Abstract
Background
Tiam1 has been identified as an oncogene and acts as an activator of GTPase Rac. Tiam1 was reported to be a promoter of cancer progression in various cancer types, while in lung adenocarcinoma, its mechanism of action is poorly understood.
Materials and Methods
Immunohistochemistry staining and Western blot assay were used to determine Tiam1 expression in lung adenocarcinoma tissues, and its association with prognosis was determined by statistical analysis. We depleted Tiam1 in both A549 and H1975 cancer cell lines. Carboxyfluorescein diacetate succinimidyl ester staining and colony formation assays were used to evaluate its impact on cell proliferation ability after depletion. Transwell migration assay and wound healing assays were performed to determine its impact on migration ability of both cell lines. Western blot assay and immunofluorescence staining were used to analyze the association between Tiam1 and epithelial–mesenchymal transition (EMT) progression. Tube formation assay and vasculogenic mimicry assay were used to show the impact of Tiam1 depletion on cancer angiogenesis.
Results
In this study, we demonstrated that Tiam1 overexpression in lung adenocarcinoma was significantly associated with advanced tumor grade and poor prognosis. In vitro assays indicated that Tiam1 depletion significantly inhibited cell proliferation, colony formation, and migration capacities in A549 and H1975 cells. Further investigations revealed that Tiam1 plays an important role in EMT program enhancement, angiogenesis, and accelerated tumor progression. Notably, Tiam1 depletion in cancer cells strongly inhibited human umbilical vein endothelial cell angiogenesis and vasculogenic mimicry capacities of both cancer cell lines.
Conclusion
Tiam1 overexpression is associated with lung adenocarcinoma progression and may indicate poor prognosis. Tiam1 accelerated tumor progression due to EMT and angiogenesis enhancement. Our data may provide a novel therapeutic target for lung adenocarcinoma.
Keywords: Tiam1, prognostic value, EMT, angiogenesis, VM, lung adenocarcinoma
Introduction
Lung cancer, a prevalent and lethal cancer worldwide,1 has become one of the leading causes of cancer therapy evolution. During the past two decades, while the trials were focusing on the methods to distinguish histological subgroups, intensified chemotherapy and radiotherapy regimens were established as the most effective treatments of lung cancer.2–4 However, the character of high metastasis rate and invasiveness of lung cancer cells contributed to its high mortality rate and poor prognosis in patients.5,6 Until lately, large amounts of data demonstrated the high efficiency of targeted therapy in suppressing tumor progression, identification of new molecular targets that contribute to lung cancer initiation and metastasis will be essential for best clinical treatment selection and outcome improvement.7–9
T-cell lymphoma invasion and metastasis-inducing factor, also known as Tiam1, was identified as a metastasis-related gene which was originally found in aggressive mice T lymphoma cells.10 Tiam1, a guanine nucleotide exchange factor, acts as the activator of the small GTPase Rac.11 Rac, a member of the Rho-like proteins (Rho, Rac, and Cdc42), plays an important role in regulating cytoskeleton dynamics and is crucial for tumor malignancies.12,13 Though the multiple cellular functions of Tiam1, via general Rho protein regulation, were elaborated,14 its controversial role in cancer progression received no final conclusion.15 Previously, perturbed expression of Tiam1 was detected in various cancer tissues, including breast, colon, ovarian, and lung.16–19 The correlation between the expression and its role in carcinogenesis was also verified.20 Moreover, it was found that the expression of Tiam1is upregulated in malignant tumors, thereby proving that Tiam1 serves as a cell migration and division enhancer.21 It is well-known that metastasis and division of cancer cells are coupled with epithelial–mesenchymal transition (EMT) acquisition, and thus Tiam1 may trigger the EMT program. Furthermore, in non-cancerous regions, Tiam1 was found to be crucial in contributing to angiogenic activities via Ras-Tiam1-Rac1-Pak1 signaling pathway.22 As angiogenesis serves as a connecting mechanism between cancer cell stemness and tumorigenesis,23 these demonstrations manifest that Tiam1 plays dual roles in cancer initiation and migration.
Herein, we aim to show that Tiam1 overexpression indicates poor prognosis in lung adenocarcinoma patients, and that Tiam1 expression is significantly related to proliferation, colonization, and migration capacities in both lung cancer cell lines, A549 and H1975. We also show that Tiam1 depletion greatly alters EMT markers’ expression and further inhibits EMT progression. Additionally, using in vitro studies, we demonstrate that Tiam1 protein level could enhance angiogenesis and vasculogenic mimicry (VM), consistent with its regulatory mechanisms operating in human tumors.
Materials and methods
Ethics statement
This study complied with the principles of the Declaration of Helsinki and was approved by the human ethics and research ethics committees of Yanbian University Medical College in China. All patients, whose tissues were used in this research, provided written informed consent. Their resected specimens were stored by our hospital and will potentially be used for scientific research. Their privacy will be maintained. The follow-up survival data were collected retrospectively through medical record analyses.
Clinical samples
Lung adenocarcinoma tissues (92 cases) with clinical features were selected from patients who underwent surgery between January 2008 and December 2011 in Shanghai Outdo Biotech Co. Ltd. and the tissue bank of Yanbian University Medical College. All specimens were taken from patients who underwent lung cancer surgery without receiving preoperative tumor therapy. Diagnosis and stage analysis were based on the Staging Manual of the American Joint Committee on Cancer, seventh edition. Clinicopathological data obtained from these 92 patients are summarized. Briefly, the 92 patients with lung adenocarcinoma included 51 males and 41 females with a mean age of 60 years. Among them, 55 were above 60 years and 37 were younger. All cases were confirmed by pathological examination. With regard to the tumor sizes, 52 cases were T1–T2 and 40 were T3–T4. According to TNM staging, 54 cases were in clinical stage I–II and 38 were in stage III–IV. Overall, 11 cases were defined as histologically well differentiated, 44 cases as moderate, and 37 cases as poorly differentiated. For lymph node (LN) metastases, 50 cases showed no LN metastases while 42 cases were LN positive. According to the fluorescence in situ hybridization assessment, 21 cases were EGFR positive and 71 cases were negative, and 10 cases were ALK positive and 82 cases were negative. None of the patients received radiochemotherapy before surgery. The 92 patients with lung adenocarcinoma were followed for 8 years or until death. The follow-up time was calculated from the day of surgery to death, or to the latest follow-up date. In addition, 88 samples from adjacent non-tumour tissues were collected from the cancer resection margin in these patients.
