Abstract
Background and objectives: blaSHV, blaTEM and blaVEB are a group of Extended-Spectrum Beta-Lactamase enzymes (ESBLs) which are able to hydrolyze Penicillins and some cephalosporin antibiotics. The present study evaluated the frequency of ESBL genes blaSHV, blaTEM and blaVEB in Acinetobacter baumannii strains isolated from nosocomial infections to outline the importance of these genes in antibiotic resistance.
Methods: One hundred Acinetobacter baumannii strains were isolated from different nosocomial infections. After antibiotic resistance evaluation with the Kirby-Bauer disc-diffusion method, the Minimum Inhibitory Concentration (MIC) of Ciprofloxacin was measured using the E-test method. Then, the ESBL producing strains were identified employing Combined Disk Methods. Finally, all isolates were evaluated with the Polymerase Chain Reaction (PCR) technique to detect the ESBL genes of interest.
Results: Out of 100 Acinetobacter baumannii isolates, 59% were ESBL positive according to the phenotypic method. The PCR assay could not detect the blaSHV and blaVEB genes in the studied isolates, but the presence of blaTEM gene was demonstrated in 42% of the strains.
Conclusion: The high resistance to most antibiotics, the high prevalence of ESBLs-producing strains and also a high prevalence of blaTEM gene in A. baumannii strains found in the current study gives cause for major concern about nosocomial infections in Iran because of the treatment complexity of these strains. Our results highlight the need for infection control measures to prevent the spread of resistant isolates, especially in hospitals.
Keywords: Acinetobacter baumannii, blaSHV, blaTEM, blaVEB, drug resistance, PCR
Zusammenfassung
Hintergrund: blaSHV, blaTEM und blaVEB gehören zur Gruppe der Extended-Spectrum Beta-Lactamase Enzyme (ESBLs) codierenden Gene, die Penicilline und einige Cephalosporine hydrolysieren können. In der vorliegenden Studie sollte die Häufigkeit dieser ESBL Gene bei Acinetobacter baumannii Stämmen, isoliert aus nosokomialen Infektionen, untersucht werden.
Methode: 100 Acinetobacter baumannii Stämme wurden bei verschiedenen nosokomialen Infektionen isoliert. Nach der Resistenzbestimmung gemäß der Agardiffusionsmethode nach Kirby Bauer wurde die Minimale Hemmkonzentration (MHK) von Ciprofloxacin mit der E-Test Methode bestimmt. Anschließend wurden die ESBL bildenden Stämme mit der Combination Disk-Methode identifiziert und abschließend die Gene mittels PCD detektiert.
Ergebnisse: 59% der Isolate waren phänotypisch ESVL positiv. Mittels PCR konnte nur das blaTEM Gen detektiert werden.
Schlussfolgerung: Die Resistenz gegen die meisten Antibiotika, die hohe Prävalenz von ESBL Bildnern und des blaTEM Gens bei Acinetobacter baumannii Stämmen ist besorgniserregend in Bezug auf die Behandlung nosokomialer Infektionen im Iran. Die Ergebnisse unterstreichen die Notwendigkeit von Infektionskontrollmaßnahmen zur Verhinderung der Ausbreitung resistenter Stämme vor allem in Krankenhäusern.
Introduction
Acinetobacter species are aerobic gram-negative microorganisms responsible for various types of infections such as pneumonia, urinary tract infection and septicemia [1]. Acinetobacter baumannii (A. baumannii) is an important opportunistic pathogen that has caused global outbreaks of nosocomial infections [2].The Acenitobacter infections have been recognized as an emerging problem (one of major cause of nosocomial infections) and appeared to be associated with high mortality rates throughout the world [3].
