Abstract
DNA mismatch repair (MMR) corrects DNA base pairing errors that occur during DNA replication. MMR catalyzes strand-specific DNA degradation and resynthesis by dynamic molecular coordination of sequential downstream pathways. The temporal and mechanistic order of molecular events is essential to insure interactions in MMR that occur over long distances on the DNA. Biophysical real-time studies of highly conserved components on mismatched DNA have shed light on the mechanics of MMR. Single-molecule imaging has visualized stochastically coordinated MMR interactions that are based on thermal fluctuation-driven motions. In this review, we describe the role of diffusivity and stochasticity in MMR beginning with mismatch recognition through strand-specific excision. We conclude with a perspective of the possible research directions that should solve the remaining questions in MMR.
Graphical Abstract
Introduction
A recent NMR analysis demonstrated that a G/T mismatch could form a Watson-Crick-like geometry by a tautomerization and/or ionization in the ~1 ms time scale [1]. The participating nucleotides temporarily converted into alternative chemical states, which allows the base-pairing error to partner into the Watson-Crick geometry. These observations suggested that a DNA polymerase might continue synthesis normally extemporaneously leaving behind such base-pair errors that have escaped recognition by the intrinsic proofreading activity [2]. Fortunately, the majority of organisms maintain a mismatch repair (MMR) system that recognizes and repairs these base-pairing errors enhancing the overall fidelity of DNA replication up to 1000-fold [3, 4]. The combination of polymerase editing and MMR systems in human cells results in less than one error per cell division [5], ultimately insuring the integrity of the genome.
The core MutS homologs (MSHs) and MutL homologs (MLH/PMSs) that accomplish MMR have been highly conserved throughout evolution. Mutations of the human MSH2 and MLH1 genes are the most frequent cause of the common hereditary cancer predisposition Lynch syndrome or hereditary non-polyposis colorectal cancer (LS/HNPCC) [6, 7]. These and partner MMR genes are also associated with a variety of sporadic colorectal, endometrial, ovarian and upper urinary tract tumors [8–10]. While genetic studies in 1960’s identified most of the players [11, 12], it was the biochemical reconstitution of E. coli MMR in the 1980’s that led to significant understanding of the MMR mechanism [13]. Since then, structural and biophysical analysis has contributed to the development of updated MMR models. Most recently, single-molecule manipulation and real-time imaging have successfully explored a variety of unresolved questions in MMR. These studies demonstrated that molecular coordination of multiple components in the right place and time is essential for efficient repair. Single-molecule imaging also clearly showed that the stochastic coordination through thermal fluctuation-driven motion controls MMR. Nevertheless, there still remain a number of unknown details in the MMR mechanics. This review contains an overview of the MMR, with a strong focus on the current challenges to future major advances. We conclude with a perspective of the research directions and the prospects that we expect to see in the MMR field in the near future and beyond.
Overview of mismatch repair processes
The central outcome of MMR is a DNA mismatch-dependent excision of the newly synthesized DNA strand that begins at a strand break that may be several thousand nucleotides from the mismatch and terminates just past the mismatch. The overall MMR reaction has been described extensively in previous review articles (see recent reviews [5, 14]). To initiate MMR, a dimer (bacteria, archaea) or heterodimer (eukaryotes) of MSH searches the genomic duplex DNA culminating in binding to the base-pair error (Fig. 1, I Mismatch Recognition). Presumably because of the increased complexity of the genome, eukaryotes have evolved two MSH heterodimers. The MSH2-MSH6 heterodimer primarily recognizes single base pair [15, 16] or small insertion/deletion nucleotides, while the MSH2-MSH3 heterodimer binds an overlapping set of small insertion/deletion nucleotides as well as significantly larger insertion/deletion loop-type (IDL) mismatches [17, 18]. Remarkably, only human MSH2 and MSH6 have been shown to cause LS/HNPCC, suggesting that single base pair mismatches or some other lesion processed by MSH2-MSH6 is responsible for tumorigenesis [19].
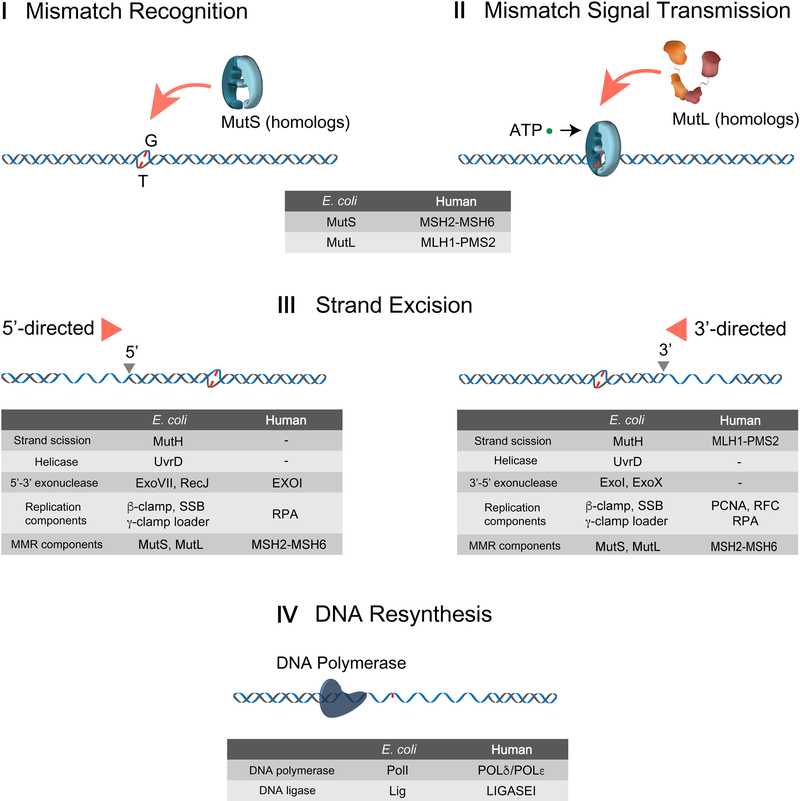
Figure 1. DNA mismatch repair processes.
I Mismatch Recognition: MutS homologs (MSHs) search and recognize the mismatch. → II Mismatch Signal Transmission: Mismatch finding is transmitted to a downstream site by MSH and MLH/PMS, for which ATP is required. → III Strand Excision: Strand excision is initiated at a 5’-end (5’-directed strand excision) or a 3’-end (3’-directed strand excision) to release the mismatch. → IV DNA Resynthesis: After the mismatch is eliminated, DNA polymerase fills the gap and Ligase joins two ends of single-stranded DNAs (ssDNAs). Components participating in each MMR process are listed on the table.
MSH proteins are members of the AAA (ATPase Associated with diverse cellular Activities) family and appear to retain at least one ADP while in search of a mismatch [20–22]. Mismatch binding triggers the dissociation of the bound ADP, allowing both subunits to bind ATP [23]. The ATP-bound MSH physically interacts with the MLH/PMS proteins, which are the first downstream MMR component (Fig.1, II Mismatch Signal Transmission) [13, 24, 25]. MLH/PMS proteins exist as a homodimer (prokaryotes and archaea) or heterodimer (eukaryotes) and contain a GHKL (DNA Gyrase, Hsp90, Histidine Kinase and MutL) ATP binding and hydrolysis motif [26]. The role of ATP binding and hydrolysis by MLH/PMS during MMR has been puzzling for decades.
