]. We analyzed the sequence related to alternative splicing in intron 6 by RegRNA2.0 (http://regrna2.mbc.nctu.edu.tw/detection.html) and selected the 437 bp of 5′intron 6 and the 446 bp of 3′intron 6 for minigene assay. A 1053 bp genomic segment of GPR126 spanning exon 6, 437 bp of 5′intron 6, 446 bp of 3′intron 6, and exon 7 sequences was synthesized and then the fragments with the ancestral or mutant allele were cloned into pCAS vector v1.0 (Figure 2(a)). All constructs were sequenced to verify that contain the correct sequence.
Figure 2.
The exon trapping vector pCAS vector v1.0 used to assay SNP function. (a) The pCAS vector v1.0 contains 2 exons (exon A, exon B) and a functional intron. Ancestral or mutant plasmids containing exon 6, 437 bp of 5′intron 6, 446 bp of 3′intron 6 and exon7, and harboring either the G or A allele was separately cloned into the Bam HI and Mlu I clone site of the pCAS vector v1.0. Exon A: 73 bp; exon B: 621 bp; exon 6: 84 bp; exon7: 86 bp. (b) Agarose gel electrophoresis of RT-PCR products. Ancestral type: rs41289839-G; Mutant type: rs41289839-G; Lane 1: Marker; Lane 2: Mock; Lane 3: 694 bp (73bp+621bp); Lane 4: 864 bp (73 bp+84 bp+86 bp+621 bp), 780 bp (73 bp+86 bp+621 bp); Lane 5: 864 bp, 780 bp. There was a significant difference in the ratio of exon6in to exon6ex between Lane 4 and Lane 5. Ratio: exon6in/exon6ex. (c) The expression levels of GPR126-exon6in in ancestral type group and mutant type group were detected by RT-qCPR.
2.4. Cell Culture and Transfection
DMEM medium contains 10% fetal bovine serum (FBS), penicillin (100 U/L), and streptomycin (100 mg/L). Human epithelial kidney 293 FT (HEK 293 FT) cells were cultured in DMEM medium and grow to approximately 70% to 80% confluence in a humidified atmosphere of 5% CO2 at 37°C. Cells were then transfected with 10 μg plasmid DNA using OPTI-MEM medium and Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer's instructions. Cells were harvested after 24 h transfection and total RNA was extracted with TRIzol (Invitrogen, USA).
2.5. Chondrogenic Differentiation
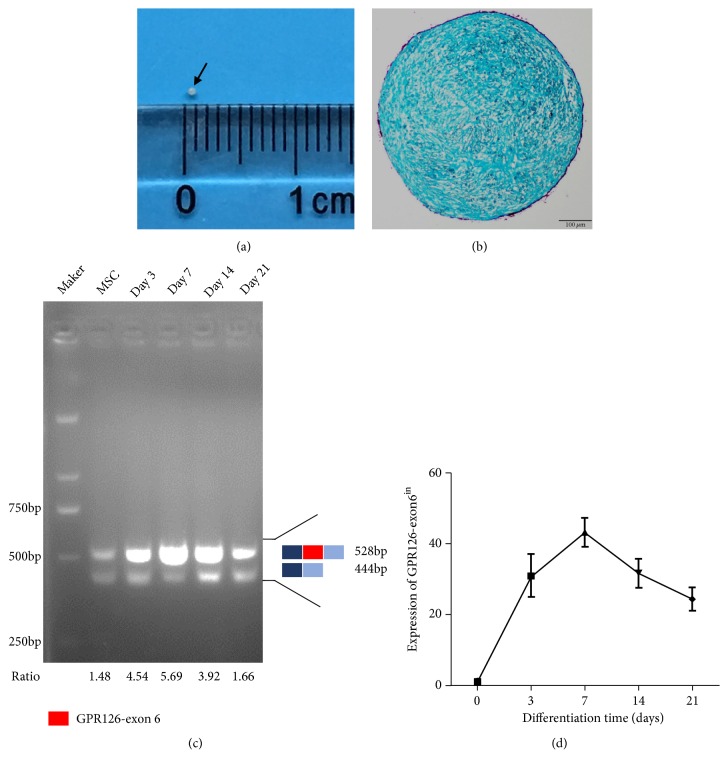
Chondrogenic differentiation of human mesenchymal stem cells (hMSCs) with rs41289839 genotype GG was applied by OriCell™ Human Mesenchymal Stem Cell Chondrogenic Differentiation Medium (Cyagen, USA). Chondrogenic medium contains high-glucose DMEM, 10 ng/mL recombinant human transforming growth factor-β3 (TGF-β3), 100 nM dexamethasone, 50 μg/mL ascorbic acid, 1 mM sodium pyruvate, 40 μg/mL proline, and ITS+ Supplement. MSCs were trypsinized, washed, and then resuspended in chondrogenic medium. 0.5 mL (2.5 × 105 cells) of the cell suspension was transferred to a 15 mL polypropylene culture tube and centrifuged at 150 g for 5 minutes at room temperature. The caps of the tubes were loosened one half turn to allow gas exchange and the tubes were incubated in a humidified atmosphere of 5% CO2 at 37°C. The medium in each tube was completely replaced every 2-3 days. Chondrogenic pellets should be cultured for 3 to 21 days.