Immunohistochemistry (IHC) staining
Sections of paraffin-embedded specimens were deparaffinized in xylene and rehydrated in graded alcohol washes. On antigen retrieval, the slides were, respectively, incubated overnight with antibodies against humanTiam1 at room temperature, followed by incubation with the secondary antibodies. Diaminobenzidine-hydrogen peroxide (Sigma-Aldrich Co.) was the chromogen, and counterstaining was carried out with 0.5% hematoxylin. Mouse IgG isotope was used as a negative control. Positive tissue sections were processed omitting the primary antibody as an additional negative control.
Evaluation of IHC staining
All slides were scored by two pathologists (ZL and SC) independently, who were blind to all clinical data. Final score established for cases with discrepancies was determined by reassessment of both pathologists with a double-headed microscope. Briefly, the IHC staining for Tiam1 was semi-quantitatively scored as “−” (negative, no or <5% positive cells), “+” (6%–50% positive cells), and “++” (<50% positive cells, considered as strongly positive). For the double scoring system taken together, “++” scored samples were considered as showing high expression of Tiam1, while “−” and “+” scored samples were considered as showing low expression of Tiam1.
Cell culture and transfection
Lung adenocarcinoma cell lines, A549 and H1975, were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). A549 cells were cultured in DMEM (Thermo Fisher Scientific, Waltham, MA, USA). H1975 cells were cultured in RPMI 1640 medium. All cell lines were supplemented with 10% FBS (Thermo Fisher Scientific) and appropriate antibiotics at 37°C in a 5% CO2 humidified incubator.
Three different Tiam1 siRNAs were purchased from RiboBio (Guangzhou, Guangdong, China). The sequence of si-RNA1 (si-Tiam1#1) was 5′-CCTCCGTACAGTA-ATTATA-3′. The sequence of si-RNA2 (si-Tiam1#2) was 5′-GGAGCTGATTTGCAAGACA-3′. The sequence of si-RNA3 (si-Tiam1#3) was 5′-GCAGTTTGCATGAGAT-GAA-3′. Control siRNA (si-control) was also used. Cells were transfected with 30 nM of siRNA by using Lipofectamine 3000 (Thermo Fisher Scientific) following the manufacturer’s instructions.
Wound healing assay
Cells seeded in six-well plates were incubated with complete tissue culture medium. After the cells became confluent, wounds were created by making scratches using a 200 µL micropipette tip. Medium was then replaced, and spontaneous cell migration was pictured by using a Nikon inverted microscope at 0, 12, and 24 hours.
Immunofluorescence staining
Cells grown on six-well culture slides were fixed with 4% paraformaldehyde for 15 minutes, and permeabilized using 0.5% Triton X-100 (CWBIO, Beijing, China). Then, cells were blocked with 3% BSA for 2 hours and incubated with Tiam1 antibody in 3% BSA at 4°C overnight. After washing three times with PBS, cells were incubated with Alexa Fluor 546-labeled secondary antibody (Thermo Fisher Scientific) in 3% BSA for 2 hours, and then analyzed by using Cytation 5 cell imaging system.
Migration assay
The migration assay was performed by using 24-well millicell (EMD Millipore, Billerica, MA, USA) with 8 µm polyethylene terephthalate (PET) membrane. Cells were seeded in the upper insert with serum-free media, while media containing 10% FBS was added to the lower chamber as chemoattractant. After 12 hours, the cells were removed from the upper surface of the filter by scraping with a cotton swab. Cells that infiltrated through the filter were fixed and stained with Giemsa. Images were then taken by Olympus BX53 microscope.
Colony formation assay
Single-cell suspension (1,000 cells per well) was seeded in six-well plates and incubated for 2 weeks. Cells were then fixed by ice-cold methanol and stained with Giemsa for 25 minutes. Then, the cells were washed with tap water for 30 minutes. Colonies (containing >50 cells) were counted directly. Statistical significance was analyzed from each of the three independent experiments.
Carboxyfluorescein diacetate succinimidyl ester (CFSE) and cell proliferation
Cell proliferation was evaluated by CellTrace™ CFSE cell proliferation kit according to the manufacturer’s protocol. Briefly, mammospheres were dissociated and incubated with PBS containing 5 µM CFSE/1×106 cells and 0.1% BSA for 15 minutes at 37°C. The reaction was stopped with cold culture medium and after washing with the medium, cells were seeded at 10,000 cells/cm2 and incubated in full culture medium for 24 hours. Mammospheres were allowed to adhere on the poly-L -lysine coated glass coverslips and were fixed with 4% paraformaldehyde for 10 minutes at room temperature. Cell nuclei were stained with DAPI (0.5 µg/mL). Coverslips were then mounted with Vectashield mounting medium and examined using Leica TCS SP5 confocal microscope (Leica Microsystems, Mannheim, Germany).
Western blot
Western blot assay was performed as previously described.24 The following primary antibodies were used: Tiam1 (Santa Cruz Biotechnology Inc.); vimentin, Snail, Slug, MMP-2, VEGF, and VEGFA (Abcam); E-cadherin and ZO-1 (Cell Signaling Technology); β-Actin (Zhongshan).
Endothelial cell tube formation assay
The 96-well tissue culture dishes were coated with 100 µL of Matrigel (BD Biosciences) and incubated at 37°C for 4 hours. Human umbilical vein endothelial cells (HUVECs) were cultured in complete medium for 12 hours and resuspended in respective complete medium at a density of 1×105 cells/mL. About 100 µL of the cell suspension was loaded onto the surface of the gelled matrix and incubated at 37°C for 4–6 hours. Tubular networks were visualized using Olympus BX53 microscope.
VM assay
The 96-well tissue culture dishes were coated with 100 µL of Matrigel (BD Biosciences) and incubated at 37°C for 4 hours. A549 cells and H1975 cells with non-targeting siRNA or si-Tiam1 were seeded onto the plates at 1×105 cells/well and VM networks were imaged by Olympus BX53 microscope over 12 hours within four randomly selected fields of view per well.
