Abstract
Purpose
The mutation status and prognostic value of PIK3CA in breast cancer were widely investigated, which showed significant difference among the patients from vast areas around the world. In this study, the frequency, distribution, bias, and burden of PIK3CA mutations and their relationships with clinicopathologic variables and prognostic significances were investigated in the breast cancer patients from Central China.
Materials and methods
Somatic mutations in exon 9 and exon 20 of PIK3CA gene were analyzed using Sanger sequencing combining with targeted next generation sequencing in 494 breast cancer patients from Central China. The correlations between PIK3CA mutations and clinicopathological characteristics and the prognostic values of multiple PIK3CA mutation statuses were evaluated.
Results
PIK3CA mutations were found in 38% of the patients and associated with estrogen receptor-positive, progesterone receptor-positive, low Ki67 labeling index, and luminal/human epidermal growth factor receptor 2-enriched subtypes. Meanwhile, the prognosis of the total patients and the patients in old diagnostic age, progesterone receptor-negative, low Ki67 labeling index, and luminal/human epidermal growth factor receptor 2-enriched subgroups was significantly related to PIK3CA mutations. Most interestingly, the distribution, bias, and burden of PIK3CA mutations were correlated with different clinical, pathological, and molecular features as well as distinct prognostic implications in multiple breast cancer subgroups.
Conclusion
The frequency, distribution, bias, and burden of PIK3CA mutations were associated with various clinical, pathological, and molecular characteristics in the breast cancer patients from Central China. These different mutation statuses can be used as potential indicators of prognosis in multiple breast cancer subgroups.
Keywords: breast cancer, PIK3CA, clinicopathological characteristics, prognosis
Introduction
Breast cancer is the most diagnosed female cancer and the fifth leading cause of cancer death, and the mortality rate is 70,700 patients every year in China.1 The incidence and mortality of this cancer in women is increasing in recent years.1 Classical therapeutic strategy for this disease in clinic is combining the loco-regional therapy (surgery and radiation) with subsequent adjuvant systemic therapy.2–4 However, as a highly heterogeneous disease, the therapeutic effect of breast cancer is determined by various classical clinical phenotypes (age at diagnosis, tumor size, stage of tumor, lymph node invasiveness, etc.) as well as intrinsic molecular subtypes (triple negative, human epidermal growth factor receptor 2 [HER2]-enriched, luminal A and luminal B subtypes).2,5 Besides, in the era of personalized medicine, genetic factors get increasing attention, which are considered as the most important aspects in tumor initiation, progression, prognosis, and drug resistance.4,6 Therefore, accurate molecular diagnosis of specific biomarkers that can respond to and monitor the therapeutic effect of breast cancer is essential.
As a major component of PI3K/AKT/mTOR signaling pathway, the activation of phosphatidylinositol-3 kinases (PI3K) which interacts with transmembrane tyrosine-kinase growth factor receptors subsequently activates AKT, mTOR, MAPK signaling pathways, and plays essential roles in multiple cellular processes including translation regulation, protein synthesis, cell metabolism, autophagy, cell adhesion, and apoptosis.7,8 Numerous studies illustrated that hyperactivation of PI3K signaling intimately was associated with pathogenesis and progression of various types of cancers.9–13 There are two most important events that could constitutively activate the PI3K signaling pathway – loss of the function of tumor suppressor phosphate and tension homology deleted on chromosome ten (PTEN) and activation mutations in p110α catalytic subunit which encoded by the PIK3CA gene.8,14 Between them, PIK3CA is one of the most prevalent mutated genes in breast cancer even though its mutation frequency varies among different investigations.15–19 According to the Catalogue of Somatic Mutations in Cancer Database (COSMIC, https://www.sanger.ac.uk), >90% of the PIK3CA mutations are located in the helical (exon 9) or kinase (exon 20) domains, including the hotspot mutations E542K, E545K in exon 9 and H1047R, H1047L in exon 20.
Therefore, multiple studies have been performed to investigate the relationship between PIK3CA mutations and clinicopathological features, prognostic value, or therapeutic relevance of breast cancer in different countries and races.15,20–22 However, the results are controversial among these different studies even those from the same country. For example, in two investigations in the patients from Italy, Buttitta et al23 showed that PIK3CA mutations were associated with HER2-negative clinicopathological subtype, and in contrast, no association was determined by Barbareschi et al.24 Harlé et al25 attempted to correlate the PIK3CA mutations with low-grade tumors in breast cancer patients from Nancy, France, while by investigating the patients from Saint-Cloud, France, Cizkova et al26 believed that there were correlations between PIK3CA mutations and estrogen receptor (ER)-positive, progesterone receptor (PR)-positive, HER2 negative, low tumor grade or small tumor size. Considering the small sample sizes, population-related peculiarities of patients, and different methods used for detection of the mutations, these phenomena might demonstrate that it is crucially important to analyze carefully these factors in more areas including a larger population with a unique method. The outcomes would help us to deeply understand breast cancer and offer us specific and predictive biomarkers to be used for breast cancer diagnosis and treatment.
In this study, we investigated the frequency, distribution, bias, and burden of PIK3CA mutations in 494 breast cancer patients from a single center located in Central China and explored their associations with clinicopathological features and disease prognosis. These data will expand the knowledge of PIK3CA mutations related to breast cancer, which can be further used to provide precision medicine strategies to the breast cancer patients in Central China.
Materials and methods
Patients
The study was conducted in accordance with the Declaration of Helsinki. After obtaining written informed consent from all the patients and approval of the Ethics Committee of the First Affiliated Hospital of USTC, 537 formalin-fixed, paraffin-embedded (FFPE) primary breast tumor tissue samples were collected at the Department of Pathology, the First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China from 2010 to 2017. Ultimately, 494 samples were eligible for our analysis and the clinical characteristics of these patients are summarized in Table S1. All the samples were obtained from female patients who did not receive preoperative treatment. Based on the American Joint Committee on Cancer TNM system (2010),27 the pathological diagnosis of each sample was made by at least three pathologists. The Nottingham Prognostic Index (NPI)28 was calculated to determine the prognosis of the patients after surgery using the following formula: NPI =0.2×tumor size (cm)+ grade (1–3)+ lymph node status (1–3). Progression-free survival (PFS) was defined as the time span between surgery date and the first relapse time of tumor, second primary tumor, death, or last follow-up.29 Among 494 patients, 303 PFS and overall survival (OS) data were collected in which 28 of them had relapsed tumor or second primary tumor and 46 patients died. The follow-up period was from 5 to 97 months with a median time of 35 months.
Molecular subtypes of breast cancer
ER, PR, and HER2 statuses were determined by immunohistochemical (IHC) staining. ER or PR was regarded as positive when >1% of tumor cells were stained based on the St. Gallen International Expert Consensus.30 HER2 was considered positive when complete or intense membrane staining was determined in >30% of tumor cells. The subtypes of samples were classified using anatomopathological classification according to St. Gallen International Expert Consensus.30 The expression of Ki67 – a cellular marker for proliferation – was also examined by IHC in all patients.
DNA preparation and targeted next generation sequencing
Tumor content of >50% was qualified through H&E staining and selected for DNA extraction. Genomic DNA was extracted from one 10 µm section using the GeneRead DNA FFPE Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The quantity and quality of extracted DNA were determined using Nanodrop spectrophotometer 2000 (Thermo Fisher Scientific, MA, USA) and Qubit 3.0 (Life Technologies, Carlsbad, CA, USA).
Genetic variants of 24 samples were screened with TruSeq Amplicon Cancer Panel (Illumina, San Diego, CA, USA) using the NextSeq 500 sequencing system (Illumina). Mutations in 212 amplicons from 48 genes were examined in these samples, including BRAF, KRAS, EGFR, TP53, NRAS, ALK, IDH1, FGFR, PTEN, RB, ATM, PIK3CA, and other important cancer-related genes. Among them, the total 20 exons of PIK3CA were targeted.
After sequencing, mapping, and alignment, single nucleotide variants (SNVs) and indels were called and annotated based on the method described by The Cancer Genome Altas.31 Variants with insufficient coverage (minimum depth of coverage <8) and low variant allele frequency (<0.03) were filtered out.
PCR amplification of the PIK3CA exon 9 and exon 20 fragments
The exon 9 and exon 20 of PIK3CA gene were amplified using the following primers: exon 9 forward 5′-TGTAAAACGACG-GCCAGTCAGAGTAACAGACTAGCTAGAGACAATG-3′, exon 9 reverse 5′-CAGGAAACAGCTATGACCAATCTC-CATTTTAGCACTTACCTGTGAC-3′, and exon 20 forward 5′-TGAGCAAGAGGCTTTGGAGTAT-3′, exon 20 reverse 5′-CCTATGCAATCGGTCTTTGC-3′. The PCR was performed in a 20 µL reaction system using FastStart Essential DNA Green Master Kit (Roche, Mannheim, Germany) with 1 µL gDNA, 1 µL forward primer (10 nmol/mL), 1 µL reverse primer (10 nmol/mL), and 10 µL FastStart Essential DNA Green Master. The PCR was carried out on a LightCycler 96 Real-Time PCR System (Roche) under the following conditions: initial denaturation at 95°C for 5 minutes, then 45 cycles of 95°C for 20 seconds, 60°C for 30 seconds, and 72°C for 30seconds.
Detection of PIK3CA mutations by Sanger sequencing
For sequence analysis, PCR products were purified by PCR Product Purification Kit (Generalbiol, Anhui, China) and subjected to bidirectional dye-terminator sequencing using M13 forward primer 5′-TGTAAAACGACGGCCAGT-3′ for exon 9 amplicon and exon 20 reverse primer 5′-CCTATG-CAATCGGTCTTTGC-3′ for exon 20 amplicon. Sequencing fragments were detected by capillary electrophoresis using the ABI 3730xl DNA analyzer (Applied Biosystems, Carlsbad, CA, USA) and then analyzed by SnapGene Viewer 4.2.6. The mutations were identified by manual review.
Statistical analysis
The relations between PIK3CA mutation statuses and clinicopathological characteristics were assessed by the Pearson chi-squared tests using SPSS software v19.0.0 (IBM, NY, USA). The HR of PIK3CA mutation as well as clinicopathological variables was calculated by the Cox proportional hazards regression model in univariate analysis. Based on the PFS in different mutation conditions, the Kaplan–Meier survival curves were drawn using GraphPad Prism software v5.01 (GraphPad, San Diego, CA, USA) and the significant differences were displayed by the log-rank test (SPSS v 19.0.0, IBM). Statistical significance was considered as P<0.05.
Results
Somatic mutations in breast cancer patients from Central China
To investigate the somatic mutations in breast cancer, 24 primary tumor samples were analyzed using targeted next generation sequencing (NGS). A mean coverage depth of 8,525× was achieved. 94.4% of amplicons were covered at >500× depth. After SNV and indel calling, a total of 93 mutations were detected, including 27 synonymous SNVs, 48 nonsynonymous SNVs, 3 stopgains, 3 splicings, 10 frameshift deletions, 1 nonframeshift deletion, and 1 frameshift insertion. The overall mutation frequency was 2.75 nonsilent mutations (range of 0–12 mutations) per sample. As shown in Figure 1, the most frequently mutated gene was TP53 (41.7%, 10 of the 24 patients) with various mutation types (missense mutation, nonsense mutation, frameshift insertion, and splicing). PIK3CA mutation was found in 33% of the samples, which ranked as the second highest mutated gene. All of the PIK3CA mutations were hotspot mutations including three E542K, three E545K, and two H1047R (Table S2). This much higher mutation rate in PIK3CA exon 9 (75%) in breast cancer was quite different from previous reports19,32–34 which attracted our attention. Besides, we examined the clinicopathological variables of these patients and found that the samples harbored exon 9 mutations were HER2-negative tumors and all belonged to luminal B molecular subtype (Table S2). Furthermore, the only two tumor-relapsed cases were also from the group with exon 9 mutations, which demonstrated that exon 9 mutations might be related to poor prognosis (Table S2). Therefore, we hypothesized that the proportion of PIK3CA exon 9 mutations was much higher in the female breast cancer patients from our area (Central China) than the other regions, and this preference was correlated with special clinicopathological characteristics and tumor prognosis.
Figure 1.
Somatic mutations of 24 significant mutated genes in 24 breast cancer patients.
Notes: The figure shows an overview of genomic alterations (right legend) in particular genes (rows) affecting individual samples (columns). The missense mutation, nonsense mutation, frame shift insertion, frame shift deletion, in frame deletion, splice site mutation, and multiple mutation are shown as green, red, purple, blue, brown, yellow, and black, respectively. Left legend denotes the percentage of mutations in 24 breast cancer samples and right plot represents the total number of mutations for each gene.
PIK3CA mutations detection in archival FFPE tissues
To verify our hypothesis, the mutations in exon 9 and exon 20 of PIK3CA gene were examined by direct sequencing in 537 primary breast tumor samples including the 24 specimens analyzed by NGS to validate the results of sequencing. Among the succeeded sequenced 494 (92%) samples, PIK3CA mutations were determined in 188 (38%) tumors, including 74 mutations in exon 9, 106 mutations in exon 20, and 8 in both exons 9 and 20 (Table 1). The hotspot mutations accounted for 74.6% of total mutations (153/205) in which E542K (p.542), E545K (p.545), and H1047R/L (p.1047) were found in 28, 34, and 91 patients, respectively, with p.1047 ranking the highest (Table 1). Seventeen patients carried two mutations and two of them had H1047R simultaneous with E542K or E545K, which had never been reported before (Table 1). Meanwhile, we characterized 52 non-hotspot mutations in which 1 silent and 5 nonsense mutations were observed in 37 patients (Table 1).
Table 1.
