Abstract
Purpose
To study the prognostic value of klotho (KL) and its promoter DNA methylation in head and neck squamous cell carcinoma (HNSCC) and to assess their associations with the autophagy gene LC3 and the RNA transferase gene NSUN2.
Materials and methods
Upper quartile normalized RNA-seq V2 RSEM values of KL mRNA and beta value for KL methylation were retrieved from The Cancer Genome Atlas HNSCC dataset. Kaplan–Meier survival curves were used to assess the associations of KL expression and methylation with patient survival; multivariate Cox proportional hazards regression models were used to estimate the HRs and their 95% CIs.
Results
There is a negative relationship between KL gene expression and its promoter DNA methylation in HNSCC. KL gene expression was positively correlated with overall survival, while KL methylation was inversely correlated with the overall survival of HNSCC patients. Furthermore, KL methylation was significantly associated with gender (P=0.012), tumor grade (P=0.0009) and tumor site (P<0.0001). Finally, HNSCC patients with high KL gene expression or low KL DNA methylation had high LC3 but low NSUN2.
Conclusion
KL methylation silenced its gene expression in HNSCC. Low KL expression and high KL methylation can be potential biomarkers for worse prognosis in HNSCC. As the downstream targets, LC3 and NSUN2 could be responsible for the KL expression in HNSCC.
Keywords: biomarkers, gene silencing, head neoplasms, neck neoplasms, predictive values, prognosis
Introduction
Head and neck cancer (HNC) is the sixth most common cancer worldwide, and about 90% of HNC cases are head and neck squamous cell carcinoma (HNSCC) cases.1 Despite the advancements in treatment, the survival of HNSCC patients has remained poor in the past 40 years, with the 5-year postoperative survival rate being <50%.2 However, if diagnosed at an early stage, the survival rate can be as high as 80%.3 Therefore, identification of biomarkers, especially at the early stage, is urgent for improvement of HNCSS patients’ survival.
Klotho (KL) is a single-transmembrane protein composed of 1,012 amino acids.4 The extracellular domain of KL can be cleaved and released into blood, urine and cerebrospinal fluid,5 where it acts as a hormone to regulate physiological processes.6 Although it was initially discovered as an antiaging gene in 1997,7 KL has recently emerged as a potent tumor suppressor in a wide variety of cancers, including breast cancer,7–9 renal cell carcinoma,10 thyroid cancer,11 lung cancer,12 pancreatic cancer,4 oral cancer and esophageal squamous cell carcinoma (ESCC),13 whereas in ovarian cancer, its expression was reported to be associated with poor prognosis.14 Compared to normal tissues or adjacent tissues, decreased KL gene expression has been consistently detected in many tumors. On the other hand, restoring KL inhibited cancer cell proliferation.4–8 Although studies of KL in HNSCC just started recently and are still very limited, the available data already demonstrated reduced KL levels in oral squamous cell carcinoma (OSCC) and ESCC.13,15 KL expression is epigenetically regulated by its promoter DNA methylation, a DNA modification frequently found in cancers.16 KL promoter hypermethylation has been reported to silence its expression and consequently leads to the loss of its tumor suppressive function.9 In fact, KL promoter hypermethylation has been found in malignancies such as HNSCC,17 breast cancer,9 colorectal cancer18 and cervical cancer.19 These findings raise the surge of evaluating KL expression and its DNA methylation as potential prognosticators in HNSCC.
Thus, the purpose of this study was to investigate the significance of KL expression and its DNA methylation in HNSCC survival. To explore the possible underlying mechanisms, we examined their associations with the autophagy-associated gene LC3 as well as the RNA methyltransferase gene NSUN2, a newly discovered prognostic marker for HNSCC.20
Materials and methods
Gene expression, DNA methylation and clinicopathologic data
The Cancer Genome Atlas HNSCC provisional dataset (www.cbioportal.org) enrolled 530 patients, of whom 522 tumor specimens were analyzed for RNA levels using RNA-seq and for DNA methylation using Illumina DNA methylation array HM450, which were conducted as previously described.21 Both upper quartile normalized RNA-seq V2 RNA-seq by expectation maximization (RSEM) values of KL expressions, and beta values of KL, which are the ratios of intensities between methylated and unmethylated alleles to estimate DNA methylation levels, as well as clinicopathologic data were retrieved.
Of the 522 patients who had available data on both RNA-seq and DNA methylation of KL, the average age was 60.9 years old (range from 19 to 90). There were 200 patients whose tumor was located at the tongue, 135 had tumor at the pharynx and 186 had tumor at other sites of head and neck. The patients, except for one without indicated gender, were composed of 384 males and 137 females. Based on the information available for race (506 patients), Caucasian patients accounted for 88.1%, African Americans 9.3%, Asians 2.2% and Native Americans 0.4%. One hundred seventeen patients had no smoking history and 391 had it. There were 284 patients with a stage IV disease, 105 patients with stage III, 98 patients with stage II and 20 patients with stage I disease. Sixty-three patients had a tumor with grade I (12.2%), 304 with grade II (58.8%) and 150 with grade III (29%). Post-radiotherapy information was available only for 183 patients, of whom 121 (66.1%) received radiotherapy after surgery, while 62 did not. The follow-up information was available for 519 patients with the average overall survival being 21.2 months (range from 0.07 to 210.8 months; Table 2).