While MSH and MLH/PMS are evolutionarily conserved from bacterial to human, the strand scission and the strand excision that initiate and drive MMR are distinct between a subset of gram-negative enteric bacteria that includes E. coli, and all other bacteria, archaea and eukaryotes [27]. In E. coli, the MutL that is recruited by ATP-bound MutS activates the MutH endonuclease, which cleaves the unmethylated GATC strand within a hemi-methylated GATC sequences (Fig. 1, III Strand Excision) [25, 28]. The mechanism associated with the distant communication between the mismatch and the hemimethylated GATC site has been highly controversial for decades [29]. Nevertheless, the MutH strand scission has been proposed to serve as an entry site for the UvrD helicase and RecJ or ExoVII single stranded DNA (ssDNA) exonuclease in the 3’→5’ direction or ExoI or ExoX ssDNA exonuclease in the 5’→3’ direction (Fig. 1, III Strand Excision) [30]. This bidirectional strand excision of the newly replicated strand terminates just past the mismatch.
In prokaryotes lacking the MutH gene, the mechanism of strand specific incision and bidirectional excision remains poorly understood. Similarly, the mechanism of bidirectional excision in eukaryotes is a puzzle since the unidirectional 5’→3’ exonuclease EXOI has been the only exonuclease linked to eukaryotic MMR (Fig. 1, III Strand Excision) [31–34]. Interestingly, the 5’→3’ MMR excision reaction does not require an MLH/PMS in vitro [35], while the MLH/PMS is essential for the 3’→5’ excision reaction. In bacteria that do not utilize MutH as well as archaea and eukaryotes the MLH/PMS protein contains a cryptic endonuclease domain [36]. The MLH/PMS endonuclease appears to play a functional role in the 3’→5’ excision reaction and is activated by the replication processivity factor β-Clamp in prokaryotes and PCNA in archaea and eukaryotes [36–38]. Mutation of conserved residues within the endonuclease motif of the S. cerevisae PMS1 and the MutL of gram-positive bacteria exhibits a mutator phenotype consistent with an MMR defect and an essential role in mismatch excision [37–39]. However, the detailed mechanics of the MLH/PMS strand-specific incision remains unknown. DNA polymerase I in E. coli and DNA polymerase δ/ε in eukaryotes resynthesize the resulting DNA gap following MMR excision and DNA ligase seals any remaining strand scissions (Fig. 1, IV DNA Resynthesis).
The mechanics of the MSH mismatch recognition
In recent decades, MSH structural and biophysical properties have been largely resolved by X-ray crystallography and a variety of single-molecule imaging studies. The structures of prokaryotic MutS and eukaryotic MSH (MSH2-MSH6) are remarkably similar [40–42]. MSH proteins that are stably bound to the mismatch appear to interrogate the unpaired nucleotide of one strand by intercalating a conserved Phe residue into the DNA. The stacking interactions between this Phe residue and adjacent DNA nucleotides is further stabilized by hydrogen bonding between Glu residues in MSH and DNA phosphates from the minor groove. This binding interface triggers a sharp 45–60° bend of DNA towards to the major groove, which appears to further stabilize the MSH-DNA binding. ATP-mediated conformational transitions from an “open” to a “closed” clamp were first visualized by electron microscopy with uranyl-acetate stained human MSH2-MSH6 [43, 44] and have been detailed by deuterium exchange mass spectrometry studies of the E. coli MutS and human MSH2-MSH6 [45, 46]. A crystal structure of the E.coli MutS resulting from the ATP-mediated conformational transitions has only recently been resolved [47]. Together these structural studies showed that MSH switches between at least two distinct conformations as originally predicted for the human MSH2-MSH6 [43, 44].
Single-molecule real-time tracking has explored the mechanical issues associated with the transition of an MSH from the mismatch search to the formation of a closed sliding clamp on the mismatched DNA [48, 49]. These studies used Thermus aquaticus (Ta) MutS containing a single Cys residue that was labeled with a fluorophore by maleimide chemistry [49]. Similar studies have been reported for the S. cerevisae or human MSH2-MSH6 that were visualized by a fluorescent quantum dot-labeled antibody or a protein fluorophore, respectively [50, 51]. Single DNA molecules immobilized on a flow-cell surface were used as targets for the MSH proteins (Fig. 2a) [52, 53]. Physical analyses of diffusion coefficient as a function of ionic strength and the use of emission polarization analysis of the fluorophore attached to TaMutS provided insight into the diffusion motions of an MSH along DNA (Fig. 2b, c, d) [52].
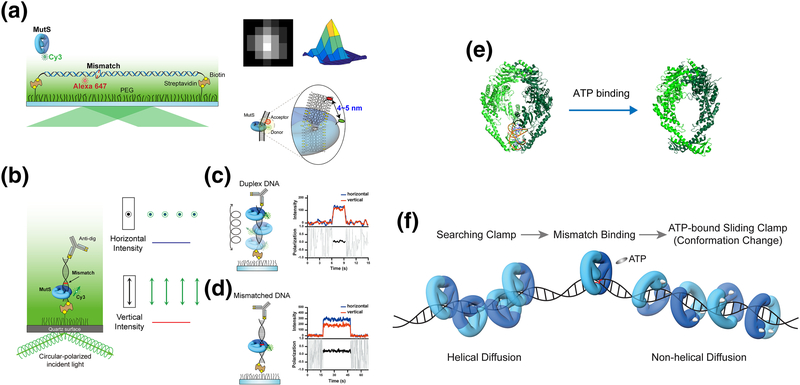
Figure 2. Conformational switch of MutS.
(a) Mismatched DNA containing a dG/dT mismatch or an unpaired thymine (dT) mismatch was stretched and immobilized on a flow-cell surface with defined hydrodynamic drag by applying a laminar flow. A single acceptor fluorophore was attached to the mismatched DNA to monitor the mismatch binding by MutS that was tagged with a single donor fluorophore using a single-molecule Förster Resonance Energy Transfer assay. The time-dependent position of MutS on the DNA was determined with an accuracy of a few nanometers using 2D Gaussian-fitted center of intensity profiles [104, 105]. (b) A schematic representation of the single-molecule fluorescence polarization system to measure the polarization of Cy3-MutS. (c) The emission signals of Cy3-MutS on duplex DNA was depolarized. (d) The emission signals of Cy3-MutS on mismatched DNA was strongly polarized, which indicates that the Cy3-MutS was trapped at the mismatch. (e) The crystal structures of E. coli MutS bound to a mismatch and ATP-bound MutS. (PDB: 1e3M [40], mismatch-bound MutS and 5akc [47], ATP-bound MutS). (f) TaMutS forms two different sliding clamps on mismatched DNA: a mismatch searching MutS clamp (~1 s) that diffuses along the DNA in a continuous contact with the DNA backbone (helical diffusion) and a highly stable (~600 s) ATP-bound sliding clamp in a discontinuous contact with the DNA backbone (non-helical diffusion).
Tracking single MSH proteins revealed that the mismatch search is initiated by nonspecific 3D diffusion-dependent DNA binding followed by 1D motion along the DNA. The lifetime on the DNA decreased while the diffusion coefficient did not vary with increasing ionic strength [52, 53]. These observations are consistent with the conclusion that the MSH moves along the DNA while in a continuous contact with the DNA backbone [54]. The rotational dynamics of MutS was determined by detecting the distinct emission anisotropic signals of the single fluorophore attached to TaMutS on duplex DNA or mismatched DNA (Fig. 2c, d). These single molecule emission polarization studies unmistakably suggested that an MSH searches for a mismatch by 1D translation-coupled rotational diffusion [55]. At physiological ionic conditions the MSH appears to search for ~1 s, which combined with its characteristic diffusion coefficient (0.034 μm2/s) suggests that it is capable of examining ~700 bp of naked DNA per event [49, 56].