PIK3CA mutation profiles in exons 9 and 20 in breast cancers (n=494)
| Exon | Nucleotide change | Codon change | Effect | Hotspot mutation | Frequency (%) | Number of mutations |
|---|---|---|---|---|---|---|
|
| ||||||
| 9 | c.1613A>G | p.Asp538Gly | Missense | No | <1 | 1 |
| 9 | c.1615C>T | p.Pro539Ser | Missense | No | <1 | 1 |
| 9 | c.1624G>A | p.Glu542Lys | Missense | Yes | 13.8 | 26 |
| 9 | c.1627A>G | p.Ile543Val | Missense | No | <1 | 1 |
| 9 | c.1633G>A | p.Glu545Lys | Missense | Yes | 16.5 | 31 |
| 9 | c.1633G>C | p.Glu545Gln | Missense | No | <1 | 1 |
| 9 | c.1634A>C | p.Glu545Ala | Missense | No | 1.6 | 3 |
| 9 | c.1634A>G | p.Glu545Gly | Missense | No | <1 | 2 |
| 9 | c.1636C>A | p.Gln546Lys | Missense | No | <1 | 1 |
| 9 | c.1637A>C | p.Gln546Pro | Missense | No | <1 | 1 |
| 9 | c.1637A>G | p.Gln546Arg | Missense | No | 1.6 | 3 |
| 9 | c.1651C>T | p.Leu551Leu | Silent | No | <1 | 1 |
| 20 | c.3120G>A | p.Met1040Ile | Missense | No | <1 | 2 |
| 20 | c.3127A>G | p.Met1043Val | Missense | No | <1 | 2 |
| 20 | c.3129G>A | p.Met1043Ile | Missense | No | <1 | 1 |
| 20 | c.3139C>T | p.His1047Tyr | Missense | No | <1 | 1 |
| 20 | c.3140A>G | p.His1047Arg | Missense | Yes | 38.3 | 72 |
| 20 | c.3140A>T | p.His1047Leu | Missense | Yes | 5.3 | 10 |
| 20 | c.3145G>C | p.Gly1049Arg | Missense | No | <1 | 1 |
| 20 | c.3146G>A | p.Gly1049Asp | Missense | No | <1 | 1 |
| 20 | c.3148G>A | p.Gly1050Ser | Missense | No | <1 | 1 |
| 20 | c.3152G>A | p.Trp1051* | Nonsense | No | <1 | 1 |
| 20 | c.3153G>A | p.Trp1051* | Nonsense | No | <1 | 1 |
| 20 | c.3154A>G | p.Thr1052Ala | Missense | No | <1 | 1 |
| 20 | c.3155C>T | p.Thr1052Ile | Missense | No | <1 | 1 |
| 20 | c.3166G>A | p.Asp1056Asn | Missense | No | <1 | 1 |
| 20 | c.3191A>G | p.Gln1064Arg | Missense | No | <1 | 2 |
| 20 | c.3194A>T | p.His1065Leu | Missense | No | <1 | 1 |
| 9 | c.[1613A>G(+)1633G>A] | p.[Asp538Gly(+)Glu545Lys] | Missense | Yes/No | <1 | 1 |
| 9 | c.[1613A>G(+)1633G>C] | p.[Asp538Gly(+)Glu545Gln] | Missense | No | <1 | 1 |
| 9+20 | c.[1624G>A(+)3140A>G] | p.[Glu542Lys(+)His1047Arg] | Missense | Yes | <1 | 1 |
| 9+20 | c.[1624G>A(+)3146G>A] | p.[Glu542Lys(+)Gly1049Asp] | Missense | Yes/No | <1 | 1 |
| 9+20 | c.[1633G>A(+)3140A>G] | p.[Glu545Lys(+)His1047Arg] | Missense | Yes | <1 | 1 |
| 9+20 | c.[1633G>A(+)3170G>A] | p.[Glu545Lys(+)Trp1057*] | Nonsense | Yes/No | <1 | 1 |
| 9+20 | c.[1636C>A(+)3166A>G] | p.[Gln546Lys(+)Glu1056Asn] | Missense | No | <1 | 1 |
| 9+20 | c.[1637A>G(+)3140A>G] | p.[Gln546Arg(+)His1047Arg] | Missense | Yes/No | <1 | 1 |
| 9+20 | c.[1637A>G(+)3155C>T] | p.[Gln546Arg(+)Thr1052Ile] | Missense | No | <1 | 1 |
| 9+20 | c.[1638G>A(+)3140A>G] | p.[Gln546Gln(+)His1047Arg] | Missense | Yes/No | <1 | 1 |
| 20 | c.[3133G>A(+)3152G>A] | p.[Asp1045Asn(+)Trp1051*] | Nonsense | No | <1 | 1 |
| 20 | c.[3137C>T(+)3140A>G] | p.[Ala1046Val(+)His1047Arg] | Missense | Yes/No | <1 | 1 |
| 20 | c.[3140A>G(+)3145G>A] | p.[His1047Arg(+)Gly1049Ser] | Missense | Yes/No | <1 | 1 |
| 20 | c.[3140A>G(+)3146G>A] | p.[His1047Arg(+)Gly1049Asp] | Missense | Yes/No | <1 | 1 |
| 20 | c.[3140A>G(+)3156A>T] | p.[His1047Arg(+)Thr1052Thr] | Missense | Yes/No | <1 | 1 |
| 20 | c.[3140A>G(+)3166G>A] | p.[His1047Arg(+)Asp1056Asn] | Missense | Yes/No | <1 | 1 |
| 20 | c.[3148G>A(+)3153G>A] | p.[Gly1050Ser(+)Trp1051*] | Nonsense | No | <1 | 1 |
| Total =188 | ||||||
In addition, we found 14 new PIK3CA mutations in exon 9 and 20 in breast cancers, which have not been reported by COSMIC. Among the newly found mutations, 8 of them were located in exon 9, 4 in exon 20, and the other 2 in both exon 9 and 20 (Table S3). Whether they have any clinical significance needs to be studied further. Thus, these new mutations were not taken for the following analyses in this study.
Association of PIK3CA gene mutations with clinicopathological data
As shown in Table 2, PIK3CA mutations were positively associated with ER-positive (P=0.016), PR-positive (P=0.007), and low Ki67 labeling index (P=0.001) tumors. Meanwhile, they were negatively correlated with triple-negative breast cancer subtype (P=0.003), but were not associated with age at diagnosis, tumor stage, lymph node status, tumor size, or HER2 status.
Table 2.
Association between PIK3CA mutations and standard clinical, pathological, and biological features of breast cancer
| Parameters | Category | n |
PIK3CA
|
Exon 9
|
Exon 20
|
p.542/545
|
p.1047
|
PIK3CA mutation
|
PIK3CA mutation burdens
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mut | P-value | Mut | P-value | Mut | P-value | Mut | P-value | Mut | P-value | Hotspot | Non- hotspot | P-value | 1 Mut | 2 Muts | P-value | |||
|
| ||||||||||||||||||
| Total | 494 | 188 (38%) | 82 (17%) | 114 (23%) | 62 (13%) | 91 (18%) | 153 (75%) | 52 (25%) | 171 (91%) | 17 (9%) | ||||||||
| Age (years) | <40 | 91 | 28 (31%) | 0.106 | 15 (16%) | 0.974 | 15 (16%) | 0.098 | 11 (12%) | 0.883 | 10 (11%) | 0.043 | 21 (66%) | 11 (34%) | 0.202 | 24 (86%) | 4 (14%) | 0.294 |
| ≥ 40 | 403 | 160 (40%) | 67 (17%) | 99 (25%) | 51 (13%) | 81 (20%) | 132 (76%) | 41 (24%) | 147 (92%) | 13 (8%) | ||||||||
| Tumor stage | I | 12 | 4 (33%) | 0.216 | 0 (0%) | 0.191 | 4 (33%) | 0.413 | 0 (0%) | 0.280 | 3 (25%) | 0.580 | 3 (75%) | 1 (25%) | 1.000 | 4 (100%) | 0 (0%) | 0.776 |
| II | 250 | 105 (42%) | 46 (18%) | 63 (25%) | 35 (14%) | 51 (20%) | 86 (75%) | 28 (25%) | 96 (91%) | 9 (9%) | ||||||||
| III | 205 | 70 (34%) | 31 (15%) | 43 (21%) | 23 (11%) | 35 (17%) | 58 (75%) | 19 (25%) | 63 (90%) | 7 (10%) | ||||||||
| Unknown | 27 | 9 | 5 | 4 | 4 | 2 | 6 | 4 | 8 | 1 | ||||||||
| Lymph node status | 0 | 229 | 86 (38%) | 0.696 | 34 (15%) | 0.543 | 54 (24%) | 0.930 | 25 (11%) | 0.637 | 41 (18%) | 0.780 | 66 (70%) | 28 (30%) | 0.199 | 78 (91%) | 8 (9%) | 0.391 |
| 1–3 | 108 | 45 (42%) | 21 (19%) | 26 (24%) | 15 (14%) | 19 (18%) | 34 (72%) | 13 (28%) | 43 (96%) | 2 (4%) | ||||||||
| >3 | 131 | 48 (37%) | 23 (18%) | 29 (22%) | 18 (14%) | 27 (21%) | 45 (83%) | 9 (17%) | 42 (88%) | 6 (12%) | ||||||||
| Unknown | 26 | 9 | 4 | 5 | 4 | 4 | 8 | 2 | 8 | 1 | ||||||||
| Tumor size (cm) | <2 | 124 | 42 (34%) | 0.523 | 19 (15%) | 0.401 | 24 (19%) | 0.209 | 14 (11%) | 0.179 | 22 (18%) | 0.917 | 36 (84%) | 7 (16%) | 0.071 | 41 (98%) | 1 (2%) | 0.074 |
| 2–5 | 315 | 125 (40%) | 57 (18%) | 72 (23%) | 45 (14%) | 58 (18%) | 103 (75%) | 34 (25%) | 113 (90%) | 12 (10%) | ||||||||
| ≥5 | 54 | 20 (37%) | 6 (11%) | 17 (31%) | 3 (6%) | 11 (20%) | 14 (58%) | 10 (42%) | 16 (80%) | 4 (20%) | ||||||||
| Unknown | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | ||||||||
| Molecular subtypes | Luminal A | 69 | 31 (45%) | 0.003 | 15 (21%) | 0.069 | 17 (25%) | 0.114 | 9 (13%) | 0.163 | 13 (19%) | 0.012 | 22 (67%) | 11 (33%) | 0.006 | 29 (94%) | 2 (6%) | 0.498 |
| Luminal B | 300 | 120 (40%) | 54 (18%) | 72 (24%) | 43 (14%) | 58 (19%) | 101 (78%) | 29 (22%) | 110 (92%) | 10 (8%) | ||||||||
| HER2+ | 66 | 27 (41%) | 10 (15%) | 18 (27%) | 8 (12%) | 17 (26%) | 25 (86%) | 4 (14%) | 25 (93%) | 2 (7%) | ||||||||
| Triple | 57 | 9 (16%) | 3 (5%) | 6 (11%) | 2 (4%) | 2 (4%) | 4 (36%) | 7 (64%) | 7 (78%) | 2 (22%) | ||||||||
| Negative | ||||||||||||||||||
| Unknown | 2 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | ||||||||
| ER | Positive | 357 | 148 (41%) | 0.014 | 68 (19%) | 0.025 | 87 (24%) | 0.260 | 57 (16%) | 0.049 | 70 (20%) | 0.334 | 127 (79%) | 33 (21%) | 0.708 | 136 (92%) | 12 (8%) | 0.670 |
| Negative | 133 | 39 (29%) | 14 (11%) | 26 (20%) | 12 (9%) | 21 (16%) | 33 (77%) | 10 (23%) | 35 (90%) | 4 (10%) | ||||||||
| Unknown | 4 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | ||||||||
| PR | Positive | 319 | 134 (42%) | 0.015 | 61 (19%) | 0.050 | 80 (25%) | 0.139 | 53 (17%) | 0.026 | 64 (20%) | 0.235 | 117 (81%) | 28 (19%) | 0.302 | 123 (92%) | 11 (8%) | 0.787 |
| Negative | 172 | 53 (31%) | 21 (12%) | 33 (19%) | 16 (9%) | 27 (16%) | 43 (74%) | 15 (26%) | 48 (91%) | 5 (9%) | ||||||||
| Unknown | 3 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | ||||||||
| HER2 | Positive | 172 | 69 (40%) | 0.519 | 26 (15%) | 0.507 | 47 (27%) | 0.111 | 22 (13%) | 0.896 | 36 (21%) | 0.303 | 58 (76%) | 18 (24%) | 0.691 | 62 (90%) | 7 (10%) | 0.564 |
| Negative | 315 | 117 (37%) | 55 (18%) | 66 (21%) | 39 (12%) | 54 (17%) | 93 (74%) | 33 (26%) | 108 (92%) | 9 (8%) | ||||||||
| Unknown | 7 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | ||||||||
| Ki67 (%) | ≤30 | 250 | 114 (46%) | 0.001 | 46 (18%) | 0.424 | 72 (29%) | 0.002 | 33 (13%) | 0.847 | 62 (25%) | 0.000 | 95 (80%) | 27 (22%) | 0.189 | 106 (93%) | 8 (7%) | 0.317 |
| >30 | 230 | 71 (31%) | 36 (16%) | 39 (17%) | 29 (13%) | 26 (11%) | 55 (70%) | 24 (30%) | 63 (89%) | 8 (11%) | ||||||||
| Unknown | 14 | 3 | 0 | 3 | 0 | 3 | 3 | 1 | 2 | 1 | ||||||||
| NPI | Good (2–3.4) | 82 | 35 (43%) | 0.671 | 15 (18%) | 0.301 | 21 (26%) | 0.662 | 11 (13%) | 0.501 | 17 (21%) | 0.788 | 28 (78%) | 8 (22%) | 0.883 | 34 (97%) | 1 (3%) | 0.392 |
| Moderate | 213 | 79 (37%) | 29 (14%) | 53 (25%) | 22 (10%) | 42 (20%) | 64 (74%) | 23 (26%) | 71 (90%) | 8 (10%) | ||||||||
| (3.4–5.4) | ||||||||||||||||||
| Poor (>5.4) | 155 | 59 (38%) | 30 (19%) | 33 (21%) | 22 (14%) | 27 (17%) | 49 (75%) | 16 (25%) | 53 (90%) | 6 (10%) | ||||||||
| Unknown | 44 | 15 | 8 | 7 | 7 | 5 | 12 | 5 | 13 | 2 | ||||||||
Note: P<0.05 was considered statistically significant and those values are shown in bold.
Abbreviations: ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; Mut, mutant; NPI, Nottingham Prognostic Index; PR, progesterone receptor.
We further investigated the relationships between clinicopathological features and PIK3CA mutation distributions, including the exon 9/20 and hotspot mutations (p.542/545 and p.1047; Table 2). ER and PR positive were also significantly correlated with mutations in exon 9 and at p.542/545. Besides, the low Ki67 index was found in patients with exon 20 and p.1047 mutations. Meanwhile, triple-negative breast cancer patients had much less p.1047 mutations (P=0.012). In addition, p.1047 mutations were significantly associated with old diagnosis age (≥40 years old; P=0.043).