The infections caused by Acinetobacter pathogens are often treated with Cephalosporins, including Ceftazidime, ceftriaxone, aminoglycosides such as tobramycin and amikacin, Carbapenems, and tetracycline. To date, however, most strains of A. baumannii have become increasingly resistant to almost all these currently available antibacterial agents due to the presence of mobile genetic elements, such as insertion sequences (ISs), plasmids, integrons and resistance islands [1], [2]. These mobile elements carry and transfer antimicrobial resistance genes to another bacterium and easily spread among various species and even genera of microorganisms. Furthermore, the transfer of these elements between chromosome and plasmid may facilitate the rapid spread of the resistance genes among different strains of bacteria. An increasing drug-resistance rate among A. baumannii strains is a major concern in hospitals worldwide [4], [5], [6]. The antimicrobial resistance in this nosocomial pathogen is mainly caused by beta-lactamase (β-lactamases) inactivating enzymes, alteration of membrane porin channels, and mutations that change cellular functions [5]. However, the most common mechanism of resistance is the production of hydrolytic enzymes of antimicrobial agents, including extended-spectrum beta-lactamases (ESBLs) that belong to Ambler classes A, D and B [7], [8], [9], [10]. ESBL-producing strains are mutant, acquired plasmid-mediated β-lactamases which demonstrate unique hydrolytic properties. They are enzymes with the potential to digest β-lactamase antibiotics, which possess Oxyimino groups such as Oxyimino-Cephalosporins. However, they are inhibited by Clavulanate and Tazobactam, β-lactamase inhibitors [4]. Clinical types of ESBL genes includes blaSHV, blaTEM, blaVEB, blaKPC, blaPER, blaBEL-1, blaBES-1, blaSFO-1, blaTLA and blaBIC that are associated with mobile genetic elements, predominantly plasmids [11]. The wide use of numerous new β-lactam antibiotics in recent decades has led to the emergence of ESBLs, which are mostly derivatives of TEM-1 (named for the patient, Temoneira) and SHV-1 (sulfhydryl variable) enzymes. They are capable of hydrolyzing a wide range of β-lactam antibiotics, except carbapenems that have been detected in Acinetobacter strains and many gram-negative bacteria [12].
Non-TEM-, non-SHV-derived ESBLs (VEB [Vietnamese extended-spectrum-beta-lactamase]) have been documented in Acinetobacter species from Europe and Asia but not yet from the Americas [11], [13]. The blaVEB-1 ESBL gene is belonging to class A β-lactamase and is located in a class 1 integron. These genetic structures are responsible for the expression of cassette-associated and mobile resistance genes, initially detected in Enterobacteriaceae and Pseudomonas aeruginosa from Southeast Asia [14], [15]. Subsequently, it has been described in clonally related A. baumannii isolates recovered during an outbreak that lasted 9 months (August 2001–April 2002) in the intensive care unit of a hospital in northern France. In these strains, the location of the blaVEB-1 gene on the chromosomes and integrons was identified [15]. This enzyme is associated with a high level of resistant to Cephems, Monobactams and Ceftazidime [16], [17]. In light of the above, the increasing rate of antibiotic consumption and its impact on treatment failure due to misuse or overuse of antibiotics by patients, and finally widespread drug-resistant strains particularly in hospitals, it is imperative to search for new means of controlling the increasing mortality rate based on the failure of drug therapy [18], [19], [20]. Regarding the growing importance of ESBLs in antibiotic resistance and its impact on treatment failure, this study was performed to evaluate the antimicrobial susceptibility patterns of clinical strains and determine the frequency of blaSHV, blaTEM and blaVEB genes in A. baumannii strains isolated from several hospitals in Tehran.
Methods
Bacterial strains
In this cross-sectional descriptive study, 100 isolated samples of A. baumannii were collected from 10 hospitals in Tehran (Imam Hoseyn, Mofid, Imam khomeyni, Shahid Labafi nazhad, Loghman, Khatamalanbya, Milad, Mostafa Khomeyni, Shariati and Motahari) from October 2014 to April 2015. The specimens were obtained from wounds, tracheae, blood, sputum, catheters, pleural fluid, urine and CSF of hospitalized patients. These isolates were collected from different hospital wards, including ICU, Burns, Internal Medicine, Emergency Medicine, Surgery, Medical, Neurological, BMT, CCU, Orthopedic and Maxillofacial Surgery wards. Replicated isolates from the same patients were excluded from the study. All isolates were identified morphologically and using conventional biochemical methods. The following phenotypic and bacteriological tests were used in this study: Gram staining, colony morphologies, McConkey’s agar, TSI, oxidase reaction, lysine decarboxylase, growth at 37°C and 45°C, hydrolyzed gelatin, citrate utilization, OF, and hemolysis on blood agar with 5% sheep blood. The obtained strains were preserved in tryptic soy broth (Merck, Darmstadt, Germany) containing 15% glycerol [21].
Antibiotic susceptibility testing
Antibiotic susceptibility testing was performed using the agar diffusion test (Kirby-Bauer antibiotic testing), as recommended by the Clinical and Laboratory Standards Institute (CLSI), with disks containing Meropenem (MEM:10 µg), Ceftazidime (CAZ: 30 µg), Gatifloxacin (GAT: 10 µg), Levofloxacin (LEV:10 µg), Piperacillin/Tazobactam (P/T: 110 µg), Ticarcillin-Clavulanate (T/C: 85 µg), and Trimetoprim-Sulfametoxazol (TMS: 25 µg) (MAST, Merseyside, U.K). A. baumannii ATCC19606 served as the control strain for antibiotic susceptibility testing [1], [22].
