Abstract
Background
Biological mechanism of prostate cancer (PCa) recurrence and progress is complex but many of the key elements are not fully understood. Polo-like kinases (Plks) represent a family of highly conserved serine–threonine kinases that play essential roles in cell cycle progression. Plk3 plays contradictory roles in different cancers. However, the roles of Plk3 in PCa remain largely unexplored.
Methods
Kaplan–Meier analysis and Cox regression analysis were performed to evaluate the relationship between Plk3 and prognosis of patients with PCa. Gene set enrichment analysis (GSEA) was conducted to evaluate proliferation and metastasis gene sets using The Cancer Genome Atlas Dataset. MTS assay, clone formation assay, cell migration, and wound healing assay were carried out to investigate biological functions of Plk3.
Results
We found that high Plk3 expression was closely correlated with poor prognosis. GSEA revealed that Plk3 was involved in proliferation and metastasis. Loss-of-function assays demonstrated that Plk3 promoted proliferation and metastasis in PCa cells in vitro.
Conclusion
We discovered that Plk3 plays a critical role in PCa, indicating that it may be a potential prognostic marker and help predict the progression, especially recurrence of PCa.
Keywords: prostate cancer, polo-like kinase 3, recurrence, proliferation, migration, GSEA
Background
Prostate cancer (PCa) is the second most common cancer and the third leading cause of cancer death among men worldwide.1 For localized PCa, radical prostatectomy (RP) or radical radiotherapy is the mature treatment option. But there is still a certain risk of recurrence after treatment. About 16%–35% of the patients needed the second-line treatment within 5 years after primary treatment.2 Prostate-specific antigen (PSA) recurrence (namely biochemical recurrence) is the sign of clinical recurrence (CR), which includes local and distant recurrences. However, only 34% of those with PSA recurrence subsequently had a CR according to Pound et al,3 which was also confirmed by Boorjian et al.4 Recently, Nini et al5 reported that among patients experiencing PSA recurrence (n=370), CR occurred in 183 patients who experienced PSA recurrence after surgery (49.5%). Among patients who experienced CR, recurrence was local and/or nodal in 56 (30.6%), retroperitoneal in 25 (13.7%), skeletal in 77 (42.1%), and visceral in 25 (13.7%). There is defect in specificity of PSA to predict CR.
Physicians predict the CR according to prostate-specific antigen-doubling time, Gleason score, clinical stage, pathological stage, nodal and margin status besides PSA, and several predictive tools have been developed to estimate the risk of relapse following the main standard treatment options for localized PCa.6 However, some patients with good prognostic features still relapsed and succumbed to the disease due to heterogeneities of PCa. We need to explore more sensitive and specific method.
Mechanistically, recurrence arises from local and/or disseminated residual cancer cells. Cancer dormancy can be separated into mechanisms that antagonize the expansion of a dividing tumor cell population (tumor mass dormancy) and mechanisms that result in tumor cell growth arrest (tumor cell dormancy or cellular dormancy).7 Polo-like kinases (Plks) including Plk1, Plk2, Plk3, Plk4, and Plk5 represent a family of highly conserved serine-threonine kinases that play essential roles in cell cycle progression8,9 and in the cellular response to different types of stress.10–12 Mitogenic stimulation of serum-starved quiescent cells with fetal calf serum resulted in a transient modification of murine Plk3, suggesting a functional change during the entry of cells into the cell cycle from quiescence.13 Accumulating evidence has revealed that Plk3 plays mysterious roles in different cancers. Although reduced in cancers of head/neck, lung, and liver,14–16 Plk3 was overexpressed in ovarian and breast cancers.17,18 A bad prognosis was correlated with the downregulation of Plk3 in patients suffering from hepatocarcinoma. In contrast, bad prognosis was linked with overexpression of Plk3 in breast and ovarian cancers. To date, however, the role of Plk3 in the PCa remains largely unexplored.
Our previous study has proved that Plk3 is upregulated in PCa compared with the normal PCa tissues and was positively correlated with the progression of PCa.19 To go deeper into the relationship between Plk3 and PCa, we studied if the expression of Plk3 correlated with prognosis of PCa according to the Kaplan–Meier method and Cox proportional hazard regression models. We also investigated the function of Plk3 in PCa cells. Our findings strongly suggest that Plk3 participates in PCa progression and may help predict the recurrence of PCa.