Compared to breast cancers with PIK3CA hotspot mutations, cancers carrying non-hotspot mutations were more likely to belong to triple-negative subtype (P=0.006) and be larger in tumor size (Table 2). When analyzed by mutation burden, cancers with two mutations were more likely to be larger in tumor size compared with cancers harbored only one mutation (Table 2).
Associations of PIK3CA gene mutations with prognosis
As shown in Table 2, when we predicted the prognosis of the patients using the NPI method, no significant association was observed between various PIK3CA mutation statuses and prognosis.
Furthermore, prognosis analysis was conducted among 303 breast cancer patients with a median follow-up of 35 months. The Cox proportional hazards model and the Kaplan–Meier survival curve were used to evaluate the correlation between PFS rate or OS rate of breast cancer patients and PIK3CA mutation statuses.
In the univariate analysis, patients with old prognostic age (P=0.034) and small tumor size (P=0.033) exhibited better PFS, while old prognostic age (P=0.025) also correlated with better OS (Table 3). However, PIK3CA mutation frequency was not statistically significantly associated with PFS (HR[95% CI]=1.257[0.732–2.160], P=0.407), OS (HR[95% CI]=1.946[0.987–3.837], P=0.055), as well as their exon 9, exon 20, and hotspot mutations (Table 3).
Table 3.
Univariate Cox analysis of the correlation between clinicopathological parameters and progression-free/overall survival of breast cancer patients
| Variables | PFS
|
OS
|
||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
|
| ||||
| Age (years) | ||||
| <40 vs ≥40 | 1.805 (1.044–3.121) | 0.034 | 2.050 (1.092–3.848) | 0.025 |
| Tumor stage | ||||
| I and II vs III | 1.692 (0.976–2.935) | 0.061 | 1.465 (0.769–2.792) | 0.245 |
| Lymph node status | ||||
| Positive vs negative | 0.764 (0.452–1.290) | 0.313 | 0.944 (0.521–1.711) | 0.850 |
| Tumor size | ||||
| <5 cm vs ≥5 cm | 0.476 (0.240–0.943) | 0.033 | 0.493 (0.229–1.062) | 0.071 |
| Molecular subtypes | ||||
| Luminal and HER2+ vs triple negative | 1.264 (0.572–2.791) | 0.563 | 0.939 (0.417–2.110) | 0.878 |
| ER | ||||
| Positive vs negative | 1.162 (0.685–1.971) | 0.579 | 1.114 (0.600–2.068) | 0.732 |
| PR | ||||
| Positive vs negative | 1.469 (0.887–2.431) | 0.135 | 1.403 (0.782–2.518) | 0.257 |
| HER2 | ||||
| Positive vs negative | 0.735 (0.433–1.246) | 0.253 | 0.737 (0.396–1.371) | 0.335 |
| Ki67 | ||||
| ≤30% vs >30% | 0.980 (0.583–1.646) | 0.939 | 0.725 (0.389–1.349) | 0.309 |
| PIK3CA mutational status | ||||
| PIK3CA mutation vs wild-type | 1.257 (0.732–2.160) | 0.407 | 1.946 (0.987–3.837) | 0.055 |
| Exon 9 mutation vs wild-type | 0.794 (0.389–1.621) | 0.527 | 1.444 (0.515–4.050) | 0.485 |
| Exon 20 mutation vs wild-type | 1.696 (0.883–3.259) | 0.113 | 1.950 (0.907–4.194) | 0.087 |
| p.542/545 mutation vs wild-type | 0.731 (0.346–1.547) | 0.413 | 1.646 (0.508–5.329) | 0.406 |
| p.1047 mutation vs wild-type | 1.662 (0.819–3.371) | 0.159 | 2.2025 (0.857–4.783) | 0.108 |
Note: P<0.05 was considered statistically significant and those values are shown in bold.
Abbreviations: ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; OS, overall survival; PFS, progression-free survival; PR, progesterone receptor.
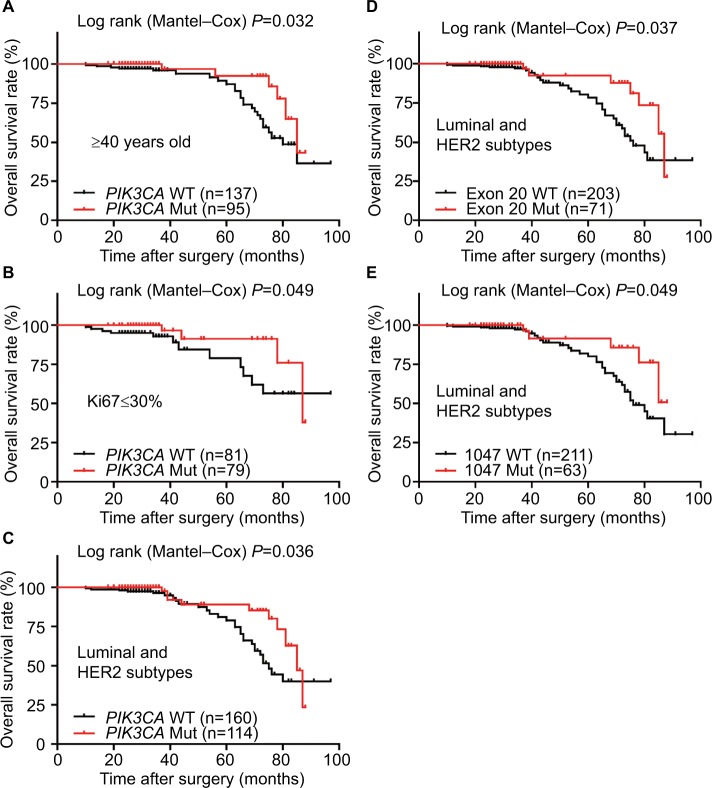
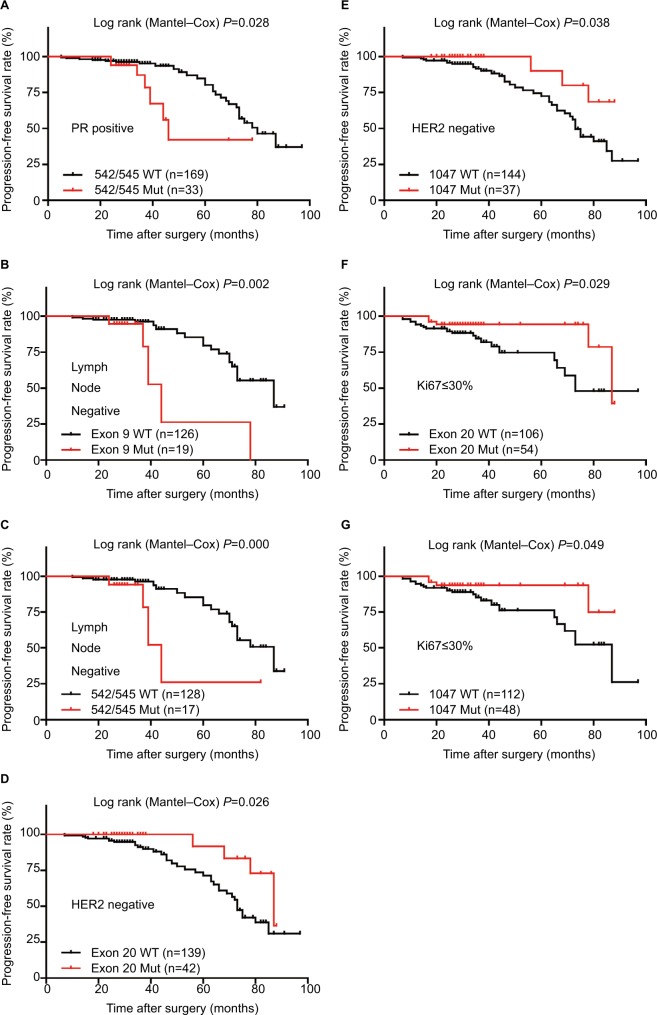
When examined by Kaplan–Meier estimate and log-rank test, the PFS of total patients with PIK3CA mutations was almost the same as wild-type patients, while the OS was significantly better in the total patients with PIK3CA mutations (Figure 2). However, there were no differences in PFS/OS between mutation and wild-type groups when examined by exon 9, exon 20, and hotspot mutations p.542/545 and p.1047 (Figures S1 and S2). Besides, PIK3CA mutations significantly improved the OS of the patients with old diagnosis age, low Ki67 labeling index, or luminal/HER2-enriched subtypes (Figure 3). Also as better prognostic effectors, exon 20 mutations as well as the hotspot p.1047 mutations were significantly associated with the PFS of the patients in HER2-negative or low Ki67 labeling index subgroups (Figure 4) and the OS of the patients diagnosed as luminal/HER2-enriched subtypes (Figure 3). In contrast, exon 9 mutations and its hotspot p.542/545 mutations were found in the patients with worse PFS, who belonged to PR-positive or lymph node-negative subgroups (Figure 4).
Figure 2.
Kaplan–Meier survival curves according to PIK3CA genotype for (A) progression-free survival and (B) overall survival of the total patients.
Abbreviations: Mut, mutation; WT, wild-type.
Figure 3.
Overall Kaplan–Meier survival curves.
Note: (A,B) OS rates of the PIK3CA mutations in ≥40 years old (A) and Ki67 labeling index ≤30% (B) subgroups; (C–E) OS rates of the luminal and HER2-enriched subtypes patients with PIK3CA (C), exon 20 (D), or p.1047 (E) mutations.
Abbreviations: HER2, human epidermal growth factor receptor 2; Mut, mutation; OS, overall survival; WT, wild-type.
Figure 4.
Progression-free Kaplan–Meier survival curves.
Note: (A) PFS rates of the PR-positive patients with p.542/545 mutations; (B,C) PFS rates of the lymph node-negative patients with exon 9 (B) or p.542/545 (C) mutations; (D,E) PFS rates of the HER2-negative patients with exon 20 (D) or p.1047 (E) mutations; (F,G) PFS rates of the Ki67 labeling index ≤30% patients with exon 20 (F) or p.1047 (G) mutations.
Abbreviations: HER2, human epidermal growth factor receptor 2; Mut, mutation; PFS, progression-free survival; PR, progesterone receptor; WT, wild-type.
When performing the univariate Cox analysis according to different clinicopathological parameters, a significant difference in PFS was observed between prognosis and exon 9 as well as p.542/545 hotspot mutations in PR-positive or lymph node-negative subgroups, exon 20 in HER2-negative or low Ki67 labeling index subgroups (Table 4). In OS, a significantly better prognosis was found in total PIK3CA and exon 20 mutations patients with luminal/HER2-enriched subtypes, while total PIK3CA mutations patients with old diagnostic age had a better OS as well (Table 4). These results were partially in accordance with the Kaplan–Meier analysis. Besides, no significance was detected between prognosis and PIK3CA mutation distribution/bias/burden under multiple other clinical, pathological, and molecular subtypes (Table S4).
Table 4.
Univariate Cox analysis of the correlation between PIK3CA mutation status and progression-free/overall survival according to different clinicopathological parameters
| Variables | PFS
|
OS
|
||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
|
| ||||
| ≥40 years old | ||||
| PIK3CA mutation vs wild-type | 1.632 (0.821–3.247) | 0.163 | 2.545 (1.042–6.215) | 0.040 |
| PR positive | ||||
| p.542/545 mutation vs wild-type | 0.397 (0.169–0.932) | 0.034 | 0.980 (0.293–3.282) | 0.974 |
| HER2 negative | ||||
| Exon 20 mutation vs wild-type | 3.039 (1.078–8.573) | 0.036 | 2.156 (0.750–6.198) | 0.154 |
| Ki67 labeling index £30% | ||||
| Exon 20 mutation vs wild-type | 2.838 (1.065–7.565) | 0.037 | 3.099 (0.869–11.047) | 0.081 |
| Lymph node negative | ||||
| Exon 9 mutation vs wild-type | 0.236 (0.085–0.655) | 0.006 | 0.444 (0.127–1.545) | 0.202 |
| p.542/545 mutation vs wild-type | 0.107 (0.029–0.403) | 0.001 | 0.401 (0.090–1.790) | 0.231 |
| Luminal and HER2 subtypes | ||||
| PIK3CA mutation vs wild–type | 1.323 (0.752–2.327) | 0.332 | 2.125 (1.032–4.375) | 0.041 |
| Exon 20 mutation vs wild-type | 1.969 (0.988–3.923) | 0.054 | 2.337 (1.024–5.331) | 0.044 |
Note: P<0.05 was considered statistically significant and those values are shown in bold.
Abbreviations: HER2, human epidermal growth factor receptor 2; PFS, progression-free survival; PR, progesterone receptor; OS, overall survival.
Discussion
To study the clinicopathological and prognostic values of PIK3CA variants in the breast cancer patients from Central China, 494 patients were investigated, and new insight into the complexity of PIK3CA mutations was provided in this research. In general, the mutation frequency (38%) in this study is relatively high as most investigations reported ~30% mutation rate using the similar detection method (Table 5).21,24,26,35–42 Interestingly, the frequency rates of PIK3CA mutations fluctuated among the studies which had been done by different groups from distinct areas of China (Table S5).43–54 This might be partially due to the sensitivity of assay methods. However, considering the controversial results from the researches around the world as well as environmental factors and lifestyles playing roles in breast cancer, we hypothesized that PIK3CA mutations and its associated factors might show diversity roles among the populations from different regions.
Table 5.