Statistical analysis
The data analysis was performed using SPSS 17.0 software and GraphPad Prism 6.0 software. Associations between Tiam1 expression and clinicopathological features were evaluated by a chi-squared test and Fisher’s exact test. The Kaplan–Meier method was used for analysis of survival curves, and statistical significance was assessed using the log-rank test. Cox proportional hazards regression model was used to examine univariate and multivariate hazard ratios for the study variables. Group comparisons for continuous data were done by Mann–Whitney U test or one-way ANOVA. Biochemical experiments were performed in triplicate. The value of P<0.05 was considered as statistically significant.
Results
Tiam1 is overexpressed in lung adenocarcinoma
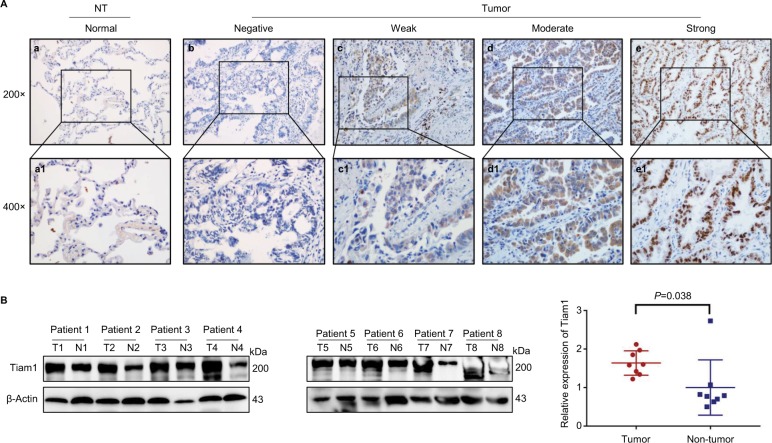
To further confirm the hypothesis and increase our understandings of its clinical significance, we performed IHC staining of specimens obtained form 92 lung adenocarcinoma patients. Consistent with previous studies, Tiam1 was detected both in the cytoplasm and the cell nuclei (Figure 1A). Here, Tiam1 positive staining ratio was only 3.4% (3/88) in the adjacent non-tumor tissues, while in tumor tissues the positive rate was 82.6% (16/92), which shows that Tiam1was aberrantly upregulated (P<0.01). Paralleled with the positive staining ratio, the strongly positive staining of Tiam1 was 2.2% (2/88) and 48.9% (45/92), respectively, which is considered as statistically significant (P<0.01) (Table 1). Western blot was performed for Tiam1 expression evaluation in eight lung adenocarcinoma patients, for both tumor and non-tumor tissues. Tissue samples from tumor and adjacent non-tumor tissues were paired. It is obvious that Tiam1 expression is highly expressed in lung adenocarcinoma tissue compared with adjacent non-tumor tissue (Figure 1B).
Figure 1.
Tiam1 protein expression in lung adenocarcinoma tissues.
Notes: (A) IHC staining for Tiam1 protein expression in lung adenocarcinoma. (a) Negative Tiam1 staining in normal lung tissues. (b) Negative Tiam1 staining in normal lung adenocarcinoma tissues. (c) Weak Tiam1 staining in lung adenocarcinoma. Tiam1 protein was mainly detected in the cytoplasm. (d) Diffuse and moderate Tiam1 staining in lung adenocarcinoma tissues. Protein signals were detected in both cytoplasm and cell nuclei. (e) Diffuse and strongly positive Tiam1 staining in lung adenocarcinoma tissues. Protein signals were revealed mainly in the nuclei. (a1–e1) Indicate higher magnification of the selected area in (a) to (e), respectively (original magnification, a–e: 200×; a1–e1: 400×). (B) Western blot analysis of Tiam1 expression value in tumor vs non-tumor tissue, and Tiam1 was significantly overexpressed in tumor tissue (P=0.0378).
Abbreviations: IHC, immunohistochemistry; NT, non-tumor; T, tumor; N, non-tumor.
Table 1.
Tiam1 expression in lung adenocarcinoma patients
| Diagnosis | No of cases | Positive cases | Positive rates (+ to ++) | Strongly positive rates (++) | ||
|---|---|---|---|---|---|---|
|
| ||||||
| − | + | ++ | ||||
|
| ||||||
| Lung adenocarcinoma | 92 | 16 | 31 | 45 | 82.6%** | 48.9%** |
| Adjacent non-tumor | 88 | 85 | 1 | 2 | 3.4% | 2.2% |
Notes: Tiam1 is significantly overexpressed in lung adenocarcinoma patients.
P<0.01 compared with the adjacent non-tumor tissues.
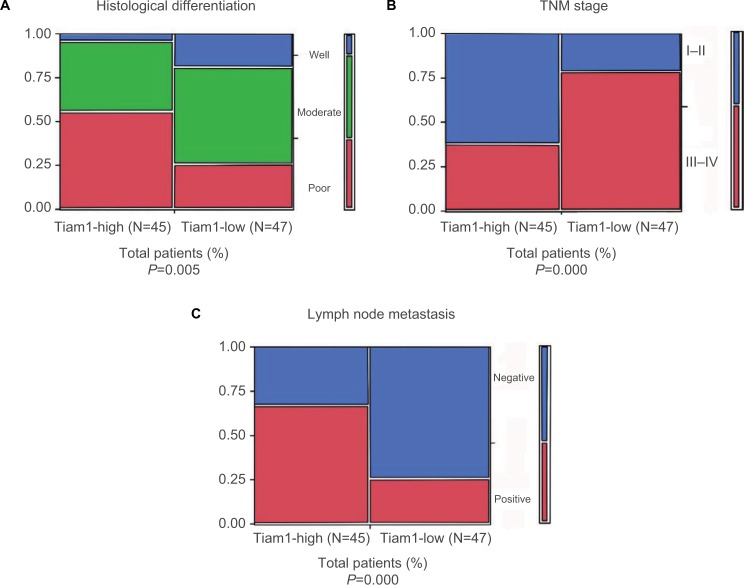
Further, to determine the correlation between Tiam1 expression and clinical features, we performed Pearson’s chi-squared test. It was evident that aberrant Tiam1 expression was correlated with histological differentiation (P=0.005), TNM stage (P<0.000), and LN metastasis (P<0.000), but not with age, gender, tumor size, EGFR, or ALK expression (Table 2, Figure 2A–C). Thus, Tiam1 overexpression in lung adenocarcinoma could indicate poor clinical stage.
