Abstract
One obstacle in diagnostic pathology is the harmonization of one drug-one diagnostic tests for programmed death ligand-1 (PD-L1). There are many challenges in accurate comparisons of diagnostic tests, such as differences in the titer of each antibody, detection system and dynamic range of visualization. Our previously developed digital immunostaining technique is highly sensitive and quantitative with the ability to quantify particles that bind in a one-to-one fashion with antibody in each cell. Determining the differences in the titer of each antibody with digital immunostaining may be beneficial for future harmonized analysis. To demonstrate the accuracy of digital immunostaining, the present study compared the number of particles with ELISA and nCounter data from five cell lines. NCI-H460 exhib-ited the highest level of PD-L1 protein, followed by A549, PC-3, NCI-H1299, and NCI-H446 cells. In addition, the PD-L1 mRNA values determined by nCounter corresponded with the order of the protein levels determined by ELISA. The present study revealed that digital immunostaining for PD-L1 was highly associated with ELISA and nCounter data. Among the four antibodies tested, the titer of all but SP142 coincided with ELISA and nCounter data. These results indicated that our digital immunostaining technique may be beneficial for future harmonized analysis.
Keywords: programmed death ligand-1, digital immunostaining, ELISA, nCounter
Introduction
We recently developed a novel quantitative immunostaining technique termed ‘digital immunostaining with phosphor-integrated dot (PID)’. This method is highly sensitive with a wide range of detection compared with conventional immunostaining methods such as 3,3′-diaminobenzidine (DAB)- or alkaline phosphatase-based visualization (1–3). Most conventional immunohistochemistry (IHC) scoring systems are dependent on the staining intensity and are semi-quantitative. Compared with conventional IHC, our digital IHC provides more objective data because we can count the number of particles that bind in a one-to-one fashion with antibody in each cell. Furthermore, our previous study of digital immunostaining of human epidermal growth factor receptor 2 protein revealed the close correlation with clinical response to trastuzumab therapy compared with that of conventional DAB staining (2).
PD-1 is a type of immune checkpoint receptor expressed on the surface of cytotoxic T-cells. The upregulation of the PD-1 receptor plays an important role in suppressing the inactivation of programmed death ligand-1 (PD-L1) (4). Recently, therapeutic antibodies to PD-1/PD-L1 have shown promising clinical results for several tumors such as melanoma, renal cancer, and non-small cell lung cancer (NSCLC) (5). There is a strong association between the efficacy of PD-1/PD-L1 antibody therapy and PD-L1 protein expression measured with DAB-based IHC (6). One of the difficulties in diagnostic pathology is the harmonization of one drug-one diagnostic tests for PD-L1. To overcome this challenge, several studies, e.g., the Blue-print project, were performed (7). The consensus was that all diagnostic kits, excluding the SP142 assay, showed robust harmonization (7). However, there are many variables to consider for accurate comparison of diagnostic tests, such differences in antibody titer, detection system, and dynamic range of visualization.
Because the estimation of the ability of each antibody test is less informative in daily practice, the ability of each diagnostic test was evaluated together with antibody titer, detection system, and visualization method. Understanding the differences in titer of each antibody used for diagnostic kits or research with digital immunostaining may be beneficial for future harmonized analysis.
Materials and methods
Cell lines and cell block construction
Five cell lines with various PD-L1 expression levels (NCI-H446, PC-3, NCI-H1299, A549, and NCI-H460) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). PC-3 and A549 cells were cultured in F-12K (ATCC 30-2004), whereas NCI-H1299, H460, and H446 cells were cultured in RPMI 1640 (ATCC 30-2001) with 10% fetal bovine serum (Life Technologies, Carlsbad, CA, USA) and 1% penicillin-streptomycin (Nacalai Tesque, Kyoto, Japan). Cells were maintained in a humidified incubator with 5% CO2 at 37°C.
A formalin-fixed, paraffin-embedded (FFPE) cell block was constructed for the five cell lines. The specimens were centrifuged for 10 min at 2,000 rpm. The resulting pellet was fixed in 10% buffered formalin for 6 h and embedded in paraffin.
ELISA
ELISA was performed according to the manufacturers instructions using a PathScan Total PD-L1 Sandwich ELISA kit (cat. no. 14784; Cell Signaling Technology, Inc., Danvers, MA, USA). The ELISA results were obtained as relative (OD), as there were no standard substances for PD-L1 in the kit.
mRNA molecular counting
Cell line-derived cell blocks were cut into 5-µm-thick pieces with a microtome. The total mRNA was extracted from 5–6 sections using a NucleoSpin total RNA FFPE kit (Machery-Nagel, Düren, Germany). The RNA concentrations were determined from the 260/280-nm OD ratio.
