Abstract
The long terminal repeat (LTR) retrotransposons Tf1 and Tf2 of Schizosaccharomyces pombe are active mobile elements of the Ty3/gypsy family. The mobilization of these retrotransposons depends on particle formation, reverse transcription and integration, processes typical of other LTR retrotransposons. However, Tf1 and Tf2 are distinct from other LTR elements in that they assemble virus-like particles from a single primary translation product, initiate reverse transcription with an unusual self-priming mechanism, and, in the case of Tf1, integrate with a pattern that favors specific promoters of RNA pol II-transcribed genes. To avoid the chromosome instability and genome damage that results from increased copy number, S. pombe applies a variety of defense mechanisms that restrict Tf1 and Tf2 activity. The mRNA of the Tf elements is eliminated by an exosome-based pathway when cells are in favorable conditions whereas nutrient deprivation triggers an RNA interference-dependent pathway that results in the heterochromatization of the elements. Interestingly, Tf1 integrates into the promoters of stress-induced genes and these insertions are capable of increasing the expression of adjacent genes. These properties of Tf1 transposition raise the possibility that Tf1 benefits cells with specific insertions by providing resistance to environmental stress.
INTRODUCTION
The fission yeast Schizosaccharomyces pombe, discovered in the late 1800s in East Africa and genetically characterized in the 1950s by Urs Leupold (97), has became a central model for studies of cell cycle, gene expression, and the complex relationship between transposable elements (TEs) and their host. Schizosaccharo-myces pombe, also known as fission yeast, can be studied with a sophisticated toolbox of molecular and genetic techniques. The haploid genome is 12.57 Mbp and encodes 5,052 genes distributed among three chromosomes (1). The complete genome sequence of the Leupold isolate revealed that TEs constitute 1.1% of the genome (2) (Table 1). All TE-related sequences in S. pombe derive from long terminal repeat (LTR) retrotransposons. The intact elements present in the Leupold strain are 13 full-length copies of the Tf2 element (3). Recombination that occurs between the LTRs of a full-length element results in solo LTRs that serve as a fossil record of TEs that are no longer present. The Leupold strain contains 249 solo LTRs or LTR fragments that are derived from nine clades of LTR retrotransposons. The youngest clades are the 35 LTR sequences from Tf2 and the 28 LTRs from Tf1, an element related to Tf2 but that is no longer present in the Leupold strain (2). Full-length Tf1 elements are present in wild isolates of S. pombe collected from different geographic regions (3). The transposition activity of Tf1 and the function of its proteins is measured by expressing a plasmid-encoded copy of Tf1 that contains neo (4, 5). Levels of Tf1 transposition correspond to amounts of G418 resistance. wtfs are another form of repeat identified in the Leupold strain (6). They are present in 25 copies and are generally 250 bp downstream of an LTR (2). Their function is unknown, but they appear to encode protein and their transcription is strongly induced during meiosis (7, 8).
TABLE 1.
Features and content in transposable elements of the Schizosaccharomyces pombe genome
| Feature | Number |
|---|---|
| Genome size (sequenced portion, excluding rDNA)a | 12.57 Mb |
| Chromosome numbera | 3 |
| Chromosome size rangea | 2.45–5.58 Mb |
| Overall GC contenta | 36.06% |
| Gene count (protein coding genes on Chr I, II and III)a | 5,123 |
| Gene count excluding dubious genes (protein coding genes on Chr I, II and III)a | 5,052 |
| Gene density (genes per Mb)a | 554.41 |
|
Transposon related sequences in strains derived from 972 | |
| Full-length Tf2 elementsb | 13 |
| Full-length Tf1 elementsb | 0 |
| Solo Tf2-related LTRsb,d | 35 |
| Solo Tf1-related LTRsb,d | 28 |
| Solo LTRs non-Tf1 nor Tf2b,d | 111 |
| Fragments related to LTRb,e | 75 |
| wtf elementsc | 25 |
THE CYCLE OF Tf1 TRANSPOSITION
The full-length Tf1 and Tf2 elements are flanked by two LTRs and encode Gag, protease (PR), reverse transcriptase (RT), and integrase (IN) (Fig. 1, Table 2). The LTR sequences of Tf1 and Tf2 share stretches of sequence identity and although the Gags of these two elements are divergent, the coding sequences for RT and IN are virtually identical (3, 9). The structure of these TEs and the function of their proteins are closely related to those of retroviruses. Indeed, the order of RT before IN in the open reading frame (ORF) and phylogenetic analysis of the coding sequences place Tf1 and Tf2 firmly within the Gypsy/Ty3 family of LTR retrotransposons, a clade that includes elements with envelope-like proteins that appear to mediate infection (10–14).
FIGURE 1.
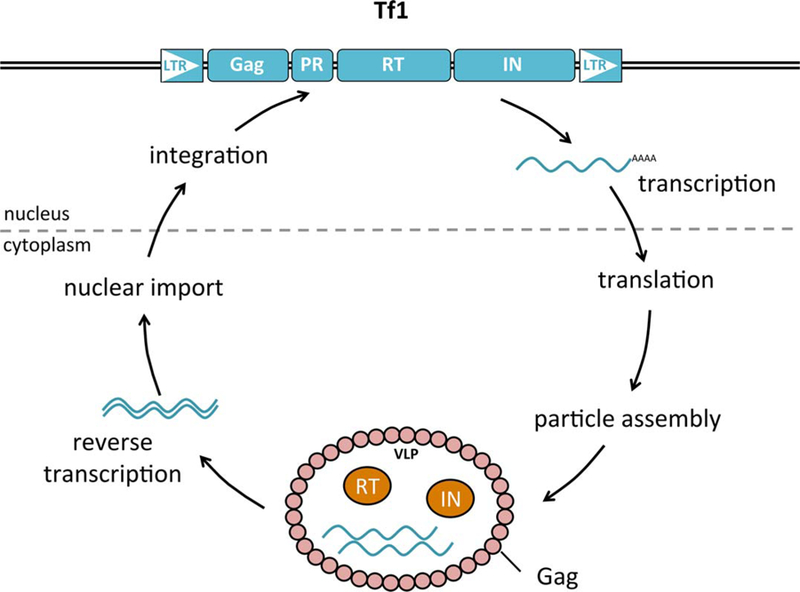
Tf1 transposition in Schizosaccharomyces pombe. Full-length Tf1 (blue rectangles) is transcribed and translated. The polyprotein assembles into particle precursors and PR processes the protein into Gag (pink), protease (PR), reverse transcriptase (RT) (orange), and integrase (IN) (orange). The mature virus-like particles (VLP) contain the processed proteins and two copies of mRNA (blue wavy lines). The RT reverse-transcribes the mRNA into double-stranded cDNA that associates with IN. Once transported into the nucleus (above dotted line), the IN integrates the cDNA at a new position in the genome. The long terminal repeats are symbolized by triangles. The protein coding sequences are represented by rectangles. doi:10.1128/microbiolspec.MDNA3-0040-2014.f1
TABLE 2.