Minimum inhibitory concentration (MIC)
The minimum inhibitory concentration (MIC) test was performed according to CLSI guidelines by the E-test method for Ciprofloxacin-resistant isolates on Mueller-Hinton agar medium. E-test-Imipenem strips were applied on the plates, and the plates were incubated at 37°C in air for 16 to 20 h [23]. Non-susceptible A. baumannii strains with MIC≥32 µg/ml were considered as Ciprofloxacin resistant [24]. A. baumannii ATCC19606 and Pseudomonas aeruginosa ATCC27853 served as controls for the Ciprofloxacin E-test method.
Phenotypic detection of ESBL-producing isolates
To detect ESBLs, all the isolates were tested employing the disk diffusion test (CDDT) containing Ceftazidime (CAZ) 30 µg and Cefotaxime (CTX) 30 µg with a combination of CAZ 30 µg+clavulanic acid (CA) 10 µg and CTX 30 µg+CA 10 µg per disc (Mast Group, Merseyside, UK). Zones of inhibition were compared with the CTX and CAZ discs alone and compared with the combined CAZ 30 µg+CA 10 µg and CTX 30 µg+CA 10 µg discs. An increase in zone diameter of ≥5 mm in the presence of clavulanic acid indicated the existence of ESBL in the test microorganism. Escherichia coli ATCC25922 and Klebsiella pneumonia ATCC700603 were used as negative and positive controls for ESBL production, respectively [25].
DNA extraction
For molecular diagnosis, the total DNA of all A. baumannii isolates was extracted by the DNA extraction kit (Bioneer Company, Korea, Cat. number K-3032-2) specifically for the given bacterial colony.
Detection of ESBL genes by PCR method
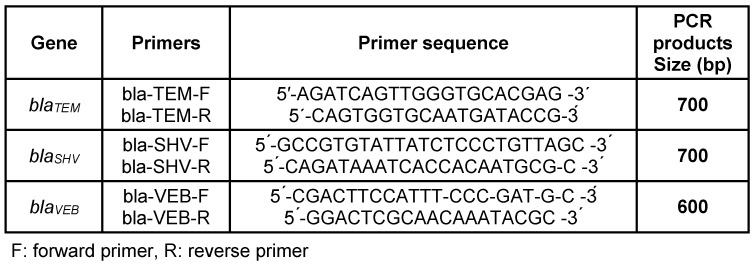
The genetic basis of the β-lactam resistance mediated by enzymatic mechanisms was investigated using the PCR method on all of the isolates phenotypically positive for ESBL. The PCR assay for detection of β-lactamase genes blaSHV, blaTEM and blaVEB was carried out using the previously reported specific oligonucleotide primers shown in Table 1 (Tab. 1). The PCR mixture contained the DNA template, Forward/Reverse primers, and master mix (Bioneer Co., Korea, Cat. number K-2016).
Table 1. Oligonucleotides used in the study for each tested gene in PCR method.
Amplification was carried out with the following thermocycling conditions. The cycling parameters used for the blaSHV gene were: 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 sec, annealing at 56°C for 30 sec, extension at 72°C for 30 sec and a final extension at 72°C for 10 min. The parameters for the blaTEM gene were: 94°C for 3 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 59°C for 1 min, extension at 72°C for 1 min and a final extension at 72°C for 10 min. The parameters used for the blaVEB gene were: 94°C for 10 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, extension at 72°C for 5 min and a final extension at 72°C for 10 min. The PCR product bands were analyzed electrophoretically on a 1% agarose gel at 100 V for 45–60 min in 1X TBE containing ethidium bromide. The results were examined under UV irradiation. The positive controls were three A. baumannii clinical strains containing blaSHV, blaTEM and blaVEB genes.