The nCounter GX Custom CodeSet was used to detect gene expression in the purified RNA. RNA was hybridized with CodeSets of 41 genes for 18 h at 65°C and processed according to the manufacturers instructions. This method directly quantifies the RNA using simultaneous hybridization processes with multiple probes.
IHC with PID
Specimens of the cell lines were cut into pieces with a thickness of 5 µm. Tissue microar-rays of NSCLC were obtained from US Biomax (Rockville, MD, USA; LC241). They consisted of six NSCLC samples, one adenocarcinoma sample (C5 core), and five squamous cell carcinoma samples (A1, B1, C1, A6, and B6 cores). Staining was performed on serial sections with each antibody.
The sections were deparaffinized, washed with distilled water, boiled in 10 mM sodium citrate buffer (pH=6.3) for 20 min, allowed to cool at 25°C for 40 min, rinsed with deionized water, and washed in PBS for 5 min. They were then incubated with the anti-rabbit monoclonal PD-L1 antibody diluted with the antibody diluent. Antibodies were diluted as follows: E1L3N (Cell Signaling Technology, Inc.) 1:800, SP142 (Spring Bioscience, Pleasanton, CA, USA) 1:100, SP263 (Ventana Medical Systems, Oro Valley, AZ, USA) 1:1, and 28-8 (Abcam, Cambridge, UK) 1:500.
Sections were incubated with a biotinylated secondary monoclonal anti-rabbit antibody (LO-RG-1, 2 µg/ml) for 30 min and with streptavidin-conjugated PID (0.03 nM) for 2 h, both at 25°C.
Measurement of PID fluorescence
Sections were irradiated at 580 nm, and the fluorescence intensity was measured using a fluorescence microscope (BX-53; Olympus Corp., Tokyo, Japan) and a CCD camera (DP73; Olympus Corp.). The PID particle number per cell was measured using the PidAnalyzer (Konica Minolta, Inc., Tokyo, Japan). Five fields were selected randomly, and over 1,000 tumor cells were counted.
Statistical analysis
The linearity of PID was determined by comparing the ELISA and nCounter data and calculated by Pearsons test. PidAnalyzer v.2.14.1.1 software (Konica-Minolta, Inc.) was used to conduct statistical analyses.
Results
Comparison between protein and mRNA expression
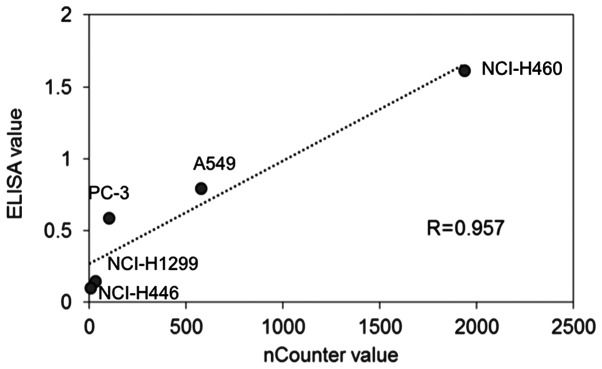
The linearity of ELISA data was determined by comparison with the nCounter data. The highest protein expression level of PD-L1 measured by ELISA was in NCI-H460 cells (1.62) followed by A549 (0.80), PC-3 (0.59), NCI-H1299 (0.15), and NCI-H446 (0.10) cells. Regarding the mRNA levels of PD-L1, the highest value measured by nCounter was in H460 cells (1937) followed by A549 (575), PC-3 (101), NCI-H1299 (31), and NCI-H446 (4) cells. The PD-L1 protein amount measured by ELISA and the mRNA counts showed a high correlation (Fig. 1: R>0.957).
Figure 1.
Line chart of PD-L1 protein measurements using ELISA and PD-L1 mRNA measurements, and the nCounter system. Linearity of PD-L1 protein measurements using ELISA and PD-L1 mRNA measurements resulted in a statistically significant correlation (Pearson's r=0.957). PD-L1, programmed death ligand-1.