Mismatch recognition halts the 1D helical diffusion of MSH at the site of the mismatch [52, 53]. These observations strongly suggest that MSH proteins identify a mismatch by a facilitated diffusion mechanism [57, 58]. Interestingly, the mismatch recognition by MSH is influenced by the sequence of mismatched nucleotides, nearest neighbor nucleotides, and lesions [16, 59, 60]. Previous studies showed that mismatched nucleotides increase DNA flexibility of the mismatched region by weakening nucleotide base stacking interactions, which can then introduce a deformation in the double-stranded DNA (dsDNA) helical structure [61–63]. The stalling of an MSH at the mismatch may result when a diffusing MSH is captured by these local structural alterations rather than specific recognition of the mismatched nucleotides [64] or base-stacking alterations [65]. The recognition of an altered configuration rather than the mismatch itself may explain why MSH proteins can be activated by a wide-range of mismatches, lesions, and insertion/deletion loop-type (IDL) structures [16–18].
In the absence of ATP the MSH binds to the mismatch for approximately 30 s [49]. Early studies demonstrated that an MSH bound to a mismatch has a higher affinity for ATP than ADP [23]. Moreover, when the MSH binds ATP, it releases the mismatch and slides off short oligonucleotides [43]. Single-molecule imaging showed that in the presence of ATP the MSH paused for ~ 3 s and then dissociated from the mismatch via 1D diffusion along the DNA. The conformational transition of an MSH searching for a mismatch to an ATP-bound MSH moving along the DNA was found to alter the 1D diffusion characteristics: ATP-bound MSH diffused with random rotation in discontinuous contact with the DNA backbone compared to 1D rotation-coupled diffusion of a searching MSH [52]. The diffusion mechanics were inferred from direct single molecule observation of the accelerated diffusion induced by the electrostatic screening of increased ionic strength as well as the depolarized emission of the single fluorophore attached to TaMutS on a DNA molecule that has an effective 1D diffusion space shorter than a helical pitch (10.5 bp) [52]. A recent structure of ATP-bound MutS that forms a loose ring around duplex DNA explains the diffusion mechanics of ATP-bound MutS (Fig. 2e), which was observed by single-molecule studies [47]. Together these observations imply that the freely diffusing sliding clamp conformation of the ATP-bound MSH facilitates the interaction with MLH/PMS and the transmission of mismatch recognition to a distant site for the initiation of the strand excision (Fig. 2f).
Distant communications of mismatch recognition
The most critical question in the mechanics of MMR has been how mismatch recognition by an MSH is communicated with a distant downstream site [29]. After the mismatch recognition by MSH, MLH associated with the MSH has been suggested to eventually activate a strand breaking or initiate the strand excision at a distant site (Fig. 1). There have been extensive experimental efforts to address the mechanism of MMR that have resulted in several different models for MMR [29]. Based on electron microscopy and ATPase studies, the ATP Hydrolysis-Dependent Translocation model envisaged that cycling hydrolysis of ATP by an MSH guides the directional movement of the MSH-MLH/PMS complex from the mismatch to the downstream target site [66, 67]. The Molecular Switch Sliding Clamp model was formulated on the observation that mismatch recognition resulted in ADP→ATP exchange much like G-protein molecular switch that ultimately formed an ATP-bound sliding clamp [23, 43]. In this model the complex of MSH and MLH/PMS was proposed to diffuse along the DNA to the strand scission site driven by thermal fluctuation [23, 25, 43, 68]. In contrast to these models proposing movement of an MSH-MLH/PMS complex from the mismatch, the Static Transactivation model was based on in silico marrying of the E.coli MutS and MutL proteins on a mismatch and then proposing a direct interaction with a MutH bound to the strand breaking site via looping of the intervening DNA [69]. This Static Transaction model has been largely dismissed since artificial blocks located on the DNA between the mismatch and the hemimethylated GATC site inhibited MutH activation; a result that suggest some DNA translocation is necessary [70].
Fluorescence imaging of MMR proteins in live S. cerevisiae cells revealed that MSH2-MSH6 was localized at the replication centers during the synthesis phase (S phase) and MLH1-PMS1 formed nuclear foci dependent on MSH2-MSH6 but rarely colocalized with MSH2-MSH6 [71]. One possible interpretation suggests that MSH2-MSH6 bound to a mismatch recruits multiple MLH1-PMS1 proteins [71]. The MLH/PMS Polymerization model proposes that a polymer chain of MLH/PMS proteins is loaded from an MSH bound to the mismatch to the distant excision-initiation site. A single-molecule FRET experiment with TaMutS and TaMutL appeared to show TaMutL trapping TaMutS at or near a mismatch after the recognition by TaMutS [72]. Similar to the S. cerevisae cellular studies, these single-molecule observations appeared consistent with multiple MutL proteins associated with MutS bound to the mismatch, leading to MutL polymerization; although an obligatory FRET requirement at the mismatch might have biased these observations [72].
On the other hand, Gorman et al. visualized individual S. cerevisiae MSH2MSH6 and MLH1-PMS1 on a λ-phage-based DNA (48. 5 kb) that contained three repetitive G/T mismatches [53]. The single-particle real-time imaging suggested that MLH1-PMS1 is associated with MSH2-MSH6 bound to the mismatch and then the complex diffuses away from the mismatch in an ATP hydrolysis-independent manner, supporting the Molecular Switch Sliding Clamp model. These studies reduce the likelihood of the ATP Hydrolysis-Dependent Translocation and the Static Transaction models. However, the MutL polymerization model could not be eliminated easily because the concentration of MLH1-PMS1 (5 ~ 20 nM) in the experimental conditions may be much lower than its physiological concentration. Moreover, Quantum-dot (~ 25 nm in diameter) probes attached to MSH2-MSH6 and MLH1-PMS1 might spatially hinder the polymerization of MLH1-PMS1 (~ 10 nm). In spite of the extensive efforts to solve these controversial issues using single-molecule real-time imaging, until recently no clear understanding of the collaborative mechanism of MSH and MLH/PMS was forthcoming in any organism.