Results
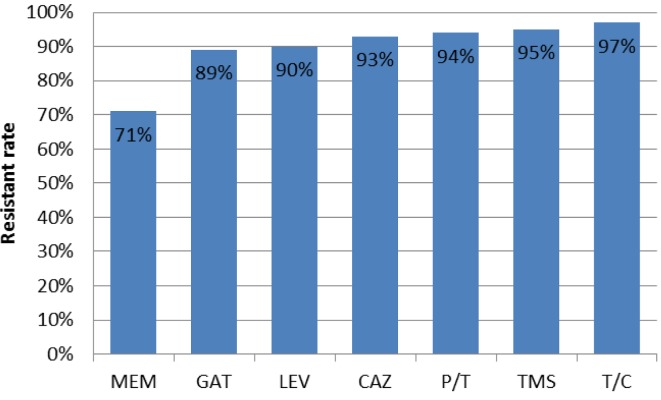
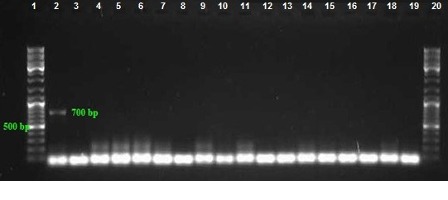
Among 100 collected strains, 57 and 43 were isolated from men and women, respectively. The majority and minority of isolates were obtained from wounds (40%) and CSF (3%), respectively (Table 2 (Tab. 2)). The highest isolation rate was associated with the ICU ward and the lowest with the orthopedic ward. As shown in Figure 1 (Fig. 1), sensitivity to Meropenem, Gatifloxacin and Levofloxacin was 29%, 11% and 10% respectively, which were most effective antibiotics against clinical isolates. The maximum rate of resistance was related to Ticarcilin-Clavelonic acid. The E-test illustrated that the MIC level of ciprofloxacin in all ciprofloxacin-resistant A. baumannii isolates (91%) was ≥32 µg/ml. In the phenotypic method, 59 (59%) isolates produced ESBL enzymes (increase of ≥7 mm in zone diameter of CAZ-clavulanic acid disks compared to CAZ disks. same value as for the CTX+clavulanic acid compared to Imipenem disks). In the molecular assay (PCR), blaSHV and blaVEB genes were not detected in any A. baumannii clinical isolates, but the presence of the blaTEM gene was demonstrated in 42% of these strains (Figure 2 (Fig. 2), Figure 3 (Fig. 3) and Figure 4 (Fig. 4)).
Table 2. Frequency of A. baumannii isolates based on the type of samples.
Figure 1. Antibiotic resistance pattern of A. baumannii isolates in this study.
Figure 2. PCR assay for detection of blaTEM gene with product size: 700 bp. Lanes 2–36: Amplified products, Lane 37: Positive control, Lanes 1 and 40: DNA ladder 100 bp, Lane 38–40: Negative control.
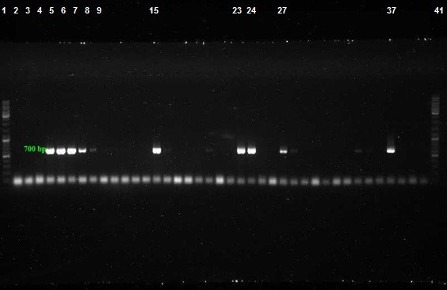
Figure 3. PCR assay for detection of blaVEB gene with product size: 600 bp. Lanes 3–17: Amplified products, Lanes 2 and 19: Positive control, Lanes 1 and 20: DNA ladder 100 bp, Lane 18: Negative control.
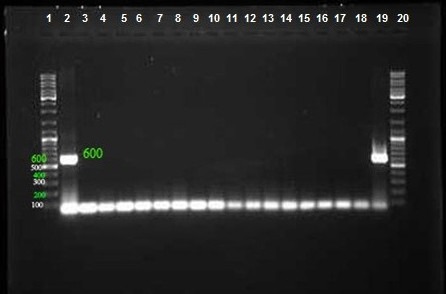
Figure 4. PCR assay for detection of blaSHV gene with product size: 700 bp. Lanes 3–18: Amplified products, Lane 2: Positive control, Lanes 1 and 20: DNA ladder 100 bp, Lane 19: Negative control.
Discussion
A. baumannii is responsible for hospital-acquired infections and has recently become one of the most important healthcare-associated infections in hospitals. Infections caused by this pathogen often leads to significant mortality and morbidity [26]. Antimicrobial resistance in A. baumannii has become a worldwide problem, especially in healthcare centers and hospitalized patients. The emergence of clinical A. baumannii isolates with diverse antibiotic resistance phenotypes leads to difficulties in treating infections caused by this pathogen [3], [27]. In recent years, A. baumannii has been commonly reported as multiple-drug resistant (MDR); the resistance rates to Imipenem, Meropenem, Ceftazidime, Piperacillin/Tazobactam, Ciprofloxacin and Gentamicin in Latin America seem to be among the world's largest [28]. In the present study, resistance to Meropenem, Gatifloxacin, Levofloxacin, Ceftazidim, Pipracilin-Tazobactam, Cotrimoxazole and Ticarcilin-Clavelonic acid were 71%, 89%, 90%, 93%, 94%, 95% and 97%, respectively. In the study conducted by Fallah et al., the resistance of A. baumannii isolates against the tested antibiotics were as follows: 95.4% to Ceftazidime, 100% to Cefotaxime, 91.7% to Meropenem, 92.6% to ciprofloxacin, 95.4% to Piperacillin/Tazobactam and 98.1% to Cotrimoxazole [11]. According to a research by Al-Agamy et al., 100% of A. baumannii isolates were resistant to Cefepime, Cefotaxime and Ceftazidime, 70% were resistant to Imipenem and 85% of isolates were resistant to Ciprofloxacin [29]. In the study carried out by Chagas et al., Ciprofloxacin, Cefepime and Piperacillin/Tazobactam showed the highest resistance rate (99.4%), followed by Ceftazidime (97.4%), Imipenem (95.5%), Meropenem (94.2%) and Ampicillin/Sulbactam (93.5%), while Sulfamethoxazole/Trimethoprim showed the highest susceptibility rate (23.9%) [30]. Safari et al. reported that the resistance rates of A. baumannii isolates were 85%, 94%, 97%, 84%, 95%, and 98% against Imipenem, Meropenem, Ciprofloxacin, Amikacin, Piperacillin/Tazobactam and Cefotaxime, respectively [31]. The results of antimicrobial resistance in other studies are very close to those of the present work, demonstrating a high prevalence of A. baumannii resistance isolates and an increase in MDR strains in recent years.