Pellets were fixed in formalin and embedded in paraffin for alcian blue stain. The slides were deparaffinized and hydrated in distilled water and stained for 30 minutes in alcian blue solution and washed in running tap water for 2 minutes and rinsed in distilled water. Images for analysis were captured and observed under light microscope. Blue staining indicated that hMSCS differentiated into chondrocytes and synthesized proteoglycans. Total RNA of pellet was extracted with TRIzol (Invitrogen, USA).
2.6. Reverse Transcription and PCR Analysis
A total of 1 μg of RNA from the transfected cells and chondrogenic differentiation pellets were reverse transcribed into cDNA using oligo(dT)18 mRNA primer and SuperScript™ II Reverse Transcriptase in a 20 μL reaction volume according to the manufacturer's instructions (Invitrogen, USA). The cDNA of transfected cells was amplified with primers: pCAS-F: 5′- GTCCGCTGACGTCGCC -3′; pCAS-R: 5′- GATCTGAGAGACTGAGGTGA-3′. The cDNA of pellets was amplified with primers: PLT-F: 5′- ACAGCTCTGCCTTGTTTGGA-3′; PLT-R: 5′- ACCTCAGGGTGACGAAGGAT-3′. PCR conditions were set as follows: an initial denaturation at 94°C for 1 min, 30 cycles at 94°C for 15 s, 58°C for 10 s, 72°C for 1 min, and a final extension at 72°C for 10 min. The PCR products were analyzed by electrophoresis on a 2% agarose gel. The signal ratio was obtained through comparing the band intensity of GPR126-exon6in transcript with the band intensity of GPR126-exon6ex transcript. The band intensity of each transcript was analyzed by Image-Pro Plus. We purified the PCR products of all bands using TIANgel Purification Kit (TIANGEN, China) and verified by Sanger sequencing.
2.7. Real-Time qPCR
The expression level of GPR126-exon6in was determined by real-time qPCR (RT-qPCR) using SYBR-Premix Ex Taq (Takara, Japan) and ABI Prism 7900HT sequence detection system (Applied Biosystems, Carlsbad, CA). The genes were amplified using specific primers and human β-actin gene was used as an endogenous control. The PCR primer sequences used were as follows: exon 6 included GPR126-transcripts (GPR126-exon6in): forward: 5′- TACACCACCCACTGTCACCA-3′, reverse: 5′- ATTCTGCCACCTTGCTCTGT-3′; β-actin: forward: 5′- ACCGAGCGCGGCTACAG-3′, reverse: 5′- CTTAATGTCACGCACGATTTCC-3′. Data were analyzed using the comparative Ct method (2-ΔΔCt). Three separate experiments were performed for each group.
2.8. Statistical Analysis
Analysis of the data was performed using SPSS version 23.0, with p value < 0.05 considered statistically significant. The Student's t-test was used to compare the difference of GPR126-exon6in expression between the ancestral type group and mutant type group. One-way analysis of variance (ANOVA) was used to analyze the differences between groups. The LSD method of multiple comparisons was used when the probability for ANOVA was statistically significant. We assessed the frequencies of the alleles and genotypes in patients and controls by χ2 test. Linkage disequilibrium analysis was carried out online using SHEsis (http://analysis.bio-x.cn) [19]. D' >0.7 and r2 >1/3 indicates strong LD between SNPs [20, 21].