Sizes of transposable elements and the molecular weights of their components in Schizosaccharomyces pombe
The transposition cycle of Tf1 begins with transcription of the full-length mRNA, which is then exported to the cytoplasm where it is translated (Fig. 1). Most retro-viruses and LTR retrotransposons encode two separate ORFs for the pol and gag proteins, and PR cleaves the polyproteins into their mature size. Mechanisms such as frame shifting that result in low levels of the Pol proteins RT and IN relative to Gag are thought to be important to allow efficient particle assembly. However, Tf1 encodes one long polyprotein. The processing activity of PR cleaves the polyprotein into the mature sized Gag, PR, RT, and IN (15). An unknown activity degrades RT and IN to produce the appropriate low levels relative to Gag that allow for particle assembly (16). Gag forms the coat of the virus-like particle, a 50-nm structure that contains at least two copies of Tf1 mRNA, RT, and IN (15, 17, 18). RT copies the mRNA into double-stranded cDNA, which is transported into the nucleus where IN mediates the integration of the cDNA fragment at a new genomic position and generates a 5-bp target site duplication (5).
As is true for the Gag proteins of retroviruses, Tf1 Gag is sufficient for the formation of virus-like particles and is required for the packaging of Tf1 mRNA and reverse transcription (18). Deletion analyses of Gag identified an N-terminal nuclear localization signal and adjacent residues that modulate the mode of import (19, 20). The import of Tf1 Gag is governed by a unique requirement for the FXFG nuclear pore factor Nup124p and mutations that compromise virus-like particle structure bypass the requirement for Nup124p (19–21). These data, together with the finding that Gag interacts directly with Nup124p, led to the model that Nup124p is required for Tf1 import because it accommodates the particle-like structure of Tf1 Gag (21). Interestingly, the role of Nup124p in particle import may be conserved in humans. Nup153, the component of the human nuclear pore complex that is most closely related to Nup124p, contains a domain that can mediate the import of Tf1 Gag in S. pombe that lack Nup124p (22, 23). Similar to the role of Nup124p in the import of Tf1 Gag, Nup153p directly engages the capsid protein of HIV-1 and is required for infectivity (24–27).
Another host factor that mediates the import of Tf1 Gag into the nucleus is the histone acetyltransferase interacting factor Pst1p and experiments indicate that acetyltransferase activity is necessary for transport of Gag into the nucleus (28).
The reverse transcription of Tf1 mRNA into full-length cDNA relies on the same intermediates and strand transfers of these intermediates as other LTR retroelements (see Mobile DNA III chapter 46, Reverse Transcription of Retroviruses and LTR Retrotransposons by S. Hughes). However, unlike typical LTR retrotransposons and retroviruses, Tf1 does not require a tRNA to initiate reverse transcription. Instead, Tf1 possesses a unique mechanism of self-priming that relies on the priming activity of the first 11 bp of the Tf1 mRNA (4, 29, 30). This 11-bp segment of Tf1 RNA anneals to the primer binding site in the mRNA and the RNaseH domain of RT cleaves the mRNA between bases 11 and 12 (Fig. 2A) (30–32). The self-primer is then used by RT to initiate minus-strand cDNA synthesis.
FIGURE 2.

Reverse transcription of Tf1 is initiated by a self-priming mechanism. (A) The 5′ end of the Tf1 mRNA anneals to the primer binding site (PBS). The first 11 nucleotides (red) are cleaved from the Tf1 mRNA (black) and prime reverse transcription towards the 5′ end of the mRNA. The scissors indicate the position of cleavage. The arrow indicates the direction of reverse transcription. (B) The 5′ end of the Tf1 mRNA folds into a complex duplex structure. The scissors indicate the position of the cleavage that liberates the self-primer (red). The position of the U5-IR stem-loop is indicated. doi:10.1128/microbiolspec.MDNA3-0040-2014.f2
Self-priming of reverse transcription requires the formation of a complex RNA structure that includes duplexes of several segments of the Tf1 mRNA (Fig. 2B) (33). One element of the RNA structure termed the U5-IR stem loop includes a downstream segment of the LTR (U5) and is required for self-priming. This structure is surprisingly similar to a feature in the mRNA of Rous sarcoma virus that is also necessary for reverse transcription (34). This structural similarity indicates that there is strong conservation in the way RT recognizes the minus-strand primer. Nevertheless, the lack of a potential tRNA primer and the formation of a duplex between the primer binding site and the 5′ end of the mRNA are hallmarks of Tf1 that are central to the self-priming mechanism. Scrutiny of many other LTR retrotransposons reveals that Tf1 and Tf2 are the founding members of a large class of elements that also use self-priming to initiate reverse transcription (35, 36). While virtually all of the self-priming elements belong to the Ty3-gypsy family of retrotransposons, one element from the Ty1-copia family also has the potential to self-prime (37).
In another twist of Tf1 reverse transcription the self-primer is not removed during reverse transcription, which results in 11 additional bases at the 3′ end of the plus strand cDNA that are complementary to the self-primer (38). Biochemical experiments with recombinant protein show that IN has an unusual processing activity that removes these 11 bases from the 3′ end of the plus strand cDNA and allows integration to occur (38, 39). The plus-strand primer of reverse transcription or polypurine tract (PPT) contains 13 ribonucleotides positioned immediately 5′ of the downstream LTR. Analysis with single nucleotide resolution of the 5′ plus-strand and 3′ minus-strand ends of the cDNA indicate the PPT consists of UUGGGGAGGGCAA (31). In vivo and biochemical experiments show that the PPT is removed by RT before reverse transcription is complete and that purified RT can remove the self-primer from the 5′ end of the minus-strand (31, 40). Interestingly, the PPT removal activity of RT is greatly stimulated by IN (40). Other biochemical assays demonstrate that Tf1 RT possesses several unusual activities such as template- independent DNA synthesis and the ability to clamp very short primer terminal sequences onto the 3′ termini of template strands (41–44).
The IN of Tf1 has many similarities with INs of retroviruses and other LTR retrotransposons including the 3′ processing activity that exposes the 3′ CA of the cDNA and the strand transfer activity that simultaneously cleaves the phosphodiester bonds in the target DNA and catalyzes formation of the covalent bond between the 3′ end of the LTR and the 5′ end of the target DNA (see Mobile DNA III chapter 45, Host Factors in Retroviral Integration and the Selection of Integration Target Sites by F. Bushman and R. Craigie) (39). The structure of retrovirus and LTR retrotransposon INs is divided into three domains that contain conserved motifs. The N-terminal domain binds zinc and contains an HHCC motif; the center domain has the catalytic residues and contains the D-D-E motif; and the C-terminal domain has DNA binding activity. Some INs in their C-terminal domain have subdomains, a GP(Y/F) domain, a chromodomain, or both (13). Tf1 possesses both the GP(Y/F) domain and the chromodomain (45). Both of these features are necessary for integration in vivo (45, 46). In vitro experiments demonstrate that the GP(Y/F) domain is necessary for strand transfer activity, binds DNA, and mediates IN multimerization (45). The chromodomains of INs are similar in structure to the chromodomains of chromatin factors and in cases of plant retrotransposons they direct binding to heterochromatin (47). The chromodomain of the fungal element Maggy directs integration to heterochromatin by interacting with histone H3 methylated at lysine 9 (47). Although no interaction has been detected between the chromodomain of Tf1 IN and histones, it does play a role in binding IN to cDNA in vivo (48).
