Abstract
Purpose
The aim of this work was to investigate the clinicopathological significance of fibrinogen gamma chain (FGG) and its biological roles during hepatocellular carcinoma (HCC) development and progression.
Methods
The expression of FGG was examined by Western blot and reverse transcription quantitative PCR in two different sample sets, including 24 or 35 pairs of HCC tumor tissues and their corresponding adjacent non-tumorous tissues. Afterward, association analysis between the expression of FGG and clinicopathological characteristics was systematically analyzed in 79 HCC patients. Subsequently, the mobility and invasiveness of SK-HEP-1 cells with FGG overexpression or knockdown were evaluated by transwell assay and wound healing assay. Additionally, the expressions of epithelial to mesenchymal transition (EMT)-associated markers were also detected in FGG overexpressed or silenced SK-HEP-1 cells.
Results
The expression of FGG was significantly increased in primary HCC tissues comparing with its corresponding adjacent non-tumorous tissues. Clinical pathological analysis demonstrated that upregulation of intracellular FGG was significantly associated with increased vascular invasion, more satellite nodules, and more advanced TNM stage, and HCC patients with stronger expression of FGG had a higher recurrence rate and correspondingly a shorter overall survival time. Meanwhile, the high expression of FGG was also proved to be an independent risk factor for disease-free survival after surgical resection. In vitro phenotype studies showed that overexpression of FGG could promote the migration and invasion in SK-HEP-1 cells; conversely, these phenotypes could be significantly inhibited by knocking down the expression of FGG. Mechanism studies indicated that FGG could promote the migration and invasion through EMT signaling pathway by regulating the expressions of Slug and ZEB1.
Conclusion
FGG played important roles in enhancing cancer cell motility and invasiveness through EMT signaling, and might serve as a potential prognostic biomarker for HCC patients.
Keywords: hepatocellular carcinoma, fibrinogen gamma chain, epithelial to mesenchymal transition, recurrence, metastasis
Introduction
According to the global cancer statistics in 2018, hepatocellular carcinoma (HCC) was considered as the sixth common malignant cancer and the fourth leading cause of cancer-related mortality worldwide, resulting in 782,000 deaths each year.1 Although surgical resection combined with chemo- or radiotherapy is the mainstay of HCC treatment and significantly improves the postoperative 5-year survival rate, the patients with advanced HCC remain showing poor outcomes.2 Based on clinical practice, recurrence and metastasis of HCC remain the major obstacle to prolong the long-term survival of postoperative patients. Hence, to further understand the recurrence/metastasis mechanisms of HCC and identify novel biomarkers for predicting its prognosis will help for improving the long-term clinical outcomes of HCC patients.
Epithelial to mesenchymal transition (EMT) is a well-known process by which epithelial cells are transformed into mesenchymal cells. EMT plays crucial roles in wound healing, embryogenesis, and malignant cancer progression. Especially for cancer progression, EMT affects almost overall characteristics of cancer, such as migration, invasion, proliferation, resistance to therapeutics, and immune escape.3,4 Additionally, increasing evidences reported that Slug and ZEB-1 are well studied transcriptional factors and important inducers of EMT, whose expression might be elevated at the early stage of EMT. Subsequently, upregulation of Slug or ZEB-1 results in decreasing the expression of epithelial markers and induction the expression of mesenchymal genes.5,6 There is no doubt that EMT also plays a vital role in HCC development. HCC cells acquire more invasive and aggressive phenotypes as they undergo EMT and gain stemness properties like cancer stem cells.7 In the meantime, downregulation of E-cadherin or overexpression of Snail and Slug in HCC tissues was confirmed to be correlated with poor prognosis of HCC patients.8
Fibrinogen is a dimeric protein composed of pairs of three polypeptide chains termed as Aα, Bβ, and γ, which are coded by three different genes, fibrinogen alpha chain (FGA), fibrinogen beta chain (FGB), and fibrinogen gamma chain (FGG), respectively. It is well known that fibrinogen plays important roles in blood coagulation, fibrinolysis, cellular and matrix interactions, inflammation, and wound healing.9 Meanwhile, numerous studies have reported that hyperfibrinogenemia as a prognostic factor10 is frequently observed in patients with malignant cancers and is closely associated with the process of tumor invasion, metastasis,11–13 angiogenesis,14 and tumor growth.15 However, the basis of how fibrinogen causing cancer progression remains unclear.
As one of three peptide chains of fibrinogen, the dys-regulation of FGG is also frequently detected in many malignant cancer types, such as HCC,16 gastric cancer,10 and prostate cancer.17 In addition, it has been proved that FGG was involved in regulating the formation and function of fibrinogen. For instance, FGG inhibits the adhesion of platelets to fibrinogen by interacting with hepatitis B spliced protein;18 Yokoyama et al discovered that C terminal region of FGG as a major integrin-binding site of fibrinogen was involved in the process of thrombus formation, angiogenesis, and inflammation.19,20 Nagata et al showed that a frameshift mutation of FGG led to hypofibrinogenemia, indicating that FGG participated in the regulation of fibrinogen secretion.21 However, these findings were based on the facts that FGG was an extracellular protein and exerted these functions as structural domains of fibrinogen. Whether FGG had biological functions inside the cells remained unclear so far.