Table 2.
Associations of KL expression and DNA methylation with clinicopathologic variables
| KL expression | KL methylation | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable | n | Low | Medium | High | P-value | P-trend | Low | Medium | High | P-value | P-trend |
| Gender | 521 | 0.271 | 0.394 | 0.012 | 0.04 | ||||||
| Female | 137 | 39 (28.5)a | 52 (38.0) | 46 (33.6) | 32 (23.4) | 56 (40.9) | 49 (35.8) | ||||
| Male | 384 | 135 (35.2) | 121 (31.5) | 128 (33.3) | 141 (36.7) | 118 (30.7) | 125 (32.6) | ||||
| Race | 506 | 0.672 | 0.390 | 0.775 | 0.736 | ||||||
| Caucasian | 446 | 146 (32.7) | 148 (33.2) | 152 (34.1) | 148 (33.2) | 152 (34.1) | 146 (32.7) | ||||
| Other | 60 | 23 (38.3) | 19 (31.7) | 18 (30.0) | 20 (33.3) | 18 (30.0) | 22 (36.7) | ||||
| Smoking | 508 | 0.282 | 0.270 | 0.288 | 0.115 | ||||||
| No | 117 | 37 (31.6) | 34 (29.1) | 46 (39.2) | 33 (28.2) | 39 (33.3) | 45 (38.5) | ||||
| Yes | 391 | 131 (33.5) | 136 (34.8) | 124 (31.7) | 137 (35.0) | 130 (33.3) | 124 (31.7) | ||||
| Stage | 507 | 0.183 | 0.039 | 0.520 | 0.141 | ||||||
| I | 20 | 3 (15.0) | 8 (40.0) | 9 (45.0) | 6 (30.0) | 8 (40.0) | 6 (30.0) | ||||
| II | 98 | 26 (26.5) | 32 (32.7) | 40 (40.8) | 29 (29.6) | 34 (34.7) | 35 (35.7) | ||||
| III | 105 | 42 (40.0) | 31 (29.5) | 32 (30.5) | 30 (28.6) | 33 (31.4) | 42 (40.0) | ||||
| IV | 284 | 97 (34.2) | 98 (34.5) | 89 (31.3) | 105 (37.0) | 93 (32.8) | 86 (30.3) | ||||
| Grade | 517 | 0.779 | 0.223 | 0.0009 | 0.0006 | ||||||
| I | 63 | 24 (38.1) | 22 (34.9) | 17 (27.0) | 17 (27.0) | 16 (25.4) | 30 (47.6) | ||||
| II | 304 | 101 (33.2) | 102 (33.6) | 101 (33.2) | 86 (28.3) | 115 (37.8) | 103 (33.9) | ||||
| III | 150 | 47 (31.3) | 49 (32.7) | 54 (36.0) | 67 (44.7) | 42 (28.0) | 41 (27.3) | ||||
| Tumor site | 521 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||||||
| Tongue | 200 | 52 (26.0) | 60 (30.0) | 88 (44.0) | 79 (39.5) | 72 (36.0) | 49 (24.5) | ||||
| Pharynx | 135 | 39 (28.9) | 52 (38.5) | 44 (32.6) | 53 (39.3) | 45 (33.3) | 37 (27.4) | ||||
| Other | 186 | 83 (44.6) | 61 (32.8) | 42 (22.6) | 41 (22.0) | 57 (30.7) | 88 (47.3) | ||||
Note:
n (%).
Statistical analyses
Statistical analyses were performed using SAS version 9.2 (SAS Institute Inc.). The overall survival in months was calculated as the time from surgery until either the event of death or last follow-up, whichever came first. Spear-man correlation was performed to analyze the correlations between KL expression and DNA methylation, NSUN2 or LC3 expression. When KL expression and DNA methylation were treated as categorical variables, they were classified into three groups: low, medium and high, based on the distribution of tertile as the cutoff values. The chi-squared test was used to analyze the associations of KL expression and DNA methylation with clinicopathologic variables. Kaplan–Meier survival curves were used to assess the associations of KL expression and DNA methylation with patient survival, and multivariate Cox proportional hazard regression models were used to estimate the HRs and their 95% CIs, after adjusting for patients’ age at diagnosis, disease stage, tumor grade, gender and smoking status. A P-value <0.05 was considered statistically significant.