Comparison of association between PIK3CA mutations and various clinicopathological features in different studies
| Authors | Year of publication | Country | No. of patients | PIK3CA mutation | Sample type | Methods | Association between PIK3CA mutations and clinicopathological characteristics |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Arsenic et al21 | 2014 | Germany | 241 | 15.8% | FTS | DS | H1047R mutation: worse overall survival |
| Barbareschi et al24 | 2007 | Italy | 163 | 27.6% | FTS | SSCP + DS | Exon 20 mutations: prolonged overall and disease-free survival; exon 9 mutations: poor prognosis for disease-free survival and overall survival |
| Bozhanov et al35 | 2010 | Bulgaria | 145 | 31.3% | FTS | SSCP + DS | PIK3CA mutations: PR positive |
| Cizkova et al26 | 2012 | France | 452 | 33.4% | FTS | DS | PIK3CA mutations: low histopathological grade, small macroscopic tumor size, ER positive, PR positive, HER2 negative, favorable metastasis- free survival |
| Kalinsky et al36 | 2009 | USA | 590 | 33% | FFPE | MA + DS | PIK3CA mutations: ER positive, PR positive, HER2 negative, low-grade tumor; exon 9 mutations: older age; exon 20 mutations: node negative |
| Liang et al37 | 2006 | Singapore | 80 | 39% | FFPE | DS | Exon 20 mutations: older age, early stage |
| López-Knowles et al38 | 2010 | Australia | 168 | 7% | FFPE | DS | No significance |
| Mangone et al39 | 2012 | Brazil | 86 | 27% | FTS | SSCP + DS | Exon 20 mutations: poor overall survival and disease-free survival |
| Maruyama | 2007 | Japan | 188 | 24.47% | FTS | DS | PIK3CA mutations: ER positive, favorable |
| et al40 | prognosis | ||||||
| Pérez-Tenorio et al41 | 2007 | Sweden | 270 | 24% | FTS | SSCP + DS | PIK3CA mutations: ER positive, small tumor size, HER2 negative, longer local recurrence- free survival |
| Azizi Tabesh et al42 | 2016 | Iran | 80 | 45% | FTS | DS | PIK3CA mutations: low grade; Exon 20 mutations: PR positive |
| Current study | 2018 | China | 494 | 38.06% | FFPE | DS | PIK3CA mutations: luminal and HER2 positive, ER positive, PR positive, low Ki67 index, better overall survival; exon 9 mutations: worse progressive-free survival; exon 20 mutations: better progressive-free survival and overall survival; H1047 mutations: older age, better progressive-free survival and overall survival; non-hotspot mutations: larger tumor size |
Abbreviations: DS, direct sequencing; ER, estrogen receptor; FFPE, formalin-fixed, paraffin-embedded tissue samples; FTS, frozen tissue samples; HER2, human epidermal growth factor receptor 2; MA, MassARRAY; PR, progesterone receptor; SSCP, single-strand conformation polymorphism.
Then, we identified a significant association of PIK3CA mutations with clinicopathological and molecular characteristics, such as luminal/HER2-enriched subtypes, ER-positive, PR-positive, and low Ki67 labeling index which were partly consistent with the literature.26,35,36,40,41,44,45,47 When we separately analyzed the mutations in exon 9 and exon 20, their differences on the relationships with the clinicopathological characteristics were identified and should be considered separately when used for disease monitoring, therapeutic effect evaluation, and prognosis prediction. Besides, the point mutation p.1047, non-hotspot mutations, and more mutation burdens related to specific clinical and biological features of breast cancer might play particular roles and need to be investigated in future.
Numerous investigators reported that PIK3CA mutations are associated with prognosis.24,26,39–41,43,44 However, this association varies and even contradict among studies. Some of them showed better prognosis of the patients with PIK3CA mutations,24,26,40 and others believed worse outcome,39,43,44,50 while many researchers did not find any prognostic significance.35,36,38,49 In our study, interesting outcomes were explored when we tested the prognostic value of each PIK3CA mutation status in the subgroups separated according to different clinicopathological parameters. Firstly, total PIK3CA mutations exhibited disparate roles between FPS and OS in subgroups (Figures 2 and 3). Secondly, both exon 9 and exon 20 mutations correlated with FPS, but in diverse subgroups (Figure 4). Thirdly, only the effect of exon 20 mutations on the OS was identified (Figure 3D). Furthermore, exon 9 and exon 20 mutations revealed completely converse roles in the prognosis (Figures 3D and 4). Moreover, the hotspot mutations were in perfect accord with their exons (Figures 3D, E and 4). These results demonstrated that the variant status of PIK3CA mutations played different roles in the prognosis of breast cancer patients in our area.
In addition, when checking our samples, we realized that 60% of tumors in our study belonged to luminal B molecular subtype, which was extremely higher than the ratio in the other studies (~30%). This phenomenon also demonstrated that breast cancer patients in our area might have some specific preferences in genetic and clinicopathological features. However, the reasons and mechanisms need to be elucidated in future.
Limitations
This study still has some limitations. Firstly, all the samples were from a single center with a relatively small sample size. Although we identified some rules in the clinicopathological features, prognostic relevance, and PIK3CA mutation preferences, the sample size in the subgroups (stage I tumors, tumors with two mutations, and the relapsed patients) was quite small which made the results not that solid. Secondly, the followup times for most patients were too short. As >80% of breast cancer patients survive for >5 years after diagnosis, longer follow-up time is needed. Moreover, the effect and association of adjuvant systemic therapy with PIK3CA mutation status were not evaluated in this study, which might influence the progression-free/overall survival rate. Furthermore, the oncogenetic mutations in other exons were not examined, which might contribute to the prognosis of the patients.
Conclusion
PIK3CA mutations were detected using Sanger sequencing combined with targeted NGS in 38% of breast cancer patients from a single hospital in Central China. Different from the other studies, 60% of breast cancer patients were diagnosed with luminal B tumors. PIK3CA mutations were associated with ER-positive, PR-positive, low Ki67 labeling index, and luminal/HER2-enriched subtypes, while exon 9, exon 20, hotspot mutations, and mutation burdens made distinct contributions. In addition, p.1047 mutations were significantly associated with older diagnosis age. Significant heterogeneity was identified in the univariable effect of PIK3CA mutation status on FPS and OS. PIK3CA mutations patients had a better OS, which was also showed in the older diagnostic age, PR-negative, low Ki67 labeling index, and luminal/HER2-enriched subgroups. As better prognostic markers, exon 20 and p.1047 hotspot mutations significantly persisted in the HER2-negative and low Ki67 labeling index subgroups (analyzed by FPS) as well as luminal/HER2-enriched subgroup (analyzed by OS). In contrast, exon 9 and p.542/545 hotspot mutations exhibited worse prognostic factors in PR-positive and lymph node-negative subgroups when assayed using FPS. Therefore, these results demonstrated that the mutation frequency, distribution, bias, and burden of PIK3CA were related to different clincopathological characteristics, prognosis, and might play different roles in breast cancer from Central China. These differences and relationships should be deeply studied and taken into consideration during disease management.
Supplementary materials
Progression-free Kaplan–Meier survival curves of the total patients.
Note: (A–D) PFS rate of total patients with exon 9 (A), p.542/545 (B), exon 20 (C), or p.1047 (D) mutations.
Abbreviations: Mut, mutation; PFS, progression-free survival; WT, wild-type.
Overall Kaplan–Meier survival curves of the total patients.
Note: (A–D) OS rate of total patients with exon 9 (A), p.542/545 (B), exon 20 (C), or p.1047 (D) mutations.
Abbreviations: Mut, mutation; OS, overall survival; WT, wild-type.
Table S1.
Clinical, pathological, and biological features of breast cancer patients
| Parameters | Category | Number of patients | Percentage | Note |
|---|---|---|---|---|
|
| ||||
| Total | 494 | 100.00 | Eligible samples for analysis in 537 samples | |
| Age (years) | <40 | 91 | 18.40 | Age range: 25–89 years old |
| ≥40 | 403 | 81.58 | Median age: 48 years old | |
| Tumor stage | I | 12 | 2.43 | |
| II | 250 | 50.61 | ||
| III | 205 | 41.50 | ||
| Unknown | 27 | 5.47 | ||
| Lymph node status | 0 | 229 | 46.36 | |
| 1–3 | 108 | 21.86 | ||
| >3 | 131 | 26.52 | ||
| Unknown | 26 | 5.26 | ||
| Tumor size (cm) | <2 | 124 | 25.10 | Tumor size range: 0.3–12 cm |
| 2–5 | 315 | 63.77 | Median size: 2.6 cm | |
| ≥5 | 54 | 10.93 | ||
| Unknown | 1 | 0.20 | ||
| Molecular subtypes | Luminal A | 69 | 13.97 | |
| Luminal B | 300 | 60.73 | ||
| HER2+ | 66 | 13.36 | ||
| Triple negative | 57 | 11.54 | ||
| Unknown | 2 | 0.40 | ||
| ER | Positive | 357 | 72.27 | |
| Negative | 133 | 26.92 | ||
| Unknown | 4 | 0.81 | ||
| PR | Positive | 319 | 64.57 | |
| Negative | 172 | 34.82 | ||
| Unknown | 3 | 0.61 | ||
| HER2 | Positive | 172 | 34.82 | |
| Negative | 315 | 63.77 | ||
| Unknown | 7 | 1.42 | ||
| Ki67 (%) | ≤30 | 250 | 50.61 | |
| >30 | 230 | 46.56 | ||
| Unknown | 14 | 2.83 | ||
| Follow-up data collected | Total | 303 | 61.34 | Follow-up period: 5–97 months |
| Relapsed or second tumor | 28 | 9.24a | Median time: 35 months | |
| Died | 46 | 15.18a | ||
Note:
Percentage of the total follow-up data collected samples.
Abbreviations: ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; PR, progesterone receptor.
Table S2.
General clinical and pathological features of breast cancers used for targeted sequencing (n=24)
| Patient ID | PIK3CA mutation | Age (years) | Tumor stage | Lymph node status | Tumor size (cm) | Molecular subtype | ER | PR | HER2 | Ki67 | PFS (Month) | Note |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| R160066 | WT | 39 | III | 8 | 3 | Luminal B | 90% | 90% | 2+ | 20% | 24 | |
| R160069 | WT | 30 | III | 12 | 3 | HER2+ | Negative | Negative | 3+ | 60% | 37 | |
| R160070 | WT | 31 | III | 0 | 6 | Luminal B | 30% | 10% | Negative | 50% | Unknown | |
| R160071 | WT | 32 | II–III | 2 | 2 | Luminal B | 70% | 40% | 2+ | 30% | 40 | |
| R160080 | WT | 35 | II | 4 | 3 | Luminal B | 50% | 50% | 1+ | 20% | Unknown | |
| R160082 | WT | 35 | Unknown | 23 | 4.5 | Luminal B | 90% | 30% | 1+ | Unknown | 43 | |
| R160088 | WT | 37 | II–III | 4 | 1.6 | Luminal B | 80% | 15% | 3+ | 20% | 44 | |
| R160090 | E542K | 37 | II–III | 1 | 2.2 | Luminal B | 95% | 5% | Negative | 60% | 35 | Relapsed |
| R160093 | WT | 39 | II–III | 0 | 3.5 | Luminal B | 95% | 95% | 2+ | 15% | 40 | |
| R160104 | WT | 43 | II | 2 | 2 | Luminal B | 100% | 100% | Negative | 30% | 22 | |
| R160107 | E545K | 43 | Unknown | 24 | 3.5 | Luminal B | 90% | 30% | 1+ | 30% | 27 | |
| R160110 | WT | 43 | II–III | 0 | 2.2 | Luminal B | 90% | 90% | 3+ | 40% | 29 | |
| R160111 | WT | 43 | III | 0 | 1 | Luminal B | 40% | 90% | 3+ | 70% | 23 | |
| R160114 | WT | 44 | II | 0 | 1.5 | Luminal B | 90% | 90% | 1+ | 60% | 23 | |
| R160123 | E545K | 45 | II | 2 | 2.3 | Luminal B | 95% | 95% | Negative | 35% | 28 | |
| R160127 | WT | 45 | II | 0 | 1 | Luminal B | 80% | 70% | Negative | 25% | 21 | |
| R160130 | WT | 45 | III | 1 | 2.5 | Triple negative | Negative | Negative | 1+ | 80% | 23 | |
| R160133 | E542K | 46 | II | 0 | 4.2 | Luminal B | 95% | 95% | Negative | 25% | Unknown | |
| R160138 | WT | 46 | III | 0 | 4 | Triple negative | Negative | Negative | Negative | 80% | 22 | |
| R160145 | E542K | 47 | II | 0 | 1.8 | Luminal B | 50% | 20% | Negative | 75% | 26 | |
| R160158 | WT | 48 | III | 6 | 1.3 | Luminal B | 30% | Negative | Negative | 60% | 28 | |
| R160161 | E545K | 49 | Unknown | 10 | 8 | Luminal B | 50% | 50% | Negative | 20% | 36 | Relapsed |
| R160164 | H1047R | 50 | II–III | 0 | 2 | Luminal B | 3% | Negative | Negative | 60% | 27 | |
| R160169 | H1047R | 51 | II | 12 | 2 | Luminal A | 90% | 30% | 1+ | 8% | 30 | |
Abbreviations: ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; PFS, progression-free survival; PR, progesterone receptor; WT, wild-type.
Table S3.
New PIK3CA mutations in exons 9 and 20 in breast cancers (n=494)
| Exon | Nucleotide change | Codon change | New mutation effect | Number of mutations |
|---|---|---|---|---|
|
| ||||
| 9 | c.1618C>G | p.Leu540Val | Missense | 1 |
| 9 | c.1621T>C | p.Ser541Pro | Missense | 1 |
| 9 | c.1628T>C | p.Ile543Thr | Missense | 1 |
| 9 | c.1628T>G | p.Ile543Ser | Missense | 1 |
| 9 | c.1629C>G | p.Ile543Met | Missense | 2 |
| 9 | c.1655G>A | p.Trp552* | Nonsense | 1 |
| 20 | c.3139_3140CA>TG | p.His1047Cys | Missense | 1 |
| 20 | c.3206G>A | p.*1069* | Silent | 1 |
| 9 | c.[1621T>C(+)1644delA] | p.[Ser541Pro(+)Lys548fs*10] | Frameshift | 1 |
| 9+20 | c.[1651C>T(+)3117C>T] | p.[Leu551Leu(+)Phe1039Phe] | Silent | 1 |
| 20 | c.[3120G>A(+)3201G>A] | p.[Met1040Ile(+)Leu1067Leu] | Silent | 1 |
| 9+20 | c.[1644A>G(+)3140A>G(+)3178C>T] | p.[Lys548Lys(+)His1047Arg(+)His1060Tyr] | Missense | 1 |
| 20 | c.[3117C>T(+)3140A>G(+)3145G>A] | p.[Phe1039Phe(+)His1047Arg(+)Gly1049Ser] | Silent | 1 |
Table S4.