SITE SELECTION OF Tf1 INTEGRATION
Integration of TEs can damage the genetic content of the host. LTR retrotransposons feature strategies to avoid disrupting coding sequences during integration. In Saccharomyces cerevisiae the IN of Ty3 interacts with components of TFIIIB to position integration one to four nucleotides upstream of RNA pol III-transcribed genes (see Mobile DNA III chapter 42, Ty3, A Position-Specific Retrotransposon in Budding Yeast by S. Sandmeyer and K. Patterson) (49). The IN of Ty5 directs integration to heterochromatin by binding Sir4, a component of heterochromatin (50). In S. pombe Tf1 avoids integrating in coding sequences by a different mechanism that is not fully understood. The choice of integration sites is thought to depend on features of Tf1 IN as well as on features of the host factors and sequence.
Existing TE sequences provide evidence of past integration events and can provide information about the integration behavior of elements. The genome sequence of S. pombe contains 13 full-length Tf2 elements and 249 solo LTRs or LTR fragments (2) (Table 1). Phylogenetic analysis of solo LTRs demonstrates that Tf1 and Tf2 are the most recently active transposable elements in S. pombe. Tf1 and Tf2 sequences are in promoter regions of RNA polymerase II-transcribed genes, in a window located 100–400 bp upstream of ORFs (2). The elements are absent from tRNA gene clusters and are greatly underrepresented in intergenic sequences between convergent genes. Although it is possible that these integrated copies persist because of selective pressure, it is now known that the positions of the existing copies result from integration directed to promoter sequences.
Studies of Tf1 integration within the S. pombe genome rely on heterologous promoters to induce the expression of Tf1 with a drug-resistance gene. Selection of cells with de novo insertions allowed a total of 27 sites to be isolated and sequenced (51). Schemes that lack selection of insertions in S. pombe are also used to generate and sequence insertion sites (52, 53). In two studies not using genetic selection, 51 and 9 insertions were sequenced. Regardless of whether selection is used, the insertions distribute throughout the genome and cluster in promoter regions upstream of ORFs (52, 53).
Unlike Tf1, Tf2 does not depend on IN for the majority of its activity, and mobilizes with a frequency 10-to 20-fold lower than Tf1 (54). When Tf2 marked with neo is expressed from a heterologous promoter 70% of events that introduce Tf2-neo into the genome result from homologous recombination between Tf2 cDNA and pre-existing copies of Tf2. This mechanism is a different strategy to avoid disrupting the host coding sequences, which has been viewed as a recycling of the integration sites. In addition to integration site recycling, the remaining 30% of mobilization events do occur through true integration events (54). Although the amino acid sequences of Tf1 and Tf2 INs are 98% identical it was possible that the low levels of Tf2 integration were due to the small number of amino acid differences. The results of domain swapping experiments between Tf1 and Tf2 coding sequences revealed that Tf2 IN is fully active. Instead, the low levels of Tf2 integration relative to Tf1 were the result of sequence differences in capsid and PR (55).
To understand what features of promoter sequences are recognized by Tf1, and to facilitate the isolation of independent insertions, integration events are isolated in target plasmids. These plasmids are targets of integration when introduced into cells that express Tf1. Transposition assays with plasmids containing a collection of genes confirm that insertions occur in promoter sequences and reveal that each promoter possesses a unique and reproducible pattern of integration sites (46). By generating a series of deletions in target plasmids, minimal sequences as short as 160 bp and 70 bp are found to function as efficient targets (46, 56). In specific cases the minimal sequences contain binding sites for transcription factors and in the case of insertions in the fbp1 promoter, binding of the transcription activator Atf1p to the promoter mediates integration (46). The minimal target sequences from the promoters do not support transcription and artificial promoters that do drive transcription are not targets of integration (46, 56). These results indicate that specific transcription factors, not general transcription machinery, may mediate integration. Although Atf1p mediates integration in the fpb1 promoter it does not contribute to integration at the promoter of ade6 and the overall frequency of genome-wide transposition is not reduced in the absence of Atf1p.
The target plasmid assay can also be used to test domains of IN for a role in positioning integration. Tf1 IN lacking the chromodomain can be expressed from a version of Tf1 that has a frameshift mutation in IN. Although this substantially reduces the frequency of transposition, integration events can be isolated in the target plasmid (48). Tf1 IN lacking the chromodomain greatly modifies the insertion sites in target plasmids containing a variety of target promoters. These data suggest that the chromodomain plays an important role in positioning integration. However, the pattern of Tf1 integration genome-wide is not altered when the chromodomain is absent (57). These data indicate that with natural genome targets, the chromodomain does not contribute to the selection of insertion sites.
The development of deep-sequencing technologies has radically increased the number of TE insertions that can be mapped genome-wide. Cells with insertions are generated by overexpressing Tf1 containing neo, extracting DNA from G418-resistant cells, and using ligation-mediated polymerase chain reaction to sequence libraries of insertions. A saturated map of Tf1 integration with reproducible numbers of events in each intergenic region can be obtained using 454 pyrosequencing (58). Insertion profiles generated in haploid and diploid strains are indistinguishable. Over 95% of 73,125 independent integration events target intergenic regions, showing again a clear preference for promoters of RNA polymerase II-transcribed genes. An improved method for sequencing Tf1 integration (described below) mapped 800,723 independent insertions and these showed the same preference for promoter regions as observed earlier (Fig. 3). Just 3.8% of the integrations occur in ORFs. The integration pattern relative to ORFs as shown in Fig. 3 is highly similar to the one of existing integration events, indicating that the distribution of pre-existing Tf elements results from integration mechanisms, not selection pressure (2).
FIGURE 3.

Distribution of the distance from Tf1 integration to the nearest open reading frame (ORF). A collection of 800,723 independent integration events was positioned according to the distance to the nearest ORF (red) (57). The x-axis represents the distance from the 5′ end and 3′ end of ORFs in bins that are 100 bp wide; distances within ORFs are divided into 15 bins of equal proportion. The y-axis represents the percentage of independent integration events positioned within the bins. The majority of integration events are located near the 5′ end of ORFs, 3.8% of all integration events targeted coding sequences. Figure reproduced with permission from ref. (57). doi:10.1128/microbiolspec.MDNA3-0040-2014.f3
The dense profiles of integration produced with deep sequencing show that a specific set of promoters receive the bulk of the insertion events (58). Approximately 20% of the intergenic sequences have higher levels of integration than would occur by random selection. Comparisons with genome-wide profiles of transcription do not identify any correlation between amounts of integration in promoters and the levels of transcription (58). Importantly, the intergenic sequences with enhanced integration are enriched with promoters of stress-induced genes (57, 58). Promoters of genes that are induced by cadmium, hydrogen peroxide, heat, methylmethane sulfonate, and sorbitol display high levels of Tf1 integration. This pattern supports the model that specific transcription factors mediate the pattern of integration. The bias favoring integration in stress-induced promoters is also seen in diploid strains, indicating that this insertion preference is the result of integration bias, not selection occurring after integration (57, 58). In contrast to the integration in promoter sequences, the 3.5% of insertion events that occur in ORFs exhibit no gene-specific bias and levels of integration in individual ORFs vary between experiments (58). This indicates that Tf1 integration may occur by two discrete mechanisms, one that directs the bulk of integration to specific promoters, and one that distributes low levels of integration randomly.
Another type of factor that may play a role in positioning integration is the DNA binding protein Sap1. This essential factor binds to DNA with sequence specificity, blocks replication forks in rDNA repeats, and is required for the imprinting and recombination mechanisms that cause mating type switching (59–64). Interestingly, Sap1 binding sites show a strong correlation with positions of Tf1 integration (65). This link with Sap1 binding sites suggests that Sap1 plays a role in integration.