Table 2.
Correlations between Tiam1 protein expression and the clinicopathological features of the lung adenocarcinoma patients
| Clinical features | No of cases (n) | Strongly positive Tiam 1 cases N (%) | χ2 | P-value |
|---|---|---|---|---|
|
| ||||
| Gender | 1.643 | 0.216 | ||
| Male | 51 | 28 (54.9) | ||
| Female | 41 | 17 (41.5) | ||
| Age (years) | 0.796 | 0.402 | ||
| ≤60 | 37 | 16 (43.2) | ||
| >60 | 55 | 29 (52.7) | ||
| Tumor size | 1.049 | 0.400 | ||
| T1–T2 | 52 | 23 (44.2) | ||
| T3–T4 | 40 | 22 (55.0) | ||
| Histological differentiation | 10.438 | 0.005** | ||
| Well | 11 | 2 (18.2) | ||
| Moderate | 44 | 18 (40.9) | ||
| Poor | 37 | 25 (67.6) | ||
| TNM stage | 15.898 | 0.000** | ||
| I–II | 54 | 17 (31.5) | ||
| III–IV | 38 | 28 (73.7) | ||
| Lymph node metastasis | 15.678 | 0.000** | ||
| N0 | 50 | 15 (30.0) | ||
| N+ | 42 | 30 (71.4) | ||
| EGFR expression | 0.131 | 0.806 | ||
| Negative | 71 | 34 (47.9) | ||
| Positive | 21 | 11 (52.4) | ||
| ALK expression | 1.996 | 0.193 | ||
| Negative | 82 | 38 (46.3) | ||
| Positive | 10 | 7 (70.0) | ||
Notes: Correlations between Tiam1 protein expression and the clinicopathological features of the lung adenocarcinoma patients. Tiam1 protein expression is strongly correlated with histological differentiation, TNM stage, and lymph node metastasis (**P<0.01).
Abbreviation: ALK, anaplastic lymphoma kinase.
Figure 2.
Association between Tiam1 expression and clinicopathological parameters of lung adenocarcinoma.
Notes: (A) Tiam1 overexpression rates in well, moderate, and poorly differentiated lung adenocarcinoma tissues. (B) Tiam1 overexpression rates in different TNM stages of lung adenocarcinoma. (C) Tiam1 overexpression rates in suspected lymph node metastasis cases. (Tiam1 expression in poorly differentiated tissues, late TNM stages, and lymph node metastasis positive lung adenocarcinoma tissues was significantly higher, respectively).
Tiam1 overexpression is correlated with poor prognosis in lung adenocarcinoma patients
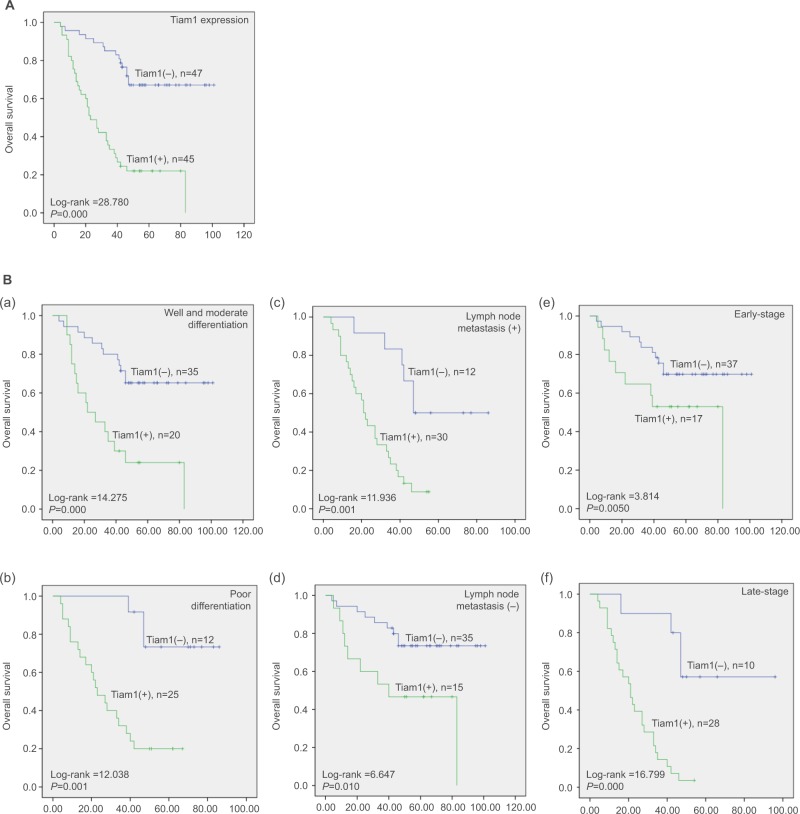
To further demonstrate the impact of Tiam1 overexpression on overall survival (OS), we utilized Kaplan–Meier survival analysis for illustration. Here, we discovered that patients with high Tiam1 expression had significantly shorter OS (P<0.001) (Figure 3A). Compared with Tiam1 low-expression cases, with regard to differentiation status (both P<0.01), LN metastasis (P=0.01, P<0.001, respectively), and early and late clinical stages (P=0.050, P=0.000, respectively), the OS of Tiam1 high-expression cases was significantly shortened (Figure 3Ba–f). Furthermore, when univariate and multivariate Cox regression analyses were applied, the data further verified that Tiam1 overexpression and TNM stages were independent risk factors for shorter OS in lung adenocarcinoma patients (Table 3).
Figure 3.
Kaplan–Meier survival curves illustrating the significance of Tiam1 expression in lung adenocarcinoma.