Methods
Cell lines and cell culture
Human PCa cell line DU145 was bought from Center of Experiment Animal of Sun Yat-sen University (Guangzhou, China) and cultured in a humidified CO2 incubator at 37°C. DU145 was cultured with RPMI 1640 (Thermo Fisher Scientific, Waltham, MA, USA) and supplemented with 10% FBS (Thermo Fisher Scientific) and 1% penicillin–streptomycin (Thermo Fisher Scientific).
siRNA transfection
siRNA oligos targeting human Plk3 (siRNA-2: GCAU-CAAGCAGGUUCACUATT, siRNA-4: GCAGAAA- GAAGAAGAGUAATT) or negative control siRNA were designed and synthesized by GenePharma (Shanghai, China). The siRNAs were transfected with RNAiMAX (Thermo Fisher Scientific) for 6–8 hours. Mock cells were cultured in Opti-MEM for 6–8 hours, but without siRNA.
Western blot
Cell lysates were prepared and subjected to immunoblot analysis of Plk3 protein. Cells (about 1×107 cells) washed twice with ice-cold PBS were lysed with RIPA lysis buffer (no. P0013B; Beyotime Biotechnology, Shanghai, China) and complete protease inhibitor (cocktail, no. B14001a; Selleck, Shanghai, China) for 30 minutes on ice and then cleared by centrifugation at 12,000 rpm at 4°C for another 30 minutes. The total protein concentration in the extracts was measured utilizing a bicinchoninic acid protein assay kit (Beyotime Biotechnology). Equal amounts of protein were separated by SDS-PAGE and transferred to a polyvinylidene fluoride membrane (EMD Millipore, Billerica, MA, USA). The membranes were blocked with 5% BSA or non-fat dry milk in a mixture tris-buffered saline (TBS) and Tween 20 for 1 hour and then probed with antibodies against Plk3 (D14F12, Rabbit mAb, 1:1,000; Cell Signaling Technology Danvers, MA, USA), β-tubulin (abs830032, 1:1,000; Absin, Shanghai, China), GAPDH (abs130609, 1:1,000; Absin), SLUG (sc-166476, 1:500; Santa Cruz Biotechnology Inc., Dallas, TX, USA), Claudin-1 (D3H7C, 1:1,000; Cell Signaling Technology and D3H7C, American) and MMP2 (sc-13594, 1:500; Santa Cruz Biotechnology Inc.). Western blotting was performed as previously described.20
Cell proliferation assay
The MTS colorimetric assay was used to screen for cell viability. PCa cells transfected with siRNA for 48 hours, reseeded in 96-well plates at 2,000 cells per well for a final volume of 100 mL and then incubated for 6 days. Cell viability was determined with the CellTiter 96 non-radioactive cell proliferation assay (MTS; Promega Corporation, Madison, WI, USA). Colony formation assay was performed as described previously.21
Cell migration assay
Cells were suspended in serum-free RPMI-1640 medium, and 5×104 cells were seeded into the upper chambers of a Tran-swell well (Corning, NY, USA) 48 hours after transfection with siRNA. The lower chamber of each well was incubated with 500 µL of RPMI-1640 medium with 10% FBS at 37°C for 24 hours. Cells were fixed and stained, and non-migratory cells in the upper chamber were removed. Migrated cells were counted in 10 random high-power fields.
Wound-healing assay
Wound-healing assays were performed to investigate migration under siRNA treatment. PCa cells were transfected with siRNAs for 48 hours. When cell density in six-well plates was ~90%, a sterile 10 µL pipette tip was used to make a linear wound. Cells were washed to remove superfluous floating cells and debris, then incubated in serum-free RPMI-1640 for an additional 72 hours. Wound healing was photographed at 0, 24, 48, and 72 hours using a microscope. The width of the wound was measured by ImageJ software (National Institutes of Health, Bethesda, MD, USA). The wound-healing ratio (%) = (wound area at 0 hour − wound area at 24, 48, and 72 hours)/wound area at 0 hour ×100%.
Apoptosis assay
PCa cells were transfected with siRNAs for 48 hours. After harvesting and washing twice with PBS, the cells were stained with annexin-V–fluorescein isothiocyanate and propidium iodide (PI) according to the manufacturer’s directions. The stained cells (106 cells) were then analyzed immediately using a FACSVerse flow cytometer (BD, Franklin Lakes, NJ, USA), and the results were expressed as a percentage of living (AnnV− [RayBiotech, Peachtree Corners, GA, USA], PI− [MP Biomedicals, Santa Ana, CA, USA]), early apoptotic (AnnV+, PI−) and late apoptotic/dead cells (AnnV+, PI+). Apoptotic rates were reported as the percentage of apoptotic cells (including early apoptotic cells and late apoptotic cells) among total cells.