Univariate Cox analysis of the correlation between PIK3CA mutation status and progression-free/overall survival according to different clinicopathological parameters
| Variables | PFS
|
OS
|
||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
|
| ||||
| <40 years old | ||||
| PIK3CA mutation vs wild-type | 0.746 (0.298–10.866) | 0.531 | 1.365 (0.463–4.022) | 0.572 |
| Exon 9 mutation vs wild-type | 0.554 (0.173–1.780) | 0.322 | 1.345 (0.298–6.077) | 0.700 |
| Exon 20 mutation vs wild-type | 1.050 (0.336–3.282) | 0.934 | 1.238 (0.339–4.519) | 0.747 |
| p.542/545 mutation vs wild-type | 0.382 (0.117–1.247) | 0.111 | 0.995 (0.221–4.470) | 0.995 |
| p.1047 mutation vs wild-type | 0.894 (0.256–3.117) | 0.860 | 1.133 (0.252–5.092) | 0.871 |
| ≥40 years old | ||||
| PIK3CA mutation vs wild-type | 1.632 (0.821–3.247) | 0.163 | 2.545 (1.042–6.215) | 0.040 |
| Exon 9 mutation vs wild-type | 1.029 (0.403–2.628) | 0.953 | 1.858 (0.440–7.837) | 0.399 |
| Exon 20 mutation vs wild-type | 1.977 (0.879–4.446) | 0.099 | 2.283 (0.874–5.962) | 0.092 |
| p.542/545 mutation vs wild-type | 1.068 (0.380–3.003) | 0.901 | 3.175 (0.431–23.373) | 0.257 |
| p.1047 mutation vs wild-type | 1.942 (0.819–4.607) | 0.132 | 2.412 (0.840–6.928) | 0.102 |
| ER negative | ||||
| PIK3CA mutation vs wild-type | 1.747 (0.589–5.181) | 0.314 | 5.336 (0.699–40.709) | 0.106 |
| Exon 9 mutation vs wild-type | 1.507 (0.202–11.232) | 0.689 | – | – |
| Exon 20 mutation vs wild-type | 1.711 (0.504–5.810) | 0.389 | 3.824 (0.500–29.247) | 0.196 |
| p.542/545 mutation vs wild-type | 1.507 (0.202–11.232) | 0.689 | – | – |
| p.1047 mutation vs wild-type | 1.497 (0.440–5.092) | 0.518 | 3.312 (0.433–25.325) | 0.248 |
| ER positive | ||||
| PIK3CA mutation vs wild-type | 1.060 (0.555–2.025) | 0.860 | 1.583 (0.736–3.403) | 0.240 |
| Exon 9 mutation vs wild-type | 0.665 (0.303–1.459) | 0.309 | 1.185 (0.410–3.425) | 0.754 |
| Exon 20 mutation vs wild-type | 1.707 (0.779–3.741) | 0.182 | 1.714 (0.727–4.041) | 0.219 |
| p.542/545 mutation vs wild-type | 0.579 (0.253–1.324) | 0.195 | 1.295 (0.390–4.295) | 0.673 |
| p.1047 mutation vs wild-type | 1.785 (0.744–4.279) | 0.194 | 1.801 (0.684–4.738) | 0.233 |
| PR negative | ||||
| PIK3CA mutation vs wild-type | 1.522 (0.657–3.526) | 0.327 | 3.198 (0.953–10.729) | 0.060 |
| Exon 9 mutation vs wild-type | 1.888 (0.451–7.908) | 0.384 | – | – |
| Exon 20 mutation vs wild-type | 1.347 (0.517–3.510) | 0.542 | 2.099 (0.623–7.070) | 0.231 |
| p.542/545 mutation vs wild-type | 1.665 (0.397–6.975) | 0.486 | – | – |
| p.1047 mutation vs wild-type | 1.159 (0.444–3.023) | 0.763 | 1.860 (0.552–6.271) | 0.317 |
| PR positive | ||||
| PIK3CA mutation vs wild-type | 0.983 (0.468–2.066) | 0.964 | 1.396 (0.582–3.353) | 0.455 |
| Exon 9 mutation vs wild-type | 0.406 (0.170–0.973) | 0.043 | 0.733 (0.246–2.187) | 0.578 |
| Exon 20 mutation vs wild-type | 2.014 (0.811–4.999) | 0.131 | 1.880 (0.686–5.1479) | 0.220 |
| p.542/545 mutation vs wild-type | 0.340 (0.134–0.863) | 0.023 | 0.786 (0.231–2.674) | 0.700 |
| p.1047 mutation vs wild-type | 2.317 (0.801–6.704) | 0.121 | 2.258 (0.664–7.677) | 0.192 |
| HER2 negative | ||||
| PIK3CA mutation vs wild-type | 1.496 (0.727–3.076) | 0.274 | 2.168 (0.885–5.314) | 0.091 |
| Exon 9 mutation vs wild-type | 0.507 (0.221–1.166) | 0.110 | 1.238 (0.373–4.112) | 0.728 |
| Exon 20 mutation vs wild-type | 3.039 (1.078–8.573) | 0.036 | 2.156 (0.750–6.198) | 0.154 |
| p.542/545 mutation vs wild-type | 0.452 (0.187–1.096) | 0.079 | 1.546 (0.366–6.526) | 0.553 |
| p.1047 mutation vs wild-type | 3.225 (0.991–10.488) | 0.052 | 2.262 (0.685–7.472) | 0.180 |
| HER2 positive | ||||
| PIK3CA mutation vs wild-type | 0.948 (0.410–2.193) | 0.900 | 1.983 (0.630–6.239) | 0.242 |
| Exon 9 mutation vs wild-type | 1.709 (0.400–7.306) | 0.470 | 1.964 (0.257–15.004) | 0.515 |
| Exon 20 mutation vs wild-type | 0.903 (0.370–2.205) | 0.823 | 2.248 (0.627–8.065) | 0.214 |
| p.542/545 mutation vs wild-type | 1.483 (0.347–6.342) | 0.595 | 1.683 (0.220–12.867) | 0.616 |
| p.1047 mutation vs wild-type | 0.873 (0.343–2.219) | 0.776 | 2.502 (0.563–11.118) | 0.228 |
| Ki67 labeling index £30% | ||||
| PIK3CA mutation vs wild-type | 1.672 (0.764–3.663) | 0.199 | 2.977 (0.955–9.281) | 0.600 |
| Exon 9 mutation vs wild-type | 0.556 (0.221–1.399) | 0.213 | 1.040 (0.233–4.637) | 0.959 |
| Exon 20 mutation vs wild-type | 2.838 (1.065–7.565) | 0.037 | 3.099 (0.869–11.047) | 0.081 |
| p.542/545 mutation vs wild-type | 0.464 (0.171–1.262) | 0.133 | 1.480 (0.192–11.378) | 0.706 |
| p.1047 mutation vs wild-type | 2.773 (0.958–8.027) | 0.060 | 3.548 (0.805–15.642) | 0.094 |
| Ki67 labeling index >30% | ||||
| PIK3CA mutation vs wild-type | 1.270 (0.546–2.951) | 0.579 | 1.716 (0.689–4.274) | 0.246 |
| Exon 9 mutation vs wild-type | 1.315 (0.398–4.343) | 0.653 | 1.941 (0.458–8.234) | 0.368 |
| Exon 20 mutation vs wild-type | 1.409 (0.492–4.039) | 0.523 | 1.611 (0.552–4.705) | 0.383 |
| p.542/545 mutation vs wild-type | 1.267 (0.384–4.186) | 0.697 | 1.881 (0.443–7.977) | 0.391 |
| p.1047 mutation vs wild-type | 1.478 (0.448–4.875) | 0.521 | 1.661 (0.493–5.597) | 0.413 |
| Tumor stage I and II | ||||
| PIK3CA mutation vs wild-type | 1.070 (0.557–2.059) | 0.838 | 1.817 (0.808–4.086) | 0.148 |
| Exon 9 mutation vs wild-type | 0.578 (0.247–1.349) | 0.205 | 0.790 (0.270–2.314) | 0.667 |
| Exon 20 mutation vs wild-type | 1.584 (0.744–3.370) | 0.233 | 2.301 (0.904–5.853) | 0.080 |
| p.542/545 mutation vs wild-type | 0.490 (0.194–1.237) | 0.131 | 0.837 (0.248–2.825) | 0.837 |
| p.1047 mutation vs wild-type | 1.572 (0.691–3.577) | 0.281 | 2.510 (0.859–7.333) | 0.092 |
| Tumor stage III | ||||
| PIK3CA mutation vs wild-type | 1.519 (0.498–4.635) | 0.463 | 2.331 (0.514–10.581) | 0.273 |
| Exon 9 mutation vs wild-type | 1.359 (0.309–5.979) | 0.685 | – | – |
| Exon 20 mutation vs wild-type | 1.867 (0.428–8.137) | 0.406 | 1.201 (0.265–5.442) | 0.812 |
| p.542/545 mutation vs wild-type | 1.225 (0.277–5.414) | 0.789 | – | – |
| p.1047 mutation vs wild-type | 1.498 (0.342–6.550) | 0.592 | 0.911 (0.200–4.149) | 0.904 |
| Lymph node negative | ||||
| PIK3CA mutation vs wild-type | 1.002 (0.423–2.372) | 0.997 | 1.626 (0.606–4.360) | 0.334 |
| Exon 9 mutation vs wild-type | 0.236 (0.085–0.655) | 0.006 | 0.444 (0.127–1.545) | 0.202 |
| Exon 20 mutation vs wild-type | 2.214 (0.742–6.612) | 0.154 | 2.399 (0.766–7.513) | 0.133 |
| p.542/545 mutation vs wild-type | 0.107 (0.029–0.403) | 0.001 | 0.401 (0.090–1.790) | 0.231 |
| p.1047 mutation vs wild-type | 3.065 (0.717–13.102) | 0.131 | 3.120 (0.712–13.673) | 0.131 |
| Lymph node positive | ||||
| PIK3CA mutation vs wild-type | 1.428 (0.681–2.996) | 0.346 | 2.279 (0.848–6.124) | 0.102 |
| Exon 9 mutation vs wild-type | 1.926 (0.586–6.328) | 0.280 | 4.403 (0.593–32.711) | 0.147 |
| Exon 20 mutation vs wild-type | 1.283 (0.558–2.951) | 0.557 | 1.669 (0.570–4.892) | 0.350 |
| p.542/545 mutation vs wild-type | 1.687 (0.514–5.540) | 0.389 | 4.059 (0.546–30.159) | 0.171 |
| p.1047 mutation vs wild-type | 1.132 (0.492–2.600) | 0.771 | 1.471 (0.502–4.309) | 0.482 |
| Tumor size <5 cm | ||||
| PIK3CA mutation vs wild-type | 1.074 (0.606–1.905) | 0.806 | 1.584 (0.784–3.200) | 0.200 |
| Exon 9 mutation vs wild-type | 0.619 (0.298–1.283) | 0.197 | 1.101 (0.387–3.129) | 0.857 |
| Exon 20 mutation vs wild-type | 1.530 (0.766–3.058) | 0.228 | 1.602 (0.730–3.516) | 0.240 |
| p.542/545 mutation vs wild-type | 0.553 (0.257–1.191) | 0.130 | 1.216 (0.372–3.983) | 0.746 |
| p.1047 mutation vs wild-type | 1.704 (0.801–3.623) | 0.166 | 1.839 (0.767–4.410) | 0.172 |
| Tumor size ≥5 cm | ||||
| PIK3CA mutation vs wild-type | 3.231 (0.407–25.614) | 0.267 | – | – |
| Exon 9 mutation vs wild-type | – | – | – | – |
| Exon 20 mutation vs wild-type | 3.231 (0.407–25.614) | 0.267 | – | – |
| p.542/545 mutation vs wild-type | – | – | – | – |
| p.1047 mutation vs wild-type | 0.286 (0.026–3.161) | 0.307 | – | – |
| Luminal and HER2 subtypes | ||||
| PIK3CA mutation vs wild-type | 1.323 (0.752–2.327) | 0.332 | 2.125 (1.032–4.375) | 0.041 |
| Exon 9 mutation vs wild-type | 0.723 (0.351–1.488) | 0.378 | 1.254 (0.443–3.550) | 0.670 |
| Exon 20 mutation vs wild-type | 1.969 (0.988–3.923) | 0.054 | 2.337 (1.024–5.331) | 0.044 |
| p.542/545 mutation vs wild-type | 0.637 (0.298–1.361) | 0.244 | 1.373 (0.421–4.478) | 0.600 |
| p.1047 mutation vs wild-type | 1.945 (0.917–4.125) | 0.083 | 2.493 (0.971–6.399) | 0.058 |
| Triple negative subtype | ||||
| PIK3CA mutation vs wild-type | 0.720 (0.083–6.247) | 0.766 | 0.720 (0.083–6.247) | 0.766 |
| Exon 9 mutation vs wild-type | – | – | – | – |
| Exon 20 mutation vs wild-type | 0.255 (0.026–2.463) | 0.237 | 0.255 (0.026–2.463) | 0.237 |
| p.542/545 mutation vs wild-type | – | – | – | – |
| p.1047 mutation vs wild-type | 0.255 (0.026–2.463) | 0.237 | 0.255(0.026–2.463) | 0.237 |
Note: P<0.05 was considered statistically significant and those values are shown in bold.
Abbreviations: ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; OS, overall survival; PFS, progression-free survival; PR, progesterone receptor.
Table S5.
Comparison of association between PIK3CA mutations and various clinicopathological features in different areas in China
| Authors | Year of publication | Area | No. of patients | PIK3CA mutation | Sample type | Methods | Association between PIK3CA mutations and clinicopathological characteristics |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Hu et al1 | 2018 | Changsha, Hunan | 68 | 33.82% | Peripheral blood | Targeted NGS | PIK3CA mutations: worse progression-free survival |
| Deng et al2 | 2019 | Chengdu, Sichun | 507 | 46.5% | FTS | Targeted NGS | PIK3CA mutations: ER positive, PR positive, HER3 negative; two or three mutations in PIK3CA: poor prognosis for overall survival |
| Chen et al3 | 2018 | Shanghai | 149 | 43.6% | FTS | Targeted NGS | PIK3CA mutations: older age, ER positive, PR positive |
| Cheng et al4 | 2017 | Luzhou, Sichun | 32 | 28.12% | FFPE | DS | PIK3CA mutations: more invasiveness lymph node, bigger tumor size |
| Yuan et al5 | 2015 | Beijing | 729 | 28.3% | FTS | DS | PIK3CA mutations: ER positive, PR positive, less response to neoadjuvant chemotherapy |
| Wang et al6 | 2015 | Xining, Qinghai | 25 | 32% | FTS | DS | Not detected |
| Liu et al7 | 2015 | Dalian, Liaoning | 80 | 32.5% | FFPE | Targeted NGS | No significance |
| Deng et al8 | 2015 | Chengdu, Sichun | 288 | 15.6% | FFPE | DS | PIK3CA mutations: poor outcome of ER-positive breast cancer |
| Zhang et al9 | 2014 | Beijing | 93 | 32.3% | FTS | xTAG liquid chip | PIK3CA mutations: patients’ clinical response to neoadjuvant chemotherapy |
| Bai et al10 | 2014 | Xi’an Shaanxi | 105 | 35.2% | FFPE | Targeted NGS | PIK3CA mutations: older age |
| Tong et al11 | 2012 | Guangzhou, Guangdong | 120 | 7.5% | FTS | MA | PIK3CA mutations: older age |
| Li et al12 | 2010 | Shanghai | 233 | 19.7% | FFPE | DS | PIK3CA mutations: high grade, ER positive, PR positive, PTEN positive |
| Current study | 2018 | Hefei, Anhui | 494 | 38.06% | FFPE | DS | PIK3CA mutations: luminal and HER2 positive, ER positive, PR positive, low Ki67 index, better overall survival; exon 9 mutations: worse progression-free survival; exon 20 mutations: better progression- free survival and overall survival; H1047 mutations: older age, better progression-free survival and overall survival; non-hotspot mutations: larger tumor size |
Abbreviations: DS, direct sequencing; ER, estrogen receptor; FFPE, formalin-fixed, paraffin-embedded tissue samples; FTS, frozen tissue samples; HER3, human epidermal growth factor receptor 3; MA, MassARRAY; NGS, next generation sequencing; PR, progesterone receptor.