In our previous study, an iTRAQ-based quantitative proteomics approach was applied to systematically compare the proteome differences among small HCC (diameter ≤3 cm), media HCC (3 cm < diameter <5 cm), large HCC (5 cm ≤ diameter <10 cm), and huge HCC (diameter ≥10 cm) in 60 pairs of tumor and non-tumor specimens for the first time. Finally, we found FGG (but not other chains of fibrinogen) was significantly upregulated in primary HCC tissues comparing with the adjacent non-cancerous tissues.22 Furthermore, the upregulation degree of FGG was significantly correlated with tumor size. Thus, we hypothesize that the FGG may also play important roles in the progression of HCC. In this study, based on the clinical characteristics of FGG and phenotype changes of FGG-manipulated HCC cells, we provide evidences that FGG, as an intracellular protein, is a potential prognostic biomarker for HCC patients and promotes the migration and invasion of HCC cells through activating EMT. These results would expand our understanding of the biological functions of FGG and might provide theoretical basis for HCC treatment with FGG as the target.
Materials and methods
Patients and specimens
To detect the expression levels of FGG in HCC patients, the fresh frozen tumor tissues and adjacent non-tumorous liver tissues obtained from HCC patients after surgical resection were subjected to protein extraction for Western blot examination (n=24 pairs) or RNA extraction for reverse transcription quantitative PCR (RT-qPCR) analysis (n=35 pairs). In addition, a total of 79 archived paraffin-embedded specimens were randomly collected from HCC patients undergoing curative resection at the Mengchao Hepatobiliary Hospital of Fujian Medical University from January 2014 to June 2016. Clinical and pathologic diagnosis of HCC met the criteria of the American Association for the Study of Liver Diseases. Tumor stage was determined according to Barcelona Clinic Liver Cancer (BCLC) staging classification and TNM Staging for Hepatocellular Cancer (eighth edition, 2017). The HCC patients enrolled in this study were divided into two groups: the patients who had recurrence/metastasis within 2 years after operation (R/M group); the patients who had no recurrence/metastasis within 2 years of operation (NR/M group). Patient follow-up was terminated on June 30, 2018. The median follow-up period was 33 months (range 5–48 months). Disease-free survival (DFS) and overall survival (OS) were considered as the primary endpoints. DFS was calculated from the operation date to the date of tumor recurrence (intrahepatic recurrence and extrahepatic metastasis). OS was defined as the time range from the date of surgical resection to the date of death or the last follow-up.
Cell lines and cell culture
The human HCC cell lines SK-HEP-1 and HEK-293T were purchased from American Type Culture Collection (ATCC) in 2014. MHCC97L was purchased from from the cell bank of the Shanghai Institute of Cell Biology (Shanghai, China) in 2017. The last cytogenetic test and authentication were performed by a commercial company, Genewiz, in March, 2017. And its STR profiles 100% matched with ATCC STR database. Cells were routinely cultured in Minimum Essential Medium (Gibco, Rockville, MD, NY, USA) for SK-HEP-1 cells or DMEM (Gibco, Thermo, Waltham, MA, USA) for MHCC97L and HEK-293T cells supplemented with 10%(V/V) FBS (Excell Bio, Shanghai, China). Cells were incubated at 37°C in a 5% CO2-humidified cell culture incubator. All the cell lines were used <30 passages from acquisition to discard.
RT-qPCR analysis of FGG expression
Total RNA was isolated from 24 pairs of fresh-frozen HCC tissues and their adjacent non-tumorous liver tissues using TransZol reagents (TransGen Biotech, Beijing, China). Afterward, the mRNA expression of FGG was detected by RT-qPCR assays as previously described22. Briefly, reverse transcription was carried out using Transcriptor First Strand cDNA Synthesis Kit (Roche, Indianapolis, IN, USA) according to the manufacturer’s instructions. Real-time PCR determination was performed with the following parameters: 45 cycles of 95°C for 15 seconds and 60°C for 60 seconds. The relative gene expression of FGG was normalized with the reference gene 18S rRNA, and calculated according to the Livak method (2−ΔΔCt). Specific primers (human FGG, forward primer sequence – TTATTGTCCAACTACCT-GTGGC; reverse primer sequence – GACTTCAAAGTA-GCAGCGTCTAT; human 18S rRNA, forward primer sequence – AGAAACGGC-TACCACATCCA, and reverse primer sequence – CACCAGACTTGCCCTCCA-3) were verified with PCR amplification efficiency, which ranked between 90% and 110%.