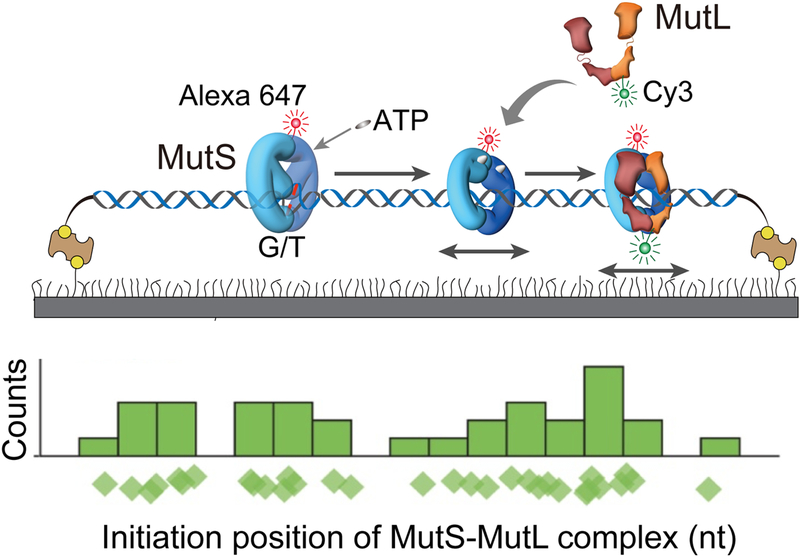
A ternary complex of MMR components was recently reconstituted on mismatched DNA using Alexa Fluor or Cyan dye (~ 3 nm)-tagged E. coli MutS, MutL and MutH [73]. The real-time tracking of individual MutS, MutL and MutH proteins on the 17.3 kb-mismatched DNA enabled visual analysis of the dynamic characteristics of each protein and intermediate interactions between these MMR proteins (Fig. 3). Remarkably, the processes of mismatch binding transmission appeared totally stochastic. In these studies, MutL was found to associate with an ATP-bound MutS sliding clamp after it is released from the mismatch (Fig. 3; The distribution of initial position of MutS-MutL). Interestingly, the MutS-MutL complex oscillates between association and dissociation states (see Liu et al. review in this special issue for details). Moreover, a previously unidentified stable MutL sliding clamp was formed that diffuses rapidly along DNA. ATP binding by MutL as well as an ATP-bound MutS was required to form the MutL sliding clamp. The MutS-MutL complex or an ATPbound MutL sliding clamp recruits MutH forming a ternary complex of MutS-MutL-MutH that is capable of searching for a target hemimethylated GATC site by 1D diffusion along DNA in a continuous contact with the DNA backbone (see Liu et al. review in this special issue for details). Interestingly, the lifetime of an ATP-bound MutL sliding clamp on DNA (~ 800 s) is four-fold longer than that of ATP-bound MutS (~ 200 s). Thus, multiple MutL sliding clamps can be loaded by an ATP-bound MutS, which may result in an excess number of MutL sliding clamps diffusing along DNA. This observation appears to explain the surplus of MLH1-PMS1 foci formed far beyond MSH2-MSH6 in live S. cerevisiae cells. These studies strongly suggested that MSH and MLH/PMS sliding clamps are capable of reaching the excision initiation site by diffusion, at least in E. coli. Such a sequential stochastic coordination guiding directional DNA mismatch repair is uniquely consistent with the Molecular Switch Sliding Clamp model for MMR (see Liu et al. this special issue).
Figure 3. Dynamics of the MutS-MutL complex [73].
E coli MutS and MutL were imaged on doubly-tethered 17.3 kb mismatched DNAs in single-molecule total internal reflection fluorescence microscopy. The initial locations MutS-MutL complexes were randomly distributed on extended DNA substrates.
Stochastic control of strand excision
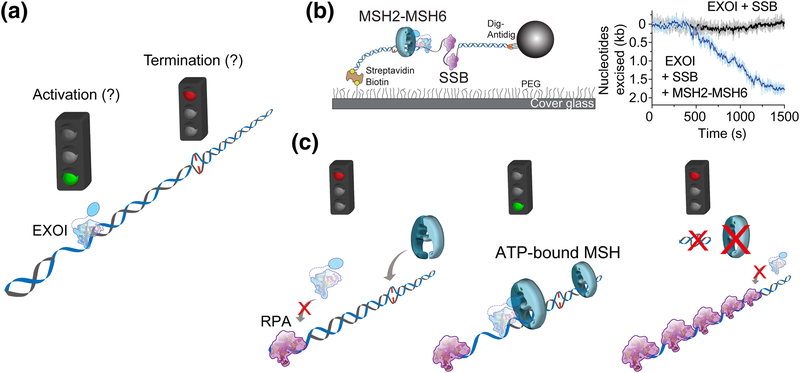
With the exception of E. coli, most of prokaryotes, archaea and eukaryotes do not require a MutH-dependent strand breaking to initiate strand excision. E.coli MMR also utilizes the UvrD helicase regardless of the excision direction. Yet there is no apparent helicase requirement for MMR that does not use MutH. Perhaps more importantly, as suggested above, the component requirements for MMR initiated from the 3’-direction are different from MMR initiated from the 5’-direction. Hypothetically, this could reflect a difference in MMR in the leading and lagging strand of a replication fork [74]. Nevertheless, the mismatch is eliminated through the excision of the newly replicated strand to just past the mismatch. However, the mechanism of how the strand excision stops is not understood (Fig. 4a). This question is general for the mechanism of strand excision in all organisms because there does not appear to be any MMR component capable of recognizing the mismatch after an ATP-bound MSH sliding clamp is formed.
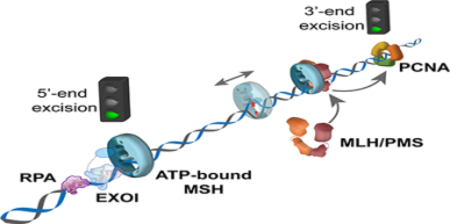
Figure 4. Eukaryotic 5’-directed excision [51].
(a) The initiation and termination of EXOI-dependent strand excision. (b) A single-molecule flow-stretching assay to visualize the strand transition of dsDNA to ssDNA [106, 107]. Time trajectories of the EXOI exonuclease activity in the presence of only single strand DNA binding proteins (SSBs) or both MSH2-MSH6 and SSB, respectively. (c) A model of 5’-directed excision reaction.
To our knowledge the human 5’-directed excision is the only MMR process that has been examined in real-time on single mismatched DNA [51]. These studies were accomplished by monitoring the production of ssDNA from dsDNA that was linked to the surface and then extended by a slow laminar-flow force on a polystyrene bead linked to the opposite DNA end (Fig. 4b) [51]. Under these conditions the production of ssDNA by MMR excision shortens the DNA length [75], which can be monitored at high resolution (~120 nt) in real-time via movement of the polystyrene bead [51].
Human EXOI spontaneously initiates 5’→3’ excision from any 5’-strand break. A single EXOI excises a single DNA strand at a rate of ~3.5 nt/s for just over 1 knt [51]. The addition of RPA dramatically inhibits EXOI 5’→3’ exonuclease activity (Fig. 4b, c; left). However, EXOI will resume 5’→3’ excision in the presence of human MSH2-MSH6 and ATP (Fig. 4b). Remarkably, the excision rate and processivity when one MSH2-MSH6 sliding clamp was present is nearly identical to EXOI alone [51]. These results are consistent with the conclusion that the ATP-bound MSH2-MSH6 merely tethers the EXOI to the 5’-end but does not alter its fundamental activity [51]. This progression appears extraordinarily similar to the tethering of polymerases by the replication processivity factors β-Clamp (bacteria) or PCNA (archaea and eukaryotes) [76, 77]. As predicted by the original biochemical studies [43] increasing the concentration of MSH2-MSH6 resulted in more sliding clamps on the mismatched DNA, and significantly longer excision tracts. Taken as a whole, these studies are consistent with a model where ATP-bound MSH2-MSH6 sliding clamp formed at the mismatch diffuses along DNA and then physically interacts with EXOI at the 5’-end, which results in the strand excision by EXOI in the presence of RPA (Fig. 4c; middle). A single MSH2-MSH6/EXOI complex may spontaneously dissociate after degrading ~1 knt. However, since multiple MSH2-MSH6 sliding clamps can be loaded at the mismatch, the next closest ATP-bound MSH2-MSH6 sliding clamp reinitiates the strand excision with another EXOI at the newly located 5’-end (Fig. 4c; middle). This animated process continues until the mismatch is removed. In the absence of a mismatch no additional MSH2-MSH6 sliding clamps may be loaded, allowing RPA to halt any further EXOI excision (Fig. 4c; right). In this model ATP-bound MSH sliding clamps provide a platform that stabilizes EXOI at the 5’-end of an ss/dsDNA junction via well-known protein-protein interaction domains [78]. Moreover, initiation and termination of the MMR strand excision are stochastically controlled by the number of ATP-bound MSH sliding clamp.