3. Results
3.1. GPR126-SNP Identification and Linkage Disequilibrium Analysis
All individuals enrolled in our study were Chinese Han. Women in the case group account for a relatively high proportion (124/7), which is characteristic of AIS disease. The mean age of AIS patient was 13.94 ± 1.93 (range, 11-18). Since AIS occurs in adolescent with immature skeleton, the age of the controls was greater than 20 years to ensure they had mature bone (36.72 ± 16.84, range, 20-75). AIS patients were diagnosed at 12.91 ± 1.31 years old. The average cobb angle of the curve was 37.40° ± 13.90 (range, 13-105°).
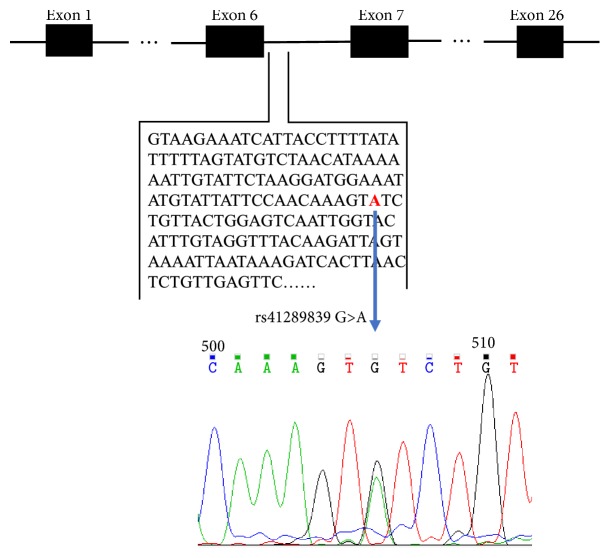
The GRP126 region was sequenced and compared to the control population. There were statistically significant differences in genotype and allele frequencies of rs41289839 (p = 1.55 × 10−3 and 2.68 × 10−3, respectively), indicating that we have newly discovered a SNP (rs41289839 G>A) at the junction of exon 6 and intron 6 associated with AIS (Table 2, Figure 1).
Table 2.
The allele and genotype frequencies of GPR126 gene polymorphisms in patients with AIS and controls.
| SNP | group | Genotype frequency | Allele frequency | ||||||
|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | p | A | G | OR(95%CI) | p | ||
| rs41289839 | AIS | 7 (0.053) | 64 (0.49) | 60 (0.46) | 1.55 × 10−3 | 78 (0.30) | 184 (0.70) | 1.86 (1.24-2.80) | 2.68 × 10−3 |
| A/G | control | 6 (0.045) | 37 (0.28) | 89 (0.67) | 49 (0.19) | 215 (0.81) | |||
| rs6570507 | AIS | 36 (0.28) | 53 (0.40) | 42 (0.32) | 4.05 × 10−3 | 125 (0.48) | 137 (0.52) | 1.89 (1.33-2.69) | 4.02 × 10−4 |
| A/G | control | 17 (0.13) | 52 (0.39) | 63 (0.48) | 86 (0.33) | 178 (0.67) | |||
Linkage disequilibrium tests: D' = 0.984, r2 = 0.461
Figure 1.
Identification of a novel SNP associated with AIS in GPR126.GPR126 gene consists of 26 exons and alternative splicing of exon 6 and exon 25 produces 4 protein-coding transcripts. A newly discovered SNP (rs41289839 G>A) at the junction of exon 6 and intron 6 was associated with AIS.
Rs6570507 was found to be associated with AIS in a large-scale genome-wide analysis [7] and this result was confirmed in our population (p = 4.05 ×10−3 and 4.02 × 10−4, respectively). Interestingly, linkage disequilibrium analysis by SHEsis showed that rs41289839 and rs6570507 were in strong LD (D' = 0.984, r2 = 0.461, Table 2).
3.2. The Newly Discovered SNP Regulates GPR126-Exon6 Splicing
Since intronic SNPs in the intron–exon boundaries have been reported to have effects on transcription factor binding sites and native splicing sites [22–24], we speculated that rs41289839 G>A may be associated with alternative splicing of exon 6. Splice prediction software HSF (http://www.umd.be/HSF3/) showed that the mutant type of this SNP may destroy the binding sites of the splice enhancement elements SPp55 and SC35 (Table 3).