References
- 1.Hu ZY, Xie N, Tian C, et al. Identifying circulating tumor DNA mutation profiles in metastatic breast cancer patients with multiline resistance. EBioMedicine. 2018;32:111–118. doi: 10.1016/j.ebiom.2018.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng L, Zhu X, Sun Y, et al. Prevalence and prognostic role of PIK3CA/AKT1 mutations in Chinese breast cancer patients. Cancer Res Treat. 2019;51(1):128–140. doi: 10.4143/crt.2017.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen L, Yang L, Yao L, et al. Characterization of PIK3CA and PIK3R1 somatic mutations in Chinese breast cancer patients. Nat Commun. 2018;9(1):1357. doi: 10.1038/s41467-018-03867-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng J, Fu S, Wei C, et al. Evaluation of PIK3CA mutations as a biomarker in Chinese breast carcinomas from Western China. Cancer Biomark. 2017;19(1):85–92. doi: 10.3233/CBM-160380. [DOI] [PubMed] [Google Scholar]
- 5.Yuan H, Chen J, Liu Y, et al. Association of PIK3CA mutation status before and after neoadjuvant chemotherapy with response to chemotherapy in women with breast cancer. Clin Cancer Res. 2015;21(19):4365–4372. doi: 10.1158/1078-0432.CCR-14-3354. [DOI] [PubMed] [Google Scholar]
- 6.Wang YL, Dai X, Li YD, et al. Study of PIK3CA, BRAF, and KRAS mutations in breast carcinomas among Chinese women in Qinghai. Genet Mol Res. 2015;14(4):14840–14846. doi: 10.4238/2015.November.18.49. [DOI] [PubMed] [Google Scholar]
- 7.Liu S, Wang H, Zhang L, et al. Rapid detection of genetic mutations in individual breast cancer patients by next-generation DNA sequencing. Hum Genomics. 2015;9(1):2. doi: 10.1186/s40246-015-0024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng L, Chen J, Zhong XR, et al. Correlation between activation of PI3K/Akt/mTOR pathway and prognosis of breast cancer in Chinese women. PLoS One. 2015;10(3):e0120511. doi: 10.1371/journal.pone.0120511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Liu M, Yang H, et al. PIK3CA mutations are a predictor of docetaxel plus epirubicin neoadjuvant chemotherapy clinical efficacy in breast cancer. Neoplasma. 2014;61(4):461–467. doi: 10.4149/neo_2014_057. [DOI] [PubMed] [Google Scholar]
- 10.Bai X, Zhang E, Ye H, et al. PIK3CA and TP53 gene mutations in human breast cancer tumors frequently detected by ion torrent DNA sequencing. PLoS One. 2014;9(6):e99306. doi: 10.1371/journal.pone.0099306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong L, Yang XX, Liu MF, et al. Mutational analysis of key EGFR pathway genes in Chinese breast cancer patients. Asian Pac J Cancer Prev. 2012;13(11):5599–5603. doi: 10.7314/apjcp.2012.13.11.5599. [DOI] [PubMed] [Google Scholar]
- 12.Li H, Zhu R, Wang L, et al. PIK3CA mutations mostly begin to develop in ductal carcinoma of the breast. Exp Mol Pathol. 2010;88(1):150–155. doi: 10.1016/j.yexmp.2009.09.016. [DOI] [PubMed] [Google Scholar]
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant Numbers: 81702954 and 81372214), the Key Technologies R&D Programme of Anhui Province (Grant Number: 1604a0802103), the Fundamental Research Funds for the Central Universities (Grant Number: WK9110000042), the Major/Innovative Program of Development Foundation of Hefei Center for Physical Science and Technology (Grant Number: 2016FXCX006), and the 100-Talent Program of Chinese Academy of Sciences.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Goldhirsch A, Wood WC, Coates AS, et al. Strategies for subtypes– dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011;22(8):1736–1747. doi: 10.1093/annonc/mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eifel P, Axelson JA, Costa J, et al. National Institutes of health consensus development conference statement: adjuvant therapy for breast cancer, November 1-3, 2000. J Natl Cancer Inst. 2001;93(13):979–989. doi: 10.1093/jnci/93.13.979. [DOI] [PubMed] [Google Scholar]
- 4.Kyrochristos ID, Peristeri DV, Ziogas DE, Lykoudis EG, Roukos DH. Precise predictive and therapeutic strategy for breast cancer. Future Oncol. 2018;14(18):1777–1780. doi: 10.2217/fon-2018-0277. [DOI] [PubMed] [Google Scholar]
- 5.Akram M, Iqbal M, Daniyal M, Khan AU. Awareness and current knowledge of breast cancer. Biol Res. 2017;50(1):33. doi: 10.1186/s40659-017-0140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van’t Veer LJ, Dai H, Van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 7.Aoki M, Fujishita T. Oncogenic roles of the PI3K/Akt/mTOR axis. Curr Top Microbiol Immunol. 2017;407:153–189. doi: 10.1007/82_2017_6. [DOI] [PubMed] [Google Scholar]
- 8.Follo MY, Manzoli L, Poli A, Mccubrey JA, Cocco L. PLC and PI3K/Akt/mTOR signalling in disease and cancer. Adv Biol Regul. 2015;57:10–16. doi: 10.1016/j.jbior.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Bahrami A, Khazaei M, Shahidsales S, et al. The therapeutic potential of PI3K/Akt/mTOR inhibitors in breast cancer: rational and progress. J Cell Biochem. 2018;119(1):213–222. doi: 10.1002/jcb.26136. [DOI] [PubMed] [Google Scholar]
- 10.Matsuoka T, Yashiro M. The role of PI3K/Akt/mTOR signaling in gastric carcinoma. Cancers. 2014;6(3):1441–1463. doi: 10.3390/cancers6031441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sathe A, Nawroth R. Targeting the PI3K/Akt/mTOR pathway in bladder cancer. Methods Mol Biol. 2018;1655:335–350. doi: 10.1007/978-1-4939-7234-0_23. [DOI] [PubMed] [Google Scholar]
- 12.Slomovitz BM, Coleman RL. The PI3K/Akt/mTOR pathway as a therapeutic target in endometrial cancer. Clin Cancer Res. 2012;18(21):5856–5864. doi: 10.1158/1078-0432.CCR-12-0662. [DOI] [PubMed] [Google Scholar]
- 13.Liao X, Morikawa T, Lochhead P, et al. Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res. 2012;18(8):2257–2268. doi: 10.1158/1078-0432.CCR-11-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polivka J, Janku F. Molecular targets for cancer therapy in the PI3K/Akt/mTOR pathway. Pharmacol Ther. 2014;142(2):164–175. doi: 10.1016/j.pharmthera.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Dirican E, Akkiprik M, Özer A. Mutation distributions and clinical correlations of PIK3CA gene mutations in breast cancer. Tumor Biol. 2016;37(6):7033–7045. doi: 10.1007/s13277-016-4924-2. [DOI] [PubMed] [Google Scholar]
- 16.Dumont AG, Dumont SN, Trent JC. The favorable impact of PIK3CA mutations on survival: an analysis of 2587 patients with breast cancer. Chin J Cancer. 2012;31(7):327–334. doi: 10.5732/cjc.012.10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan H, Li C, Xiang Q, et al. PIK3CA mutations and their response to neoadjuvant treatment in early breast cancer: a systematic review and meta-analysis. Thorac Cancer. 2018;9(5):571–579. doi: 10.1111/1759-7714.12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zardavas D, Phillips WA, Loi S. PIK3CA mutations in breast cancer: reconciling findings from preclinical and clinical data. Breast Cancer Res. 2014;16(1):201. doi: 10.1186/bcr3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zardavas D, Te Marvelde L, Milne RL, et al. Tumor PIK3CA genotype and prognosis in early-stage breast cancer: a pooled analysis of individual patient data. J Clin Oncol. 2018;36(10):981–990. doi: 10.1200/JCO.2017.74.8301. [DOI] [PubMed] [Google Scholar]
- 20.Ademuyiwa FO, Tao Y, Luo J, Weilbaecher K, Ma CX. Differences in the mutational landscape of triple-negative breast cancer in African Americans and Caucasians. Breast Cancer Res Treat. 2017;161(3):491–499. doi: 10.1007/s10549-016-4062-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arsenic R, Lehmann A, Budczies J, et al. Analysis of PIK3CA mutations in breast cancer subtypes. Appl Immunohistochem Mol Morphol. 2014;22(1):50–56. doi: 10.1097/pdm.0b013e318297afea. [DOI] [PubMed] [Google Scholar]
- 22.Benvenuti S, Frattini M, Arena S, et al. PIK3CA cancer mutations display gender and tissue specificity patterns. Hum Mutat. 2008;29(2):284–288. doi: 10.1002/humu.20648. [DOI] [PubMed] [Google Scholar]
- 23.Buttitta F, Felicioni L, Barassi F, et al. PIK3CA mutation and histological type in breast carcinoma: high frequency of mutations in lobular carcinoma. J Pathol. 2006;208(3):350–355. doi: 10.1002/path.1908. [DOI] [PubMed] [Google Scholar]
- 24.Barbareschi M, Buttitta F, Felicioni L, et al. Different prognostic roles of mutations in the helical and kinase domains of the PIK3CA gene in breast carcinomas. Clin Cancer Res. 2007;13(20):6064–6069. doi: 10.1158/1078-0432.CCR-07-0266. [DOI] [PubMed] [Google Scholar]
- 25.Harlé A, Lion M, Lozano N, et al. Analysis of PIK3CA exon 9 and 20 mutations in breast cancers using PCR-HRM and PCR-ARMS: correlation with clinicopathological criteria. Oncol Rep. 2013;29(3):1043–1052. doi: 10.3892/or.2013.2229. [DOI] [PubMed] [Google Scholar]
- 26.Cizkova M, Susini A, Vacher S, et al. PIK3CA mutation impact on survival in breast cancer patients and in ERα, Pr and ERBB2-based subgroups. Breast Cancer Res. 2012;14(1):R28. doi: 10.1186/bcr3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edge SB, Compton CC. The American Joint Committee on cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17(6):1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 28.Galea MH, Blamey RW, Elston CE, Ellis IO. The Nottingham prognostic index in primary breast cancer. Breast Cancer Res Treat. 1992;22(3):207–219. doi: 10.1007/BF01840834. [DOI] [PubMed] [Google Scholar]
- 29.Dancey JE, Dodd LE, Ford R, et al. Recommendations for the assessment of progression in randomised cancer treatment trials. Eur J Cancer. 2009;45(2):281–289. doi: 10.1016/j.ejca.2008.10.042. [DOI] [PubMed] [Google Scholar]
- 30.Curigliano G, Burstein HJ, Winer EP, et al. De-escalating and escalating treatments for early-stage breast cancer: the St. Gallen international expert consensus Conference on the primary therapy of early breast cancer 2017. Ann Oncol. 2017;28(8):1700–1712. doi: 10.1093/annonc/mdx308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sobhani N, Roviello G, Corona SP, et al. The prognostic value of PI3K mutational status in breast cancer: a meta-analysis. J Cell Biochem. 2018;119(6):4287–4292. doi: 10.1002/jcb.26687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thirumal Kumar D, George Priya Doss C. Role of E542 and E545 missense mutations of PIK3CA in breast cancer: a comparative computational approach. J Biomol Struct Dyn. 2017;35(12):2745–2757. doi: 10.1080/07391102.2016.1231082. [DOI] [PubMed] [Google Scholar]
- 34.Filipenko ML, Os’kina NA, Oskorbin IA, et al. Association between the prevalence of somatic mutations in PIK3CA gene in tumors and clinical and morphological characteristics of breast cancer patients. Bull Exp Biol Med. 2017;163(2):250–254. doi: 10.1007/s10517-017-3777-z. [DOI] [PubMed] [Google Scholar]