Results
Association of KL expression and its DNA methylation with clinicopathologic features in patients
In the analyzed 522 tumor samples, the average expression level of KL was 11.4 Fragments Per Kilobase of transcript per Million mapped reads (FPKM) ranging from 0 to 1,422 FPKM; the average DNA methylation level was 0.36 ranging from 0.03 to 0.80 (Table 1). A significant negative correlation was observed between KL DNA methylation and its expression. The correlation coefficient was −0.29 (95% CI: −0.36 to −0.20, P<0.0001; Table 1).
Table 1.
Spearman correlation between KL expression and DNA methylation
| Variable | n | Mean | SD | Median | Range |
|---|---|---|---|---|---|
| KL expression | 522 | 23.6 | 71.5 | 11.4 | 0–1,422 |
| KL DNA methylation | 522 | 0.36 | 0.16 | 0.37 | 0.03–0.80 |
| Correlation coefficient (95% CI) | 522 | −0.29 | (−0.36, −0.20) | ||
| P-value | <0.0001 | ||||
Table 2 shows the associations of KL expression and its DNA methylation with clinicopathologic variables. To limit variation, we first classified patients into three categories: low, medium and high, based on the tertile distribution of KL expression and its methylation, respectively. Although patients with more advanced stage of disease tended to have lower KL expression (P-trend =0.039), KL expression levels between disease stages were not statistically different (P=0.183). However, KL expression was found to be significantly associated with tumor sites (P<0.0001). Patients with tumor on tongue tended to have a high KL expression, while tumor on other sites beyond the tongue and pharynx tended to have a low KL expression. No statistically significant associations were found between KL expression with other variables such as tumor grade, race, gender and smoking status.
KL DNA methylation was found to be significantly associated with gender (P=0.012), tumor grade (P=0.0009) and tumor site (P<0.0001). Female patients tended to have a higher KL methylation than males (P-trend =0.04). Patients with higher tumor grade (poor differentiation) tended to have a lower KL methylation, while those with lower tumor grade tended to have a higher KL methylation (P-trend =0.0006). In addition, it was found that patients with a disease on tongue tended to have a lower KL methylation, while those with a tumor located beyond the tongue and pharynx tended to have a higher KL methylation (P-trend <0.0001). No significant associations were found between KL methylation with other variables, such as race (P=0.775), smoking status (P=0.288) and disease stage (P=0.520).
Associations of KL expression and methylation with patient survival
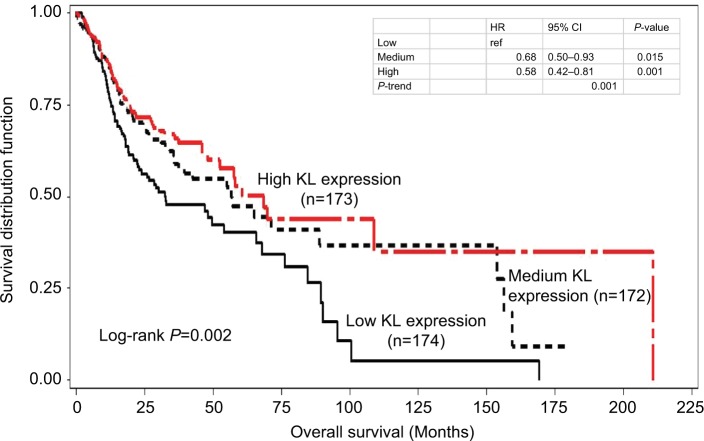
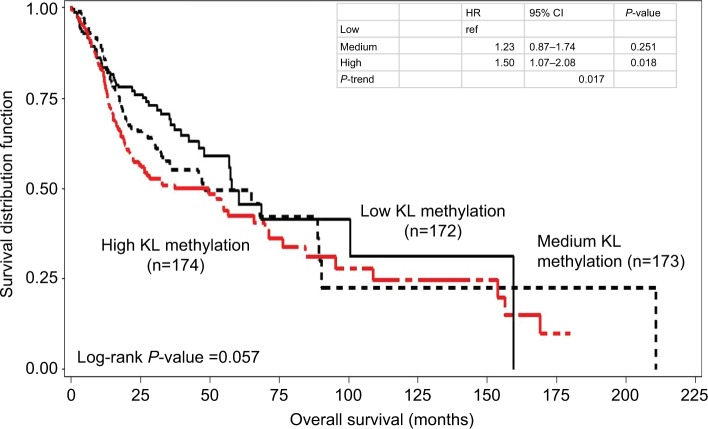
Kaplan–Meier survival curve analyses stratified by either KL expression or KL methylation levels were first performed to examine the associations of KL expression and DNA methylation with overall survival. The results showed that patients with high KL expression had significantly superior overall survival in comparison to those with low KL expression (log-rank P=0.002; Figure 1). The survival curve for patients with low KL expression was clearly separated from the curves for those with either medium or high expression. The medians of overall survival were 68.4 months (95% CI: 42.3–210.8 months) for those with high expression, 56.9 months (95% CI: 35.8–153.8 months) for those with medium expression and 32.6 months (95% CI: 22.3–65.8 months) for those with low expression. The difference in overall survival between those with high and low expression was 25.8 months. The HRs for death were 0.58 (95% CI: 0.42–0.81) for those with high vs low expression and 0.68 (95% CI: 0.50–0.93) for those with medium vs low expression. Conversely, patients with high KL DNA methylation had inferior overall survival in comparison to those with low methylation (log-rank P=0.057; Figure 2). The medians of overall survival were 57.9 months (95% CI: 47.9–159.5 months) for those with low methylation, 48.2 months (95% CI: 32.5–89.3 months) for those with medium methylation and 37.2 months (95% CI: 22.2–65.8 months) for those with high methylation. Patients with low KL DNA methylation had 20.7 months longer overall survival than those with high methylation. The HRs were 1.23 with 95% CI ranging from 0.87 to 1.74 (P=0.251) for patients with medium vs low methylation and 1.50 with 95% CI ranging from 1.07 to 2.08 (P=0.018) for those with high vs low methylation. There was a statistically significant trend (P-trend =0.017).