Notes: (A) OS rate of patients with high and low Tiam1 expression. (B) (a, b) High Tiam1 expression was associated with poor OS in well, moderate, or poorly differentiated lung adenocarcinoma cases; (c, d) high Tiam1 expression was associated with poor OS in patients with or without lymph node metastasis; (e, f) high Tiam1 expression was associated with poor OS in early- or late-stage lung adenocarcinoma cases.
Abbreviation: OS, overall survival.
Table 3.
Univariate and multivariate survival analyses of the clinicopathological features of lung adenocarcinoma patients
| Characteristics | B | SE | Wald | HR | 95% CI
|
P-value | |
|---|---|---|---|---|---|---|---|
| Lower limit | Upper limit | ||||||
|
| |||||||
| Univariate survival analysis | |||||||
| Gender | 0.137 | 0.212 | 0.418 | 1.147 | 0.757 | 1.737 | 0.518 |
| Age (years) | 0.213 | 0.218 | 0.960 | 1.238 | 0.808 | 1.896 | 0.327 |
| Tumor size | 0.512 | 0.220 | 5.397 | 1.668 | 1.083 | 2.569 | 0.020* |
| Histological differentiation | 0.338 | 0.157 | 4.629 | 1.403 | 1.031 | 1.909 | 0.031* |
| TNM stage | 0.914 | 0.225 | 16.526 | 2.495 | 1.606 | 3.877 | 0.000** |
| Lymph node metastasis | 0.85 | 0.222 | 14.666 | 2.339 | 1.514 | 3.612 | 0.000** |
| EGFR expression | 0.017 | 0.250 | 0.005 | 1.017 | 0.623 | 1.660 | 0.946 |
| ALK expression | 0.550 | 0.340 | 2.616 | 1.733 | 0.890 | 3.375 | 0.106 |
| Tiam 1 expression | 1.026 | 0.223 | 21.141 | 2.789 | 1.801 | 4.318 | 0.000** |
| Multivariate survival analysis | |||||||
| Tumor size | 0.203 | 0.238 | 0.722 | 1.225 | 0.767 | 1.954 | 0.395 |
| Histological differentiation | 0.104 | 0.177 | 0.347 | 1.110 | 0.785 | 1.570 | 0.556 |
| TNM stage | 0.629 | 0.256 | 6.046 | 1.876 | 1.136 | 3.098 | 0.014* |
| Lymph node metastasis | 0.502 | 0.260 | 3.731 | 1.652 | 0.993 | 2.750 | 0.053 |
| Tiam 1 expression | 0.827 | 0.238 | 12.048 | 2.286 | 1.433 | 3.647 | 0.001** |
Notes: Univariate and multivariate survival analyses of the clinicopathological features of lung adenocarcinoma patients. Tiam1 overexpression in lung adenocarcinoma could indicate poor clinical outcome (*P<0.05; **P<0.01). The statistical analysis was performed using the Cox proportional hazard regression model.
Abbreviations: SE, standard error; ALK, anaplastic lymphoma kinase.
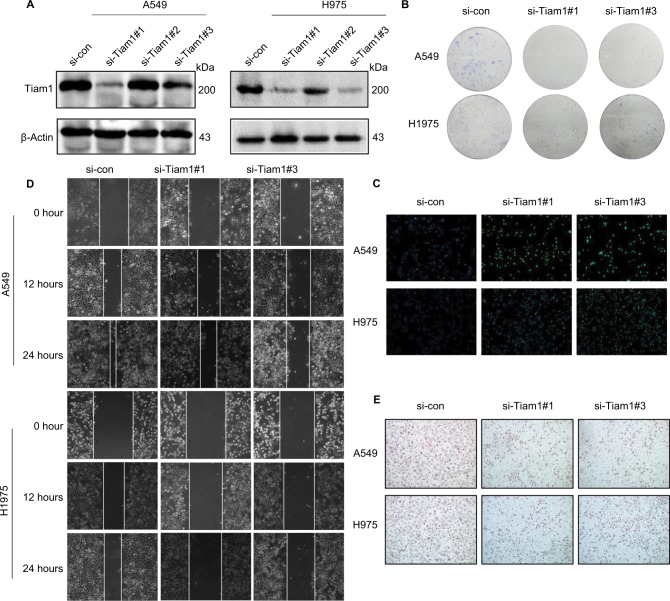
Tiam1 depletion reduces cancer cell proliferation, colonization, and migration capacities
Since our previous study demonstrated that Tiam1 could serve as a prognostic factor for lung adenocarcinoma patients, we intended to analyze whether depletion of Tiam1 affects malignant tumor progression. Both lung adenocarcinoma cell lines, A549 and H1975, were transfected with non-targeting siRNA as si-control or three different Tiam1-specific lentiviral si-RNAs (Figure 4A). Tiam1 was significantly depleted by si-Tiam1 sequence #1 and #3, and sequence #2 showed only a slight depletion effect on Tiam1. Therefore, si-Tiam1 sequence #1 and #2 were chosen for further investigation. Corresponding with the silencing effect of Tiam1, the reduction on cell proliferation and colonization capacities were demonstrated by colony formation assay and CFSE staining (Figure 4B, C).
Figure 4.
Tiam1 depletion reduces cancer cell proliferation, colonization, and migration capacities.
Notes: (A) Tiam1 protein expression was inhibited by Tiam1-specific siRNAs in A549 and H1975 cancer cell lines, compared with si-control, as verified by Western blot analysis. (B) Colony formation assay of A549 and H1975 cancer cell lines. (C) CFSE incorporation for evaluation of proliferation capacities of A549 and H1975 cancer cell lines (original magnification, 100×). (D) Wound healing assay to detect the effects of si-Tiam1 on A549 and H1975 cancer cell motilities. (E) Transwell migration assay of si-Tiam1 cells in both A549 and H1975 cancer cell lines (original magnification, 100×).
Abbreviation: CFSE, carboxyfluorescein diacetate succinimidyl ester.
We next focused on the functionalities of Tiam1 on cancer cell migration. As expected, Tiam1 depletion significantly attenuated the cancer cell migration capacities, according to the wound healing and transwell assays (Figure 4D, E).