Comparison of PD-L1 protein expression between ELISA and digital immunostaining with PID
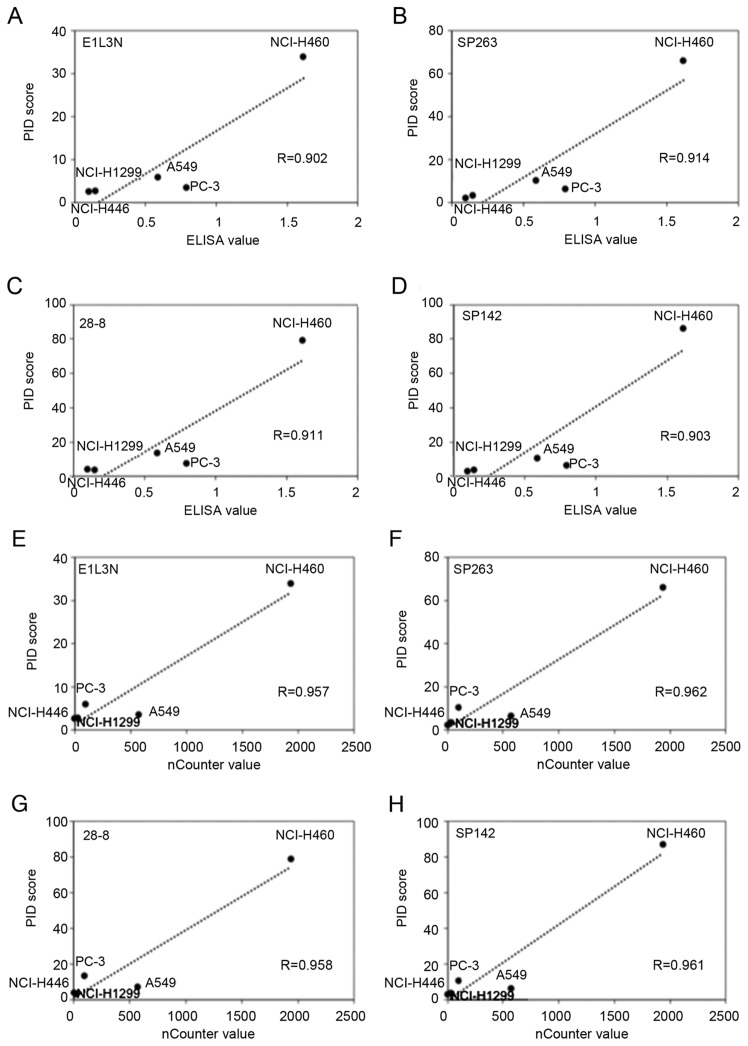
The average PID score was determined by fluorescence microscopy (Fig. 2). The highest concordance between the PID score and ELISA was observed for SP263 (r=0.914) followed by 28-8 (r=0.911), SP142 (r=0.903), and E1L3N (r=0.902; Fig. 3A-D). Comparison of the PD-L1 score (PID count per cell) with the ELISA levels revealed a high correlation (R>0.90) for all primary antibodies. Furthermore, there was no difference in the PD-L1 expression tendency for each antibody.
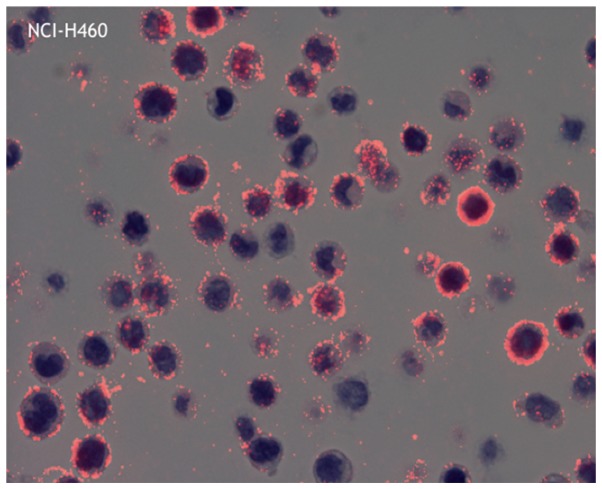
Figure 2.
Representative image of phosphor-integrated dot staining of the PD-L1 antibody using 28-8 (cell line: NCI-H446). The average dot contained 78.8 individual dots (max: 96.6 and min: 63.4). Magnification, ×400. PD-L1, programmed death ligand-1.
Figure 3.