- 35.Bozhanov SS, Angelova SG, Krasteva ME, et al. Alterations in p53, BRCA1, ATM, PIK3CA, and HER2 genes and their effect in modifying clinicopathological characteristics and overall survival of Bulgarian patients with breast cancer. J Cancer Res Clin Oncol. 2010;136(11):1657–1669. doi: 10.1007/s00432-010-0824-9. [DOI] [PubMed] [Google Scholar]
- 36.Kalinsky K, Jacks LM, Heguy A, et al. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res. 2009;15(16):5049–5059. doi: 10.1158/1078-0432.CCR-09-0632. [DOI] [PubMed] [Google Scholar]
- 37.Liang X, Lau QC, Salto-Tellez M, Putti TC, Loh M, Sukumar S. Mutational hotspot in exon 20 of PIK3CA in breast cancer among Singapore Chinese. Cancer Biol Ther. 2006;5(5):544–548. doi: 10.4161/cbt.5.5.2656. [DOI] [PubMed] [Google Scholar]
- 38.López-Knowles E, O’Toole SA, McNeil CM, et al. PI3K pathway activation in breast cancer is associated with the basal-like phenotype and cancer-specific mortality. Int J Cancer. 2010;126(5):1121–1131. doi: 10.1002/ijc.24831. [DOI] [PubMed] [Google Scholar]
- 39.Mangone FR, Bobrovnitchaia IG, Salaorni S, Manuli E, Nagai MA. PIK3CA exon 20 mutations are associated with poor prognosis in breast cancer patients. Clinics. 2012;67(11):1285–1290. doi: 10.6061/clinics/2012(11)11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maruyama N, Miyoshi Y, Taguchi T, Tamaki Y, Monden M, Noguchi S. Clinicopathologic analysis of breast cancers with PIK3CA mutations in Japanese women. Clin Cancer Res. 2007;13(2Pt 1):408–414. doi: 10.1158/1078-0432.CCR-06-0267. [DOI] [PubMed] [Google Scholar]
- 41.Pérez-Tenorio G, Alkhori L, Olsson B, et al. PIK3CA mutations and PTEN loss correlate with similar prognostic factors and are not mutually exclusive in breast cancer. Clin Cancer Res. 2007;13(12):3577–3584. doi: 10.1158/1078-0432.CCR-06-1609. [DOI] [PubMed] [Google Scholar]
- 42.Azizi Tabesh G, Izadi P, Fereidooni F, Emami Razavi AN, Tavakkoly Bazzaz J. The high frequency of PIK3CA mutations in Iranian breast cancer patients. Cancer Invest. 2017;35(1):36–42. doi: 10.1080/07357907.2016.1247455. [DOI] [PubMed] [Google Scholar]
- 43.Hu ZY, Xie N, Tian C, et al. Identifying circulating tumor DNA mutation profiles in metastatic breast cancer patients with multiline resistance. EBioMedicine. 2018;32:111–118. doi: 10.1016/j.ebiom.2018.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng L, Zhu X, Sun Y, et al. Prevalence and prognostic role of PIK3CA/AKT1 mutations in Chinese breast cancer patients. Cancer Res Treat. 2019;51(1):128–140. doi: 10.4143/crt.2017.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen L, Yang L, Yao L, et al. Characterization of PIK3CA and PIK3R1 somatic mutations in Chinese breast cancer patients. Nat Commun. 2018;9(1):1357. doi: 10.1038/s41467-018-03867-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng J, Fu S, Wei C, et al. Evaluation of PIK3CA mutations as a biomarker in Chinese breast carcinomas from Western China. Cancer Biomark. 2017;19(1):85–92. doi: 10.3233/CBM-160380. [DOI] [PubMed] [Google Scholar]
- 47.Yuan H, Chen J, Liu Y, et al. Association of PIK3CA mutation status before and after neoadjuvant chemotherapy with response to chemotherapy in women with breast cancer. Clin Cancer Res. 2015;21(19):4365–4372. doi: 10.1158/1078-0432.CCR-14-3354. [DOI] [PubMed] [Google Scholar]
- 48.Wang YL, Dai X, Li YD, et al. Study of PIK3CA, BRAF, and KRAS mutations in breast carcinomas among Chinese women in Qinghai. Genet Mol Res. 2015;14(4):14840–14846. doi: 10.4238/2015.November.18.49. [DOI] [PubMed] [Google Scholar]
- 49.Liu S, Wang H, Zhang L, et al. Rapid detection of genetic mutations in individual breast cancer patients by next-generation DNA sequencing. Hum Genomics. 2015;9(1):2. doi: 10.1186/s40246-015-0024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng L, Chen J, Zhong XR, et al. Correlation between activation of PI3K/Akt/mTOR pathway and prognosis of breast cancer in Chinese women. PLoS One. 2015;10(3):e0120511. doi: 10.1371/journal.pone.0120511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Liu M, Yang H, et al. PIK3CA mutations are a predictor of docetaxel plus epirubicin neoadjuvant chemotherapy clinical efficacy in breast cancer. Neoplasma. 2014;61(4):461–467. doi: 10.4149/neo_2014_057. [DOI] [PubMed] [Google Scholar]
- 52.Bai X, Zhang E, Ye H, et al. PIK3CA and TP53 gene mutations in human breast cancer tumors frequently detected by ion torrent DNA sequencing. PLoS One. 2014;9(6):e99306. doi: 10.1371/journal.pone.0099306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tong L, Yang XX, Liu MF, et al. Mutational analysis of key EGFR pathway genes in Chinese breast cancer patients. Asian Pac J Cancer Prev. 2012;13(11):5599–5603. doi: 10.7314/apjcp.2012.13.11.5599. [DOI] [PubMed] [Google Scholar]
- 54.Zhu Li H, Wang RL, et al. PIK3CA mutations mostly begin to develop in ductal carcinoma of the breast. Exp Mol Pathol. 2010;88(1):150–155. doi: 10.1016/j.yexmp.2009.09.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Progression-free Kaplan–Meier survival curves of the total patients.
Note: (A–D) PFS rate of total patients with exon 9 (A), p.542/545 (B), exon 20 (C), or p.1047 (D) mutations.
Abbreviations: Mut, mutation; PFS, progression-free survival; WT, wild-type.
Overall Kaplan–Meier survival curves of the total patients.
Note: (A–D) OS rate of total patients with exon 9 (A), p.542/545 (B), exon 20 (C), or p.1047 (D) mutations.
Abbreviations: Mut, mutation; OS, overall survival; WT, wild-type.
Table S1.
Clinical, pathological, and biological features of breast cancer patients
| Parameters | Category | Number of patients | Percentage | Note |
|---|---|---|---|---|
|
| ||||
| Total | 494 | 100.00 | Eligible samples for analysis in 537 samples | |
| Age (years) | <40 | 91 | 18.40 | Age range: 25–89 years old |
| ≥40 | 403 | 81.58 | Median age: 48 years old | |
| Tumor stage | I | 12 | 2.43 | |
| II | 250 | 50.61 | ||
| III | 205 | 41.50 | ||
| Unknown | 27 | 5.47 | ||
| Lymph node status | 0 | 229 | 46.36 | |
| 1–3 | 108 | 21.86 | ||
| >3 | 131 | 26.52 | ||
| Unknown | 26 | 5.26 | ||
| Tumor size (cm) | <2 | 124 | 25.10 | Tumor size range: 0.3–12 cm |
| 2–5 | 315 | 63.77 | Median size: 2.6 cm | |
| ≥5 | 54 | 10.93 | ||
| Unknown | 1 | 0.20 | ||
| Molecular subtypes | Luminal A | 69 | 13.97 | |
| Luminal B | 300 | 60.73 | ||
| HER2+ | 66 | 13.36 | ||
| Triple negative | 57 | 11.54 | ||
| Unknown | 2 | 0.40 | ||
| ER | Positive | 357 | 72.27 | |
| Negative | 133 | 26.92 | ||
| Unknown | 4 | 0.81 | ||
| PR | Positive | 319 | 64.57 | |
| Negative | 172 | 34.82 | ||
| Unknown | 3 | 0.61 | ||
| HER2 | Positive | 172 | 34.82 | |
| Negative | 315 | 63.77 | ||
| Unknown | 7 | 1.42 | ||
| Ki67 (%) | ≤30 | 250 | 50.61 | |
| >30 | 230 | 46.56 | ||
| Unknown | 14 | 2.83 | ||
| Follow-up data collected | Total | 303 | 61.34 | Follow-up period: 5–97 months |
| Relapsed or second tumor | 28 | 9.24a | Median time: 35 months | |
| Died | 46 | 15.18a | ||
Note:
Percentage of the total follow-up data collected samples.
Abbreviations: ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; PR, progesterone receptor.
Table S2.
General clinical and pathological features of breast cancers used for targeted sequencing (n=24)
| Patient ID | PIK3CA mutation | Age (years) | Tumor stage | Lymph node status | Tumor size (cm) | Molecular subtype | ER | PR | HER2 | Ki67 | PFS (Month) | Note |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| R160066 | WT | 39 | III | 8 | 3 | Luminal B | 90% | 90% | 2+ | 20% | 24 | |
| R160069 | WT | 30 | III | 12 | 3 | HER2+ | Negative | Negative | 3+ | 60% | 37 | |
| R160070 | WT | 31 | III | 0 | 6 | Luminal B | 30% | 10% | Negative | 50% | Unknown | |
| R160071 | WT | 32 | II–III | 2 | 2 | Luminal B | 70% | 40% | 2+ | 30% | 40 | |
| R160080 | WT | 35 | II | 4 | 3 | Luminal B | 50% | 50% | 1+ | 20% | Unknown | |
| R160082 | WT | 35 | Unknown | 23 | 4.5 | Luminal B | 90% | 30% | 1+ | Unknown | 43 | |
| R160088 | WT | 37 | II–III | 4 | 1.6 | Luminal B | 80% | 15% | 3+ | 20% | 44 | |
| R160090 | E542K | 37 | II–III | 1 | 2.2 | Luminal B | 95% | 5% | Negative | 60% | 35 | Relapsed |
| R160093 | WT | 39 | II–III | 0 | 3.5 | Luminal B | 95% | 95% | 2+ | 15% | 40 | |
| R160104 | WT | 43 | II | 2 | 2 | Luminal B | 100% | 100% | Negative | 30% | 22 | |
| R160107 | E545K | 43 | Unknown | 24 | 3.5 | Luminal B | 90% | 30% | 1+ | 30% | 27 | |
| R160110 | WT | 43 | II–III | 0 | 2.2 | Luminal B | 90% | 90% | 3+ | 40% | 29 | |
| R160111 | WT | 43 | III | 0 | 1 | Luminal B | 40% | 90% | 3+ | 70% | 23 | |
| R160114 | WT | 44 | II | 0 | 1.5 | Luminal B | 90% | 90% | 1+ | 60% | 23 | |
| R160123 | E545K | 45 | II | 2 | 2.3 | Luminal B | 95% | 95% | Negative | 35% | 28 | |
| R160127 | WT | 45 | II | 0 | 1 | Luminal B | 80% | 70% | Negative | 25% | 21 | |
| R160130 | WT | 45 | III | 1 | 2.5 | Triple negative | Negative | Negative | 1+ | 80% | 23 | |
| R160133 | E542K | 46 | II | 0 | 4.2 | Luminal B | 95% | 95% | Negative | 25% | Unknown | |
| R160138 | WT | 46 | III | 0 | 4 | Triple negative | Negative | Negative | Negative | 80% | 22 | |
| R160145 | E542K | 47 | II | 0 | 1.8 | Luminal B | 50% | 20% | Negative | 75% | 26 | |
| R160158 | WT | 48 | III | 6 | 1.3 | Luminal B | 30% | Negative | Negative | 60% | 28 | |
| R160161 | E545K | 49 | Unknown | 10 | 8 | Luminal B | 50% | 50% | Negative | 20% | 36 | Relapsed |
| R160164 | H1047R | 50 | II–III | 0 | 2 | Luminal B | 3% | Negative | Negative | 60% | 27 | |
| R160169 | H1047R | 51 | II | 12 | 2 | Luminal A | 90% | 30% | 1+ | 8% | 30 | |
Abbreviations: ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; PFS, progression-free survival; PR, progesterone receptor; WT, wild-type.
Table S3.
New PIK3CA mutations in exons 9 and 20 in breast cancers (n=494)
| Exon | Nucleotide change | Codon change | New mutation effect | Number of mutations |
|---|---|---|---|---|
|
| ||||
| 9 | c.1618C>G | p.Leu540Val | Missense | 1 |
| 9 | c.1621T>C | p.Ser541Pro | Missense | 1 |
| 9 | c.1628T>C | p.Ile543Thr | Missense | 1 |
| 9 | c.1628T>G | p.Ile543Ser | Missense | 1 |
| 9 | c.1629C>G | p.Ile543Met | Missense | 2 |
| 9 | c.1655G>A | p.Trp552* | Nonsense | 1 |
| 20 | c.3139_3140CA>TG | p.His1047Cys | Missense | 1 |
| 20 | c.3206G>A | p.*1069* | Silent | 1 |
| 9 | c.[1621T>C(+)1644delA] | p.[Ser541Pro(+)Lys548fs*10] | Frameshift | 1 |
| 9+20 | c.[1651C>T(+)3117C>T] | p.[Leu551Leu(+)Phe1039Phe] | Silent | 1 |
| 20 | c.[3120G>A(+)3201G>A] | p.[Met1040Ile(+)Leu1067Leu] | Silent | 1 |
| 9+20 | c.[1644A>G(+)3140A>G(+)3178C>T] | p.[Lys548Lys(+)His1047Arg(+)His1060Tyr] | Missense | 1 |
| 20 | c.[3117C>T(+)3140A>G(+)3145G>A] | p.[Phe1039Phe(+)His1047Arg(+)Gly1049Ser] | Silent | 1 |
Table S4.