Table 3.
Threshold scores of exonic splice enhancer (ESE) motifs associated with rs41289839 G>A.
| Linked SR protein | Reference Motif (value 0-100) | Variation |
|---|---|---|
| SRp55 | agtgtc (74.12) | Site broken -100 |
| Sc35 | gtctgtta (75.78) | Site broken -100 |
Next, we constructed a minigene expression system and transfected it into 293FT cells (Figure 2(a)). The results of minigene assay demonstrated that rs41289839 G>A will inhibit the inclusion of exon 6 during alternative splicing, leading to a decreased expression level of exon 6-included transcript (GPR126-exon6in) relative to the exon6 excluded transcript (GPR126-exon6ex) (Figures 2(b) and 2(c)).
3.3. GPR126-Exon 6 Has an Important Role in Cartilage Development
We performed an in vitro stem cell differentiation experiment to explore the role of exon 6 in cartilage development. Human mesenchymal stem cells were successfully differentiated into chondrocytes (Figures 3(a) and 3(b)). Reverse transcription PCR and real-time qPCR of chondrogenic pellets showed that GPR126-exon6in has a high expression level relative to GPR126-exon6ex during chondrogenic differentiation of hMSCs, suggesting that exon 6 may have important functions in cartilage development (Figures 3(c) and 3(d)). It indicated that a decreased inclusion of exon 6 caused by rs41289839 G>A may lead to cartilage malformation, which may be the cause of AIS.
Figure 3.
Inclusion of GPR126-exon6 during chondrogenic differentiation. (a) Chondrogenic differentiation of hMSCs for 21d. Black arrow: a chondrogenic pellet. (b) Alcian blue stain of chondrogenic pellet. (c) RT-PCR assay with primer PLT-F and PLT-R to monitor alternative splicing patterns of GPR126-exon6 during chondrogenic differentiation. Ratio: exon6in/exon6ex. (d) The expression levels of GPR126-exon6in during chondrogenic differentiation were detected by RT-qCPR.
4. Discussion
Alternative splicing is associated with multiple diseases such as spinal muscular atrophy [25], breast cancer [26], ovarian cancer [27], prostate cancer [28], and colon cancer [29]. Some studies have shown that alternative splicing is associated with intracellular localization and function of protein. The leptin gene can be translated into an isoform located in the nucleus and a cytoplasmic isoform, the former being a transcription factor and the latter having phosphatase activity [30, 31]. Typically the erythropoietin receptor is a membrane protein, but one of its splicing isoforms is a soluble protein [32]. The ability of proteins to bind to other proteins will change due to alternative splicing. An insulin splicing isoform (exon 11 skips) showed abnormally high affinity for IGF-II [33]. In addition, single nucleotide polymorphisms play an important role in exon splicing. CCND1 gene polymorphism (A870G) modulates alternative splicing and allows expression of an alternative cyclin D1 transcript (transcript cyclin D1b) which is correlated to the risk and/or severity of disease or drug response across a range of malignancies, including lung cancer [34]. A SNP in KLF6 intron generates a novel functional SRp40 DNA binding site and increases three alternatively spliced KLF6 isoforms [35] that are associated with prostate cancer [36] and lung cancer [37].
The etiology and pathogenesis of AIS are still uncertain [38]. Several familial surveys of idiopathic scoliosis provided strong evidence that genetic factors play a major role [39]. SNPs of ESR1 [40], ESR2 [41], MATN1 [42], MTNR1B [43], TPH1 [44], IL-6, and MMP-3 [45] have been reported to be related to AIS. Recently, a GWAS study found that GPR126 may also be a predisposing gene and rs6570507 of GPR126 was the most significantly associated SNP with AIS in Japanese [7]. In our study, we for the first time provided evidence for strong association of rs41289839 G>A with AIS susceptibility. Further functional analysis revealed that the newly discovered SNP regulates the splicing of GPR126-exon6.