Tiam1 depletion inhibits cancer cell EMT progression
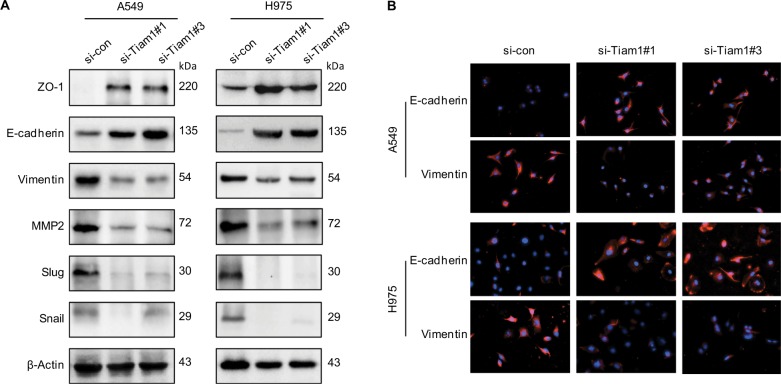
Given that EMT activation program causes plasticity of cancer cells and may enhance continuous adaptions to tumor microenvironment,25,26 and considering the crucial role of Tiam1 in lung adenocarcinoma progression from initiation to metastasis, we intend to investigate further the interplay of Tiam1 with EMT progression of cancer cells. Hence, Western blot analysis was displayed to reflect the alteration of EMT markers. Epithelial markers E-cadherin and ZO-1 were upregulated, while mesenchymal markers including vimentin, MMP2, Slug, and Snail were significantly downregulated, which were paralleled with Tiam1 expression (Figure 5A). To further confirm our findings, immunofluorescence staining was applied. Consistent with the previous data, Tiam1 depletion led to an increase in E-cadherin expression and decrease in vimentin (Figure 5B). These data demonstrated that Tiam1 expression is associated with EMT progression in lung adenocarcinoma cells.
Figure 5.
Correlation between Tiam1 overexpression and cancer cell EMT progression.
Notes: (A) Relative expression levels of EMT markers in A549 and H1975 cancer cells with si-control or si-Tiam1, respectively. (B) IF staining for EMT markers in A549 and H1975 cancer cells, where cells were transfected with si-control and si-Tiam1 (400×).
Abbreviations: EMT, epithelial–mesenchymal transition; IF, immunofluorescence.
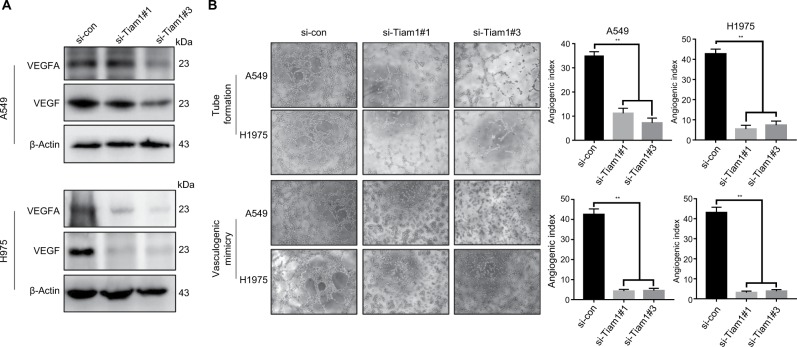
Tiam1 depletion reduces angiogenesis and cancer cell VM capacities
According to previous studies, Tiam1 affects the expression of various genes,15 and lots of data had demonstrated that angiogenesis is considered to be crucial for the metastasis and progression of cancer. Notably, cell signaling modulated by Tiam1 could influence endothelial cell migration, according to Lee et al.22 We therefore performed Western blot analysis, and the findings indicate that the expression of angiogenesis-associated markers, VEGF and VEGFA, was significantly decreased by Tiam1 depletion (Figure 6A). To validate the Tiam1 expression in angiogenesis and its association in vitro, endothelial tube formation assay for HUVECs and VM assay for both cancer cell lines were performed. Tiam1 depletion significantly inhibited the microtubule formation capacities of HUVECs, as well as in VM assay, Tiam1 depletion inhibited the formation of VM networks in both A549 and H1975 cells (Figure 6B). Altogether, our findings indicated that increased Tiam1 expression could enhance angiogenesis and further accelerate tumorigenesis and tumor progression.
Figure 6.
Tiam1 depletion reduces angiogenesis and cancer cell VM capacities.
Notes: (A) Relative expression levels of VEGF and VEGFA in A549 and H1975 cells with si-control or si-Tiam1, respectively. (B) Endothelial tube formation assay was performed with HUVECs incubated in culture medium of A549 and H1975 cancer cells 48 hours after transfection with the si-control and si-Tiam1. The VM assay was performed with A549 and H1975 cancer cells with si-control or si-Tiam1, respectively (original magnification, 100×).
Abbreviations: HUVECs, human umbilical vein endothelial cells; VM, vasculogenic mimicry.
Discussion
Herein, we described the key role of the malignant tumor enhancer Tiam1. According to our findings, Tiam1 is aberrantly overexpressed in lung adenocarcinoma, and was significantly associated with patients’ clinical outcome. High Tiam1 expression led to poor histological differentiation, later clinical stages, LN metastasis, and shortened OS. In the in vitro study, Tiam1 depletion reduced the proliferation, colony formation, and migration capacities of lung adenocarcinoma cancer cell lines A549 and H1975. These data suggested that Tiam1 may serve a role in cancer progression promotion from initiation to metastasis. Consistent with previous findings, Tiam1 was highly expressed in various tumors and was demonstrated as a novel therapeutic biomarker for head and neck squamous cell carcinoma.27–29 Thus, in this study, univariate and multivariate Cox regression analyses investigated the value of Tiam1 in prognosis prediction. We verified that abnormal high expression of Tiam1 could be an independent biomarker of poor prognosis in lung adenocarcinoma patients.