Gene Set Enrichment Analysis (GSEA)
GSEA software was downloaded from Broad Institute (http://www.broadinstitute.org/gsea/index.jsp). The high group and low group were classified according to the average of mRNA expression of Plk3 because the mRNA expression obeys the normal distribution for its large sample. Significantly enriched gene sets were identified, which produced FDR q-value <0.05.
The Cancer Genome Atlas (TCGA) data mining
Patients’ clinical profiles in the TCGA prostate adenocarcinoma cohort22 are available at TCGA or cBioPortal (TCGA).23,24
Statistical analysis
SPSS 22.0 software (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. Student’s t-tests were used to analyze the association of Plk3 mRNA expression with clinicopathological characteristics. Disease-free survivals were analyzed using the Kaplan–Meier method, and differences were assessed using the log-rank test. Univariate analysis comparisons and multivariate survival comparisons were performed using Cox proportional hazard regression models. The RRs of death were expressed as adjusted HRs and corresponding 95% CIs. One-way ANOVA was used to analyze the difference between the control and treatment group. Differences were statistically significant when P<0.05.
Results
Plk3 serves as an independent prognostic factor for the disease-free survival of PCa patients
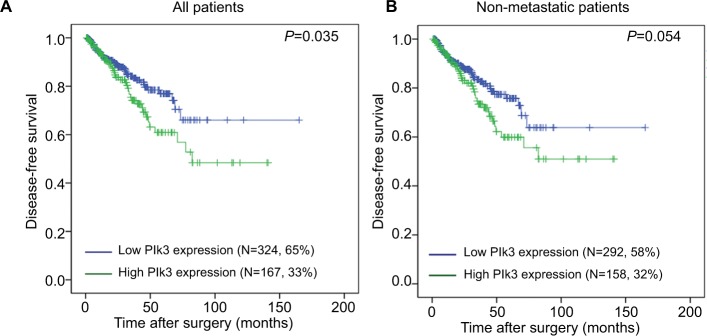
The association of Plk3 expression with the disease-free survival time of PCa patients was analyzed by Kaplan–Meier plots using the TCGA database. As shown in Figure 1A and B, the disease-free survival of PCa patients (all patients) with high Plk3 expression was significantly shorter than that with low Plk3 expression (P=0.035), whereas it is not significant in non-metastatic patients (P=0.054). Then, we carried out univariate analysis using age, Gleason score, tumor stage, lymph node stage, distant metastasis, PSA, and Plk3 expression. The univariate analysis indicated that Gleason score, tumor stage, lymph node stage, PSA, and Plk3 expression (HR 1.855, 95% CI 1.048–3.285; P=0.034) were significant prognostic factors for disease-free survival in patients with PCa (Table 1). Multivariate analysis using Cox proportional hazards model revealed that high Plk3 expression and tumor stage were significant independent prognostic factors in PCa (P=0.025, P<0.001, respectively) (Table 1).
Figure 1.
The disease-free survival rates of all patients and non-metastatic patients.
Notes: (A) All patients (low Plk3 expression [N=324, 65%], high Plk3 expression [N=167, 33%]) and (B) non-metastatic patients (low Plk3 expression [N=292, 58%], high Plk3 expression [N=158, 32%]) with prostate cancer were compared according to low- and high-Plk3 status. Statistical significance was determined using the log-rank test.
Abbreviation: Plk3, polo-like kinase 3.
Table 1.