Recent efforts to characterize patterns of Tf1 integration include a new method of sequencing that maps significantly more insertion events than previous techniques. This method, termed the serial number system, provides more integration data by distinguishing events that occur at the same nucleotide position as independent (57). Tf1 expression plasmids are induced in cultures of S. pombe to generate large numbers of insertions. But in contrast to previous strategies, diverse libraries of expression plasmids are used that contain 8-bp serial numbers consisting of random nucleotides embedded near the downstream end of the LTR. Insertions that occur at the same nucleotide position and orientation have different serial numbers and can be identified as independent events because the sequence reads that reveal the position of integration also include the serial number sequence. With this method as many as 455,000 insertion events can be sequenced from a single culture of S. pombe (57). The integration data from the serial number system have the added dimension of tabulating numbers of insertions at each nucleotide coordinate, creating profiles of peaks, spikes, and flat terrains. Comparison of integration profiles between independent experiments shows very high reproducibility. The tabulation of the number of integration events at individual positions reveals that 75% of all the integration occurs at just 35% of the insertion positions. The integration positions with the highest number of repeated insertions have strong nucleotide preferences, traces of which are observed at the 35% of the positions where 75% of the events occur (57).
HOST DEFENSES AGAINST Tf ELEMENTS IN S. POMBE
Unrestricted transposition is detrimental to the host. Even when integration is directed to gene-free regions the over-accumulation of TEs is a burden on the DNA replication of the host and reduces genome stability. As a result, host organisms such as S. pombe have evolved a variety of defense mechanisms that inhibit TE activity (Table 3) (66, 67). RNA interference (RNAi) is a highly conserved system that inhibits the expression of TEs either by degrading their mRNA or by assembling TE sequences into heterochromatin (68, 69). In S. pombe, RNAi is mediated by the Argonaute family protein Ago1, the RNA cleaving factor Dcr1, and the RNA-dependent RNA polymerase Rdr1 (70, 71). In vertebrates, insects, nematodes, and plants, RNAi strongly inhibits the expression of TEs (72). Surprisingly, deletion of ago1, dcr1, or rdr1 resulted only in slight increases in Tf2 expression and did not affect the expression of genes adjacent to LTRs, suggesting that RNAi has a relatively minor role in inhibiting the transcription of TEs in S. pombe (73). The chromatin features that determine heterochromatin-mediated silencing, histone H3 methylated on lysine 9 and the HP1 homolog swi6, are not associated with Tf sequences (74). Further-more, deep sequencing of small interfering RNAs (siRNAs) does not detect Tf-derived sequences (74–76). Interestingly, another yeast from the Schizosaccharomyces genus, Schizosaccharomyces japonicus, possesses an extensive population of LTR retrotransposons and mRNAs of these elements are processed into siRNAs (77).
TABLE 3.
Host factors that restrict transposable element activity
| Host defense machinery | Host factors / Function | Regulatory impact on Tf elements | Refs |
|---|---|---|---|
| Exosome | Pla1: canonical polyA polymerase | Elimination of transcripts via the exosome | (78) |
| Red1: RNA surveillance factor | (96) | ||
| Pab2: polyA binding protein | |||
| Rrp6: exosome subunit | |||
| Mtl1: component of Red1 associated MTREC complex | (79) | ||
| RNAi | Nrl1: splicing factor binding protein | Activation of RNAi response through recognition of cryptic introns | (79) |
| Under exosome deficiency or nutrient starvation | Clr4: histone methyltransferase | Activation of RNAi response through histone methylation | (78) |
| Ago1: argonaute, RNA binding | Processing of mRNA into siRNAs and the formation of heterochromatin | (78) | |
| Dcr1: dicer, ribonuclease | |||
| Rdr1: RNA-directed RNA polymerase | |||
| Swi6: HP1 chromodomain protein | Heterochromatinization | (74) | |
| Nucleosome deposition by histone chaperone complex HIRA | Hip1: hira histone chaperone | Inhibition of transcription through nucleosome deposition | (80,81) |
| Hip3: HIRA interacting protein | |||
| Slm9: HIRA protein | |||
| CENP-B proteins and associated factors | Abp1/Cbp1: CENP-B homolog | Inhibition of transcription | (86,87) |
| Cbh1: CENP-B homolog | Restriction of homologous recombination | ||
| Cbh2: CENP-B homolog | Recruitment of HDACs Clr3 and Clr6 | ||
| Clr3: histone deacetylase | Inhibition of transcription through histon hypoacetylation | (73,86,87) | |
| Clr6: histone deacetylase | |||
| Set1: histone methyltransferase | Inhibition of sense and antisense transcription | (87) | |
| Ku70: Ku domain protein, DNA binding | Clustering into Tf-bodies | (88) | |
| Ku80: Ku domain protein, DNA binding |
Although the RNAi machinery of S. pombe does not appear to limit TE activity under optimal conditions, cells with defects in nuclear exosome activity possess a strong RNAi response that processes Tf2 mRNA into siRNAs and assembles Tf2 sequences into heterochromatin (Table 3) (78). The targeting of Tf2s by RNAi relies on the recognition of a cryptic intron by Nrl1, a protein associated with splicing factors (79). Splicing of the cryptic intron is predicted to remove 20 amino acids from RT near the N terminus of the protein. The incorporation of Tf2 into heterochromatin when cells lack the exosome demonstrates that there is competition between the exosome and RNAi for Tf2 mRNA. Under conditions of nutrient deprivation, such as low glucose, the competition is altered such that Tf2 mRNAs are processed into siRNAs and Tf2 sequences assemble into heterochromatin even when cells possess functional exosomes (78).
The transcription apparatus of the host inhibits the expression of genes that are not necessary during vegetative growth in ideal conditions. The histone chaperone complex Histone Regulator A (HIRA) deposits nucleosomes independent of DNA replication and this prevents the transcription of many genes when cells grow in ideal conditions (80). Interestingly, HIRA also plays a key role in inhibiting TE expression in S. pombe. Cells lacking either of the two HIRA factors, Hip1 or Slm9, express high levels of Tf2 mRNA and noncoding transcripts that initiate at solo LTRs (80, 81).
Another class of proteins that inhibit the expression of TEs in S. pombe is the centromere protein B (CENP-B) proteins. CENP-B factors are a conserved set of proteins that bind DNA repeats in many species from S. pombe to humans (82). Interestingly, CENP-B proteins are thought to have evolved from an ancient pogo-like DNA transposon (83, 84). In human cells CENP-B binds to a 17-bp sequence present in pericentromeric α-satellite repeats (85). The three CENP-B homologs in S. pombe are Abp1/Cbp1, Cbh1 and Cbh2. Together, these proteins bind to specific sequences in the LTRs throughout the genome (86). The CENP-B proteins, in particular, Abp1, greatly inhibit the transcription of Tf1, Tf2 and genes adjacent to LTRs (Table 3) (86). The reduced transcription can result in inhibition of Tf1 integration events. Abp1 inhibits transcription of Tf2 in part by recruiting the histone deacetylases Clr3 and Clr6. In addition, the histone H3K4 methyltransferase Set1 cooperates with Abp1 to regulate the sense and antisense transcription of Tf2 (87). Importantly, Abp1-mediated inhibition of Tf2 transcription is independent of the repressive function of the HIRA complex (86). If CENP-B proteins are derived from ancient DNA transposons it is interesting to speculate that the DNA-binding activity of the transposase evolved to recognize sequences within LTR retrotransposons and this resulted in an inhibitory activity that was co-opted by the host to control TEs.