For further investigation, we proposed that impact of Tiam1 on cancer growth and metastasis may result in EMT progression. As expected, we pictured a comprehensive appearance of EMT progression driven by Tiam1. The ablation of Tiam1 significantly attenuated the expression of mesenchymal markers vimentin, MMP2, Slug, and Snail. Immunofluorescence staining further confirmed the alternations in E-cadherin and vimentin expression. These findings indicated that Tiam1 is strongly associated with EMT progression. Additionally, according to Liu et al, Tiam1 promotes cancer progression by modulating EMT via Wnt/β-catenin signaling pathway.27 Moreover, Buongiorno et al confirmed that Tiam1 enhancement on Rac1 and Wnt activation increased β-catenin nuclear translocation and finally promoted cancer cell migration and invasion.30 These results indicated that the Rac1/Wnt/β-catenin signaling is involved in the metastasis promotion effects of Tiam1. Tiam1 is widely demonstrated as a cytoplasmic protein, but recently, Tiam1 was found to be localized in the nucleus.31 Tiam1 is downregulated by multiple factors or miRNAs which finally inhibited its promotional activity on tumorigenesis.32,33 We believe that Tiam1 regulation and lung adenocarcinoma tumorigenesis enhancement need further investigation.
Notably, another essential aspect of tumor initiation and progression is angiogenesis. Angiogenesis is considered to be crucial for the metastasis and progression of cancer and is involved in the carcinogenesis of lung cancer.34 As reports showed that Tiam1 could influence endothelial cell migration,22 we utilized endothelial tube formation assay to investigate its role in angiogenesis. HUVECs cultured in the presence of supernatant of Tiam1 depletion cells strongly attenuated their capacity of tube formation, and Western blot assay also showed that both markers of angiogenesis, VEGF and VEGFA, were downregulated. Moreover, VM describes the ability of tumor cells to acquire endothelial cell behavior that enables to generate tumor-derived vascular networks,35 and the transcriptional signature of VM shares components with EMT in tumor plasticity during metastasis.36 We show that Tiam1 contributes to VM network formation of lung adenocarcinoma cells and further enhances cancer cell initiation and migration. Consistent with previous findings that VM contributes to tumor progression and EMT in non- small-cell lung cancer,37,38 we discovered that Tiam1 depletion significantly inhibited the VM networks generated by both A549 and H1975 cells. Overall, our data suggest that Tiam1 may contribute to VM formation in vitro and further enhance tumor progression.
Conclusion
Taken together, we indicated that Tiam1 is aberrantly overex-pressed in lung adenocarcinoma patients and is significantly associated with poor clinical outcome. Tiam1 serves as an oncogene in lung adenocarcinoma progression via EMT and angiogenesis enhancement. Thus, Tiam1 may be a promising biomarker for the prognosis of lung adenocarcinoma patients and may serve as a potential therapeutic target for lung adenocarcinoma.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (no 31760313), and The Funds of Changbai Mountain Scholar Project and Key Laboratory of the Science and Technology Department of Jilin Province (no 20170622007JC).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Aupérin A, Le Péchoux C, Rolland E, et al. Meta-analysis of concomitant versus sequential radiochemotherapy in locally advanced non-small-cell lung cancer. J Clin Oncol. 2010;28(13):2181–2190. doi: 10.1200/JCO.2009.26.2543. [DOI] [PubMed] [Google Scholar]
- 3.Curran WJ, Jr, Paulus R, Langer CJ, et al. Sequential vs. concurrent chemoradiation for stage III non-small cell lung cancer: randomized phase III trial RTOG 9410. J Natl Cancer Inst. 2011;103(19):1452–1460. doi: 10.1093/jnci/djr325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly K, Chansky K, Gaspar LE, et al. Phase III trial of maintenance gefitinib or placebo after concurrent chemoradiotherapy and docetaxel consolidation in inoperable stage III non-small-cell lung cancer: SWOG S0023. J Clin Oncol. 2008;26(15):2450–2456. doi: 10.1200/JCO.2007.14.4824. [DOI] [PubMed] [Google Scholar]
- 5.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 6.Wang X, Adjei AA. Lung cancer and metastasis: new opportunities and challenges. Cancer Metastasis Rev. 2015;34(2):169–171. doi: 10.1007/s10555-015-9562-4. [DOI] [PubMed] [Google Scholar]
- 7.Velizheva NP, Rechsteiner MP, Valtcheva N, et al. Targeted next-generation-sequencing for reliable detection of targetable rearrangements in lung adenocarcinoma-a single center retrospective study. Pathol Res Pract. 2018;214(4):572–578. doi: 10.1016/j.prp.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monterisi S, Lo Riso P, Russo K, et al. HOXB7 overexpression in lung cancer is a hallmark of acquired stem-like phenotype. Oncogene. 2018;37(26):3575–3588. doi: 10.1038/s41388-018-0229-9. [DOI] [PubMed] [Google Scholar]
- 9.Kim J, McMillan E, Kim HS, et al. XPO1-dependent nuclear export is a druggable vulnerability in KRAS-mutant lung cancer. Nature. 2016;538(7623):114–117. doi: 10.1038/nature19771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Habets GG, Scholtes EH, Zuydgeest D, et al. Identification of an invasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for Rho-like proteins. Cell. 1994;77(4):537–549. doi: 10.1016/0092-8674(94)90216-x. [DOI] [PubMed] [Google Scholar]
- 11.Michiels F, Habets GG, Stam JC, van der Kammen RA, Collard JG. A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature. 1995;375(6529):338–340. doi: 10.1038/375338a0. [DOI] [PubMed] [Google Scholar]
- 12.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9(9):690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 13.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279(5350):509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 14.Welch HC. Regulation and function of P-Rex family Rac-GEFs. Small GTPases. 2015;6(2):49–70. doi: 10.4161/21541248.2014.973770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boissier P, Huynh-do U. The guanine nucleotide exchange factor Tiam1: a Janus-faced molecule in cellular signaling. Cell Signal. 2014;26(3):483–491. doi: 10.1016/j.cellsig.2013.11.034. [DOI] [PubMed] [Google Scholar]