PD-L1 protein and mRNA levels. Linearity of PD-L1 protein measurements using ELISA and digital immunostaining with PID resulted in a statistically significant correlation for all antibodies. (A) E1L3N: r=0.902, (B) SP263: r=0.914, (C) 28-8: r=0.911 and (D) SP142: r=0.903. Linearity of PD-L1 mRNA measurements using the nCounter system and digital immunostaining with PID resulted in a statistically significant correlation for all antibodies. (E) E1L3N: r=0.957, (F) SP263: r=0.962, (G) 28-8: r=0.958 and (H) SP142: r=0.961. PD-L1, programmed death ligand-1; PID, phosphor-integrated dot.
Comparison of PD-L1 mRNA expression between nCounter and digital immunostaining with PID
The highest concordance between the PID score and nCounter value was observed for SP263 (r=0.962) followed by SP142 (r=0.961), 28-8 (r=0.958), and E1L3N (r=0.957; Fig. 3E-H). Comparison of the PD-L1 score (PID count per cell) with the nCounter levels revealed a high correlation for all primary antibodies. Furthermore, there was no difference in the PD-L1 expression tendency for each antibody.
PD-L1 expression in NSCLC specimen
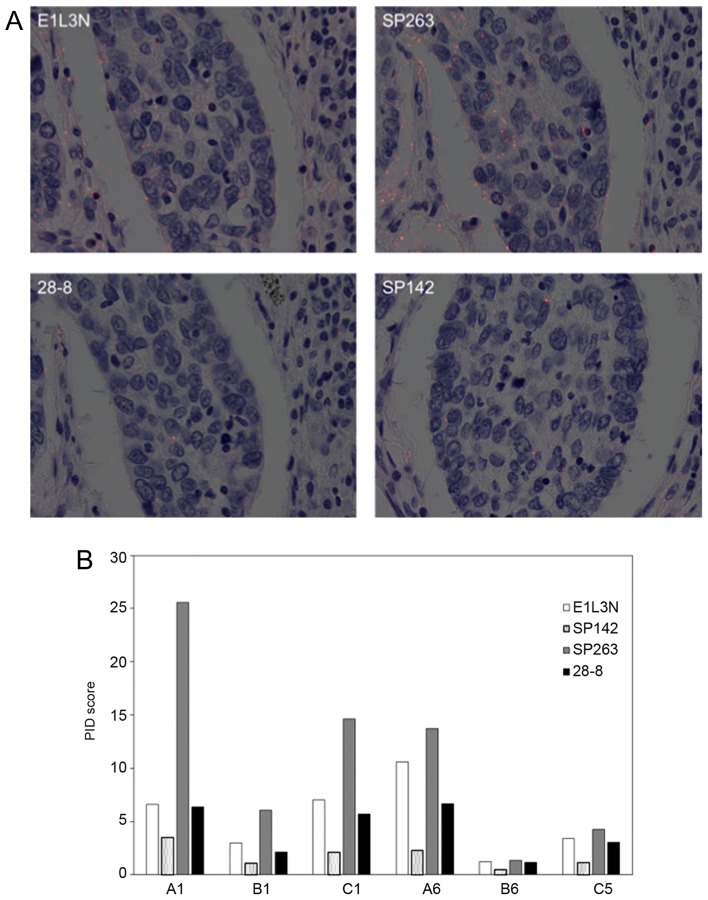
The representative images of PID for NSCLC are shown in Fig. 4. All but one core (B8) showed a similar tendency in average PID score using four primary antibodies: The score of SP263 was highest and decreased in the order of E1L3N, 28-8, and SP142.
Figure 4.
PD-L1 expression and PID scores. (A) PD-L1 expression of each antibody for squamous cell carcinoma (core A1). The average red dot of E1L3N contained 6.6 dots (max: 8.5 and min: 4.5); SP263, 25.6 dots (max: 32.1 and min: 18.0); 28-8, 6.3 dots (max: 8.1 and min: 5.2); and SP142, 3.5 dots (max: 5.2 and min: 0.8). Magnification, ×400. (B) Bar chart of the PID scores of non-small cell lung cancer specimens. D-L1, pro-grammed death ligand-1; PID, phosphor-integrated dot.
Discussion
The present study revealed that digital immunostaining of PD-L1 expression was highly correlated with protein expression measured by ELISA and quantitative mRNA data obtained using an nCounter system. Based on digital immunostaining data of each antibody of PD-L1 in NSCLC specimens, the number of dots per cell was the lowest for SP142 among the four antibodies.