Western blot analysis
Cells and fresh-frozen tumor tissue samples were lysed in ice-cold RIPA buffer (Beyotime Biotechnology, Shanghai, China) containing protease inhibitor cocktail (Roche). Protein extraction was subsequently quantified by bicinchoninic acid assay according to the manufacturer’s instruction (TransGen Biotech, Beijing, China). Equal amounts of protein samples were separated in 12% SDS-PAGE and transferred onto nitrocellulose membranes. After blocking with 5% skim milk, the membranes were probed with primary antibodies against FGG (Santa Cruz Biotechnology, Dallas, TX, USA), N-cadherin (Cell Signaling Technology, Beverly, MA, USA), Slug (Cell Signaling Technology), ZEB-1 (Cell Signaling Technology) or β-actin (Santa Cruz Biotechnology) overnight at 4°C. All these primary antibodies were diluted by 1,000 folds. Following wash, the membranes were incubated with the appropriate horseradish peroxidase-conjugated secondary antibody (1:5,000 dilution; Santa Cruz Biotechnology) for 1 hours at room temperature. Later, the blots were detected by chemiluminescence and visualized by the ChemiDoc MP imaging system (Bio-Rad, Hercules, CA, USA). Quantification of blots was carried out by densitometry and normalized by β-actin.
Immunohistochemical analysis
Immunohistochemical analysis was performed as previously described23 with slight modification. Briefly, FGG was immunohistochemically detected using a commercial IHC staining kit (Maixin Bio, Fuzhou, Fujian, China) according to the manufacturer’s instructions. The slides were incubated with the antibody against FGG (1:100 dilution; Santa Cruz Biotechnology) overnight at 4°C. The tumor expression of FGG was evaluated by two independent pathologists in a blinded manner, and discrepancies were resolved by consensus. Images were visualized using an Olympus BX40 microscope (Olympus Co., Tokyo, Japan). Ten random fields were selected for each section. The intensity of staining was defined as follows: 0, no visible staining; 1, weak staining (a faint cytoplasmic immunopositivity); 2, moderate staining (a clear cytoplasmic immunopositivity); and 3, strong staining (a pronounced cytoplasmic immunopositivity equivalent to that of the hepatic cells). And the percentage of positive-staining cells was graded as 0 (<5%), 1 (5%–25%), 2 (26%–50%), 3 (51%–75%), and 4 (>75%). The final immunoreactivity score was calculated by multiplying proportion and intensity scores. Each case was considered as negative if the final score was 0–1, weakly positive (score: 2–4), moderate positive (score: 5–7), or strongly positive (score: ≥8)
Establishment of FGG overexpression cells
The full-length human FGG (GenBank accession number NM_000509.5) was synthesized by GENEWIZ, a commercial company and cloned into a lentiviral vector pCDH-CMV-MCS-EF1-copGFP. Then the recombinant lentiviral particles for FGG overexpression were produced and concentrated as previously reported.24 In brief, 7.5 µg candidate plasmid was co-transfected with 5 µg pLP1, 3 µg pLP2, and 3 µg VSV-G into 107 of 293 T cells using the Lipofectamine 3000 (Invitrogen, Waltham, MA, USA) in 10 cm petri dish. After 20 hours incubation, the culture medium was replaced. Following that, the supernatant was collected at 48 and 72 hours of incubation time. Lentiviral particles were concentrated by 200-folds with ultracentrifugation for 1.5 hours at 100,000 g, 4°C. The concentrated lentivirus was used to infect SK-HEP-1 cells in the presence of 2 µg/mL Polybrene (Santa Cruz Biotechnology). Cells infected with empty vector were used as negative controls. Modified cells with FGG overexpression were selected with 2 µg/mL puromycin for 2 weeks.
RNA interference
Small interfering RNA duplexes against FGG, Slug, and ZEB-1 were synthesized by RiboBio Co., Ltd (Guangzhou, Guangdong, China). The siRNA sequences are provided in Table S1. After trial experiment, si RNA duplexes: siSlug-3 and siZEB-1-3 were finally selected to knockdown the expression of Slug and ZEB-1. siRNAs were transfected into the SK-HEP-1 cells using Lipofectamine 3000 according to the manufacturers’ protocol. The transfected cells were incubated at 37°C in 5% CO2 for 24 or 48 hours before Western-blot detection or phenotype analysis.
Wound healing assay
Culture-Insert (Ibidi, Munich, Germany) with 2 cell culture reservoirs was used to this experiment according to the manufacturer’s protocol. Namely, SK-HEP-1 cells with or without FGG overexpression were seeded into each side of the reservoirs at a density of 7×104 cells. After 12 hours of incubation, the molds were removed and the cells were washed twice with PBS to remove the un-adherent cells. Afterwards, the adherent cells were cultured and photographed at 0 hour, 12 and 24 hours.
Cell migration and invasion assay
The migration and invasion assays were carried out using transwell units with 8 µm pore (Corning, Brooklyn, NY, USA), as previously described.25 Briefly, 104 of modified SK-HEP-1 cells were suspended in 200 µL serum-free medium and loaded into the upper chamber in the migration assay. In addition, the lower chamber was filled with 500 µL of complete medium (10% FBS). After 18-hour incubation, the migrating cells were fixed with 4% formaldehyde for 30 minutes and stained with 0.5% crystal violet for another 30 min. The cells on the upper surface of the membrane were wiped off with a cotton swab. Following wash, the cells that had passed through the membrane were visualized and counted under a microscope (Zeiss GmbH, Jena, Germany) in five different fields at 200-folds magnification. On the other hand, for the invasion assay, only two different procedures were undertaken: the upper chamber was pre-coated with 100 µL of matrigel and more cells (2×104 cells) were subsequently loaded into the upper chamber.