- 16.Stebel A, Brachetti C, Kunkel M, Schmidt M, Fritz G. Progression of breast tumors is accompanied by a decrease in expression of the Rho guanine exchange factor Tiam1. Oncol Rep. 2009;21(1):217–222. [PubMed] [Google Scholar]
- 17.Minard ME, Herynk MH, Collard JG, Gallick GE. The guanine nucleotide exchange factor Tiam1 increases colon carcinoma growth at metastatic sites in an orthotopic nude mouse model. Oncogene. 2005;24(15):2568–2573. doi: 10.1038/sj.onc.1208503. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Liang S, Jin H, Xu C, Ma D, Lu X. Tiam1, negatively regulated by miR-22, miR-183 and miR-31, is involved in migration, invasion and viability of ovarian cancer cells. Oncol Rep. 2012;27(6):1835–1842. doi: 10.3892/or.2012.1744. [DOI] [PubMed] [Google Scholar]
- 19.Wang HM, Wang J. Expression of Tiam1 in lung cancer and its clinical significance. Asian Pac J Cancer Prev. 2012;13(2):613–615. doi: 10.7314/apjcp.2012.13.2.613. [DOI] [PubMed] [Google Scholar]
- 20.Rygiel TP, Mertens AE, Strumane K, van der Kammen R, Collard JG. The Rac activator Tiam1 prevents keratinocyte apoptosis by controlling ROS-mediated ERK phosphorylation. J Cell Sci. 2008;121(Pt 8):1183–1192. doi: 10.1242/jcs.017194. [DOI] [PubMed] [Google Scholar]
- 21.Ellenbroek SI, Iden S, Collard JG. The Rac activator Tiam1 is required for polarized protrusional outgrowth of primary astrocytes by affecting the organization of the microtubule network. Small GTPases. 2012;3(1):4–14. doi: 10.4161/sgtp.19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SH, Kunz J, Lin SH, Yu-Lee LY. 16-kDa prolactin inhibits endothelial cell migration by down-regulating the Ras-Tiam1-Rac1-Pak1 signaling pathway. Cancer Res. 2007;67(22):11045–11053. doi: 10.1158/0008-5472.CAN-07-0986. [DOI] [PubMed] [Google Scholar]
- 23.Fantozzi A, Gruber DC, Pisarsky L, et al. VEGF-mediated angiogenesis links EMT-induced cancer stemness to tumor initiation. Cancer Res. 2014;74(5):1566–1575. doi: 10.1158/0008-5472.CAN-13-1641. [DOI] [PubMed] [Google Scholar]
- 24.Lin Z, Bazzaro M, Wang MC, Chan KC, Peng S, Roden RB. Combination of proteasome and HDAC inhibitors for uterine cervical cancer treatment. Clin Cancer Res. 2009;15(2):570–577. doi: 10.1158/1078-0432.CCR-08-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brabletz T. To differentiate or not--routes towards metastasis. Nat Rev Cancer. 2012;12(6):425–436. doi: 10.1038/nrc3265. [DOI] [PubMed] [Google Scholar]
- 26.Diepenbruck M, Christofori G. Epithelial-mesenchymal transition (EMT) and metastasis: yes, no, maybe? Curr Opin Cell Biol. 2016;43:7–13. doi: 10.1016/j.ceb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 27.Liu L, Wu B, Cai H, et al. Tiam1 promotes thyroid carcinoma metastasis by modulating EMT via Wnt/β-catenin signaling. Exp Cell Res. 2018;362(2):532–540. doi: 10.1016/j.yexcr.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 28.Zhu G, Fan Z, Ding M, et al. An EGFR/PI3K/AKT axis promotes accumulation of the Rac1-GEF Tiam1 that is critical in EGFR-driven tumorigenesis. Oncogene. 2015;34(49):5971–5982. doi: 10.1038/onc.2015.45. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Ding Y, Huang J, et al. miR-141 suppresses the migration and invasion of HCC cells by targeting Tiam1. PLoS One. 2014;9(2):e88393. doi: 10.1371/journal.pone.0088393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buongiorno P, Pethe VV, Charames GS, Esufali S, Bapat B. Rac1 GTPase and the Rac1 exchange factor Tiam1 associate with Wntresponsive promoters to enhance beta-catenin/TCF-dependent transcription in colorectal cancer cells. Mol Cancer. 2008;7:73. doi: 10.1186/1476-4598-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diamantopoulou Z, White G, Fadlullah MZH, et al. Tiam1 antagonizes TAZ/YAP both in the destruction complex in the cytoplasm and in the nucleus to inhibit invasion of intestinal epithelial cells. Cancer Cell. 2017;31(5):e626:621–634. doi: 10.1016/j.ccell.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen G, Lu L, Liu C, Shan L, Yuan D. MicroRNA-377 suppresses cell proliferation and invasion by inhibiting Tiam1 expression in hepatocellular carcinoma. PLoS One. 2015;10(3):e0117714. doi: 10.1371/journal.pone.0117714. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Huang H, Fan L, Zhan R, Wu S, Niu W. Expression of microRNA-10a, microRNA-342-3p and their predicted target gene Tiam1 in extranodal NK/T-cell lymphoma, nasal type. Oncol Lett. 2016;11(1):345–351. doi: 10.3892/ol.2015.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ellis PM. Anti-angiogenesis in personalized therapy of lung cancer. Adv Exp Med Biol. 2016;893:91–126. doi: 10.1007/978-3-319-24223-1_5. [DOI] [PubMed] [Google Scholar]
- 35.Maniotis AJ, Folberg R, Hess A, et al. Vascular channel formation by human melanoma cells in vivo and in vitro: vasculogenic mimicry. Am J Pathol. 1999;155(3):739–752. doi: 10.1016/S0002-9440(10)65173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hendrix MJ, Seftor EA, Hess AR, Seftor RE. Vasculogenic mimicry and tumourcell plasticity: lessons from melanoma. Nat Rev Cancer. 2003;3(6):411–421. doi: 10.1038/nrc1092. [DOI] [PubMed] [Google Scholar]
- 37.Zhou X, Gu R, Han X, Wu G, Liu J. Cyclin-dependent kinase 5 controls vasculogenic mimicry formation in non-small cell lung cancer via the FAK-AKT signaling pathway. Biochem Biophys Res Commun. 2017;492(3):447–452. doi: 10.1016/j.bbrc.2017.08.076. [DOI] [PubMed] [Google Scholar]
- 38.Yao L, Zhang D, Zhao X, et al. Dickkopf-1-promoted vasculogenic mimicry in non-small cell lung cancer is associated with EMT and development of a cancer stem-like cell phenotype. J Cell Mol Med. 2016;20(9):1673–1685. doi: 10.1111/jcmm.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]