An MLH/PMS complex is not essential for the 5’-directed excision reaction in eukaryotic MMR [79, 80]. However, the addition of human MLH1-PMS2 to these single molecule reactions appears to alter excision termination to just past the mismatch similar to previous studies [35]. The molecular mechanism of how MLH1-PMS2 inhibits runaway excision by the MSH2-MSH6 and EXOI complex during MMR is unknown. One clue to this mechanism was the observation that MLH1-PMS2 appears to modulate the number of ATP-bound MSH2-MSH6 sliding clamps capable of initiating the EXOI excision reaction [51]. However this observation cannot rule out the possibility of regulated loading of MSH2-MSH6 at the mismatch or the titration of an excision component such as EXOI independent of the mismatch by MLH1-PMS2 [78]. Importantly, the regulatory role of MLH1-PMS2 in 5’-directed excision is likely to be significantly different from its catalytic role of its intrinsic endonuclease on 3’-directed excision [36]. Stochastic control of human 5’-MMR termination past the mismatch by ATP-bound MSH sliding clamp may easily be applied to 3’-directed MMR excision as well as strand excision in the other organisms. In all these cases it would be the number of MSH sliding clamps loaded at the mismatch that would determine the initiation and extent of MMR excision.
Concluding Perspectives
Single molecule imaging studies are capable of visualizing molecular dynamics in real-time with nm accuracy. Moreover, these studies may detect intermediate and rare populations of molecules that help decipher the biophysical mechanics. These advantages have facilitated our understanding of a variety of biological problems. For the last ten years, MMR mechanics has been explored by various single-molecule techniques such as single-molecule FRET [49, 72, 81, 82], fluorescence single-particle tracking [50, 52, 53, 73, 83], flow-stretching analysis [51, 81], and force spectroscopy [84–86]. They have successfully revealed the detailed mechanics of mismatch searching/recognition, mismatch signal transmission by MSH and MLH/PMS to a distant site, and the termination of strand excision. However, there remain a number of important questions in MMR including the detailed mechanics of the 3’-directed MMR excision.
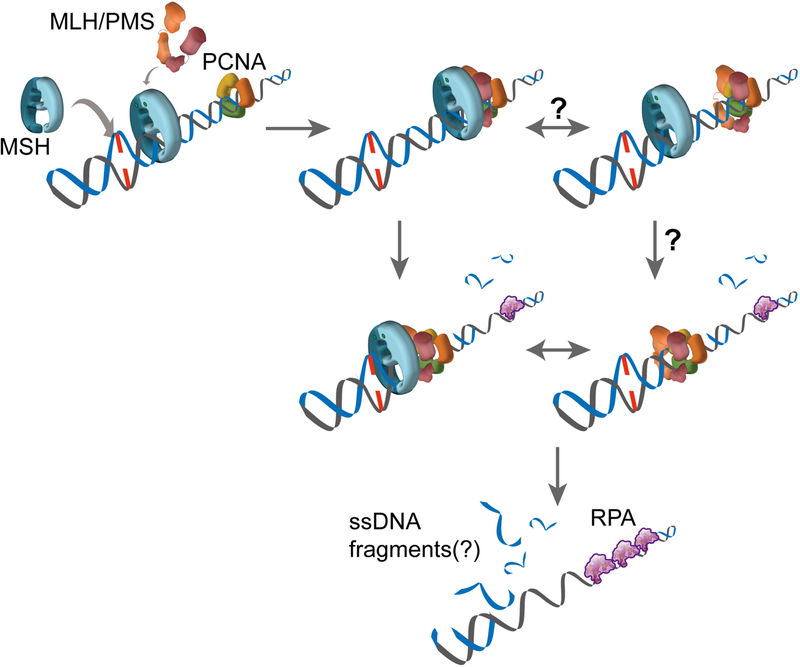
Perhaps the most puzzling process in 3’-directed strand excisions is the activation of the cryptic MLH/PMS endonuclease that depends on mismatch recognition and a 3’-end (Fig. 1; III Strand Excision). Based on the Molecular Switch Sliding Clamp model, a hypothetical model for 3’-MMR excision may be considered where the complex of ATP-bound MSH and MLH/PMS sliding clamps diffuses along DNA and then interacts with PCNA (Fig. 5). PCNA may be associated with the 3’-end of the leading strand or it may diffuse along the template (Fig. 5) [87]. This interaction is proposed to activate the cryptic MLH/PMS endonuclease activity to cleave the newly replicated DNA strand [36–38]. We imagine that this activation results in the production of short ssDNA fragments that might be released by thermal energy and/or digested from a 5’-end by EXOI. As with the 5’-directed strand excision reactions, these processes are likely to be iterative and stochastic utilizing multiple complexes of ATP-bound MSH and MLH/PMS sliding clamps until the mismatch is released.
Figure 5. Eukaryotic 3’-directed excision.
The EXOI-independent strand excision is likely to be performed by the following sequential interactions: ATP-bound MSH → MLH/PMS sliding clamp → MLH/PMS endonuclease activity triggered by PCNA → Multiple strand scissions on the newly replicated strand until the mismatch is removed → Short fragments of ssDNA are released.
Regardless of the hypothetical model, the mechanics of this reaction requires substantial additional studies to resolve the important biophysical questions. How does MLH/PMS recognize and incise the newly replicated strand? Is the ternary complex of MSH-MLH/PMS-PCNA required to activate MLH/PMS endonuclease? Can the MLH/PMS sliding clamp alone trigger its endonuclease activity? Moreover, whether the MSH-MLH/PMS complex continuously interacts with PCNA or MSH delivers one or multiple MLH/PMS to PCNA during this process is also unknown. The interface of PCNA interacting with PCNA Interacting protein (PIP) motifs is in the direction of the synthesis [88, 89]. Since the mismatch is located on the opposite side of the PIP motif-coupling surface, the orientation between MLH/PMS and PCNA must be resolved. These questions are likely to only be solved by single-molecule studies. Clearly successful reconstitution of 3’-directed MMR on single DNA molecules should finally solve the three-step processes of mismatch recognition, mismatch signal transmission, and strand excision.
Finally, to fully understand the mechanics of MMR, it is crucial to study MMR coupled with DNA replication in living cells. Fluorescent protein-fused MMR components have been visualized in living bacterial [90–94] and eukaryotic cells [71, 95–97]. However, these studies did not achieve sufficient resolution to image individual proteins. Recently, individual MutS fused to the photactivatable fluorescent protein (PAmcheery1) was successfully visualized in living B. subtilis cells using photoactivated localization microscopy [98]. Yet, at the current stage of single-molecule fluorescence imaging in living cells, it is difficult to visualize the MMR processes in detail during DNA replication. Recent advances in single-molecule imaging techniques and the endogenous gene tagging with fluorescent proteins using CRISPR/Cas9 for single-molecule imaging in cells [99] available for probing MMR function should enable visualization of MMR function within live cells in many organisms. Single-molecule MMR studies in live cells will likely provide the spatiotemporal organization of MMR and additional insights into the kinetics and function of MMR.