The mobility of LTR retrotransposons can include efficient homologous recombination between transposon cDNA and existing copies of the TE in the host genome. This recycling of insertion sites is the primary mode of action for Tf2 and it is thought to repair damaged copies of the TE (54). The CENP-B proteins not only defend S. pombe by inhibiting Tf2 transcription, they also restrict the homologous recombination between cDNA and existing Tf2s (86).
An intriguing feature of the CENP-B proteins is that they play a role in organizing the Tf2 sequences into one to three subnuclear foci called Tf bodies (86). The clustering of Tf bodies also requires the Ku heterodimer complex of Ku70 and Ku80 and the condensin complex (88). In a chain of interactions, Abp1 recruits the Ku complex to Tf2 and LTR sequences and in turn Ku recruits condensin. Tf bodies are dynamic and disassociate during S-phase and after DNA damage when histone H3 Lys56 acetylation inhibits Ku recruitment (88). Although it is clear that Tf2 elements and presumably solo LTRs organize into clusters in the nucleus, and that this must have significant impact on the three-dimensional structure of the genome, the function of Tf bodies remains to be determined. Although the Ku complex and the dimerization domain of Abp1 are required for the formation of Tf bodies they are not required for restricting Tf2 transcription (86).
High copy number is a feature of TEs that inflict harm on the host by reducing genome stability by way of chromosome rearrangements including duplications and deletions. The LTRs in S. pombe impair DNA replication by blocking fork progression, a property that results from the binding of Sap1 to LTRs (65). Although the function of this replication block is not clear, it also occurs in rDNA repeats and has the intriguing feature of occurring in only one orientation. In the absence of the three CENP-B factors the stalled replication forks collapse and this leads to a high frequency of double-stranded chromosomal breaks that results in lethality (65, 89).
THE BIOLOGICAL IMPACT OF TRANSPOSITION IN S. POMBE
The relationship between TEs and the host is often described as one of genetic conflict where each evolves machinery to combat the other. The examples of host mechanisms that reduce the expression of Tf2 described above clearly show that S. pombe has methods to combat TE activity. However, the relation is far more complex, in part because the propagation of TEs is entirely dependent on the host. In addition, TEs have been proposed to benefit the host greatly by providing regulatory sequences and by reorganizing the genome in response to environmental stress (90). Examples of TE insertions that alter the expression of adjacent genes are observed in many organisms and include Saccharomyces cerevisiae in which copies of Ty1 or one representative copy of Ty3 increase expression of tRNA genes (91, 92). Genome-wide studies of RNA in S. pombe reveal examples of Tf2 LTRs that regulate adjacent genes. The LTRs function as transcription start sites for mRNAs that read through neighboring genes. When cells are grown in low oxygen conditions, the transcription activator Sre1 binds to Tf2 LTRs at the sequence 5′-ATCGTACCAT-3′, and induces transcription of Tf2 (93). Importantly, solo Tf2 LTRs are also bound by Sre1 and this generates transcripts that initiate in the LTR and read through into downstream genes (Fig. 4A). However, the Sre1 binding sequence does not exist in Tf1 LTRs and Sre1 does not bind or regulate Tf1 LTRs. To determine whether Tf1 insertion can alter the expression of adjacent genes a target plasmid was studied that contains a Tf1 insertion in the intergenic sequence between the genes bub1 and ade6. Northern blots show that the Tf1 insertion increases the expression of both bub1 and ade6 (Fig. 4B) (46).
FIGURE 4.

Impact of Tf elements on gene expression. Tf element integration can activate the expression of adjacent genes. (A) Tf2 long terminal repeat (LTR) (blue) possesses a motif ATCGTACCAT bound by the transcription factor Sre1 (green), which activates the transcription of oxygen-dependent genes such as amt3. Pol II (pink); RNA polymerase II. (B) Sequences within the Tf1 LTR substitute for the elements in the ade6 promoter that are disrupted by Tf1 integration. Tf1 integration results in a 2.1- to 3.6-fold increase in ade6 expression. TF (grey): transcription factor. (C) Tf1 integration near the heat shock gene ssa1 acts as an enhancer of transcription, through a mechanism that may involve the transcription activation factor Atf1 (red). A motif TGACGT similar to the sequence bound by Atf1 is within the Tf1 LTR (blue). doi:http://10.1128/microbiolspec.MDNA3-0040-2014.f4
To understand the biological impact of TE insertion it is important to know what fraction of transposition events alter the expression of neighboring genes. In general, this is not known because insertions studied are initially identified using phenotypes that result from altered expression. To gauge what fraction of Tf1 insertions alter gene expression of adjacent genes an unbiased collection of integration events can be studied. A systematic analysis of 14 strains with single Tf1 insertions reveals that 40% of the insertions increase expression of adjacent genes (94). Surprisingly, none of the insertions reduce the expression of adjacent genes. The insertions that increase expression of adjacent genes do so by introducing a transcriptional enhancer. This results in higher amounts of transcripts that initiate from the original start site (Fig. 4C). The genes that are induced by the Tf1 enhancer turn out to be the genes that, in the absence of Tf1, are induced by heat shock and oxidative stress (94). The Tf1 promoter itself is activated by heat shock and oxidative stress, suggesting that the Tf1 enhancer synergizes with stress response promoters to increase the transcription of genes adjacent to Tf1 (94).
As more information accumulates about the properties of Tf1 it becomes clear that this transposable element is wired into a network of stress response mechanisms. First, Tf1 transcription increases substantially when cells are exposed to heat and oxidative stress (94). This suggests a mechanism that activates transposition in response to stress. Second, the profile of Tf1 integration shows a pattern of insertion sites that clearly favors stress response promoters (57, 58). Third, the expression of stress response genes is increased by adjacent copies of Tf1 (94). Together, these features suggest the intriguing possibility that Tf1 integration is a mechanism that is induced by stress and efficiently generates mutations that have the potential to improve survival of individual cells. While this scenario may sound compelling, much must be done to test it. Does environmental stress actually induce Tf1 integration? Can Tf1 integration provide a competitive benefit to cells exposed to environmental stress? If Tf1 integration does improve survival of individual cells, is the frequency of cells with this outcome any greater than the spontaneous rate of resistance mutations that are selected during stress. Once these questions are answered it will be clear whether Tf1 possesses specific mechanisms that provide direct benefit to cells exposed to stress.
CONCLUSIONS
The strains of S. pombe studied in laboratories around the world all originated from an individual isolate that contained 13 Tf2 LTR retrotransposons together with about 250 single LTRs. Although these 13 retrotransposons represent the only TEs present in the laboratory strains, it is important to remember that these sequences are a single snapshot of the dynamic and polymorphic mobile collection that exist in wild populations of S. pombe. The 250 single LTRs in the laboratory strain are evidence of many different families of LTR retrotransposons including Tf1, the highly active TE that was isolated from a wild strain. The static nature of Tf2 elements in laboratory strains could be a result of the inhibitory functions of CENP-Bs, the exosome, and the heterochromatization of Tf2 that occurs under nutrient deprivation. However, conditions of environmental stress overcome the mechanisms that restrict these TEs and result in induction of Tf1 and Tf2 mRNA. Once Tf1 integrates into new positions it can overcome host mechanisms that inhibit its own expression and increase transcription of adjacent genes. Because integration often occurs next to stress response genes we propose that the ability of Tf1 to induce the expression of adjacent genes may be beneficial to the host when exposed to environmental stress. This idea is consistent with the observation that environmental stress induces Tf1 and Tf2 transcription, a process that may lead to de novo insertions. The mechanisms that restrict TE activity and the ability of Tf1 and Tf2 to escape the restrictions are vivid examples of the conflict that exists between the TEs and S. pombe. The possibility that Tf1 integration may be beneficial to the host may explain the balance that has been struck between restriction and mobility.