Statistical analysis
All data were analyzed using SPSS statistical software (version 19.0, SPSS Inc., Chicago, IL, USA) or Prism statistical software (GraphPad version 6.01, San Diego, CA, USA). The results are presented as mean ± SD. In the analysis of clinical data, the comparisons of two groups were carried out by two-tailed paired Student’s t-test. Otherwise, other type of comparisons between two groups was used by two-tailed unpaired Student’s t-test. The correlation between FGG expression and clinicopathologic features was examined by Pearson’s chi-squared test. The cumulative recurrence and survival probability were evaluated using the Kaplan–Meier method, and log-rank test. The Cox proportional hazards model was used to determine the independent risk factors of recurrence and survival based on the variables selected in univariate analysis. P<0.05 was considered as statistically significant.
Ethics statement
The collection of clinical samples was approved by the ethics committee of Mengchao Hepatobiliary Hospital of Fujian Medical University and all patients signed an informed consent in accordance with the WMA Declaration of Helsinki 2013.
Results
FGG is upregulated in primary HCC tissues
In our previous study, iTRAQ-based proteomic strategy was used to screen HCC-associated protein, whose expression altered along with the tumor deterioration. Finally, we found that the expression of FGG, but not other chains of fibrinogen, significantly correlated with HCC progression. Thus, we would like to further evaluate the prognostic potential in HCC patients and investigate the biological functions in HCC cells.
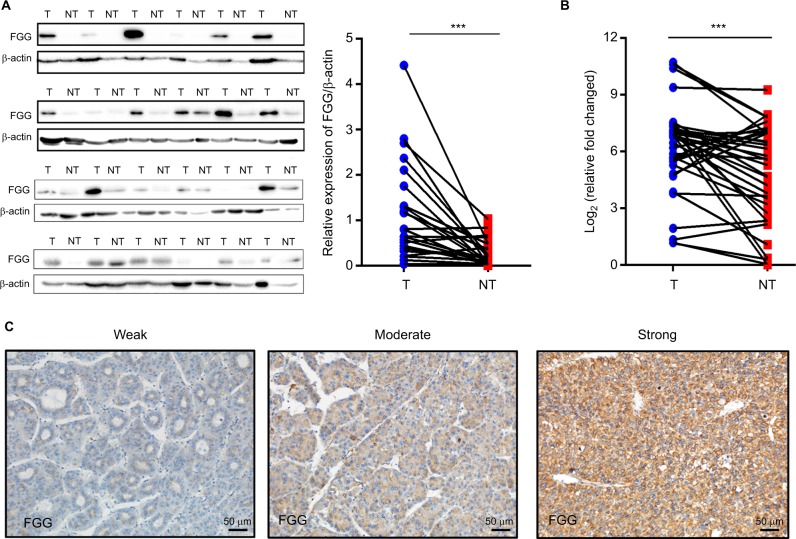
The expression of FGG in 24 pairs of HCC specimens and their adjacent non-tumorous liver samples were detected at the protein level, firstly. As shown in Figure 1A, the expression of FGG in HCC tissues was significantly higher than that in their paired non-malignant tissues (P=0.0005, paired t-test). Quantitative analysis of FGG protein suggested that 87.5% (21/24 samples) of HCC samples showed upregulated level of FGG expression compared with their paired non-malignant tissues. To further confirm this observation, the FGG mRNA expression levels were further examined by RT-qPCR in another sample set containing 35 pairs of HCC and their corresponding non-tumor tissues. Similarly, FGG mRNA expression was also significantly upregulated in most HCC specimens (77.1%, 27/35 samples; P=0.0018, paired t-test; Figure 1B), which was consistent with previous report.26 Overall, these results clearly revealed that the expression of FGG was significantly increased at both the mRNA and protein levels in HCC tissues compared with corresponding non-malignant tissues.
Figure 1.
FGG is significantly upregulated in HCC tumor tissues.
Notes: (A) Western blot analysis of FGG expression in 24 pairs of HCC tumor tissues and their adjacent non-tumorous tissues; P=0.0005, paired t-test. (B) RT-qPCR analysis of FGG mRNA expression in another sample set containing 35 pairs of HCC tumor samples and their non-tumorous tissues; P=0.0018, paired t-test. (C) Different FGG expression levels in HCC tumor tissues. FGG expression was categorized into three groups: weak – weak positive staining (1+); moderate – moderate positive staining (2+); and strong – strong positive staining (3+). ***P<0.001.
Abbreviations: FGG, fibrinogen gamma chain; HCC, hepatocellular carcinoma; NT, non-tumor sample; T, tumor sample; RT-qPCR, reverse transcription quantitative PCR.