Prognostic value of Plk3 expression for the disease-free survival by Cox proportional hazards model
| Variables | Disease-free survival
|
|
|---|---|---|
| HR (95% CI) | P-value | |
|
| ||
| Univariate analysis | ||
| Age (≥60 vs <60 years) | 1.488 (0.968–2.287) | 0.070 |
| Gleason score (>7 vs =7 vs < 7) | 9.464 (1.319–67.921) | 0.025* |
| Tumor stage (T2–T4 vs T1) | 2.972 (1.732–5.102) | <0.001*** |
| Lymph node stage (N1 vs N 0) | 1.801 (1.111–2.920) | 0.017 |
| Distant metastasis (M1 vs M0) | 0.049 (0–2488134.592) | 0.740 |
| PSA (≥4 vs <4) | 3.410 (1.644–7.075) | 0.001** |
| Plk3 expression (high vs low) | 1.855 (1.048–3.285) | 0.034* |
| Multivariate analysis | ||
| Gleason score (>7 vs = 7 vs < 7) | 5.709 (0.789–41.328) | 0.085 |
| Tumor stage (T2–T4 vs T1) | 2.763 (1.1607–4.751) | <0.00024*** |
| Plk3 expression (high vs low) | 1.933 (1.091–3.639) | 0.025* |
Note:
P<0.05,
P<0.01, and
P<0.001. Statically significant P-values are shown in bold.
Abbreviations: Plk3, polo-like kinase 3; PSA, prostate-specific antigen.
Downregulation of Plk3 reduces proliferation and metastasis of PCa
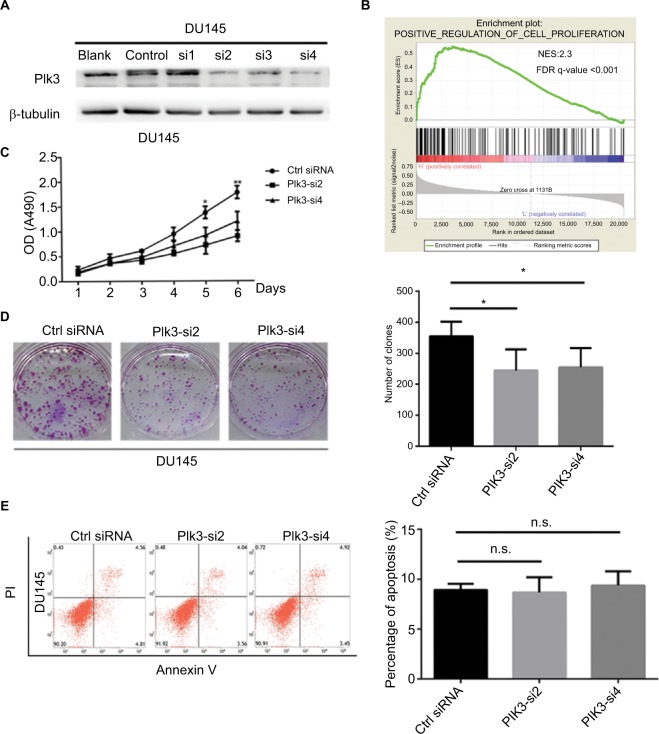
We suppressed Plk3 in PCa cells via siRNA transfection to study the role of Plk3 in PCa. We designed four siRNAs and screened their effectiveness with Western blotting. As shown in Figure 2, Plk3 was remarkably reduced in DU145 cells transfected with the Plk3 siRNA2 and siRNA4 as compared with those transfected with control siRNA (Figure 2A). We initially classified gene expression profiles of PCa obtained from TCGA (498 PCa tissues) into 2 groups according to the Plk3 expression level (high and low) using the average as the cutoff to detect biological function of Plk3 using GSEA. The list of genes expressed in Plk3 high expression groups was highly enriched in genes involved in proliferation when assessed through GSEA (Figure 2B). The ten top differential expression genes (according to rank metric score) are shown in Table 2. To validate the bioinformatics analysis result, we performed MTS assay and clone formation assay to determine the biological functions of Plk3. Plk3 knockdown remarkably reduced DU145 proliferation as compared with control (Figure 2C). Consistently, Plk3 knockdown cells formed obviously fewer colonies than control (Figure 2D). Collectively, these results indicate that Plk3 plays a key role in proliferation of PCa. Given that a previous publication has shown the implication of Plk3 in the induction of apoptosis in PCa cell line LNCap,25 we assessed the effect of Plk3 knockdown on apoptosis of DU145 according to FCM. We found that Plk3 knockdown does not affect the apoptosis of DU145 obviously (Figure 2E).
Figure 2.
Plk3 knockdown reduces prostate cancer cell proliferation.
Notes: (A) Western blotting verification of si-Plk3 knockdown efficiency of DU145 cells. (B) GSEA of proliferation-associated gene set in Plk3 high (H) and low (L) expression groups. The samples were classified into low (mRNA expression <136) or high (mRNA expression >136) based on the average. (C) MTS assay evaluation of influence of Plk3 knockdown on DU145 viability. (D) Colony formation assay determining the effect of Plk3 knockdown in DU145 cells. (E) FCM assay evaluation of influence of Plk3 knockdown on DU145 apoptosis. The data are presented as mean ± SD. *P<0.05 and **P<0.01.