Univariate Cox analysis of the correlation between PIK3CA mutation status and progression-free/overall survival according to different clinicopathological parameters
| Variables | PFS
|
OS
|
||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
|
| ||||
| <40 years old | ||||
| PIK3CA mutation vs wild-type | 0.746 (0.298–10.866) | 0.531 | 1.365 (0.463–4.022) | 0.572 |
| Exon 9 mutation vs wild-type | 0.554 (0.173–1.780) | 0.322 | 1.345 (0.298–6.077) | 0.700 |
| Exon 20 mutation vs wild-type | 1.050 (0.336–3.282) | 0.934 | 1.238 (0.339–4.519) | 0.747 |
| p.542/545 mutation vs wild-type | 0.382 (0.117–1.247) | 0.111 | 0.995 (0.221–4.470) | 0.995 |
| p.1047 mutation vs wild-type | 0.894 (0.256–3.117) | 0.860 | 1.133 (0.252–5.092) | 0.871 |
| ≥40 years old | ||||
| PIK3CA mutation vs wild-type | 1.632 (0.821–3.247) | 0.163 | 2.545 (1.042–6.215) | 0.040 |
| Exon 9 mutation vs wild-type | 1.029 (0.403–2.628) | 0.953 | 1.858 (0.440–7.837) | 0.399 |
| Exon 20 mutation vs wild-type | 1.977 (0.879–4.446) | 0.099 | 2.283 (0.874–5.962) | 0.092 |
| p.542/545 mutation vs wild-type | 1.068 (0.380–3.003) | 0.901 | 3.175 (0.431–23.373) | 0.257 |
| p.1047 mutation vs wild-type | 1.942 (0.819–4.607) | 0.132 | 2.412 (0.840–6.928) | 0.102 |
| ER negative | ||||
| PIK3CA mutation vs wild-type | 1.747 (0.589–5.181) | 0.314 | 5.336 (0.699–40.709) | 0.106 |
| Exon 9 mutation vs wild-type | 1.507 (0.202–11.232) | 0.689 | – | – |
| Exon 20 mutation vs wild-type | 1.711 (0.504–5.810) | 0.389 | 3.824 (0.500–29.247) | 0.196 |
| p.542/545 mutation vs wild-type | 1.507 (0.202–11.232) | 0.689 | – | – |
| p.1047 mutation vs wild-type | 1.497 (0.440–5.092) | 0.518 | 3.312 (0.433–25.325) | 0.248 |
| ER positive | ||||
| PIK3CA mutation vs wild-type | 1.060 (0.555–2.025) | 0.860 | 1.583 (0.736–3.403) | 0.240 |
| Exon 9 mutation vs wild-type | 0.665 (0.303–1.459) | 0.309 | 1.185 (0.410–3.425) | 0.754 |
| Exon 20 mutation vs wild-type | 1.707 (0.779–3.741) | 0.182 | 1.714 (0.727–4.041) | 0.219 |
| p.542/545 mutation vs wild-type | 0.579 (0.253–1.324) | 0.195 | 1.295 (0.390–4.295) | 0.673 |
| p.1047 mutation vs wild-type | 1.785 (0.744–4.279) | 0.194 | 1.801 (0.684–4.738) | 0.233 |
| PR negative | ||||
| PIK3CA mutation vs wild-type | 1.522 (0.657–3.526) | 0.327 | 3.198 (0.953–10.729) | 0.060 |
| Exon 9 mutation vs wild-type | 1.888 (0.451–7.908) | 0.384 | – | – |
| Exon 20 mutation vs wild-type | 1.347 (0.517–3.510) | 0.542 | 2.099 (0.623–7.070) | 0.231 |
| p.542/545 mutation vs wild-type | 1.665 (0.397–6.975) | 0.486 | – | – |
| p.1047 mutation vs wild-type | 1.159 (0.444–3.023) | 0.763 | 1.860 (0.552–6.271) | 0.317 |
| PR positive | ||||
| PIK3CA mutation vs wild-type | 0.983 (0.468–2.066) | 0.964 | 1.396 (0.582–3.353) | 0.455 |
| Exon 9 mutation vs wild-type | 0.406 (0.170–0.973) | 0.043 | 0.733 (0.246–2.187) | 0.578 |
| Exon 20 mutation vs wild-type | 2.014 (0.811–4.999) | 0.131 | 1.880 (0.686–5.1479) | 0.220 |
| p.542/545 mutation vs wild-type | 0.340 (0.134–0.863) | 0.023 | 0.786 (0.231–2.674) | 0.700 |
| p.1047 mutation vs wild-type | 2.317 (0.801–6.704) | 0.121 | 2.258 (0.664–7.677) | 0.192 |
| HER2 negative | ||||
| PIK3CA mutation vs wild-type | 1.496 (0.727–3.076) | 0.274 | 2.168 (0.885–5.314) | 0.091 |
| Exon 9 mutation vs wild-type | 0.507 (0.221–1.166) | 0.110 | 1.238 (0.373–4.112) | 0.728 |
| Exon 20 mutation vs wild-type | 3.039 (1.078–8.573) | 0.036 | 2.156 (0.750–6.198) | 0.154 |
| p.542/545 mutation vs wild-type | 0.452 (0.187–1.096) | 0.079 | 1.546 (0.366–6.526) | 0.553 |
| p.1047 mutation vs wild-type | 3.225 (0.991–10.488) | 0.052 | 2.262 (0.685–7.472) | 0.180 |
| HER2 positive | ||||
| PIK3CA mutation vs wild-type | 0.948 (0.410–2.193) | 0.900 | 1.983 (0.630–6.239) | 0.242 |
| Exon 9 mutation vs wild-type | 1.709 (0.400–7.306) | 0.470 | 1.964 (0.257–15.004) | 0.515 |
| Exon 20 mutation vs wild-type | 0.903 (0.370–2.205) | 0.823 | 2.248 (0.627–8.065) | 0.214 |
| p.542/545 mutation vs wild-type | 1.483 (0.347–6.342) | 0.595 | 1.683 (0.220–12.867) | 0.616 |
| p.1047 mutation vs wild-type | 0.873 (0.343–2.219) | 0.776 | 2.502 (0.563–11.118) | 0.228 |
| Ki67 labeling index £30% | ||||
| PIK3CA mutation vs wild-type | 1.672 (0.764–3.663) | 0.199 | 2.977 (0.955–9.281) | 0.600 |
| Exon 9 mutation vs wild-type | 0.556 (0.221–1.399) | 0.213 | 1.040 (0.233–4.637) | 0.959 |
| Exon 20 mutation vs wild-type | 2.838 (1.065–7.565) | 0.037 | 3.099 (0.869–11.047) | 0.081 |
| p.542/545 mutation vs wild-type | 0.464 (0.171–1.262) | 0.133 | 1.480 (0.192–11.378) | 0.706 |
| p.1047 mutation vs wild-type | 2.773 (0.958–8.027) | 0.060 | 3.548 (0.805–15.642) | 0.094 |
| Ki67 labeling index >30% | ||||
| PIK3CA mutation vs wild-type | 1.270 (0.546–2.951) | 0.579 | 1.716 (0.689–4.274) | 0.246 |
| Exon 9 mutation vs wild-type | 1.315 (0.398–4.343) | 0.653 | 1.941 (0.458–8.234) | 0.368 |
| Exon 20 mutation vs wild-type | 1.409 (0.492–4.039) | 0.523 | 1.611 (0.552–4.705) | 0.383 |
| p.542/545 mutation vs wild-type | 1.267 (0.384–4.186) | 0.697 | 1.881 (0.443–7.977) | 0.391 |
| p.1047 mutation vs wild-type | 1.478 (0.448–4.875) | 0.521 | 1.661 (0.493–5.597) | 0.413 |
| Tumor stage I and II | ||||
| PIK3CA mutation vs wild-type | 1.070 (0.557–2.059) | 0.838 | 1.817 (0.808–4.086) | 0.148 |
| Exon 9 mutation vs wild-type | 0.578 (0.247–1.349) | 0.205 | 0.790 (0.270–2.314) | 0.667 |
| Exon 20 mutation vs wild-type | 1.584 (0.744–3.370) | 0.233 | 2.301 (0.904–5.853) | 0.080 |
| p.542/545 mutation vs wild-type | 0.490 (0.194–1.237) | 0.131 | 0.837 (0.248–2.825) | 0.837 |
| p.1047 mutation vs wild-type | 1.572 (0.691–3.577) | 0.281 | 2.510 (0.859–7.333) | 0.092 |
| Tumor stage III | ||||
| PIK3CA mutation vs wild-type | 1.519 (0.498–4.635) | 0.463 | 2.331 (0.514–10.581) | 0.273 |
| Exon 9 mutation vs wild-type | 1.359 (0.309–5.979) | 0.685 | – | – |
| Exon 20 mutation vs wild-type | 1.867 (0.428–8.137) | 0.406 | 1.201 (0.265–5.442) | 0.812 |
| p.542/545 mutation vs wild-type | 1.225 (0.277–5.414) | 0.789 | – | – |
| p.1047 mutation vs wild-type | 1.498 (0.342–6.550) | 0.592 | 0.911 (0.200–4.149) | 0.904 |
| Lymph node negative | ||||
| PIK3CA mutation vs wild-type | 1.002 (0.423–2.372) | 0.997 | 1.626 (0.606–4.360) | 0.334 |
| Exon 9 mutation vs wild-type | 0.236 (0.085–0.655) | 0.006 | 0.444 (0.127–1.545) | 0.202 |
| Exon 20 mutation vs wild-type | 2.214 (0.742–6.612) | 0.154 | 2.399 (0.766–7.513) | 0.133 |
| p.542/545 mutation vs wild-type | 0.107 (0.029–0.403) | 0.001 | 0.401 (0.090–1.790) | 0.231 |
| p.1047 mutation vs wild-type | 3.065 (0.717–13.102) | 0.131 | 3.120 (0.712–13.673) | 0.131 |
| Lymph node positive | ||||
| PIK3CA mutation vs wild-type | 1.428 (0.681–2.996) | 0.346 | 2.279 (0.848–6.124) | 0.102 |
| Exon 9 mutation vs wild-type | 1.926 (0.586–6.328) | 0.280 | 4.403 (0.593–32.711) | 0.147 |
| Exon 20 mutation vs wild-type | 1.283 (0.558–2.951) | 0.557 | 1.669 (0.570–4.892) | 0.350 |
| p.542/545 mutation vs wild-type | 1.687 (0.514–5.540) | 0.389 | 4.059 (0.546–30.159) | 0.171 |
| p.1047 mutation vs wild-type | 1.132 (0.492–2.600) | 0.771 | 1.471 (0.502–4.309) | 0.482 |
| Tumor size <5 cm | ||||
| PIK3CA mutation vs wild-type | 1.074 (0.606–1.905) | 0.806 | 1.584 (0.784–3.200) | 0.200 |
| Exon 9 mutation vs wild-type | 0.619 (0.298–1.283) | 0.197 | 1.101 (0.387–3.129) | 0.857 |
| Exon 20 mutation vs wild-type | 1.530 (0.766–3.058) | 0.228 | 1.602 (0.730–3.516) | 0.240 |
| p.542/545 mutation vs wild-type | 0.553 (0.257–1.191) | 0.130 | 1.216 (0.372–3.983) | 0.746 |
| p.1047 mutation vs wild-type | 1.704 (0.801–3.623) | 0.166 | 1.839 (0.767–4.410) | 0.172 |
| Tumor size ≥5 cm | ||||
| PIK3CA mutation vs wild-type | 3.231 (0.407–25.614) | 0.267 | – | – |
| Exon 9 mutation vs wild-type | – | – | – | – |
| Exon 20 mutation vs wild-type | 3.231 (0.407–25.614) | 0.267 | – | – |
| p.542/545 mutation vs wild-type | – | – | – | – |
| p.1047 mutation vs wild-type | 0.286 (0.026–3.161) | 0.307 | – | – |
| Luminal and HER2 subtypes | ||||
| PIK3CA mutation vs wild-type | 1.323 (0.752–2.327) | 0.332 | 2.125 (1.032–4.375) | 0.041 |
| Exon 9 mutation vs wild-type | 0.723 (0.351–1.488) | 0.378 | 1.254 (0.443–3.550) | 0.670 |
| Exon 20 mutation vs wild-type | 1.969 (0.988–3.923) | 0.054 | 2.337 (1.024–5.331) | 0.044 |
| p.542/545 mutation vs wild-type | 0.637 (0.298–1.361) | 0.244 | 1.373 (0.421–4.478) | 0.600 |
| p.1047 mutation vs wild-type | 1.945 (0.917–4.125) | 0.083 | 2.493 (0.971–6.399) | 0.058 |
| Triple negative subtype | ||||
| PIK3CA mutation vs wild-type | 0.720 (0.083–6.247) | 0.766 | 0.720 (0.083–6.247) | 0.766 |
| Exon 9 mutation vs wild-type | – | – | – | – |
| Exon 20 mutation vs wild-type | 0.255 (0.026–2.463) | 0.237 | 0.255 (0.026–2.463) | 0.237 |
| p.542/545 mutation vs wild-type | – | – | – | – |
| p.1047 mutation vs wild-type | 0.255 (0.026–2.463) | 0.237 | 0.255(0.026–2.463) | 0.237 |
Note: P<0.05 was considered statistically significant and those values are shown in bold.
Abbreviations: ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; OS, overall survival; PFS, progression-free survival; PR, progesterone receptor.
Table S5.
Comparison of association between PIK3CA mutations and various clinicopathological features in different areas in China
| Authors | Year of publication | Area | No. of patients | PIK3CA mutation | Sample type | Methods | Association between PIK3CA mutations and clinicopathological characteristics |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Hu et al1 | 2018 | Changsha, Hunan | 68 | 33.82% | Peripheral blood | Targeted NGS | PIK3CA mutations: worse progression-free survival |
| Deng et al2 | 2019 | Chengdu, Sichun | 507 | 46.5% | FTS | Targeted NGS | PIK3CA mutations: ER positive, PR positive, HER3 negative; two or three mutations in PIK3CA: poor prognosis for overall survival |
| Chen et al3 | 2018 | Shanghai | 149 | 43.6% | FTS | Targeted NGS | PIK3CA mutations: older age, ER positive, PR positive |
| Cheng et al4 | 2017 | Luzhou, Sichun | 32 | 28.12% | FFPE | DS | PIK3CA mutations: more invasiveness lymph node, bigger tumor size |
| Yuan et al5 | 2015 | Beijing | 729 | 28.3% | FTS | DS | PIK3CA mutations: ER positive, PR positive, less response to neoadjuvant chemotherapy |
| Wang et al6 | 2015 | Xining, Qinghai | 25 | 32% | FTS | DS | Not detected |
| Liu et al7 | 2015 | Dalian, Liaoning | 80 | 32.5% | FFPE | Targeted NGS | No significance |
| Deng et al8 | 2015 | Chengdu, Sichun | 288 | 15.6% | FFPE | DS | PIK3CA mutations: poor outcome of ER-positive breast cancer |
| Zhang et al9 | 2014 | Beijing | 93 | 32.3% | FTS | xTAG liquid chip | PIK3CA mutations: patients’ clinical response to neoadjuvant chemotherapy |
| Bai et al10 | 2014 | Xi’an Shaanxi | 105 | 35.2% | FFPE | Targeted NGS | PIK3CA mutations: older age |
| Tong et al11 | 2012 | Guangzhou, Guangdong | 120 | 7.5% | FTS | MA | PIK3CA mutations: older age |
| Li et al12 | 2010 | Shanghai | 233 | 19.7% | FFPE | DS | PIK3CA mutations: high grade, ER positive, PR positive, PTEN positive |
| Current study | 2018 | Hefei, Anhui | 494 | 38.06% | FFPE | DS | PIK3CA mutations: luminal and HER2 positive, ER positive, PR positive, low Ki67 index, better overall survival; exon 9 mutations: worse progression-free survival; exon 20 mutations: better progression- free survival and overall survival; H1047 mutations: older age, better progression-free survival and overall survival; non-hotspot mutations: larger tumor size |
Abbreviations: DS, direct sequencing; ER, estrogen receptor; FFPE, formalin-fixed, paraffin-embedded tissue samples; FTS, frozen tissue samples; HER3, human epidermal growth factor receptor 3; MA, MassARRAY; NGS, next generation sequencing; PR, progesterone receptor.