Production of extended spectrum β-lactamases (ESBLs) is one of the most important resistance mechanisms of A. baumannii strains. A high prevalence of ESBL-enzyme-producing A. baumannii strains has been documented by various studies around the world [32], [33], [34], [35]. In our study, of 59 (59%) ESBL-positive A. baumannii strains tested by PCR method, the blaTEM gene was detected in 42% of them, while blaSHV and blaVEB genes were not found in these isolates. In a study conducted by Fallah et al., 84.2% of the investigated A. baumannii strains were ESBL positive according to the combined disk diffusion test; the blaVEB gene was found in 39.5% of these isolates [11]. In the study carried out by Pasterán et al. (2006) that investigated 21 ESBL-producing A. baumannii strains, the blaVEB gene was presented in 10 (47.6%) isolates [13]. In a study conducted by Thapa et al., out of 37 A. baumannii clinical isolates, the blaVEB was detected in 7 (9%) strains [14]. The disparate results between the current study and other research regarding the prevalence of the blaVEB gene may be due to the small sample size in the present study and/or different geographical locations.
In the study by Chaudhary et al., of 250 A. baumannii isolates, 209 (83.6%) were ESBL producers. Among the ESBL genes, the prevalence of blaTEM varied from 82% to 87%, followed by blaSHV (67–78%) in all ESBL-producing isolates [1]. In contrast to our study, Chaudhary et al. showed a high prevalence of the blaSHV gene, but the prevalence of the blaTEM gene was similar to our results in terms of prevalence rate. In research conducted by AL-Thahab et al., 83.8% of A. baumannii isolates were positive in the ESBL test. Resistance to ampicillin among ESBL positive strains was 100%. Among these isolates, 25% and 8.3% harbored blaSHV and blaTEM genes, respectively, but the blaVEB gene was not found in any of the strains [36]. These findings show that despite the high prevalence of ESBL-positive A. baumannii isolates, the spread of blaSHV and blaTEM genes in these isolates is very low. On the other hand, in the study by Asadollahi et al. on the 23 strains of A. baumannii isolated from burn-wound infections, the frequency of blaTEM and blaSHV genes was 43.4% and 4.3%, respectively [37]. The results of these studies closely resemble those of the present study. In the study by Ramoul et al., the presence of ESBLs in 23 A. baumannii strains was confirmed by the E-test. PCR assay detected blaTEM in three strains, but blaSHV was not observed in any of these isolates [22]. In a study by Al-Agamy et al., E-test strips for ESBLs were applied to 40 isolates, of which 30 (75%) strains yielded a positive result. The most prevalent ESBL gene was blaTEM, which was detected in 35 (87.5%) isolates; however, blaSHV and blaVEB genes were not found [29]. Moreover, in the study by Lopes et al., the PCR technique performed to detect beta-lactamase genes in 50 isolates showed that all isolates harbored the blaTEM gene, while blaSHV and blaVEB genes were not found in any of the strains [38]. The results of these studies regarding the prevalence of blaTEM, blaTEM and blaVEB ESBL genes were similar to our results. This indicates that the prevalence of blaTEM gene is higher than that of blaVEB and blaTEM genes in different regions. In a study by Koo et al., 35 MDR A. baumannii isolates were examined, and 7 antibiotic resistance gene determinants were investigated. They were unable to detect blaTEM, blaSHV and blaVEB beta-lactamase genes [39]. ESBL genes are commonly located on class 1 integrons and are mostly plasmid mediated, and thus transfer easily to other bacteria. This genetic transfer among bacteria causes more rapid spread of ESBL-producing strains. Therefore, the results of different studies can vary due to different regions in which the studies were performed, varied prevalence of pathogenic strains carrying resistance genes especially in hospitals, abusing/overusing of antimicrobial drugs by patients, and study design in term of methodology and sample size [5].