In general, mRNA and protein expression levels are not always closely correlated (8); however, based on ELISA and the nCounter tests, they showed a strong correlation in the five cell lines we evaluated. Therefore, the PD-L1 mRNA expression was tightly linked to the protein expression. Digital immunostaining data of PD-L1 were highly correlated with ELISA data as well as nCounter values. The nCounter system utilizes a novel digital color-coded barcode technology that is based on direct measurement of gene expression without RT-PCR and offers high levels of precision and sensitivity (<1 copy per cell). Each barcode is attached to a single target-specific probe corresponding to a gene of interest. Thus, the digital immunostaining data were sufficiently robust for further analysis.
Although the absolute signal values of PID were different among the four antibodies, similar linearity between the PD-L1 protein and mRNA expression was observed for the five cell lines. The results were similar to those of the Blueprint project (7) which found lower sensitivity of SP142 than 22C3, 28-8, and SP263. Digital IHC for PD-L1 in NSCLC specimens in the current study also revealed that SP142 had the lowest value. However, the expression pattern was almost the same as that of the other antibodies. There are many potential reasons for the lower expression level of SP142, such as different IHC staining methods, different kits used, truncation of PD-L1 protein, or different affinity for PD-L1 protein. For PD-L1 diagnostic agents, as various sensitizing reagents are used after the secondary antibody reaction (9) it is difficult to precisely estimate the affinity of the primary antibody.
Recent analysis revealed that disruption of the PD-L1 3′-untranslated region in mice ena-bles immune evasion of EG7-OVA tumor cells with elevated PD-L1 expression (10). In the case of disruption of the PD-L1 3′-untranslated region, PD-L1 expression was detected only with an antibody directed against the extracellular domain (28–8) but not the intracellular domain (E1L3N, SP142, and SP263). The current digital imaging method using PID can compare the affinities of antibodies in a digital fashion, as the reaction system after the reaction of the secondary antibody is the same. Thus, the differences in the PID score are considered to reflect the affinity of the primary antibody more accurately. Based on our data, the most reasonable explanation for the lower score of SP142 is its lower affinity for the PD-L1 antigen.
In clinical settings, only a PD-L1 diagnostic agent with the antibody 22C3 has been approved for a companion diagnostic kit for administration of pembrolizumab (6,11). We attempted to determine the utility of 22C3, but it was difficult to directly compare the 22C3 antibody and other antibodies because the 22C3 antibody was raised against mice, whereas E1L3N, SP142, SP263, and 28-8 were raised against rabbits. Although the differences between mouse and rabbit secondary detection systems in PID staining can be adjusted for by manipulating the secondary antibody system, it is difficult to accurately quantify the PID dots. Our data also indicate that the capability of the E1L3N antibody, which is mainly used for research (12–15) rather than as a complementary or companion diagnostic antibody, was equivalent to that of 28-8 and SP263.
Another benefit of digital immunostaining compared with immunofluorescence analysis, which is dependent on fluorescence intensity, is the lack of influence from fading fluorescence signals over time. Although digital immunostaining also requires fluorescence microscopy, the technique is independent of the fluorescence intensity; thus, fluorescence fade does not affect dot quantitation. In addition, we can simultaneously examine cell morphology and the number of dots as well as superimpose DAB staining with the semi transmissive mode.
In conclusion, the results show that digital immunostaining of PD-L1 provides a sensitive, quantitative, accurate, and robust assay for measurement of protein expression in FFPE cell lines and human tissue. Our developed digital immunostaining assay allows for direct correlation between the number of dots and amount of biomarker present in the tissue. Thus, it is potentially a valuable tool for comparing the titer of different antibodies such as in the Blueprint project as well as patient selection in daily practice.
Acknowledgements
The authors would like to thank Ms. Aoi Nozawa and Ms. Yoriko Ikehata (Konica-Minolta, Inc., Tokyo, Japan) for their technical assistance.
Funding
The present study was funded by The Yasuko Shiozaki Grant awarded by the Alumni Association of Kansai Medical University (Osaka, Japan; grant no. 2017-27).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors contributions
KT designed the study and drafted the manuscript. KN, MT, HO and YN analyzed and interpreted the data regarding the cell lines. TF, MY, KS, HY and HI performed the digital immunostaining examinations, and assisted in the preparation of the manuscript. TF was a major contributor in writing the manuscript. All authors contributed to data collection and interpretation, critically revised the manuscript for important intellectual content, and read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors Kenji Nishikawa, Masaru Takahashi, Hisatake Okada and Yasushi Nakano are affiliated with Konica-Minolta, Inc., (Tokyo, Japan). The software PidAnalyzer v.2.14.1.1 used for data analysis in the present study was supplied by this company. The authors declare that they have no competing interests.