Abbreviations: Ctrl, control; FCM, flow cytometry; GSEA, Gene Set Enrichment Analysis; n.s., no significance; Plk3, polo-like kinase 3.
Table 2.
GeneSet: POSITIVE_REGULATION_OF_CELL_PROLIFERATION
| Probe | Description | Rank in gene list | Rank metric score | Running ES | Core enrichment | |
|---|---|---|---|---|---|---|
| 1 | S1PR2 | 9,294 | 32 | 0.615 | 0.0207 | Yes |
| 2 | LIF | 3,976 | 53 | 0.576 | 0.0405 | Yes |
| 3 | CDKN1A | 1,026 | 69 | 0.557 | 0.0600 | Yes |
| 4 | FOSL1 | 8,061 | 78 | 0.547 | 0.0794 | Yes |
| 5 | CSF1 | 1,435 | 81 | 0.546 | 0.0990 | Yes |
| 6 | SERTAD1 | 29,950 | 105 | 0.523 | 0.1168 | Yes |
| 7 | CD86 | 942 | 122 | 0.512 | 0.1346 | Yes |
| 8 | IL15 | 3,600 | 192 | 0.477 | 0.1485 | Yes |
| 9 | FLT3LG | 2,323 | 214 | 0.467 | 0.1643 | Yes |
| 10 | FGF18 | 8,817 | 333 | 0.435 | 0.1743 | Yes |
Abbreviation: ES, enrichment score.
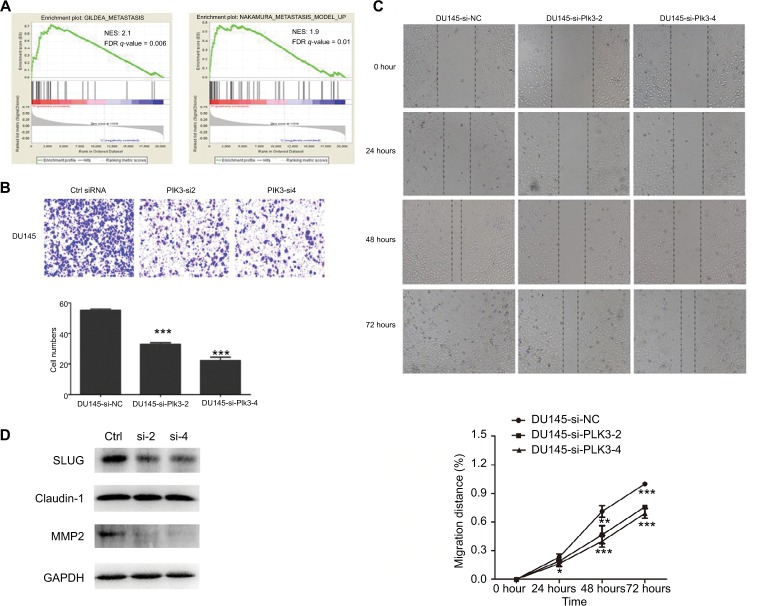
Given that high expression of Plk3 was correlated with tumor invasion, lymph node metastasis, and distant metastasis in our previous work, initially, metastasis gene sets were significantly enriched in the Plk3 high expression group when performing GSEA, which indicated that Plk3 was involved in metastasis of PCa (Figure 3A). The ten top differential expression genes (according to rank metric score) in each GSEA are shown in Tables 3 and 4. Additionally, we carried out migration and wound healing assay. Compared with control, Plk3 depletion significantly decreased migration of DU145 (Figure 3B and C). Furthermore, we detected the expression of metastasis-associated protein. We found that Plk3 knockdown reduced expression of SLUG and MMP2 (Figure 3D).
Figure 3.
Plk3 knockdown inhibits prostate cancer cell migration
Notes: (A) GSEA of metastasis-associated gene sets in the Plk3 high (H) and low (L) expression groups. The samples were as low (mRNA expression <136) or high (mRNA expression <136) based on the average. (B) Migration assay measurement of influence of Plk3 knockdown on DU145 mobility. (C) Wound healing assay measurement of influence of Plk3 knockdown on DU145 mobility. (D) Western blot evaluation of influence of Plk3 knockdown on metastasis-associated protein. The results are presented as mean ± SD. *P<0.05,**P<0.01, and ***P<0.001.