Figure 1.
Kaplan–Meier overall survival curves stratified by KL expression levels.
Notes: In comparison to low expression, high KL expression had significantly superior overall survival (log-rank P=0.002). The HR was 0.58 with 95% CI ranging from 0.42 to 0.81 (P=0.001) for high vs low, while for medium vs low, the HR was 0.68 with 95% CI ranging from 0.50 to 0.93 (P=0.015). There was a statistically significant trend (P-trend =0.001).
Figure 2.
Kaplan–Meier overall survival curves stratified by KL DNA methylation levels.
Notes: High KL DNA methylation had inferior overall survival than low methylation (log-rank P=0.057). The HR was 1.50 with 95% CI ranging from 1.07 to 2.08 (P=0.018) for high vs low, while for medium vs low, the HR was 1.23 with 95% CI ranging from 0.87 to 1.74 (P=0.251). There was a statistically significant trend (P-trend =0.017).
To further validate the Kaplan–Meier survival curve results, we performed multivariate Cox proportional hazard models with the adjustment of potential confounding variables, including patient age at diagnosis, disease stage, tumor grade, gender and smoking status. Again, the multivariate Cox models showed that patients with high KL expression had a lower risk of death than those with low expression (Table 3). The adjusted HRs were 0.69 (95% CI: 0.50–0.95) for medium vs low (P=0.025) and 0.59 (95% CI: 0.42–0.83) for high vs low (P=0.003). A significantly decreased trend existed (P=0.002). In contrast, high KL DNA methylation significantly increased the risk of death in these patients. The adjusted HRs were 1.24 (95% CI: 0.86–1.78) for medium vs low methylation (P=0.248) and 1.52 (95% CI: 1.07–2.17) for high vs low methylation (P=0.019). There was a significantly increased trend in the risk of death with increased KL DNA methylation (P-trend =0.018).
Table 3.
Associations of KL expression and DNA methylation with the risk of death
| Death | |||
|---|---|---|---|
| Variable | HRa | 95% CI | P-value |
| KL expression | |||
| Low | |||
| Medium | 0.69 | 0.50–0.95 | 0.025 |
| High | 0.59 | 0.42–0.83 | 0.003 |
| P-trend | 0.002 | ||
| Age (per 5 years) | 1.08 | 1.01–1.16 | 0.020 |
| Stage | 1.08 | 0.92–1.26 | 0.370 |
| Grade | 1.05 | 0.85–1.30 | 0.640 |
| Gender (male vs female) | 0.79 | 0.58–1.08 | 0.145 |
| Smoking (smoker vs nonsmoker) | 1.19 | 0.83–1.69 | 0.347 |
| KL methylation | |||
| Low | |||
| Medium | 1.24 | 0.86–1.78 | 0.248 |
| High | 1.52 | 1.07–2.17 | 0.019 |
| P-trend | 0.018 | ||
| Age (per 5 years) | 1.09 | 1.02–1.16 | 0.013 |
| Stage | 1.11 | 0.95–1.30 | 0.202 |
| Grade | 1.06 | 0.86–1.31 | 0.593 |
| Gender (male vs female) | 0.82 | 0.60–1.13 | 0.223 |
| Smoking (smoker vs nonsmoker) | 1.20 | 0.84–1.71 | 0.313 |
Note:
HR was estimated in multivariate Cox proportional hazard regression analysis.