Taken together, the single-molecule studies described above strongly indicate that MMR is coordinated by the random diffusive motions of multiple MMR components. However, conditions with purified proteins and constructed DNA molecules in vitro are almost certainly different from the environments inside cells where DNA binding proteins such as histone octamers may place barriers to the diffusive motion of MMR components. For example, purified MMR proteins and nuclear extracts failed to repair the mismatches in a chromatin structure [100, 101]. These observations suggest that a compact chromatin may inhibit the mismatch recognition by MSH and/or the communications of mismatch recognition with the excision initiation site. However, during DNA replication nucleosomes are temporarily dissembled near the replication fork where MMR appears to be at least partly tethered by PCNA and is modulated by histone posttranslational modifications [102]. Interestingly, ATP-bound MSH sliding clamps have been found to disassemble nucleosomes [103]. This observation suggests that there may be temporary free space along the DNA near the mismatch that could be available for MMR in cells. Clearly, understanding the coordination of MMR by 1D thermal diffusion of MSH and MLH-PMS proteins in the cellular environment is an important avenue of future MMR investigations.
Research Highlight.
MSH (MutS homologs) bound to a mispaired nucleotide switches its conformation to a sliding clamp by ATP binding.
Freely diffusing ATP-bound MSH sliding clamp recruits MLH/PMS (MutL homologs).
The diffusing complex of ATP-bound MSH and MLH/PMS activates strand excision at a distant strand break.
ATP-bound MSH sliding clamp is a key player for the initiation and termination of DNA mismatch repair.
The stochastic coordination by thermal diffusion of mismatch repair components eventually results in robust repair events.
Acknowledgements
We thank the editors for inviting us to this exciting project entitled “Plasticity of Multi-Protein Complexes”. This research was supported by Global Research Lab Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Science and ICT NRF-2017K1A1A2013241 (J.-B.L.) and NIH grant CA67007 (R.F.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Kimsey IJ, Szymanski ES, Zahurancik WJ, Shakya A, Xue Y, Chu CC, et al. Dynamic basis for dG*dT misincorporation via tautomerization and ionization. Nature. 2018;554:195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Park J, Jergic S, Jeon Y, Cho WK, Lee R, Dixon NE, et al. Dynamics of Proofreading by the E. coli Pol III Replicase. Cell Chem Biol. 2018;25:57–66 e4. [DOI] [PubMed] [Google Scholar]
- [3].Harfe BD, Jinks-Robertson S. Mismatch repair proteins and mitotic genome stability. Mutat Res. 2000;451:151–67. [DOI] [PubMed] [Google Scholar]
- [4].Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–33. [DOI] [PubMed] [Google Scholar]
- [5].Kunkel TA, Erie DA. Eukaryotic Mismatch Repair in Relation to DNA Replication. Annu Rev Genet. 2015;49:291–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–38. [DOI] [PubMed] [Google Scholar]
- [7].Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, et al. Mutations of a mutS Homolog in Hereditary Nonpolyposis Colorectal Cancer. Cell. 1993;75:1215–25. [DOI] [PubMed] [Google Scholar]
- [8].Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–61. [DOI] [PubMed] [Google Scholar]
- [9].Nicolaides NC, Papadopoulos N, Liu B, Wei YF, Carter KC, Ruben SM, et al. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371:75–80. [DOI] [PubMed] [Google Scholar]
- [10].Papadopoulos N, Nicolaides NC, Wei Y-F, Ruben SM, Carter KC, Rosen CA, et al. Mutation of a mutL Homolog in Hereditary Colon Cancer. Science. 1994;263:1625–9. [DOI] [PubMed] [Google Scholar]
- [11].Holliday RA. A mechanism for gene conversion in fungi. Genet Res. 1964;5:282–304. [DOI] [PubMed] [Google Scholar]
- [12].Witkin EM. Pure clones of lactose negative mutants obtained in Escherichia coli after treatment with 5-bromouracil. J Mol Biol. 1964;8:610–3. [DOI] [PubMed] [Google Scholar]
- [13].Lahue RS, Au KG, Modrich P. DNA mismatch correction in a defined system. Science. 1989;245:160–4. [DOI] [PubMed] [Google Scholar]
- [14].Fishel R Mismatch repair. J Biol Chem. 2015;290:26395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Marsischky GT, Kolodner RD. Biochemical characterization of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 complex and mispaired bases in DNA. J Biol Chem. 1999;274:26668–82. [DOI] [PubMed] [Google Scholar]
- [16].Mazurek A, Johnson CN, Germann MW, Fishel R. Sequence context effect for hMSH2-hMSH6 mismatch-dependent activation. Proc Natl Acad Sci USA. 2009;106:4177–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wilson T, Guerrette S, Fishel R. Dissociation of mismatch recognition and ATPase activity by hMSH2-hMSH3. J Biol Chem. 1999;274:21659–64. [DOI] [PubMed] [Google Scholar]
- [18].Harrington JM, Kolodner RD. Saccharomyces cerevisiae Msh2-Msh3 acts in repair of base-base mispairs. Mol Cell Biol. 2007;27:6546–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fishel R Signaling mismatch repair in cancer. Nature Med. 1999;5:1239–41. [DOI] [PubMed] [Google Scholar]
- [20].Lamers MH, Winterwerp HH, Sixma TK. The alternating ATPase domains of MutS control DNA mismatch repair. EMBO J. 2003;22:746–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mazur DJ, Mendillo ML, Kolodner RD. Inhibition of Msh6 ATPase activity by mispaired DNA induces a Msh2(ATP)-Msh6(ATP) state capable of hydrolysis-independent movement along DNA. Mol Cell. 2006;22:39–49. [DOI] [PubMed] [Google Scholar]
- [22].Heinen CD, Cyr JL, Cook C, Punja N, Sakato M, Forties RA, et al. Human MSH2 (hMSH2) protein controls ATP processing by hMSH2-hMSH6. J Biol Chem. 2011;286:40287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gradia S, Acharya S, Fishel R. The human mismatch recognition complex hMSH2hMSH6 functions as a novel molecular switch. Cell. 1997;91:995–1005. [DOI] [PubMed] [Google Scholar]
- [24].Plotz G, Welsch C, Giron-Monzon L, Friedhoff P, Albrecht M, Piiper A, et al. Mutations in the MutSalpha interaction interface of MLH1 can abolish DNA mismatch repair. Nucleic Acids Res. 2006;34:6574–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Acharya S, Foster PL, Brooks P, Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol Cell. 2003;12:233–46. [DOI] [PubMed] [Google Scholar]
- [26].Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci. 2000;25:24–8. [DOI] [PubMed] [Google Scholar]
- [27].Putnam CD. Evolution of the methyl directed mismatch repair system in Escherichia coli. DNA Repair (Amst). 2016;38:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Spampinato C, Modrich P. The MutL ATPase is required for mismatch repair. J Biol Chem. 2000;275:9863–9. [DOI] [PubMed] [Google Scholar]