High FGG expression was closely associated with aggressive clinicopathological features of HCC patients
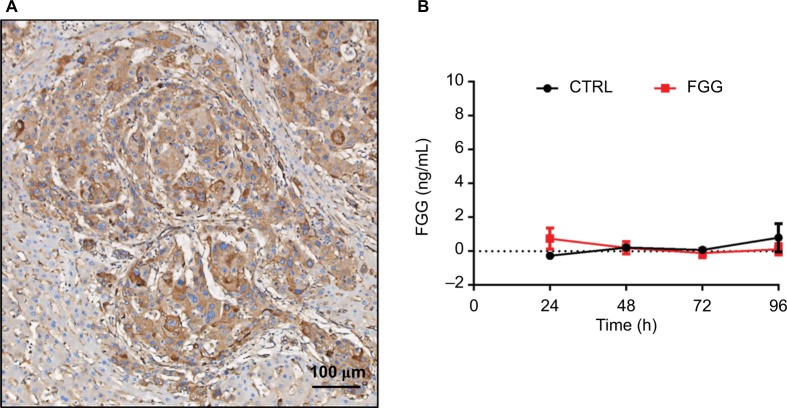
To evaluate the clinical significance of FGG, the expression of FGG was assessed in 79 paraffin-embedded HCC samples by tissue microarray. Representative photomicrographs of HCC tissues with weak positive, moderate positive, or strong positive immunohistochemical staining against FGG are shown in Figure 1C. According to the immunohistochemical results, FGG staining could be observed in all HCC specimens, including 31 weakly positive staining, 23 moderate positive staining, and 25 strong positive staining. Because fibrinogen was mainly produced by liver,27 therefore, it is not surprising that little specimens were identified as negative. In addition, FGG staining was primarily observed in the cytoplasm of HCC cells (Figure 5A), but not extracellular matrix, indicating that FGG might play roles in the HCC cells either.
Figure 5.
FGG exerts its regulatory function in an intracellular manner.
Note: (A) FGG is majorly located in HCC cells based on the observation of HCC immunohistochemical specimen. (B) ELISA assay of extracellular FGG.
Abbreviations: CTRL, control; FGG, fibrinogen gamma chain; HCC, hepatocellular carcinoma.
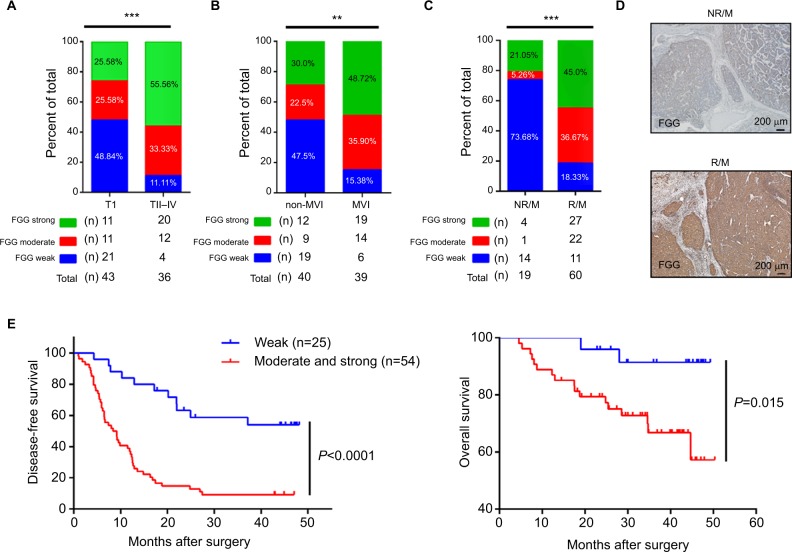
Afterwards, the correlation analysis between FGG expression and the clinical characteristics of the HCC patients was performed in the present study. Data were distributed into two groups, including: weakly positive group (n=25) and moderate and strong positive group (n=54), based on the immunohistochemical score of each slide. The results suggested that the higher expression of FGG was significantly associated with vascular invasion (both macro- and microvascular invasion, P=0.014 and P=0.002, respectively), satellite nodules (P=0.001), and more advanced TNM stage (P<0.001; Table 1, Figure 2A and B). When the enrolled patients were re-grouped, according to whether metastasis/recurrence or not, we found that the patients with metastasis or recurrence of HCC usually subject to higher expression of FGG in tumor tissues (3*2 chi-square test p<0.001, Figure 2C, 2D).
Table 1.