AIS may be caused by abnormal cartilage development. Some researchers found that the imbalanced expression of sox9, collagen II, collagen X, and aggrecan on both sides of the spine may be the cause of scoliosis [46, 47]. Cartilage oligomeric matrix protein (COMP) played a role in maintaining the structural integrity of cartilage. Gerdhem found that COMP was lower expressed in idiopathic scoliosis children [48]. SHP2 plays important roles in cartilage development, and deletion of SHP2 in the cartilage causes scoliosis in mice [49]. On the other hand, some studies have shown that GPR126 plays a role in chondrocyte proliferation and cartilage formation. Sox9 is a chondrogenesis related factor, and studies have shown that the deletion of Sox9 leads to a decreased expression of GPR126 in the intervertebral disc of mice [50]. Later, some researchers found that GPR126 is highly expressed in cartilage of human and mice [7]. Deletion of GPR126 in mice cartilage caused “split” spine deformity [51]. In our study, we found that the expression level of GPR126-exon6in transcript was higher than that of GPR126-exon6ex transcript during chondrogenic differentiation of hMSCs, suggesting that exon 6 may have important functions in cartilage development. This result may explain how SNP affects cartilage development and finally causes scoliosis.
Linkage disequilibrium analysis in our study showed that there was a strong correlation between rs41289839 and rs6570507. In fact, the relationship between genetic marker and disease is complex. Some genetic markers correlate with the pathogenesis and pathology of the disease, and some have strong linkage disequilibrium with pathogenic sites [52]. We speculated that the newly discovered SNP is likely to be the pathogenic site of some AIS patients and rs6570507 is a “marker” for rs41289839.
This is a limited research because we did not detect GPR126 expression patterns in spine cartilage tissue of AIS patients. It is difficult to collect cartilage samples of AIS patients in posterior spinal orthopedic surgery. In the next work, we will collect spinal cartilage tissue in anterior spinal surgery with informed consent to define the expression pattern of GPR126 in AIS patients. Furthermore, there is currently no research explaining how GPR126 transcripts with/without exon 6 regulate cartilage development of spine. We will overexpress GPR126-exon6in transcript or GPR126-exon6ex transcript in hMSCs and apply chondrogenic differentiation to explore the function of exon 6 in cartilage development.
5. Conclusions
In conclusion, we examined the genetic association between GPR126 and AIS risk in Chinese populations, and the intronic SNP rs41289839 G>A was found to be significantly associated with AIS in Chinese populations. Further functional analysis revealed that the newly discovered SNP regulates alternative splicing of GPR126-exon6 and even cartilage development. These results could be helpful in understanding the etiology of AIS and developing drugs for AIS.
Abbreviations
- GPR126:
G protein-coupled receptor 126
- AIS:
Adolescent idiopathic scoliosis
- SNP:
Single nucleotide polymorphism
- LD:
Linkage disequilibrium
- hMSCs:
Human mesenchymal stem cells
- ESR1:
Estrogen receptor 1
- ESR2:
Estrogen receptor 2
- MATN1:
Matrilin 1
- MTNR1B:
Melatonin receptor 1B
- TPH1:
Tryptophan hydroxylase 1
- IL-6:
Interleukin-6
- MMP-3:
Matrix metalloproteinase-3.
Contributor Information
Rui Gao, Email: rgaospine@163.com.
Xuhui Zhou, Email: xhzhouspine@163.com.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Mehta M. H. The conservative management of juvenile idiopathic scoliosis. Acta Orthopaedica Belgica. 1992;58(supplement 1):91–97. [PubMed] [Google Scholar]
- 2.Gao W., Peng Y., Liang G., et al. Association between common variants near LBX1 and adolescent idiopathic scoliosis replicated in the chinese han population. PLoS ONE. 2013;8, article e53234 doi: 10.1371/journal.pone.0053234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinstein S. L. Natural history. The Spine Journal. 1999;24(24):2592–2600. doi: 10.1097/00007632-199912150-00006. [DOI] [PubMed] [Google Scholar]