Associations of KL expression and its methylation with LC3 and NSUN2
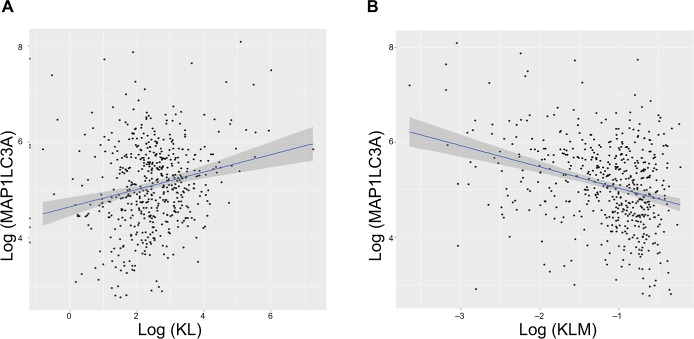
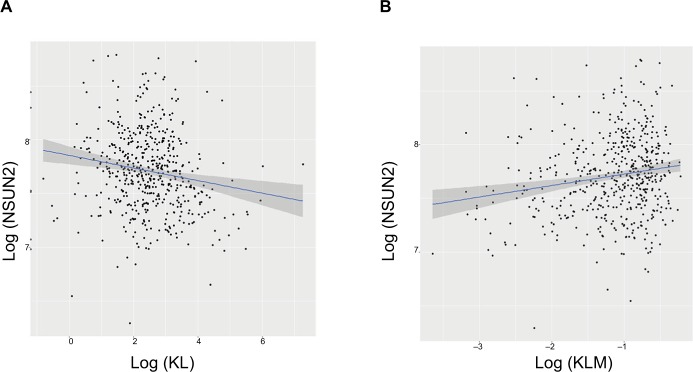
To explore the possible mechanisms underlying the prognostic factors of KL expression and its DNA methylation in HNSCC survival, we examined their associations with the expression of LC3, an autophagy-related gene, and NSUN2, a tRNA methyltransferase gene, both of which have been shown to be critically involved in HNSCC. KL expression was positively associated with LC3 expression (correlation coefficient =0.17, P=0.0003; Figure 3A), whereas KL methylation was negatively associated with LC3 expression (correlation coefficient =−0.27, P=1.01e-12; Figure 3B). On the other hand, KL expression was found to be negatively associated with NSUN2 expression (correlation coefficient =−0.16, P=0.0002; Figure 4A) and KL methylation to be positively related to NSUN2 expression (correlation coefficient =0.15, P=0.004; Figure 4B).
Figure 3.
Associations of KL expression and methylation with LC3 gene expression.
Note: Scatter plots and linear regression line (95% CI in shadow) show that high KL expression was positively associated with LC3, an autophagy gene (correlation coefficient =0.17, P=0.0003) (A), and that KL methylation was negatively associated with LC3 (correlation coefficient =−0.27, P=1.01e–12) (B).
Abbreviations: KL, klotho expression; KLM, klotho methylation.
Figure 4.
Associations of KL expression and methylation with NSUN2 gene expression.
Note: Scatter plots and linear regression line (95% CI in shadow) show that high KL expression was negatively associated with NSUN2 (correlation coefficient =−0.16, P=0.0002) (A), and that KL methylation was positively associated with NSUN2 (correlation coefficient =0.15, P=0.004) (B).
Discussion
To our knowledge, this is the first study to evaluate the prognostic value of KL in predominantly Caucasian HNSCC patients (88.1%) with a relatively large sample size (522 available data). We found an obvious negative correlation between KL expression and its DNA methylation, suggesting that KL DNA methylation leads to the silencing of its expression in HNSCC. Consistent with this inverse relationship, we discovered that patients with either high KL expression or low KL DNA methylation had lower risk of death and a superior overall survival, suggesting KL and its DNA methylation can be potential indicators for prognosis in HNSCC. Moreover, this research revealed significant associations of KL expression and its DNA methylation with NSUN2 and LC3, two reported prognosis indicators for HNCSS.
KL is an antiaging hormone and essentially involved in longevity and cellular homeostasis. It has been reported that KL, as a tumor suppressor, downregulates insulin growth factor (IGF) signaling pathway that is involved in tumorigenesis and progression of many human cancers, including renal cell carcinoma,10 breast cancer,22 lung cancer,9,23 pancreatic cancer4 and HNC.9,12,13,15,24 Compared to normal tissues, tumor tissues usually have a dramatically reduced KL expression.10,19,25 Previous studies have reported reduced KL protein levels in ESCC and OSCC.15,17 Furthermore, decreased KL protein levels usually parallel with increased DNA methylation in its promoter region and are associated with poor prognosis for a variety of cancers, such as hepatocellular cancer,24 gastric cancer26 and ESCC.15 In vitro cell line experiments suggest that restoring KL expression or the treatment with soluble KL protein suppresses cancer cell proliferation.19,25
In addition to downregulation of IGF signaling, KL may also exert its tumor suppressive function by antagonizing the Wnt/β-catenin signaling pathway,27,28 thereby leading to the suppression of oncogenes, such as c-myc and cyclin D1, which are abnormally activated in tumors. Furthermore, downregulation of the fibroblast growth factor pathway-mediated phosphorylation of Akt and ERK1/2 has been attributed to the tumor suppressive effect of KL in pancreatic cancer.4 KL has also been reported to inhibit TGF-β1-induced epithelial–mesenchymal transition (EMT) by directly binding to TGF-β receptor.29 Our previous study revealed that high expression of TWIST1, a key regulator in EMT, was associated with poor survival in HNSCC.30 In this study, we observed a significantly positive relationship between KL mRNA levels and the overall survival of HNSCC patients. For the patients with high KL expression, the overall survival is generally 25.8 months longer than in patients with low KL expression. This finding adds more evidence to support KL as a tumor suppressor in the context of human cancers.