Acknowledgments
FUNDING
This research was supported by the Intramural Research Program of the NIH from the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
REFERENCES
- 1.Wood V, Harris MA, McDowall MD, Rutherford K, Vaughan BW, Staines DM, Aslett M, Lock A, Bahler J, Kersey PJ, Oliver SG. 2011. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res 40:D695–D699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowen NJ, Jordan I, Epstein J, Wood V, Levin HL. 2003. Retrotransposons and their recognition of pol ii promoters: a comprehensive survey of the transposable elements derived from the complete genome sequence of Schizosaccharomyces pombe. Genome Res 13:1984–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levin HL, Weaver DC, Boeke JD. 1990. Two related families of retrotransposons from Schizosaccharomyces pombe [published erratum appears in Mol Cell Biol 1991;11(4):2334]. Mol Cell Biol 10:6791–6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levin HL. 1995. A novel mechanism of self-primed reverse transcription defines a new family of retroelements. Mol Cell Biol 15:3310–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levin HL, Boeke JD. 1992. Demonstration of retrotransposition of the Tf1 element in fission yeast. EMBO J 11:1145–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lespinet O, Wolf YI, Koonin EV, Aravind L. 2002. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res 12:1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mata J, Lyne R, Burns G, Bahler J. 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet 32:143–147. [DOI] [PubMed] [Google Scholar]
- 8.Watanabe T, Miyashita K, Saito TT, Yoneki T, Kakihara Y, Nabeshima K, Kishi YA, Shimoda C, Nojima H. 2001. Comprehensive isolation of meiosis-specific genes identifies novel proteins and unusual non-coding transcripts in Schizosaccharomyces pombe. Nucleic Acids Res 29:2327–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weaver DC, Shpakovski GV, Caputo E, Levin HL, Boeke JD. 1993. Sequence analysis of closely related retrotransposon families from fission yeast. Gene 131:135–139. [DOI] [PubMed] [Google Scholar]
- 10.Song SU, Kurkulos M, Boeke JD, Corces VG. 1997. Infection of the germ line by retroviral particles produced in the follicle cells: a possible mechanism for the mobilization of the gypsy retroelement of Drosophila. Development 124:2789–2798. [DOI] [PubMed] [Google Scholar]
- 11.Song SU, Gerasimova T, Kurkulos M, Boeke JD, Corces VG. 1994. An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev 8:2046–2057. [DOI] [PubMed] [Google Scholar]
- 12.Teysset L, Burns JC, Shike H, Sullivan BL, Bucheton A, Terzian C. 1998. A moloney murine leukemia virus-based retroviral vector pseudotyped by the insect retroviral gypsy envelope can infect Drosophila cells. J Virol 72:853–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malik HS, Eickbush TH. 1999. Modular evolution of the integrase domain in the Ty3/Gypsy class of LTR retrotransposons. J Virol 73:5186–5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malik HS, Henikoff S, Eickbush TH. 2000. Poised for contagion: evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res 10:1307–1318. [DOI] [PubMed] [Google Scholar]
- 15.Levin HL, Weaver DC, Boeke JD. 1993. Novel gene expression mechanism in a fission yeast retroelement: Tf1 proteins are derived from a single primary translation product [published erratum appears in EMBO J 1994;13:1494]. EMBO J 12:4885–4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atwood A, Lin JH, Levin HL. 1996. The retrotransposon Tf1 assembles virus-like particles that contain excess Gag relative to integrase because of a regulated degradation process. Mol Cell Biol 16:338–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haag AL, Lin JH, Levin HL. 2000. Evidence for the packaging of multiple copies of Tfl mRNA into particles and the trans priming of reverse transcription. J Virol 74:7164–7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teysset L, Dang VD, Kim MK, Levin HL. 2003. A long terminal repeat-containing retrotransposon of Schizosaccharomyces pombe expresses a Gag-like protein that assembles into virus-like particles which mediate reverse transcription. J Virol 77:5451–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dang VD, Levin HL. 2000. Nuclear import of the retrotransposon Tf1 is governed by a nuclear localization signal that possesses a unique requirement for the FXFG nuclear pore factor Nup124p. Mol Cell Biol 20:7798–7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim MK, Claiborn KC, Levin HL. 2005. The long terminal repeat-containing retrotransposon Tf1 possesses amino acids in Gag that regulate nuclear localization and particle formation. J Virol 79:9540–9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balasundaram D, Benedik MJ, Morphew M, Dang VD, Levin HL. 1999. Nup124p is a nuclear pore factor of Schizosaccharomyces pombe that is important for nuclear import and activity of retrotransposon Tf1. Mol Cell Biol 19:5768–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sistla S, Pang JV, Wang CX, Balasundaram D. 2007. Multiple conserved domains of the nucleoporin Nup124p and its orthologs Nup1p and Nup153 are critical for nuclear import and activity of the fission yeast Tf1 retrotransposon. Mol Biol Cell 18:3692–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varadarajan P, Mahalingam S, Liu P, Ng SB, Gandotra S, Dorairajoo DS, Balasundaram D. 2005. The functionally conserved nucleoporins Nup124p from fission yeast and the human Nup153 mediate nuclear import and activity of the Tf1 retrotransposon and HIV-1 Vpr. Mol Biol Cell 16:1823–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matreyek KA, Yucel SS, Li X, Engelman A. 2013. Nucleoporin NUP153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity. PLoS Pathog 9:e1003693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matreyek KA, Engelman A. 2013. Viral and cellular requirements for the nuclear entry of retroviral preintegration nucleoprotein complexes. Viruses 5:2483–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koh Y, Wu X, Ferris AL, Matreyek KA, Smith SJ, Lee K, KewalRamani VN, Hughes SH, Engelman A. 2013. Differential effects of human immunodeficiency virus type 1 capsid and cellular factors nucleoporin 153 and LEDGF/p75 on the efficiency and specificity of viral DNA integration. J Virol 87:648–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matreyek KA, Engelman A. 2011. The requirement for nucleoporin NUP153 during human immunodeficiency virus type 1 infection is determined by the viral capsid. J Virol 85:7818–7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dang VD, Benedik MJ, Ekwall K, Choi J, Allshire RC, Levin HL. 1999. A new member of the sin3 family of corepressors is essential for cell viability and required for retroelement propagation in fission yeast. Mol Cell Biol 19:2351–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levin HL. 1997. It’s prime time for reverse transcriptase. Cell 88:5–8. [DOI] [PubMed] [Google Scholar]