Conclusions
The results of this study reveal an alarming percentage of ESBL-producing strains occurring in Iran. ESBL-producing isolates have emerged as a major challenge, arising from overuse of expanded-spectrum Cephalosporins in hospitals and nursing homes. The high level of resistance to most antibiotics and high prevalence of blaTEM genes in this study indicate the trend to increasing antibiotic resistance in our country and the complexity of treating infections caused by this organism. This study helps understand both the need for more caution in antibiotic consumption and the alarming rate of resistance. Future studies focus on investigation of other ESBL genes. It is necessary to seek a means of monitoring the ESBLs in healthcare settings to prevent the spread of resistant strains and facilitate the selection of appropriate antibiotics for patient treatment.
Abbreviations
ESBLs: Extended-Spectrum Beta-Lactamase enzymes
PCR: Polymerase Chain Reaction
MDR: Multiple-drug resistant
ISs: Insertion sequences
TSI: Triple Sugar Iron
OF: Oxidative-fermentative
MEM: Meropenem
CAZ: Ceftazidime
GAT: Gatifloxacin
LEV: Levofloxacin
P/T: Piperacillin/Tazobactam
T/C: Ticarcillin-clavulanate
TMS: Trimetoprim-sulfametoxazol
CLSI: Clinical and Laboratory Standards Institute
MIC: Minimum Inhibitory Concentration
CDDT: combination disk diffusion test
TBE: Tris/Borate/EDTA
Notes
Acknowledgments
The authors extend their appreciation to the personnel of Iran and Shahed University of Medical Sciences for their cooperation and their technical assistance in this work.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chaudhary M, Payasi A. Molecular Characterization and Antimicrobial Susceptibility Study of Acinetobacter baumannii Clinical Isolates from Middle East, African and Indian Patients. J Proteomics Bioinform. 2012;5(11):265–269. doi: 10.4172/jpb.1000248. [DOI] [Google Scholar]
- 2.Zhou H, Zhang T, Yu D, Pi B, Yang Q, Zhou J, Hu S, Yu Y. Genomic analysis of the multidrug-resistant Acinetobacter baumannii strain MDR-ZJ06 widely spread in China. Antimicrob Agents Chemother. 2011 Oct;55(10):4506–4512. doi: 10.1128/AAC.01134-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bialvaei AZ, Kouhsari E, Salehi-Abargouei A, Amirmozafari N, Ramazanzadeh R, Ghadimi-Daresajini A, Sedighi M. Epidemiology of multidrug-resistant Acinetobacter baumannii strains in Iran: a systematic review and meta-analysis. J Chemother. 2017 Dec;29(6):327–337. doi: 10.1080/1120009X.2017.1338377. [DOI] [PubMed] [Google Scholar]
- 4.Varkey Dr, Balaji V, Abraham J. Molecular Characterisation of Extended Spectrum Beta Lactamase Producing Strains From Blood Sample. International Journal of Pharmacy & Pharmaceutical Sciences. 2014;6(3):276–278. [Google Scholar]
- 5.Zhu L, Yan Z, Zhang Z, Zhou Q, Zhou J, Wakeland EK, Fang X, Xuan Z, Shen D, Li QZ. Complete genome analysis of three Acinetobacter baumannii clinical isolates in China for insight into the diversification of drug resistance elements. PLoS ONE. 2013;8(6):e66584. doi: 10.1371/journal.pone.0066584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozen N, Ergani A, Naas T, Öğünç D, Gültekin M, Colak D, et al. Outbreak of carbapenem-resistant Acinetobacter baumannii producing the carbapenemase OXA-58 in Turkey. The Open Antimicrobial Agents Journal. 2009;1(1):1–8. doi: 10.2174/1876518100901010001. [DOI] [Google Scholar]
- 7.Fallah F, Taherpour A, Vala MH, Hashemi A. Global Spread of New Delhi mettallo-beta-lactamase-1 (NDM-1) Archives of Clinical Infectious Diseases. 2012;6(4):171–177. [Google Scholar]