Abbreviations: Ctrl, control; GSEA, Gene Set Enrichment Analysis; h, hours; Plk3, polo-like kinase 3.
Table 3.
GeneSet: NAKAMURA_METASTASIS_MODEL_UP
| Probe | Description (from dataset) | Rank in gene list | Rank metric score | Running ES | Core enrichment | |
|---|---|---|---|---|---|---|
| 1 | DUSP5 | 1,847 | 22 | 0.652 | 0.0748 | Yes |
| 2 | FOSL1 | 8,061 | 78 | 0.547 | 0.1357 | Yes |
| 3 | SLC16A3 | 9,123 | 164 | 0.488 | 0.1884 | Yes |
| 4 | TUBB6 | 84,617 | 208 | 0.470 | 0.2410 | Yes |
| 5 | LAPTM5 | 7,805 | 288 | 0.445 | 0.2889 | Yes |
| 6 | HCLS1 | 3,059 | 336 | 0.433 | 0.3370 | Yes |
| 7 | ADCY3 | 109 | 485 | 0.404 | 0.3768 | Yes |
| 8 | GRK5 | 2,869 | 683 | 0.374 | 0.4107 | Yes |
| 9 | GJB3 | 2,707 | 736 | 0.368 | 0.4510 | Yes |
| 10 | CMTM3 | 123,920 | 838 | 0.354 | 0.4873 | Yes |
Abbreviation: ES, enrichment score.
Table 4.
GeneSet: GILDEA_METASTASIS
| Probe | Description (from dataset) | Rank in gene list | Rank metric score | Running ES | Core enrichment | |
|---|---|---|---|---|---|---|
| 1 | SRGN | 5,552 | 17 | 0.664 | 0.0885 | Yes |
| 2 | IFI16 | 3,428 | 70 | 0.555 | 0.1606 | Yes |
| 3 | IGFBP4 | 3,487 | 171 | 0.485 | 0.2209 | Yes |
| 4 | GBP1 | 2,633 | 248 | 0.455 | 0.2784 | Yes |
| 5 | TIMP3 | 7,078 | 765 | 0.365 | 0.3022 | Yes |
| 6 | RAC2 | 5,880 | 814 | 0.357 | 0.3479 | Yes |
| 7 | PRKCDBP | 112,464 | 956 | 0.339 | 0.3866 | Yes |
| 8 | KRT7 | 3,855 | 1046 | 0.331 | 0.4268 | Yes |
| 9 | SPARC | 6,678 | 1140 | 0.322 | 0.4656 | Yes |
| 10 | VAMP5 | 10,791 | 1184 | 0.317 | 0.5062 | Yes |
Discussion
Primary curative procedures such as RP are well-established therapeutic options in the management of localized PCa. Jeong et al26 reported that for patients undergoing RP with pT2 and pT3a PCa, the 10-year overall survival (OS) was over 90.0%. For patients with pT3b and lymph node-positive disease, the 10-year OS was 82.8% and 76.2%, respectively. Despite technical improvements, there is still a significant risk of cancer recurrence after therapy. Not all patients with PSA recurrence after RP develop CR. More sensitive and specific markers are needed to predict the recurrence of PCa. Previously, we discovered that Plk3, upregulated in PCa tissues, was significantly associated with aggressive clinicopathological parameters of PCa. The results show that the overexpression of Plk3 is significantly associated with advanced pathological grade, clinical stage, lymph node metastasis, and distant metastasis. As opposed to cancers of lung, head/neck, and liver,14–16 Plk3 was upregulated in cancers of ovary and breast.17,18 We hypothesized that whether Plk3 plays an oncogenic or tumor suppressive role is associated with sex hormone.