- 30.Levin HL. 1996. An unusual mechanism of self-primed reverse transcription requires the RNase H domain of reverse transcriptase to cleave an RNA duplex. Mol Cell Biol 16:5645–5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atwood-Moore A, Ejebe K, Levin HL. 2005. Specific recognition and cleavage of the plus-strand primer by reverse transcriptase. J Virol 79: 14863–14875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hizi A. 2008. The reverse transcriptase of the Tf1 retrotransposon has a specific novel activity for generating the RNA self-primer that is functional in cDNA synthesis. J Virol 82:10906–10910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin JH, Levin HL. 1997. A complex structure in the mRNA of Tf1 is recognized and cleaved to generate the primer of reverse transcription. Genes Dev 11:270–285. [DOI] [PubMed] [Google Scholar]
- 34.Lin JH, Levin HL. 1998. Reverse transcription of a self-primed retrotransposon requires an RNA structure similar to the U5-IR stem-loop of retroviruses. Mol Cell Biol 18:6859–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin JH, Levin HL. 1997. Self-primed reverse transcription is a mechanism shared by several LTR-containing retrotransposons [letter]. RNA 3:952–953. [PMC free article] [PubMed] [Google Scholar]
- 36.Butler M, Goodwin T, Simpson M, Singh M, Poulter R. 2001. Vertebrate LTR retrotransposons of the Tf1/Sushi group. J Mol Evol 52:260–274. [DOI] [PubMed] [Google Scholar]
- 37.SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, Bennetzen JL. 1996. Nested retrotransposons in the intergenic regions of the maize genome [see comments]. Science 274:765–768. [DOI] [PubMed] [Google Scholar]
- 38.Atwood-Moore A, Yan K, Judson RL, Levin HL. 2006. The self primer of the long terminal repeat retrotransposon Tf1 is not removed during reverse transcription. J Virol 80:8267–8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hizi A, Levin HL. 2005. The integrase of the long terminal repeat-retrotransposon tf1 has a chromodomain that modulates integrase activities. J Biol Chem 280:39086. [DOI] [PubMed] [Google Scholar]
- 40.Herzig E, Voronin N, Hizi A. 2012. The removal of RNA primers from DNA synthesized by the reverse transcriptase of the retrotransposon Tf1 is stimulated by Tf1 integrase. J Virol 86:6222–6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oz-Gleenberg I, Herzig E, Voronin N, Hizi A. 2012. Substrate variations that affect the nucleic acid clamp activity of reverse transcriptases. FEBS J 279:1894–1903. [DOI] [PubMed] [Google Scholar]
- 42.Oz-Gleenberg I, Herschhorn A, Hizi A. 2011. Reverse transcriptases can clamp together nucleic acids strands with two complementary bases at their 3′-termini for initiating DNA synthesis. Nucleic Acids Res 39:1042–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oz-Gleenberg I, Herzig E, Hizi A. 2012. Template-independent DNA synthesis activity associated with the reverse transcriptase of the long terminal repeat retrotransposon Tf1. FEBS J 279:142–153. [DOI] [PubMed] [Google Scholar]
- 44.Kirshenboim N, Hayouka Z, Friedler A, Hizi A. 2007. Expression and characterization of a novel reverse transcriptase of the LTR retrotransposon Tf1. Virology 366:263–276. [DOI] [PubMed] [Google Scholar]
- 45.Ebina H, Chatterjee AG, Judson RL, Levin HL. 2008. The GP(Y/F) domain of TF1 integrase multimerizes when present in a fragment, and substitutions in this domain reduce enzymatic activity of the full-length protein. J Biol Chem 283:15965–15974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leem YE, Ripmaster TL, Kelly FD, Ebina H, Heincelman ME, Zhang K, Grewal SIS, Hoffman CS, Levin HL. 2008. Retrotransposon Tf1 is targeted to pol II promoters by transcription activators. Mol Cell 30:98–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao X, Hou Y, Ebina H, Levin HL, Voytas DF. 2008. Chromo- domains direct integration of retrotransposons to heterochromatin. Genome Res 18:359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chatterjee AG, Leem YE, Kelly FD, Levin HL. 2009. The chromodomain of Tf1 integrase promotes binding to cDNA and mediates target site selection. J Virol 83:2675–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qi X, Sandmeyer S. 2012. In vitro targeting of strand transfer by the Ty3 retroelement integrase. J Biol Chem 287:18589–18595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dai J, Xie W, Brady TL, Gao J, Voytas DF. 2007. Phosphorylation regulates integration of the yeast Ty5 retrotransposon into heterochromatin. Molecular Cell 27:289–299. [DOI] [PubMed] [Google Scholar]
- 51.Behrens R, Hayles J, Nurse P. 2000. Fission yeast retrotransposon Tf1 integration is targeted to 5′ ends of open reading frames. Nucleic Acids Res 28:4709–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singleton TL, Levin HL. 2002. A long terminal repeat retrotransposon of fission yeast has strong preferences for specific sites of insertion. Eukaryotic Cell 1:44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cherry KE, Hearn WE, Seshie OY, Singleton TL. 2014. Identification of Tf1 integration events in S. pombe under nonselective conditions. Gene 542:221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoff EF, Levin HL, Boeke JD. 1998. Schizosaccharomyces pombe retrotransposon Tf2 mobilizes primarily through homologous cDNA recombination. Mol Cell Biol 18:6839–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoff EKF. 1997. Dissertation, Johns Hopkins University, Baltimore, MD. [Google Scholar]
- 56.Majumdar A, Chatterjee AG, Ripmaster TL, Levin HL. 2011. The determinants that specify the integration pattern of retrotransposon Tf1 in the fbp1 promoter of Schizosaccharomyces pombe. J Virol 85:519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chatterjee AG, Esnault C, Guo Y, Hung S, McQueen PG, Levin HL. 2014. Serial number tagging reveals a prominent sequence preference of retrotransposon integration. Nucleic Acids Res 42:8449–8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo Y, Levin HL. 2010. High-throughput sequencing of retrotransposon integration provides a saturated profile of target activity in Schizosaccharomyces pombe. Genome Res 20:239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arcangioli B, Copeland TD, Klar AJ. 1994. Sap1, a protein that binds to sequences required for mating-type switching, is essential for viability in Schizosaccharomyces pombe. Mol Cell Biol 14:2058–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arcangioli B, Klar AJ. 1991. A novel switch-activating site (SAS1) and its cognate binding factor (SAP1) required for efficient mat1 switching in Schizosaccharomyces pombe. EMBO J 10:3025–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Lahondes R, Ribes V, Arcangioli B. 2003. Fission yeast Sap1 protein is essential for chromosome stability. Eukaryot Cell 2:910–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mejia-Ramirez E, Sanchez-Gorostiaga A, Krimer DB, Schvartzman JB, Hernandez P. 2005. The mating type switch-activating protein Sap1 Is required for replication fork arrest at the rRNA genes of fission yeast. Mol Cell Biol 25:8755–8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krings G, Bastia D. 2005. Sap1p binds to Ter1 at the ribosomal DNA of Schizosaccharomyces pombe and causes polar replication fork arrest. J Biol Chem 280:39135–39142. [DOI] [PubMed] [Google Scholar]
- 64.Noguchi C, Noguchi E. 2007. Sap1 promotes the association of the replication fork protection complex with chromatin and is involved in the replication checkpoint in Schizosaccharomyces pombe. Genetics 175: 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zaratiegui M, Vaughn MW, Irvine DV, Goto D, Watt S, Bahler J, Arcangioli B, Martienssen RA. 2011. CENP-B preserves genome integrity at replication forks paused by retrotransposon LTR. Nature 469:112–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fedoroff NV. 2012. Presidential address. Transposable elements, epigenetics, and genome evolution. Science 338:758–767. [DOI] [PubMed] [Google Scholar]