References
- 1.Gonda K, Miyashita M, Watanabe M, Takahashi Y, Goda H, Okada H, Nakano Y, Tada H, Amari M, Ohuchi N. Development of a quantitative diagnostic method of estrogen receptor expression levels by immunohistochemistry using organic fluorescent material-assembled nanoparticles. Biochem Biophys Res Commun. 2012;426:409–414. doi: 10.1016/j.bbrc.2012.08.105. [DOI] [PubMed] [Google Scholar]
- 2.Gonda K, Watanabe M, Tada H, Miyashita M, Takahashi-Aoyama Y, Kamei T, Ishida T, Usami S, Hirakawa H, Kakugawa Y, et al. Quantitative diagnostic imaging of cancer tissues by using phosphor-integrated dots with ultra-high brightness. Sci Rep. 2017;7:7509. doi: 10.1038/s41598-017-06534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamaki S, Yanagimoto H, Tsuta K, Ryota H, Kon M. PD-L1 expression in pancreatic ductal adenocarcinoma is a poor prognostic factor in patients with high CD8+ tumor-infiltrating lymphocytes: Highly sensitive detection using phosphor-integrated dot staining. Int J Clin Oncol. 2017;22:726–733. doi: 10.1007/s10147-017-1112-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16:275–287. doi: 10.1038/nrc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe S, et al. KEYNOTE-024 Investigators: Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375:1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 7.Hirsch FR, McElhinny A, Stanforth D, Ranger-Moore J, Jansson M, Kulangara K, Richardson W, Towne P, Hanks D, Vennapusa B, et al. PD-L1 immunohistochemistry assays for lung cancer: Results from phase 1 of the Blueprint PD-L1 IHC assay comparison project. J Thorac Oncol. 2017;12:208–222. doi: 10.1016/j.jtho.2016.11.2228. [DOI] [PubMed] [Google Scholar]
- 8.Chen G, Gharib TG, Huang CC, Taylor JM, Misek DE, Kardia SL, Giordano TJ, Iannettoni MD, Orringer MB, Hanash SM, et al. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics. 2002;1:304–313. doi: 10.1074/mcp.M200008-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Skaland I, Nordhus M, Gudlaugsson E, Klos J, Kjellevold KH, Janssen EA, Baak JP. Evaluation of 5 different labeled polymer immunohistochemical detection systems. Appl Immunohistochem Mol Morphol. 2010;18:90–96. doi: 10.1097/PAI.0b013e3181b0eaad. [DOI] [PubMed] [Google Scholar]
- 10.Kataoka K, Shiraishi Y, Takeda Y, Sakata S, Matsumoto M, Nagano S, Maeda T, Nagata Y, Kitanaka A, Mizuno S, et al. Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature. 2016;534:402–406. doi: 10.1038/nature18294. [DOI] [PubMed] [Google Scholar]
- 11.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L, et al. KEYNOTE-001 Investigators Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372:2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 12.Mori H, Kubo M, Yamaguchi R, Nishimura R, Osako T, Arima N, Okumura Y, Okido M, Yamada M, Kai M, et al. The combination of PD-L1 expression and decreased tumor-infiltrating lymphocytes is associated with a poor prognosis in triple-negative breast cancer. Oncotarget. 2017;8:15584–15592. doi: 10.18632/oncotarget.14698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ameratunga M, Asadi K, Lin X, Walkiewicz M, Murone C, Knight S, Mitchell P, Boutros P, John T. PD-L1 and tumor infiltrating lymphocytes as prognostic markers in resected NSCLC. PLoS One. 2016;11:e0153954. doi: 10.1371/journal.pone.0153954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Twyman-Saint Victor C, Rech AJ, Maity A, Rengan R, Pauken KE, Stelekati E, Benci JL, Xu B, Dada H, Odorizzi PM, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature. 2015;520:373–377. doi: 10.1038/nature14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLaughlin J, Han G, Schalper KA, Carvajal-Hausdorf D, Pelekanou V, Rehman J, Velcheti V, Herbst R, LoRusso P, Rimm DL. Quantitative assessment of the heterogeneity of PD-L1 expression in non-small-cell lung cancer. JAMA Oncol. 2016;2:46–54. doi: 10.1001/jamaoncol.2015.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.