- [29].Kolodner RD, Mendillo ML, Putnam CD. Coupling distant sites in DNA during DNA mismatch repair. Proc Natl Acad Sci USA. 2007;104:12953–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Viswanathan M, Lovett ST. Single-strand DNA-specific exonucleases in Escherichia coli - roles in repair and mutation avoidance. Genetics. 1998;149:7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tishkoff D, Johnson AW, Kolodner R. Molecular and genetic analysis of the gene encoding the Saccharomyces cerevisiae strand exchange protein SEP1. Mol Cell Biol. 1991;11:2593–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Schmutte C, Marinescu RC, Sadoff MM, Guerrette S, Overhauser J, Fishel R. Human exonuclease I interacts with the mismatch repair protein hMSH2. Cancer Res. 1998;58:4537–42. [PubMed] [Google Scholar]
- [33].Genschel J, Bazemore LR, Modrich P. Human exonuclease I is required for 5’ and 3’ mismatch repair. J Biol Chem. 2002;277:13302–11. [DOI] [PubMed] [Google Scholar]
- [34].Wei K, Clark AB, Wong E, Kane MF, Mazur DJ, Parris T, et al. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 2003;17:603–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, et al. Reconstitution of 5’-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. [DOI] [PubMed] [Google Scholar]
- [36].Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. [DOI] [PubMed] [Google Scholar]
- [37].Kadyrov FA, Holmes SF, Arana ME, Lukianova OA, O’Donnell M, Kunkel TA, et al. Saccharomyces cerevisiae MutLalpha is a mismatch repair endonuclease. J Biol Chem. 2007;282:37181–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Pillon MC, Lorenowicz JJ, Uckelmann M, Klocko AD, Mitchell RR, Chung YS, et al. Structure of the endonuclease domain of MutL: unlicensed to cut. Mol Cell. 2010;39:145–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Smith CE, Mendillo ML, Bowen N, Hombauer H, Campbell CS, Desai A, et al. Dominant mutations in S. cerevisiae PMS1 identify the Mlh1-Pms1 endonuclease active site and an exonuclease 1-independent mismatch repair pathway. PLoS Genet. 2013;9:e1003869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G × T mismatch. Nature. 2000;407:711–7. [DOI] [PubMed] [Google Scholar]
- [41].Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–10. [DOI] [PubMed] [Google Scholar]
- [42].Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, Beese LS. Structure of the human MutSalpha DNA lesion recognition complex. Mol Cell. 2007;26:579–92. [DOI] [PubMed] [Google Scholar]
- [43].Gradia S, Subramanian D, Wilson T, Acharya S, Makhov A, Griffith J, et al. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol Cell. 1999;3:255–61. [DOI] [PubMed] [Google Scholar]
- [44].Heinen CD, Wilson T, Mazurek A, Berardini M, Butz C, Fishel R. HNPCC mutations in hMSH2 result in reduced hMSH2-hMSH6 molecular switch functions. Cancer Cell. 2002;1:469–78. [DOI] [PubMed] [Google Scholar]
- [45].Mendillo ML, Putnam CD, Mo AO, Jamison JW, Li S, Woods VL Jr., et al. Probing DNA- and ATP-mediated conformational changes in the MutS family of mispair recognition proteins using deuterium exchange mass spectrometry. J Biol Chem. 2010;285:13170–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Shell SS, Putnam CD, Kolodner RD. The N terminus of Saccharomyces cerevisiae Msh6 is an unstructured tether to PCNA. Mol Cell. 2007;26:565–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Groothuizen FS, Winkler I, Cristovao M, Fish A, Winterwerp HH, Reumer A, et al. MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA. Elife. 2015;4:e06744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lee JB, Cho WK, Park J, Jeon Y, Kim D, Lee SH, et al. Single-molecule views of MutS on mismatched DNA. DNA Repair (Amst). 2014;20:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Jeong C, Cho WK, Song KM, Cook C, Yoon TY, Ban C, et al. MutS switches between two fundamentally distinct clamps during mismatch repair. Nat Struct Mol Biol. 2011;18:379–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Gorman J, Chowdhury A, Surtees JA, Shimada J, Reichman DR, Alani E, et al. Dynamic basis for one-dimensional DNA scanning by the mismatch repair complex Msh2-Msh6. Mol Cell. 2007;28:359–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Jeon Y, Kim D, Martin-Lopez JV, Lee R, Oh J, Hanne J, et al. Dynamic control of strand excision during human DNA mismatch repair. Proc Natl Acad Sci USA. 2016;113:3281–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Cho WK, Jeong C, Kim D, Chang M, Song KM, Hanne J, et al. ATP alters the diffusion mechanics of MutS on mismatched DNA. Structure. 2012;20:1264–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Gorman J, Wang F, Redding S, Plys AJ, Fazio T, Wind S, et al. Single-molecule imaging reveals target-search mechanisms during DNA mismatch repair. Proc Natl Acad Sci USA. 2012;109:E3074–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–48. [DOI] [PubMed] [Google Scholar]
- [55].Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Blainey PC, Luo G, Kou SC, Mangel WF, Verdine GL, Bagchi B, et al. Nonspecifically bound proteins spin while diffusing along DNA. Nat Struct Mol Biol. 2009;16:1224–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Bauer M, Metzler R. Generalized facilitated diffusion model for DNA-binding proteins with search and recognition states. Biophys J. 2012;102:2321–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].von Hippel PH, Berg OG. Facilitated target location in biological systems. [Review] [43 refs]. J Biol Chem. 1989;264:675–8. [PubMed] [Google Scholar]
- [59].Jones M, Wagner R, Radman M. Repair of a mismatch is influenced by the base composition of the surrounding nucleotide sequence. Genetics. 1987;115:605–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Wang H, Lawrence CW, Li GM, Hays JB. Specific binding of human MSH2.MSH6 mismatch-repair protein heterodimers to DNA incorporating thymine- or uracil-containing UV light photoproducts opposite mismatched bases. J Biol Chem. 1999;274:16894–900. [DOI] [PubMed] [Google Scholar]
- [61].Kool ET. Hydrogen bonding, base stacking, and steric effects in DNA replication. Annu Rev Bioph Biom. 2001;30:1–22. [DOI] [PubMed] [Google Scholar]
- [62].Rossetti G, Dans PD, Gomez-Pinto I, Ivani I, Gonzalez C, Orozco M. The structural impact of DNA mismatches. Nucleic Acids Res. 2015;43:4309–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Vafabakhsh R, Ha T. Extreme Bendability of DNA Less than 100 Base Pairs Long Revealed by Single-Molecule Cyclization. Science. 2012;337:1097–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. [DOI] [PubMed] [Google Scholar]
- [65].Yang W Poor base stacking at DNA lesions may initiate recognition by many repair proteins. DNA Repair (Amst). 2006;5:654–66. [DOI] [PubMed] [Google Scholar]
- [66].Allen DJ, Makhov A, Grilley M, Taylor J, Thresher R, Modrich P, et al. MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 1997;16:4467–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Blackwell LJ, Bjornson KP, Modrich P. DNA-dependent activation of the hMutS alpha ATPase. J Biol Chem. 1998;273:32049–54. [DOI] [PubMed] [Google Scholar]
- [68].Fishel R, Acharya S, Berardini M, Bocker T, Charbonneau N, Cranston A, et al. Signaling Mismatch Repair: the mechanics of an adenosine-nucleotide molecular switch. Cold Spring Harbor Symp Quant Biol. 2000;65:217–24. [DOI] [PubMed] [Google Scholar]
- [69].Junop MS, Obmolova G, Rausch K, Hsieh P, Yang W. Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol Cell. 2001;7:1–12. [DOI] [PubMed] [Google Scholar]