Associations between the expression of FGG and clinicopathological characteristics of 79 hepatocellular carcinoma patients
| Clinicopathological variables | FGG expression, IHC
|
Chi-squared value | P-value | |
|---|---|---|---|---|
| Weak (n=25) | Moderate/strong (n=54) | |||
|
| ||||
| Age (years) | ||||
| <55 | 11 | 25 | 0.036 | 0.849 |
| ≥55 | 14 | 29 | ||
| Gender | ||||
| Male | 24 | 51 | 0.086 | 1.000 |
| Female | 1 | 3 | ||
| Tumor size (cm) | ||||
| ≤5 | 16 | 22 | 2.546 | 0.111 |
| >5 | 9 | 32 | ||
| Satellite nodules | ||||
| Yes | 1 | 22 | 11.177 | 0.001** |
| No | 24 | 32 | ||
| Tumor encapsulation | ||||
| Absent | 15 | 24 | 1.654 | 0.198 |
| Present | 10 | 30 | ||
| Macrovascular invasion | ||||
| Yes | 0 | 12 | 6.551 | 0.014* |
| No | 25 | 42 | ||
| Microvascular invasion | ||||
| Yes | 6 | 33 | 9.416 | 0.002*** |
| No | 19 | 21 | ||
| AFP (ng/mL) | ||||
| <400 | 5 | 12 | 0.050 | 0.823 |
| ≥400 | 20 | 42 | ||
| HBV DNA (cps/mL) | ||||
| <500 | 14 | 21 | 2.028 | 0.154 |
| ≥500 | 11 | 33 | ||
| BCLC stage | ||||
| 0–A | 22 | 38 | 2.908 | 0.088 |
| B–D | 3 | 16 | ||
| TNM stage | ||||
| I | 21 | 22 | 12.893 | <0.001*** |
| II–IV | 4 | 32 | ||
| Fibrinogen levels (g/L) | ||||
| <4.0 | 18 | 46 | 1.931 | 0.219 |
| ≥4.0 | 7 | 8 | ||
Note:
P<0.05,
P<0.01,
P<0.001.
Abbreviations: AFP, a-fetoprotein; BCLC, Barcelona Clinic Liver Cancer staging; FGG, fibrinogen gamma chain; IHC, immunohistochemistry.
Figure 2.
Correlation analysis of FGG overexpression, clinicopathological features, and survival.
Notes: (A) HCC tumor tissues at TNM II, III, and IV stage displayed higher FGG expression level. 3*2 chi-squared test P=0.001. (B) Higher FGG expression level was significantly correlated with microvascular invasion. 3*2 chi-squared test P=0.009. (C) HCC tumor tissues in NR/M group displayed lower FGG expression level. 3*2 chi-squared test P<0.001. (D) Representative immunohistochemical staining of FGG expression in R/M or NR/M HCC tumor tissues. (E) Kaplan–Meier curves revealed an association between higher FGG expression and higher recurrence rate as well as the shorter overall survival time of patients with HCC. **P<0.01, ***P<0.001.
Abbreviations: FGG, fibrinogen gamma chain; HCC, hepatocellular carcinoma; MVI, microvascular invasion; NR/M, non-recurrence or metastasis; R/M, recurrence or metastasis; TI, TNM stage I; T II–IV, TNM stage II to IV.
Kaplan–Meier analysis and Cox’s multivariate proportional hazards model were subsequently used to systematically calculate the effect of FGG expression on DFS and OS. Kaplan– Meier analysis showed that patients with lower expression of FGG always had a longer DFS (P<0.0001, Figure 2E) and OS (P=0.015, Figure 2E) than those with higher expression of FGG postoperatively. Meanwhile, univariate analysis demonstrated that many parameters, including tumor size, tumor encapsulation, microvascular invasion, macrovascular invasion, satellite nodules, TNM stage, BCLC stage, and the expression of FGG, were prognostic variables for DFS in HCC patients (Table 2). Similarly, most of these parameters, such as tumor size, microvascular invasion, macrovascular invasion, TNM stage, BCLC stage, and the expression of FGG were prognostic variables for OS (Table 2). Multivariate analysis confirmed that higher expression of FGG was an independent poor prognostic factor for DFS (P=0.027) of patients with HCC (Table 2) after curative resection. Totally, these observations indicated that the upregulation of FGG in HCC cells may contribute to the recurrence or metastasis of HCC.
Table 2.