Plk3 participates in multiple biological processes crucial for PCa development including proliferation, apoptosis, and angiogenesis.27 The disease-free survival of PCa patients (all patients) with high Plk3 expression was significantly shorter than those with low Plk3 expression (P=0.035), whereas it is not significant in non-metastatic patients (P=0.054). It implied that Plk3 expression in PCa may be related to metastasis of PCa. GSEA proved that Plk3 correlated with the expression of proliferation and metastasis genes. In this study, we proved that Plk3 knockdown reduced proliferation of PCa cell in vitro. It may inhibit the proliferation of Plk3 but did not affect the apoptosis of PCa cell obviously. Supporting out findings, Zimmerman et al reported that Plk3 is required for entry into the S phase,28 and Chase et al13 showed that Plk3 may participate in the G2/M phase. It is noted that Plk3 knockdown can promote proliferation in human hepatocellular carcinoma with downregulated Plk3 as we said earlier.16 According to the clinicopathological parameter, correlation analysis and biological function, we found that Plk3 depletion impeded the metastasis of DU145. It is the first time to report that Plk3 knockdown decreased the mobility of cancer cell. However, Juntermanns et al29 found that Plk3 is associated with decreased tumor cell migration in cholangiocarcinoma, suggesting that the effect of Plk3 on migration differs among cancers.
Conclusion
It is our novel discovery that Plk3 serves as an independent prognostic factor for the disease-free survival of PCa patients. Moreover, Plk3 augments PCa cell proliferation and metastasis. Therefore, Plk3 may be the potential biomarker for PCa and may help predict the recurrence of PCa or gain better effect when combined with PSA.
Acknowledgments
In this study, the work of Hai Huang was supported by the National Natural Science Foundation of China (No: 81472382 and 81672550); the National Natural Science Foundation of China for Young Scientists Grant (No: 81101947); the Guangdong Province Natural Science Foundation (No: 2014A030313079); the Fundamental Research Funds for the Central Universities (No: 14ykpy19); Guangdong Province Science and Technology for Social Development Project (No: 2013B021800107 and 2017A020215018); Guangzhou City in 2015 scientific research projects (201510010298); International Science and technology cooperation project of Guangdong province science and technology plan (No: 2016A050502020); and Guangzhou International Science and Technology Cooperation Program (No: 201807010087). Also, the work of Zhenghui Guo was supported by National Natural Science Foundation of China (No: 81772733); Guangdong Province Science and Technology for Social Development Project (No: 2014A020212018, 2016A020215011); and Guangzhou science and technology plan (No: 201707010371). Also, the work of Yiming Lai was supported by the National Natural Science Foundation of China for Young Scientists Grant (No: 81802527). All these study sponsors have no roles in the study design, collection, analysis, and interpretation of data.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Bott SR. Management of recurrent disease after radical prostatectomy. Prostate Cancer Prostatic Dis. 2004;7(3):211–216. doi: 10.1038/sj.pcan.4500732. [DOI] [PubMed] [Google Scholar]
- 3.Pound CR, Partin AW, Eisenberger MA, Chan DW, Pearson JD, Walsh PC. Natural history of progression after PSA elevation following radical prostatectomy. JAMA. 1999;281(17):1591–1597. doi: 10.1001/jama.281.17.1591. [DOI] [PubMed] [Google Scholar]
- 4.Boorjian SA, Thompson RH, Tollefson MK, Rangel LJ, Bergstralh EJ, Blute ML, Karnes RJ. Long-term risk of clinical progression after biochemical recurrence following radical prostatectomy: the impact of time from surgery to recurrence. Eur Urol. 2011;59(6):893–899. doi: 10.1016/j.eururo.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 5.Nini A, Gandaglia G, Fossati N, et al. Patterns of clinical recurrence of node-positive prostate cancer and impact on long-term survival. Eur Urol. 2015;68(5):777–784. doi: 10.1016/j.eururo.2015.04.035. [DOI] [PubMed] [Google Scholar]