- [70].Pluciennik A, Modrich P. Protein roadblocks and helix discontinuities are barriers to the initiation of mismatch repair. Proc Natl Acad Sci USA. 2007;104:12709–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Hombauer H, Campbell CS, Smith CE, Desai A, Kolodner RD. Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell. 2011;147:1040–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Qiu R, DeRocco VC, Harris C, Sharma A, Hingorani MM, Erie DA, et al. Large conformational changes in MutS during DNA scanning, mismatch recognition and repair signalling. EMBO J. 2012;31:2528–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Liu J, Hanne J, Britton BM, Bennett J, Kim D, Lee JB, et al. Cascading MutS and MutL sliding clamps control DNA diffusion to activate mismatch repair. Nature. 2016;539:583–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Lujan SA, Williams JS, Pursell ZF, Abdulovic-Cui AA, Clark AB, McElhinny SAN, et al. Mismatch Repair Balances Leading and Lagging Strand DNA Replication Fidelity. Plos Genet. 2012;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Bustamante C, Bryant Z, Smith SB. Ten years of tension: single-molecule DNA mechanics. Nature. 2003;421:423–7. [DOI] [PubMed] [Google Scholar]
- [76].Tanner NA, Hamdan SM, Jergic S, Loscha KV, Schaeffer PM, Dixon NE, et al. Single-molecule studies of fork dynamics in Escherichia coli DNA replication. Nat Struct Mol Biol. 2008;15:998. [DOI] [PubMed] [Google Scholar]
- [77].Stillman B DNA polymerases at the replication fork in eukaryotes. Mol Cell. 2008;30:259–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Schmutte C, Sadoff MM, Shim KS, Acharya S, Fishel R. The interaction of DNA mismatch repair proteins with human exonuclease I. J Biol Chem. 2001;276:33011–8. [DOI] [PubMed] [Google Scholar]
- [79].Bowen N, Smith CE, Srivatsan A, Willcox S, Griffith JD, Kolodner RD. Reconstitution of long and short patch mismatch repair reactions using Saccharomyces cerevisiae proteins. Proc Natl Acad Sci USA. 2013;110:18472–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Genschel J, Modrich P. Mechanism of 5’-directed excision in human mismatch repair. Mol Cell. 2003;12:1077–86. [DOI] [PubMed] [Google Scholar]
- [81].Park J, Jeon Y, In D, Fishel R, Ban C, Lee JB. Single-molecule analysis reveals the kinetics and physiological relevance of MutL-ssDNA binding. PLoS One. 2010;5:e15496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Sass LE, Lanyi C, Weninger K, Erie DA. Single-molecule FRET TACKLE reveals highly dynamic mismatched DNA-MutS complexes. Biochemistry. 2010;49:3174–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Gorman J, Plys AJ, Visnapuu ML, Alani E, Greene EC. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat Struct Mol Biol. 2010;17:932–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Jiang J, Bai L, Surtees JA, Gemici Z, Wang MD, Alani E. Detection of high-affinity and sliding clamp modes for MSH2-MSH6 by single-molecule unzipping force analysis. Mol Cell. 2005;20:771–81. [DOI] [PubMed] [Google Scholar]
- [85].Sacho EJ, Kadyrov FA, Modrich P, Kunkel TA, Erie DA. Direct visualization of asymmetric adenine-nucleotide-induced conformational changes in MutL alpha. Mol Cell. 2008;29:112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Jiang Y, Marszalek PE. Atomic force microscopy captures MutS tetramers initiating DNA mismatch repair. EMBO J. 2011;30:2881–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Hayner JN, Douma LG, Bloom LB. The interplay of primer-template DNA phosphorylation status and single-stranded DNA binding proteins in directing clamp loaders to the appropriate polarity of DNA. Nucleic Acids Res. 2014;42:10655–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Zhang SJ, Zeng XR, Zhang P, Toomey NL, Chuang RY, Chang LS, et al. A Conserved Region in the Amino-Terminus of DNA-Polymerase-Delta Is Involved in Proliferating Cell Nuclear Antigen-Binding. J Biol Chem. 1995;270:7988–92. [DOI] [PubMed] [Google Scholar]
- [89].De March M, Merino N, Barrera-Vilarmau S, Crehuet R, Onesti S, Blanco FJ, et al. Structural basis of human PCNA sliding on DNA. Nat Commun. 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Elez M, Murray AW, Bi LJ, Zhang XE, Matic I, Radman M. Seeing mutations in living cells. Curr Biol. 2010;20:1432–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Elez M, Radman M, Matic I. Stoichiometry of MutS and MutL at unrepaired mismatches in vivo suggests a mechanism of repair. Nucleic Acids Res. 2012;40:3929–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Lenhart JS, Pillon MC, Guarne A, Simmons LA. Trapping and visualizing intermediate steps in the mismatch repair pathway in vivo. Mol Microbiol. 2013;90:680–98. [DOI] [PubMed] [Google Scholar]
- [93].Simmons LA, Davies BW, Grossman AD, Walker GC. beta clamp directs localization of mismatch repair in Bacillus subtilis. Mol Cell. 2008;29:291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Smith BT, Grossman AD, Walker GC. Visualization of mismatch repair in bacterial cells. Mol Cell. 2001;8:1197–206. [DOI] [PubMed] [Google Scholar]
- [95].Campbell CS, Hombauer H, Srivatsan A, Bowen N, Gries K, Desai A, et al. Mlh2 Is an Accessory Factor for DNA Mismatch Repair in Saccharomyces cerevisiae. Plos Genet. 2014;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Goellner EM, Smith CE, Campbell CS, Hombauer H, Desai A, Putnam CD, et al. PCNA and Msh2-Msh6 Activate an Mlh1-Pms1 Endonuclease Pathway Required for Exo1-Independent Mismatch Repair. Mol Cell. 2014;55:291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Hong Z, Jiang J, Hashiguchi K, Hoshi M, Lan L, Yasui A. Recruitment of mismatch repair proteins to the site of DNA damage in human cells. J Cell Sci. 2008;121:3146–54. [DOI] [PubMed] [Google Scholar]
- [98].Liao Y, Schroeder JW, Gao B, Simmons LA, Biteen JS. Single-molecule motions and interactions in live cells reveal target search dynamics in mismatch repair. Proc Natl Acad Sci USA. 2015;112:E6898–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Li F, Mao GG, Tong D, Huang J, Gu LY, Yang W, et al. The Histone Mark H3K36me3 Regulates Human DNA Mismatch Repair through Its Interaction with MutS alpha. Cell. 2013;153:590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Li F, Tian L, Gu LY, Li GM. Evidence That Nucleosomes Inhibit Mismatch Repair in Eukaryotic Cells. J Biol Chem. 2009;284:33056–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Li F, Ortega J, Gu LY, Li GM. Regulation of mismatch repair by histone code and posttranslational modifications in eukaryotic cells. DNA Repair (Amst). 2016;38:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Javaid S, Manohar M, Punja N, Mooney A, Ottesen JJ, Poirier MG, et al. Nucleosome remodeling by hMSH2-hMSH6. Mol Cell. 2009;36:1086–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–5. [DOI] [PubMed] [Google Scholar]
- [106].van Oijen AM, Blainey PC, Crampton DJ, Richardson CC, Ellenberger T, Xie XS. Single-molecule kinetics of lambda exonuclease reveal base dependence and dynamic disorder. Science. 2003;301:1235–8. [DOI] [PubMed] [Google Scholar]
- [107].Lee JB, Hite RK, Hamdan SM, Xie XS, Richardson CC, van Oijen AM. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–4. [DOI] [PubMed] [Google Scholar]