Univariate and multivariate analysis of recurrence or survival associated factors in 79 hepatocellular carcinoma patients
| Variables | Recurrence/metastasis | Survival | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||
|
| ||||||||
| HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | |
|
| ||||||||
| Gender (female vs male) | 0.649 (0.203–2.078) | 0.467 | 0.820 (0.109–6.158) | 0.847 | ||||
| Age (≥55 vs <55 years) | 0.802 (0.483–1.331) | 0.394 | 1.214 (0.488–3.022) | 0.676 | ||||
| Tumor size (>5 vs ≤5 cm) | 1.722 (1.028–2.883) | 0.039* | 0.958 (0.473–1.940) | 0.905 | 5.820 (1.690–20.036) | 0.005** | 4.273 (0.897–20.355) | 0.068 |
| Tumor encapsulation (present vs absent) | 1.949 (1.165–3.260) | 0.011* | 1.415 (0.798–2.509) | 0.235 | 1.977 (0.777–5.033) | 0.153 | ||
| Microvascular invasion (yes vs no) | 1.836 (1.101–3.060) | 0.02* | 0.822 (0.407–1.659) | 0.584 | 2.697 (1.024–7.104) | 0.045* | 0.717 (0.159–3.226) | 0.664 |
| Macrovascular invasion (yes vs no) | 4.353 (2.250–8.423) | <0.001*** | 2.723 (0.856–8.666) | 0.09 | 5.874 (2.242–15.386) | <0.001*** | 4.549 (0.509–40.658) | 0.175 |
| Satellite nodules (yes vs no) | 4.453 (2.477–8.005) | <0.001*** | 2.456 (1.222–4.936) | 0.012* | 2.464 (0.973–6.237) | 0.057 | ||
| AFP (≥400 ng⁄mL vs <400 ng⁄mL) | 1.032 (0.567–1.880) | 0.918 | 1.663 (0.631–4.385) | 0.304 | ||||
| HBV-DNA (≥500 cps/mL vs <500 cps/mL) | 1.243 (0.743–2.078) | 0.407 | 1.760 (0.668–4.632) | 0.253 | ||||
| Fibrosis stage (IV vs I–III) | 1.366 (0.803–2.323) | 0.25 | 1.393 (0.529–3.667) | 0.502 | ||||
| TNM stage (II–III vs I) | 2.684 (1.597–4.509) | <0.001*** | 1.271 (0.605–2.671) | 0.527 | 3.559 (1.335–9.483) | 0.011* | 0.971 (0.244–3.862) | 0.967 |
| BCLC stage (B–D vs 0–A) | 2.389 (1.361–4.193) | 0.002** | 0.726 (0.253–2.081) | 0.551 | 3.919 (1.570–9.779) | 0.003** | 0.689 (0.085–5.571) | 0.727 |
| Fibrinogen levels (≥4.0 g/L vs <4.0 g/L) | 0.738 (0.374–1.456) | 0.380 | 1.978 (0.750–5.214) | 0.168 | ||||
| FGG (strong and moderate vs weak) | 4.477 (2.298–8.721) | <0.001*** | 3.162 (1.460–6.848) | 0.004** | 5.191 (1.190–22.650) | 0.028* | 2.827 (0.505–15.836) | 0.237 |
Note:
P<0.05,
P<0.01,
P<0.001.
Abbreviations: AFP, a-fetoprotein; BCLC, Barcelona Clinic Liver Cancer staging; FGG, fibrinogen gamma chain.
FGG promoted migration and invasion of HCC cells by activating EMT
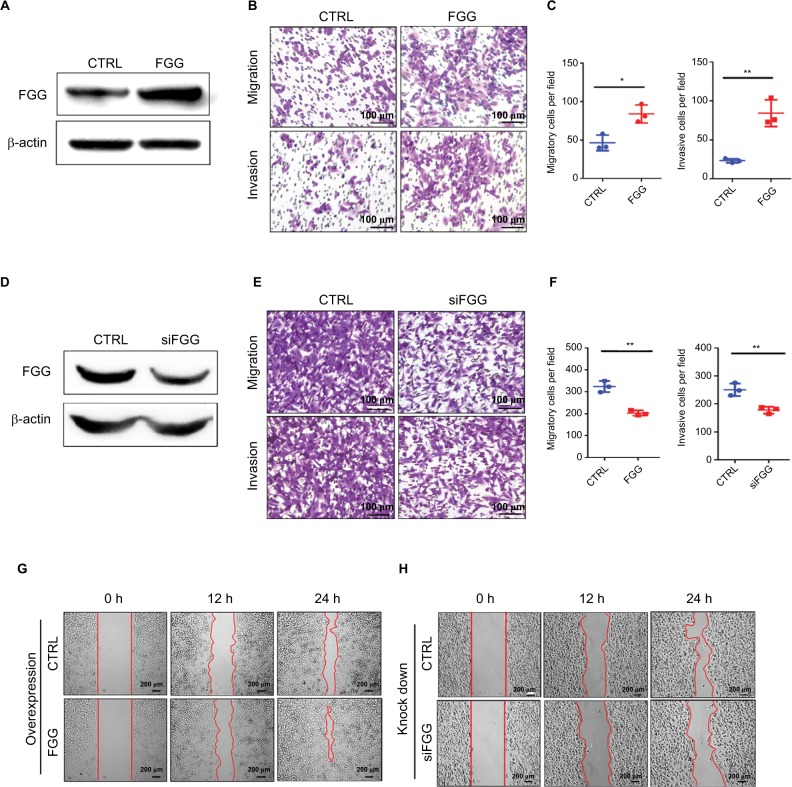
To clearly elucidate our clinicopathological results, we investigated whether FGG could affect the migration and invasion of HCC cells in vitro. At first, overexpression or knockdown of FGG was confirmed by Western blot (Figure 3A and D). Afterwards transwell assay revealed that the migration and invasion of HCC cells were significantly enhanced, when FGG was overexpressed in SK-HEP-1 cells (Figure 3B and C). A consistent result was obtained in MHCC97L cells with FGG overexpression (Figure S1). Conversely, downregulation of FGG suppressed the migration and invasion of HCC cells (Figure 3E and F). Similarly, wound healing assays showed that overexpression of FGG significantly increased the capacity of SK-HEP-1 cells to close the scratched “wound” (Figure 3G), while the motility was attenuated in FGG knockdown cells (Figure 3H). These data suggested that FGG promoted the invasion and metastasis of HCC cells in vitro, which was in well agreement with our clinical analysis.