Due to the enriched CpG sites (aka CpG island) in the promoter region, KL expression can be epigenetically silenced by DNA methylation,9,25 and the demethylation agent 5-aza-2′-deoxycytidine restores KL expression.26,31 As expected, in this study, we found a significantly negative correlation between KL DNA methylation and expression in HNSCC tissues. Consistently, we also observed that the correlation of KL methylation with disease survival was always inversely parallel to that of KL expression. That is, lower KL methylation was associated with lower risk of death and superior overall survival. These findings are consistent with previous reports showing that tumor tissues usually had higher KL methylation compared to adjacent normal tissues,9,18,26 and that high KL methylation was associated with poor prognosis and severity of several human cancers.18,24,32 For example, Rubinek et al reported that breast cancer patients with high KL methylation had poor prognosis.9 A similar result of high KL methylation and poor prognosis was reported in hepatocellular carcinoma.24 In tumor tissues from OSCC patients, DNA methyltransferase 3a was found to be higher and the KL protein level was lower than in normal tissues.13 Together, these data suggest that DNA methylation of KL plays an important role in the progression of HNSCC.33,34 However, a research done in HNSCC samples from Saudi Arabia reported that in HNSCC patients with moderately differentiated tumors, KL methylation was related to better survival rate, compared to that in those without KL methylation.17
Autophagy is critical for maintaining cellular homeostasis by the degradation of misfolded proteins and damaged organelles. In cancers, autophagy can play dual roles as tumor promotor or suppressor depends on the tumor stages. At the early stage, autophagy-mediated cell survival may contribute to cancer etiology and progression, while at a later stage, activation of autophagy can be antitumorigenic.35 In oral tongue squamous cell carcinoma (OTSCC) patients, there is evidence that the expression of autophagy-related gene beclin-1 was decreased in the tumor tissue and correlated with poor prognosis. In addition, overexpression of this autophagy gene inhibited proliferation and clonogenicity of OTSCC cells.36 LC3 is a specific marker for the autophagosome. In this study, we found that either high KL expression or low KL DNA methylation correlated positively with LC3 level in HNSCC. KL has been reported to exert its tumor suppressive function by activation of autophagy in several cancers.37,38 Our data thus suggest a link between LC3 and KL in HNSCC.
NSUN2 is an RNA methyltransferase involved in cell proliferation and differentiation.39,40 NSUN2 is a direct target of MYC,41 and its overexpression has been reported to contribute to the development of several cancers, such as breast cancer42 and HNSCC.20 Moreover, high NSUN2 level is positively associated with poor prognosis in HNSCC.20 Recently, it was discovered that NSUN2 is also a transcription target of the Lef1/β-catenin complex,39 a well-known downstream pathway inhibited by KL.43 In agreement with these reports, we found that NSUN2 was negatively correlated with KL expression but positively correlated with KL methylation in HNSCC. To our knowledge, this is the first study to demonstrate a relationship between KL and NSUN2 in HNSCC.
Conclusion
This study points out that KL gene expression and its promoter DNA methylation are potential prognostic markers for HNSCCs. Moreover, our findings also suggest correlations of KL with both LC3 and NSUN2 in HNSCC. Further studies on the roles of LC3 and NSUN2 as downstream targets regulated by KL in HNSCC are warranted.
Acknowledgments
The results shown in this manuscript are based upon the data generated by The Cancer Genome Atlas Research Network http://cancergenome.nih.gov/. This work was funded by the National Natural Science Foundation of China (grant number 81600805).