- 67.Levin HL, Moran JV. 2011. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet 12:615–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wilson RC, Doudna JA. 2013. Molecular mechanisms of RNA interference. Annu Rev Biophys 42:217–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guzzardo PM, Muerdter F, Hannon GJ. 2013. The piRNA pathway in flies: highlights and future directions. Curr Opin Genet Dev 23:44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lejeune E, Allshire RC. 2011. Common ground: small RNA programming and chromatin modifications. Curr Opin Cell Biol 23:258–265. [DOI] [PubMed] [Google Scholar]
- 71.Reyes-Turcu FE, Grewal SI. 2012. Different means, same end-heterochromatin formation by RNAi and RNAi-independent RNA processing factors in fission yeast. Curr Opin Genet Dev 22:156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Slotkin RK, Martienssen R. 2007. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8:272–285. [DOI] [PubMed] [Google Scholar]
- 73.Hansen KR, Burns G, Mata J, Volpe TA, Martienssen RA, Bahler J, Thon G. 2005. Global effects on gene expression in fission yeast by silencing and RNA interference machineries. Mol Cell Biol 25:590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI. 2005. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37:809–819. [DOI] [PubMed] [Google Scholar]
- 75.Halic M, Moazed D. 2010. Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell 140:504–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zaratiegui M, Castel SE, Irvine DV, Kloc A, Ren J, Li F, de Castro E, Marin L, Chang AY, Goto D, Cande WZ, Antequera F, Arcangioli B, Martienssen RA. 2011. RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature 479:135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, Young SK, Furuya K, Guo Y, Pidoux A, Chen HM, Robbertse B, Goldberg JM, Aoki K, Bayne EH, Berlin AM, Desjardins CA, Dobbs E, Dukaj L, Fan L, FitzGerald MG, French C, Gujja S, Hansen K, Keifenheim D, Levin JZ, Mosher RA, Müller CA, Pfiffner J, Priest M, Russ C, Smialowska A, Swoboda P, Sykes SM, Vaughn M, Vengrova S, Yoder R, Zeng Q, Allshire R, Baulcombe D, Birren BW, Brown W, Ekwall K, Kellis M, Leatherwood J, Levin H, Margalit H, Martienssen R, Nieduszynski CA, Spatafora JW, Friedman N, Dalgaard JZ, Baumann P, Niki H, Regev A, Nusbaum C. 2011. Comparative functional genomics of the fission yeasts. Science 332:930–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yamanaka S, Mehta S, Reyes-Turcu FE, Zhuang F, Fuchs RT, Rong Y, Robb GB, Grewal SI. 2013. RNAi triggered by specialized machinery silences developmental genes and retrotransposons. Nature 493:557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee NN, Chalamcharla VR, Reyes-Turcu F, Mehta S, Zofall M, Balachandran V, Dhakshnamoorthy J, Taneja N, Yamanaka S, Zhou M, Grewal SI. 2013. Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance. Cell 155: 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Anderson HE, Wardle J, Korkut SV, Murton HE, Lopez-Maury L, Bahler J, Whitehall SK. 2009. The fission yeast HIRA histone chaperone is required for promoter silencing and the suppression of cryptic antisense transcripts. Mol Cell Biol 29:5158–5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Greenall A, Williams ES, Martin KA, Palmer JM, Gray J, Liu C, Whitehall SK. 2006. Hip3 interacts with the HIRA proteins Hip1 and Slm9 and is required for transcriptional silencing and accurate chromosome segregation. J Biol Chem 281:8732–8739. [DOI] [PubMed] [Google Scholar]
- 82.Casola C, Hucks D, Feschotte C. 2008. Convergent domestication of pogo-like transposases into centromere-binding proteins in fission yeast and mammals. Mol Biol Evol 25:29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smit AF. 1996. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev 6:743–748. [DOI] [PubMed] [Google Scholar]
- 84.Tudor M, Lobocka M, Goodell M, Pettitt J, O’Hare K. 1992. The pogo transposable element family of Drosophila melanogaster. Mol Gen Genet 232:126–134. [DOI] [PubMed] [Google Scholar]
- 85.Masumoto H, Nakano M, Ohzeki J. 2004. The role of CENP-B and alpha-satellite DNA: de novo assembly and epigenetic maintenance of human centromeres. Chromosome Res 12:543–556. [DOI] [PubMed] [Google Scholar]
- 86.Cam HP, Noma K, Ebina H, Levin HL, Grewal SIS. 2008. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature 451:431–U432. [DOI] [PubMed] [Google Scholar]
- 87.Lorenz DR, Mikheyeva IV, Johansen P, Meyer L, Berg A, Grewal SI, Cam HP. 2012. CENP-B cooperates with Set1 in bidirectional transcriptional silencing and genome organization of retrotransposons. Mol Cell Biol 32:4215–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tanaka A, Tanizawa H, Sriswasdi S, Iwasaki O, Chatterjee AG, Speicher DW, Levin HL, Noguchi E, Noma K. 2012. Epigenetic regulation of condensin-mediated genome organization during the cell cycle and upon DNA damage through histone H3 lysine 56 acetylation. Mol Cell 48:532–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baum M, Clarke L. 2000. Fission yeast homologs of human CENP-B have redundant functions affecting cell growth and chromosome segregation. Mol Cell Biol 20:2852–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McClintock B. 1984. The significance of responses of the genome to challenge. Science 226:792–801. [DOI] [PubMed] [Google Scholar]
- 91.Bolton EC, Boeke JD. 2003. Transcriptional interactions between yeast tRNA genes, flanking genes and Ty elements: a genomic point of view. Genome Res 13:254–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kinsey PT, Sandmeyer SB. 1991. Adjacent pol II and pol III promoters: transcription of the yeast retrotransposon Ty3 and a target tRNA gene. Nucleic Acids Res 19:1317–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sehgal A, Lee CY, Espenshade PJ. 2007. SREBP controls oxygen-dependent mobilization of retrotransposons in fission yeast. PLoS Genet 3:e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Feng G, Leem YE, Levin HL. 2013. Transposon integration enhances expression of stress response genes. Nucleic Acids Res 41:775–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, Bowman S, Brooks K, Brown D, Brown S, Chillingworth T, Churcher C, Collins M, Connor R, Cronin A, Davis P, Feltwell T, Fraser A, Gentles S, Goble A, Hamlin N, Harris D, Hidalgo J, Hodgson G, Holroyd S, Hornsby T, Howarth S, Huckle EJ, Hunt S, Jagels K, James K, Jones L, Jones M, Leather S, McDonald S, McLean J, Mooney P, Moule S, Mungall K, Murphy L, Niblett D, Odell C, Oliver K, O’Neil S, Pearson D, Quail MA, Rabbinowitsch E, Rutherford K, Rutter S, Saunders D, Seeger K, Sharp S, Skelton J, Simmonds M, Squares R. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415:871–880. [DOI] [PubMed] [Google Scholar]
- 96.Sugiyama T, Sugioka-Sugiyama R. 2011. Red1 promotes the elimination of meiosis-specific mRNAs in vegetatively growing fission yeast. EMBO J 30:1027–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Leupold U. 1993. The origins of Schizosaccharomyces pombe genetics. In: Hall MN, Linder P. eds. The Early Days of Yeast Genetics New York: Cold Spring Harbor Laboratory Press; p 125–128. [Google Scholar]