- 4.Hadley M. N. Spine update: genetics of familial idiopathic scoliosis. Spine. 2000;25:2416–2418. doi: 10.1097/00007632-200009150-00024. [DOI] [PubMed] [Google Scholar]
- 5.Miller N. H. Genetics of familial idiopathic scoliosis. Clinical Orthopaedics and Related Research. 2002;(401):60–64. doi: 10.1097/00003086-200208000-00009. [DOI] [PubMed] [Google Scholar]
- 6.Miller N. H. Genetics of familial idiopathic scoliosis. Clinical Orthopaedics and Related Research. 2007;462:6–10. doi: 10.1097/BLO.0b013e318126c062. [DOI] [PubMed] [Google Scholar]
- 7.Kou I., Takahashi Y., Johnson T. A., et al. Genetic variants in GPR126 are associated with adolescent idiopathic scoliosis. Nature Genetics. 2013;45(6):676–679. doi: 10.1038/ng.2639. [DOI] [PubMed] [Google Scholar]
- 8.Xu J.-F., Yang G.-H., Pan X.-H., et al. Association of GPR126 gene polymorphism with adolescent idiopathic scoliosis in Chinese populations. Genomics. 2015;105(2):101–107. doi: 10.1016/j.ygeno.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 9.Qin X., Xu L., Xia C., et al. Genetic variant of GPR126 Gene is functionally associated with adolescent idiopathic scoliosis in chinese population. The Spine Journal. 2017;42(19):E1098–E1103. doi: 10.1097/BRS.0000000000002123. [DOI] [PubMed] [Google Scholar]
- 10.Liu G., Liu S., Lin M., et al. Genetic polymorphisms of GPR126 are functionally associated with PUMC classifications of adolescent idiopathic scoliosis in a Northern Han population. Journal of Cellular and Molecular Medicine. 2018;22(3):1964–1971. doi: 10.1111/jcmm.13486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stehlik C., Kroismayr R., Dorfleutner A., Binder B. R., Lipp J. VIGR - A novel inducible adhesion family G-protein coupled receptor in endothelial cells. FEBS Letters. 2004;569(1-3):149–155. doi: 10.1016/j.febslet.2004.05.038. [DOI] [PubMed] [Google Scholar]
- 12.Kouwenhoven J.-W. M., Castelein R. M. The pathogenesis of adolescent idiopathic scoliosis: review of the literature. The Spine Journal. 2008;33(26):2898–2908. doi: 10.1097/BRS.0b013e3181891751. [DOI] [PubMed] [Google Scholar]
- 13.Monk K. R., Oshima K., Jörs S., Heller S., Talbot W. S. Gpr126 is essential for peripheral nerve development and myelination in mammals. Development. 2011;138(13):2673–2680. doi: 10.1242/dev.062224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lettre G., Jackson A. U., Gieger C., et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nature Genetics. 2008;40(5):584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soranzo N., Rivadeneira F., Chinappen-Horsley U., et al. Meta-analysis of genome-wide scans for human adult stature identifies novel loci and associations with measures of skeletal frame size. PLoS Genetics. 2009;5 doi: 10.1371/journal.pgen.1000445.e1000445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sovio U., Bennett A. J., Millwood I. Y., et al. Genetic determinants of height growth assessed longitudinally from infancy to adulthood in the northern finland birth cohort 1966. PLoS Genetics. 2009;5 doi: 10.1371/journal.pgen.1000409.e1000409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao J., Li M., Bradfield J. P., et al. The role of height-associated loci identified in genome wide association studies in the determination of pediatric stature. BMC Medical Genetics. 2010;11, article 96 doi: 10.1186/1471-2350-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaildrat P., Killian A., Martins A., Tournier I., Frébourg T., Tosi M. Use of splicing reporter minigene assay to evaluate the effect on splicing of unclassified genetic variants. Methods in Molecular Biology. 2010;653:249–257. doi: 10.1007/978-1-60761-759-4_15. [DOI] [PubMed] [Google Scholar]
- 19.Shi Y. Y., He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Research. 2005;15(2):97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 20.Gabriel S. B., Schaffner S. F., Nguyen H., et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 21.Ardlie K. G., Kruglyak L., Seielstad M. Patterns of linkage disequilibrium in the human genome. Nature Reviews Genetics. 2002;3(4):299–309. doi: 10.1038/nrg777. [DOI] [PubMed] [Google Scholar]