- 8.Jawad A, Seifert H, Snelling AM, Heritage J, Hawkey PM. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J Clin Microbiol. 1998 Jul;36(7):1938–1941. doi: 10.1128/jcm.36.7.1938-1941.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonnin RA, Potron A, Poirel L, Lecuyer H, Neri R, Nordmann P. PER-7, an extended-spectrum beta-lactamase with increased activity toward broad-spectrum cephalosporins in Acinetobacter baumannii. Antimicrob Agents Chemother. 2011 May;55(5):2424–2427. doi: 10.1128/AAC.01795-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khoshnood S, Heidary M, Mirnejad R, Bahramian A, Sedighi M, Mirzaei H. Drug-resistant gram-negative uropathogens: A review. Biomed Pharmacother. 2017 Oct;94:982–994. doi: 10.1016/j.biopha.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Fallah F, Noori M, Hashemi A, Goudarzi H, Karimi A, Erfanimanesh S, Alimehr S. Prevalence of bla NDM, bla PER, bla VEB, bla IMP, and bla VIM Genes among Acinetobacter baumannii Isolated from Two Hospitals of Tehran, Iran. Scientifica (Cairo) 2014;2014:245162. doi: 10.1155/2014/245162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Czobor I, Gheorghe I, Banu O, Velican A, Lazar V, Mihaescu G, et al. ESBL genes in Multi Drug Resistant Gram negative strains isolated in a one year survey from an Intensive Care Unit in Bucharest, Romania. Romanian Biotechnological Letters. 2014;19(4):9553–9560. [Google Scholar]
- 13.Pasterán F, Rapoport M, Petroni A, Faccone D, Corso A, Galas M, Vázquez M, Procopio A, Tokumoto M, Cagnoni V. Emergence of PER-2 and VEB-1a in Acinetobacter baumannii Strains in the Americas. Antimicrob Agents Chemother. 2006 Sep;50(9):3222–3224. doi: 10.1128/AAC.00284-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thapa B, Tribuddharat C, Srifuengfung S, Dhiraputra C. Class 1 integron element in Thai Acinetobacter baumannii reveals a linkage to the European clone I. International Journal of Infection and Microbiology. 2012;1(1):24–28. doi: 10.3126/ijim.v1i1.6715. [DOI] [Google Scholar]
- 15.Naas T, Coignard B, Carbonne A, Blanckaert K, Bajolet O, Bernet C, Verdeil X, Astagneau P, Desenclos JC, Nordmann P French Nosocomial Infection Early Warning Investigation and Surveillance Network. VEB-1 Extended-spectrum beta-lactamase-producing Acinetobacter baumannii, France. Emerging Infect Dis. 2006 Aug;12(8):1214–1222. doi: 10.3201/eid1208.051547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alikhani MY, Tabar ZK, Mihani F, Kalantar E, Karami P, Sadeghi M, et al. Antimicrobial resistance patterns and prevalence of blaPER-1 and blaVEB-1 genes among ESBL-producing Pseudomonas aeruginosa isolates in West of Iran. Jundishapur Journal of Microbiology. 2014;7(1):e8888. doi: 10.5812/jjm.8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aubert D, Poirel L, Chevalier J, Leotard S, Pages JM, Nordmann P. Oxacillinase-mediated resistance to cefepime and susceptibility to ceftazidime in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2001 Jun;45(6):1615–1620. doi: 10.1128/AAC.45.6.1615-1620.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sunenshine RH, Wright MO, Maragakis LL, Harris AD, Song X, Hebden J, Cosgrove SE, Anderson A, Carnell J, Jernigan DB, Kleinbaum DG, Perl TM, Standiford HC, Srinivasan A. Multidrug-resistant Acinetobacter infection mortality rate and length of hospitalization. Emerging Infect Dis. 2007 Jan;13(1):97–103. doi: 10.3201/eid1301.060716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eliopoulos GM, Maragakis LL, Perl TM. Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clinical infectious diseases. 2008;46(8):1254–1263. doi: 10.1086/529198. [DOI] [PubMed] [Google Scholar]
- 20.Tuon FF, Penteado-Filho SR, Amarante D, Andrade MA, Borba LA. Mortality rate in patients with nosocomial Acinetobacter meningitis from a Brazilian hospital. Braz J Infect Dis. 2010 Sep-Oct;14(5):437–440. doi: 10.1590/S1413-86702010000500003. [DOI] [PubMed] [Google Scholar]
- 21.Abdi-Ali A, Hendiani S, Mohammadi P, Gharavi S. Assessment of Biofilm Formation and Resistance to Imipenem and Ciprofloxacin among Clinical Isolates of Acinetobacter baumannii in Tehran. Jundishapur Journal of Microbiology. 2014;7(1):e8606. doi: 10.5812/jjm.8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramoul A, Hammami S, Dekhil M, Aimiri S, Slim A, Boutiba-Ben Boubaker I. Phenotypic and genotypic characterization of clinical multidrug resistant Acinetobacter baumannii from Algerian intensive care units. Afr J Microbiol Res. 2013;7(10):868–874. doi: 10.5897/AJMR12.1495. [DOI] [Google Scholar]