Footnotes
Author contributions
PW designed the study. YZ, XC and QC collected and analyzed the TCGA data. YZ, LW and XZ wrote the manuscript. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Braakhuis BJ, Leemans CR, Visser O. Incidence and survival trends of head and neck squamous cell carcinoma in the Netherlands between 1989 and 2011. Oral Oncol. 2014;50(7):670–675. doi: 10.1016/j.oraloncology.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Lefebvre JL. Current clinical outcomes demand new treatment options for SCCHN. Ann Oncol. 2005;16(suppl_6):vi7–vi12. doi: 10.1093/annonc/mdi452. [DOI] [PubMed] [Google Scholar]
- 4.Abramovitz L, Rubinek T, Ligumsky H, et al. KL1 internal repeat mediates Klotho tumor suppressor activities and inhibits bFGF and IGF-I signaling in pancreatic cancer. Clin Cancer Res. 2011;17(13):4254–4266. doi: 10.1158/1078-0432.CCR-10-2749. [DOI] [PubMed] [Google Scholar]
- 5.Imura A, Iwano A, Tohyama O, et al. Secreted Klotho protein in sera and CSF: implication for post-translational cleavage in release of Klotho protein from cell membrane. FEBS Lett. 2004;565(1–3):143–147. doi: 10.1016/j.febslet.2004.03.090. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi Y, Kuro-O M, Ishikawa F. Aging mechanisms. Proc Natl Acad Sci U S A. 2000;97(23):12407–12408. doi: 10.1073/pnas.210382097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuro-O M, Matsumura Y, Aizawa H, et al. Mutation of the mouse Klotho gene leads to a syndrome resembling ageing. Nature. 1997;390(6655):45–51. doi: 10.1038/36285. [DOI] [PubMed] [Google Scholar]
- 8.Wolf I, Levanon-Cohen S, Bose S, et al. Klotho: a tumor suppressor and a modulator of the IGF-1 and FGF pathways in human breast cancer. Oncogene. 2008;27(56):7094–7105. doi: 10.1038/onc.2008.292. [DOI] [PubMed] [Google Scholar]
- 9.Rubinek T, Shulman M, Israeli S, et al. Epigenetic silencing of the tumor suppressor Klotho in human breast cancer. Breast Cancer Res Treat. 2012;133(2):649–657. doi: 10.1007/s10549-011-1824-4. [DOI] [PubMed] [Google Scholar]
- 10.Gigante M, Lucarelli G, Divella C, et al. Soluble serum αKlotho is a potential predictive marker of disease progression in clear cell renal cell carcinoma. Medicine. 2015;94(45):e1917. doi: 10.1097/MD.0000000000001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai D, Wang Q, Li X, Liu J, Ma X, Xu W. Klotho inhibits human follicular thyroid cancer cell growth and promotes apoptosis through regulation of the expression of stanniocalcin-1. Oncol Rep. 2016;35(1):552–558. doi: 10.3892/or.2015.4358. [DOI] [PubMed] [Google Scholar]
- 12.Usuda J, Ichinose S, Ishizumi T, et al. Klotho predicts good clinical outcome in patients with limited-disease small cell lung cancer who received surgery. Lung Cancer. 2011;74(2):332–337. doi: 10.1016/j.lungcan.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Adhikari BR, Uehara O, Matsuoka H, et al. Immunohistochemical evaluation of Klotho and DNA methyltransferase 3a in oral squamous cell carcinomas. Med Mol Morphol. 2017;50(3):155–160. doi: 10.1007/s00795-017-0156-9. [DOI] [PubMed] [Google Scholar]
- 14.Lu L, Katsaros D, Wiley A, de la Longrais IA, Puopolo M, Yu H. Klotho expression in epithelial ovarian cancer and its association with insulin-like growth factors and disease progression. Cancer Invest. 2008;26(2):185–192. doi: 10.1080/07357900701638343. [DOI] [PubMed] [Google Scholar]
- 15.Tang X, Fan Z, Wang Y, et al. Expression of Klotho and β-catenin in esophageal squamous cell carcinoma, and their clinicopathological and prognostic significance. Dis Esophagus. 2016;29(3):207–214. doi: 10.1111/dote.12289. [DOI] [PubMed] [Google Scholar]
- 16.Schuebel KE, Chen W, Cope L, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3(9):1709–1723. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alsofyani AA, Alsiary RA, Samkari A, et al. Prognostic potential of KLOTHO and SFRP1 promoter methylation in head and neck squamous cell carcinoma. J Appl Genet. 2017;58(4):459–465. doi: 10.1007/s13353-017-0404-7. [DOI] [PubMed] [Google Scholar]
- 18.Perveez M, Ajaz M, Afroze D. Promoter hypermethylation of KLOTHO; an anti-senescence related gene in colorectal cancer patients of Kashmir valley. Mol Biol Res Commun. 2015;4(4):217–224. [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J, Jeong DJ, Kim J, et al. The anti-aging gene KLOTHO is a novel target for epigenetic silencing in human cervical carcinoma. Mol Cancer. 2010;9(1):109. doi: 10.1186/1476-4598-9-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu L, Zhu G, Zeng H, Xu Q, Holzmann K. High tRNA transferase NSUN2 gene expression is associated with poor prognosis in head and neck squamous carcinoma. Cancer Invest. 2018;36(4):246–253. doi: 10.1080/07357907.2018.1466896. [DOI] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ligumsky H, Rubinek T, Merenbakh-Lamin K, et al. Tumor suppressor activity of Klotho in breast cancer is revealed by structure–function analysis. Mol Cancer Res. 2015;13(10):1398–1407. doi: 10.1158/1541-7786.MCR-15-0141. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Chen L, Huang G, et al. Klotho sensitizes human lung cancer cell line to cisplatin via PI3k/Akt pathway. PLoS One. 2013;8(2):e57391. doi: 10.1371/journal.pone.0057391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie B, Zhou J, Yuan L, et al. Epigenetic silencing of Klotho expression correlates with poor prognosis of human hepatocellular carcinoma. Hum Pathol. 2013;44(5):795–801. doi: 10.1016/j.humpath.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 25.Pan J, Zhong J, Gan LH, et al. Klotho, an anti-senescence related gene, is frequently inactivated through promoter hypermethylation in colorectal cancer. Tumour Biol. 2011;32(4):729–735. doi: 10.1007/s13277-011-0174-5. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Wang X, Wang X, et al. Klotho is silenced through promoter hypermethylation in gastric cancer. Am J Cancer Res. 2011;1(1):111–119. [PMC free article] [PubMed] [Google Scholar]
- 27.Kurosu H, Yamamoto M, Clark JD, et al. Suppression of aging in mice by the hormone Klotho. Science. 2005;309(5742):1829–1833. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H, Fergusson MM, Castilho RM, et al. Augmented Wnt signaling in a mammalian model of accelerated aging. Science. 2007;317(5839):803–806. doi: 10.1126/science.1143578. [DOI] [PubMed] [Google Scholar]
- 29.Doi S, Zou Y, Togao O, et al. Klotho inhibits transforming growth factor-beta1 (TGF-beta1) signaling and suppresses renal fibrosis and cancer metastasis in mice. J Biol Chem. 2011;286(10):8655–8665. doi: 10.1074/jbc.M110.174037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Y, Zhang W, Wang P. Smoking and gender modify the effect of TWIST on patient survival in head and neck squamous carcinoma. Oncotarget. 2017;8(49):85816–85827. doi: 10.18632/oncotarget.20682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai KD, Lee WX, Chen W, et al. Upregulation of PRMT6 by LPS suppresses Klotho expression through interaction with NF-κB in glomerular mesangial cells. J Cell Biochem. 2018;119(4):3404–3416. doi: 10.1002/jcb.26511. [DOI] [PubMed] [Google Scholar]
- 32.Yang W, Wang X, Li X, et al. The specific methylation characteristics of cancer related genes in Chinese colorectal cancer patients. Tumour Biol. 2014;35(8):8267–8279. doi: 10.1007/s13277-014-2100-0. [DOI] [PubMed] [Google Scholar]
- 33.Demokan S, Dalay N. Role of DNA methylation in head and neck cancer. Clin Epigenetics. 2011;2(2):123–150. doi: 10.1007/s13148-011-0045-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Worsham MJ, Stephen JK, Chen KM, et al. Delineating an epigenetic continuum in head and neck cancer. Cancer Lett. 2014;342(2):178–184. doi: 10.1016/j.canlet.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sannigrahi MK, Singh V, Sharma R, Panda NK, Khullar M. Role of autophagy in head and neck cancer and therapeutic resistance. Oral Dis. 2015;21(3):283–291. doi: 10.1111/odi.12254. [DOI] [PubMed] [Google Scholar]
- 36.Hu Z, Zhong Z, Huang S, et al. Decreased expression of Beclin-1 is significantly associated with a poor prognosis in oral tongue squamous cell carcinoma. Mol Med Rep. 2016;14(2):1567–1573. doi: 10.3892/mmr.2016.5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shu G, Xie B, Ren F, et al. Restoration of Klotho expression induces apoptosis and autophagy in hepatocellular carcinoma cells. Cell Oncol. 2013;36(2):121–129. doi: 10.1007/s13402-012-0118-0. [DOI] [PubMed] [Google Scholar]
- 38.Xie B, Zhou J, Shu G, et al. Restoration of Klotho gene expression induces apoptosis and autophagy in gastric cancer cells: tumor suppressive role of Klotho in gastric cancer. Cancer Cell Int. 2013;13(1):18. doi: 10.1186/1475-2867-13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blanco S, Kurowski A, Nichols J, Watt FM, Benitah SA, Frye M. The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 2011;7(12):e1002403. doi: 10.1371/journal.pgen.1002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hussain S, Benavente SB, Nascimento E, et al. The nucleolar RNA methyltransferase Misu (NSun2) is required for mitotic spindle stability. J Cell Biol. 2009;186(1):27–40. doi: 10.1083/jcb.200810180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frye M, Watt FM. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr Biol. 2006;16(10):971–981. doi: 10.1016/j.cub.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 42.Frye M, Dragoni I, Chin SF, et al. Genomic gain of 5p15 leads to overexpression of Misu (NSUN2) in breast cancer. Cancer Lett. 2010;289(1):71–80. doi: 10.1016/j.canlet.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 43.Chang B, Kim J, Jeong D, et al. Klotho inhibits the capacity of cell migration and invasion in cervical cancer. Oncol Rep. 2012;28(3):1022–1028. doi: 10.3892/or.2012.1865. [DOI] [PubMed] [Google Scholar]