- 22.Moyer R. A., Wang D., Papp A. C., et al. Intronic polymorphisms affecting alternative splicing of human dopamine D2 receptor are associated with cocaine abuse. Neuropsychopharmacology. 2011;36(4):753–762. doi: 10.1038/npp.2010.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rigo F., Martinson H. G. Functional coupling of last-intron splicing and 3′-end processing to transcription in vitro: the poly(A) signal couples to splicing before committing to cleavage. Molecular and Cellular Biology. 2008;28(2):849–862. doi: 10.1128/MCB.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D., Guo Y., Wrighton S. A., Cooke G. E., Sadee W. Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. The Pharmacogenomics Journal. 2011;11(4):274–286. doi: 10.1038/tpj.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorson C. L., Hahnen E., Androphy E. J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proceedings of the National Acadamy of Sciences of the United States of America. 1999;96(11):6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albert H., Santos S., Battaglia E., Brito M., Monteiro C., Bagrel D. Differential expression of CDC25 phosphatases splice variants in human breast cancer cells. Clinical Chemistry and Laboratory Medicine. 2011;49(10):1707–1714. doi: 10.1515/CCLM.2011.635. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y., Chan D. W., Liu V. W. S., Chiu P. M., Ngan H. Y. S. Differential functions of growth factor receptor-bound protein 7 (GRB7) and its variant GRB7v in ovarian carcinogenesis. Clinical Cancer Research. 2010;16(9):2529–2539. doi: 10.1158/1078-0432.CCR-10-0018. [DOI] [PubMed] [Google Scholar]
- 28.Haile S., Sadar M. D. Androgen receptor and its splice variants in prostate cancer. Cellular and Molecular Life Sciences. 2011;68(24):3971–3981. doi: 10.1007/s00018-011-0766-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Usher P. A., Sieuwerts A. M., Bartels A., et al. Identification of alternatively spliced TIMP-1 mRNA in cancer cell lines and colon cancer tissue. Molecular Oncology. 2007;1(2):205–215. doi: 10.1016/j.molonc.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han G.-S., Carman G. M. Characterization of the human LPIN1-encoded phosphatidate phosphatase isoforms. The Journal of Biological Chemistry. 2010;285(19):14628–14638. doi: 10.1074/jbc.M110.117747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang P., Takeuchi K., Csaki L. S., Reue K. Lipin-1 phosphatidic phosphatase activity modulates phosphatidate levels to promote peroxisome proliferator-activated receptor γ (PPARγ) gene expression during adipogenesis. The Journal of Biological Chemistry. 2012;287(5):3485–3494. doi: 10.1074/jbc.M111.296681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khankin E. V., Mutter W. P., Tamez H., Yuan H.-T., Karumanchi S. A., Thadhani R. Soluble erythropoietin receptor contributes to erythropoietin resistance in end-stage renal disease. PLoS ONE. 2010;5, article e9246 doi: 10.1371/journal.pone.0009246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belfiore A., Frasca F., Pandini G., Sciacca L., Vigneri R. Insulin receptor isoforms and insulin receptor/insulin-like growth factor receptor hybrids in physiology and disease. Endocrine Reviews. 2009;30(6):586–623. doi: 10.1210/er.2008-0047. [DOI] [PubMed] [Google Scholar]
- 34.Gautschi O., Ratschiller D., Gugger M., Betticher D. C., Heighway J. Cyclin D1 in non-small cell lung cancer: A key driver of malignant transformation. Lung Cancer. 2007;55(1):1–14. doi: 10.1016/j.lungcan.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 35.Narla G., DiFeo A., Reeves H. L., et al. A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Research. 2005;65(4):1213–1222. doi: 10.1158/0008-5472.CAN-04-4249. [DOI] [PubMed] [Google Scholar]
- 36.Narla G., Difeo A., Yao S., et al. Targeted inhibition of the KLF6 splice variant, KLF6 SV1, suppresses prostate cancer cell growth and spread. Cancer Research. 2005;65(13):5761–5768. doi: 10.1158/0008-5472.CAN-05-0217. [DOI] [PubMed] [Google Scholar]