- 23.Al-Agamy MH, Shibl AM, Ali MS, Khubnani H, Radwan HH, Livermore DM. Distribution of β-lactamases in carbapenem-non-susceptible Acinetobacter baumannii in Riyadh, Saudi Arabia. J Glob Antimicrob Resist. 2014 Mar;2(1):17–21. doi: 10.1016/j.jgar.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Howe RA, Andrews JM BSAC Working Party on Susceptibility Testing. BSAC standardized disc susceptibility testing method (version 11) J Antimicrob Chemother. 2012 Dec;67(12):2783–2784. doi: 10.1093/jac/dks391. [DOI] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial disk diffusion susceptibility tests,19th approved standard, M100-S19. Wayne P: CLSI; 2009. [Google Scholar]
- 26.Alsan M, Klompas M. Acinetobacter baumannii: An Emerging and Important Pathogen. J Clin Outcomes Manag. 2010 Aug;17(8):363–369. [PMC free article] [PubMed] [Google Scholar]
- 27.Manchanda V, Sanchaita S, Singh N. Multidrug resistant acinetobacter. J Glob Infect Dis. 2010 Sep;2(3):291–304. doi: 10.4103/0974-777X.68538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008 Jul;21(3):538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al-Agamy MH, Khalaf NG, Tawfick MM, Shibl AM, El Kholy A. Molecular characterization of carbapenem-insensitive Acinetobacter baumannii in Egypt. Int J Infect Dis. 2014 May;22:49–54. doi: 10.1016/j.ijid.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Chagas TP, Carvalho KR, de Oliveira Santos IC, Carvalho-Assef AP, Asensi MD. Characterization of carbapenem-resistant Acinetobacter baumannii in Brazil (2008-2011): countrywide spread of OXA-23-producing clones (CC15 and CC79) Diagn Microbiol Infect Dis. 2014 Aug;79(4):468–472. doi: 10.1016/j.diagmicrobio.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 31.Safari M, Saidijam M, Bahador A, Jafari R, Alikhani MY. High prevalence of multidrug resistance and metallo-beta-lactamase (MβL) producing Acinetobacter baumannii isolated from patients in ICU wards, Hamadan, Iran. J Res Health Sci. 2013 Sep;13(2):162–167. [PubMed] [Google Scholar]
- 32.Sinha M, Srinivasa H, Macaden R. Antibiotic resistance profile & extended spectrum beta-lactamase (ESBL) production in Acinetobacter species. Indian J Med Res. 2007 Jul;126(1):63–67. [PubMed] [Google Scholar]
- 33.Aiken AM, Mturi N, Njuguna P, Mohammed S, Berkley JA, Mwangi I, et al. Risk and causes of paediatric hospital-acquired bacteraemia in Kilifi District Hospital, Kenya: a prospective cohort study. The Lancet. 2011;378(9808):2021–2027. doi: 10.1016/S0140-6736(11)61622-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamzeh AR, Al Najjar M, Mahfoud M. Prevalence of antibiotic resistance among Acinetobacter baumannii isolates from Aleppo, Syria. Am J Infect Control. 2012 Oct;40(8):776–777. doi: 10.1016/j.ajic.2011.09.019. [DOI] [PubMed] [Google Scholar]
- 35.Shareek P, Sureshkumar D, Ramasubramanian V, Ghafur KA, Thirunarayanan M. Antibiotic sensitivity pattern of blood isolates of Acinetobacter species in a tertiary care hospital: A retrospective analysis. Am J Infect Dis. 2012;8(1):65–69. doi: 10.3844/ajidsp.2012.65.69. [DOI] [Google Scholar]
- 36.AL-Thahab AAL. Molecular Detection of Extended-Spectrum Beta-Lactamases in Clinical Isolates of Acinetobacterbaumannii. J Biol Agric Healthc. 2013;3(7):32–38. [Google Scholar]
- 37.Asadollahi P, Akbari M, Soroush S, Taherikalani M, Asadollahi K, Sayehmiri K, Maleki A, Maleki MH, Karimi P, Emaneini M. Antimicrobial resistance patterns and their encoding genes among Acinetobacter baumannii strains isolated from burned patients. Burns. 2012 Dec;38(8):1198–1203. doi: 10.1016/j.burns.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 38.Lopes BS, Gallego L, Amyes SG. Multi-drug resistance profiles and the genetic features of Acinetobacter baumannii isolates from Bolivia. J Infect Dev Ctries. 2013 Apr 17;7(4):323–328. doi: 10.3855/jidc.2711. [DOI] [PubMed] [Google Scholar]
- 39.Koo SH, Kwon KC, Cho HH, Sung JY. Genetic basis of multidrug-resistant Acinetobacter baumannii clinical isolates from three university hospitals in Chungcheong Province, Korea. Korean J Lab Med. 2010 Oct;30(5):498–506. doi: 10.3343/kjlm.2010.30.5.498. [DOI] [PubMed] [Google Scholar]