- 6.Shariat SF, Kattan MW, Vickers AJ, Karakiewicz PI, Scardino PT. Critical review of prostate cancer predictive tools. Future Oncol. 2009;5(10):1555–1584. doi: 10.2217/fon.09.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer. 2007;7(11):834–846. doi: 10.1038/nrc2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruinsma W, Aprelia M, García-Santisteban I, Kool J, Xu YJ, Medema RH. Inhibition of Polo-like kinase 1 during the DNA damage response is mediated through loss of Aurora A recruitment by Bora. Oncogene. 2017;36(13):1840–1848. doi: 10.1038/onc.2016.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCoy RC, Demko Z, Ryan A, et al. Common variants spanning PLK4 are associated with mitotic-origin aneuploidy in human embryos. Science. 2015;348(6231):235–238. doi: 10.1126/science.aaa3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu D, Dai W, Li C. Polo-like kinase 3, hypoxic responses, and tumorigenesis. Cell Cycle. 2017;16(21):2032–2036. doi: 10.1080/15384101.2017.1373224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng S, Wang H, Jia C, et al. MicroRNA-146a induces lineage-negative bone marrow cell apoptosis and senescence by targeting polo-like kinase 2 expression. Arterioscler Thromb Vasc Biol. 2017;37(2):280–290. doi: 10.1161/ATVBAHA.116.308378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrysik Z, Bernstein WZ, Deng L, et al. The novel mouse polo-like kinase 5 responds to DNA damage and localizes in the nucleolus. Nucleic Acids Res. 2010;38(9):2931–2943. doi: 10.1093/nar/gkq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chase D, Feng Y, Hanshew B, Winkles JA, Longo DL, Ferris DK. Expression and phosphorylation of fibroblast-growth-factor-inducible kinase (Fnk) during cell-cycle progression. Biochem J. 1998;333(Pt 3):655–660. doi: 10.1042/bj3330655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li B, Ouyang B, Pan H, et al. Prk, a cytokine-inducible human protein serine/threonine kinase whose expression appears to be down-regulated in lung carcinomas. J Biol Chem. 1996;271(32):19402–19408. doi: 10.1074/jbc.271.32.19402. [DOI] [PubMed] [Google Scholar]
- 15.Dai W, Li Y, Ouyang B, et al. PRK, a cell cycle gene localized to 8p21, is downregulated in head and neck cancer. Genes Chromosomes Cancer. 2000;27(3):332–336. doi: 10.1002/(sici)1098-2264(200003)27:3<332::aid-gcc15>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 16.Pellegrino R, Calvisi DF, Ladu S, et al. Oncogenic and tumor suppressive roles of polo-like kinases in human hepatocellular carcinoma. Hepatology. 2010;51(3):857–868. doi: 10.1002/hep.23467. [DOI] [PubMed] [Google Scholar]
- 17.Weichert W, Denkert C, Schmidt M, et al. Polo-like kinase isoform expression is a prognostic factor in ovarian carcinoma. Br J Cancer. 2004;90(4):815–821. doi: 10.1038/sj.bjc.6601610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weichert W, Kristiansen G, Winzer KJ, et al. Polo-like kinase isoforms in breast cancer: expression patterns and prognostic implications. Virchows Arch. 2005;446(4):442–450. doi: 10.1007/s00428-005-1212-8. [DOI] [PubMed] [Google Scholar]
- 19.Lin C, Lai Y, Peng S. Expression and clinical significance of Polo-like kinase 3 in prostate cancer. Lingnan Modern Clinics in Surgery. 2018;18:133–136. [Google Scholar]
- 20.Chen X, Xie W, Gu P, et al. Upregulated WDR5 promotes proliferation, self-renewal and chemoresistance in bladder cancer via mediating H3K4 trimethylation. Sci Rep. 2015;5:8293. doi: 10.1038/srep08293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Gu P, Xie R, et al. Heterogeneous nuclear ribonucleo-protein K is associated with poor prognosis and regulates proliferation and apoptosis in bladder cancer. J Cell Mol Med. 2017;21(7):1266–1279. doi: 10.1111/jcmm.12999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research Network The molecular taxonomy of primary prostate cancer. Cell. 2015;163(4):1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thangavel C, Boopathi E, Ciment S, et al. The retinoblastoma tumor suppressor modulates DNA repair and radioresponsiveness. Clin Cancer Res. 2014;20(21):5468–5482. doi: 10.1158/1078-0432.CCR-14-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeong BC, Chalfin HJ, Lee SB, et al. The relationship between the extent of extraprostatic extension and survival following radical prostatectomy. Eur Urol. 2015;67(2):342–346. doi: 10.1016/j.eururo.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 27.Helmke C, Becker S, Strebhardt K. The role of Plk3 in oncogenesis. Oncogene. 2016;35(2):135–147. doi: 10.1038/onc.2015.105. [DOI] [PubMed] [Google Scholar]
- 28.Zimmerman WC, Erikson RL. Polo-like kinase 3 is required for entry into S phase. Proc Natl Acad Sci U S A. 2007;104(6):1847–1852. doi: 10.1073/pnas.0610856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juntermanns B, Sydor S, Kaiser GM, et al. Polo-like kinase 3 is associated with improved overall survival in cholangiocarcinoma. Liver Int. 2015;35(11):2448–2457. doi: 10.1111/liv.12839. [DOI] [PubMed] [Google Scholar]