- 37.DiFeo A., Feld L., Rodriguez E., et al. A functional role for KLF6-SV1 in lung adenocarcinoma prognosis and chemotherapy response. Cancer Research. 2008;68(4):965–970. doi: 10.1158/0008-5472.CAN-07-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pollak L., Shlamkovic N., Minewicz A., Mirovsky Y. Otolith dysfunction as a possible cause for the development of idiopathic scoliosis. Journal of Pediatric Orthopaedics. 2013;33(3):293–297. doi: 10.1097/BPO.0b013e31827c0643. [DOI] [PubMed] [Google Scholar]
- 39.Inoue M., Minami S., Nakata Y., et al. Association between estrogen receptor gene polymorphisms and curve severity of idiopathic scoliosis. The Spine Journal. 2002;27(21):2357–2362. doi: 10.1097/00007632-200211010-00009. [DOI] [PubMed] [Google Scholar]
- 40.Wu J., Qiu Y., Zhang L., Sun Q., Qiu X., He Y. Association of estrogen receptor gene polymorphisms with susceptibility to adolescent idiopathic scoliosis. The Spine Journal. 2006;31(10):1131–1136. doi: 10.1097/01.brs.0000216603.91330.6f. [DOI] [PubMed] [Google Scholar]
- 41.Zhang H., Lu S., Tang M., et al. Association of estrogen receptor β gene polymorphisms with susceptibility to adolescent idiopathic scoliosis. The Spine Journal. 2009;34(8):760–764. doi: 10.1097/BRS.0b013e31818ad5ac. [DOI] [PubMed] [Google Scholar]
- 42.Chen Z., Tang N. L. S., Cao X., et al. Promoter polymorphism of matrilin-1 gene predisposes to adolescent idiopathic scoliosis in a Chinese population. European Journal of Human Genetics. 2009;17(4):525–532. doi: 10.1038/ejhg.2008.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu X. S., Tang N. L. S., Yeung H. Y., et al. Melatonin receptor 1B (MTNR1B) gene polymorphism is associated with the occurrence of adolescent idiopathic scoliosis. The Spine Journal. 2007;32(16):1748–1753. doi: 10.1097/BRS.0b013e3180b9f0ff. [DOI] [PubMed] [Google Scholar]
- 44.Wang H., Wu Z., Zhuang Q., et al. Association study of tryptophan hydroxylase 1 and arylalkylamine n-acetyltransferase polymorphisms with adolescent idiopathic scoliosis in han chinese. The Spine Journal. 2008;33(20):2199–2203. doi: 10.1097/BRS.0b013e31817c03f9. [DOI] [PubMed] [Google Scholar]
- 45.Aulisa L., Papaleo P., Pola E., et al. Association between IL-6 and MMP-3 gene polymorphisms and adolescent idiopathic scoliosis: a case-control study. The Spine Journal. 2007;32(24):2700–2702. doi: 10.1097/brs.0b013e31815a5943. [DOI] [PubMed] [Google Scholar]
- 46.Li Q.-Y., Zhong G.-B., Liu Z.-D., Lao L.-F. Effect of asymmetric tension on biomechanics and metabolism of vertebral epiphyseal plate in a rodent model of scoliosis. Orthopaedic Surgery. 2017;9(3):311–318. doi: 10.1111/os.12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y.-J., Yu H.-G., Zhou Z.-H., Guo Q., Wang L.-J., Zhang H.-Q. Leptin receptor metabolism disorder in primary chondrocytes from adolescent idiopathic scoliosis girls. International Journal of Molecular Sciences. 2016;17 doi: 10.3390/ijms17071160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gerdhem P., Topalis C., Grauers A., Stubendorff J., Ohlin A., Karlsson K. M. Serum level of cartilage oligomeric matrix protein is lower in children with idiopathic scoliosis than in non-scoliotic controls. European Spine Journal. 2015;24:256–261. doi: 10.1007/s00586-014-3691-2. [DOI] [PubMed] [Google Scholar]
- 49.Kim H. K. W., Aruwajoye O., Sucato D., et al. Induction of SHP2 deficiency in chondrocytes causes severe scoliosis and kyphosis in mice. The Spine Journal. 2013;38(21):E1307–E1312. doi: 10.1097/BRS.0b013e3182a3d370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henry S. P., Liang S., Akdemir K. C., De Crombrugghe B. The postnatal role of Sox9 in cartilage. Journal of Bone and Mineral Research. 2012;27(12):2511–2525. doi: 10.1002/jbmr.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karner C. M., Long F., Solnica-Krezel L., Monk K. R., Gray R. S. Gpr126/Adgrg6 deletion in cartilage models idiopathic scoliosis and pectus excavatum in mice. Human Molecular Genetics. 2015;24(15):4365–4373. doi: 10.1093/hmg/ddv170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cardon L. R., Bell J. I. Association study designs for complex diseases. Nature Reviews Genetics. 2001;2(2):91–99. doi: 10.1038/35052543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.