Figure 3.
FGG promotes HCC cell migration and invasion.
Notes: (A, D) Western blot results showing that FGG was overexpressed or knocked down in SK-Hep-1 cells. (B) Representative images reflect that overexpression of FGG enhances the migration and invasion of SK-HEP-1 cells. (E) Knockdown of FGG suppresses the migration and invasion of HCC cells. (C, F) Quantitative analysis of migration and invasion data of SK-HEP-1 cells with FGG overexpression (C) or knockdown (F). (G) Wound healing assays show that overexpression of FGG promoted the migration of SK-HEP-1 cells. (H) Wound healing assays show that downregulation of FGG inhibited the migration of SK-HEP-1 cells. Representative images of wound healing assays were taken from 0 to 24 h after the scratch was made. All experiments were independently repeated at least three times. Data are shown as mean ± SD. *P<0.05, **P<0.01.
Abbreviations: CTRL, control; FGG, fibrinogen gamma chain; HCC, hepatocellular carcinoma; siFGG, knockdown of FGG with small interfering RNA duplexes.
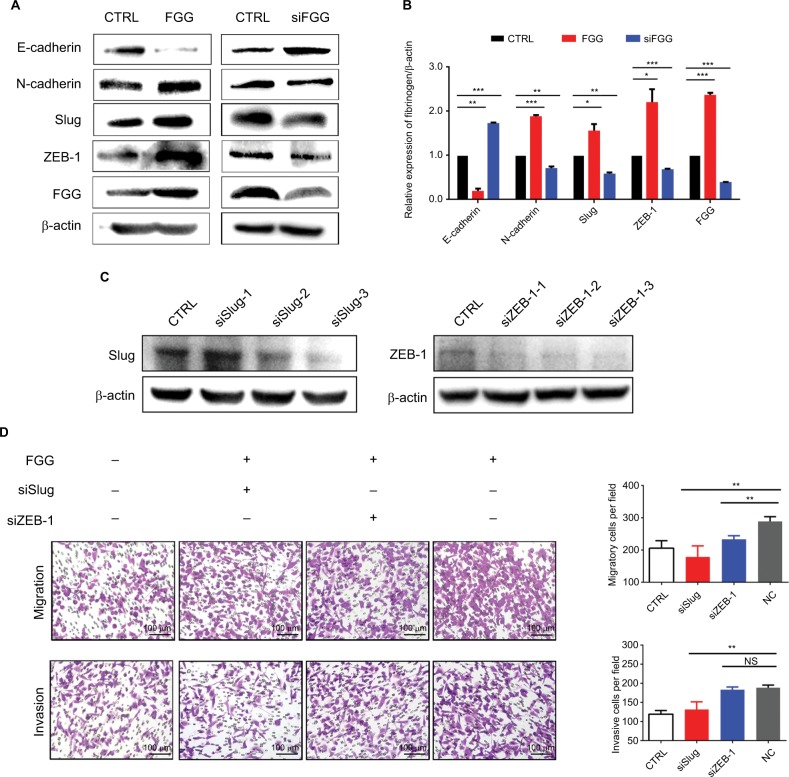
It is well known that EMT as a key process dramatically promotes the ability of cellular motility and invasiveness frequently causing cancer metastasis.28 During EMT, proteins involved in cell junctions, such as E-cadherin are downregulated, while mesenchymal markers, such as N-cadherin are upregulated.29 This conversion is believed to be triggered by many transcriptional factors with multiple zinc finger structure, including Slug, Snail1, and ZEB-1.3,30 Consequently, we speculated that FGG may activate the EMT process on the basis of our results obtained from phenotype study. The expressions of EMT markers and EMT-related transcription factors were examined by Western blot. The results showed that upregulating FGG in SK-HEP-1 cells significantly reduced the expression of epithelial marker (E-cadherin) but obviously increased the expression of mesenchymal marker (N-cadherin, Figure 4A and B) at protein level. Meanwhile, Slug and ZEB-1 were significantly upregulated to around 2-folds either. The opposite results obtained in FGG knockdown cells ideally supported these findings (Figure 4A and B). To prove that EMT was involved in the regulation of FGG, EMT inducer Slug and ZEB-1 were knocked down in SK-Hep-1 cells with FGG overexpression by their specific siRNA (Figure 4C). The phenotype assay showed that the ability of FGG promoting the migration and invasion of HCC cells was obviously attenuated by the knockdown of Slug (Figure 4D). Similarly,the knockdown of ZEB-1 inhibited the migration in HCC cells with FGG overexpression, although not the invasion. Overall, FGG promoted the migration and invasion of HCC by EMT induced by Slug and partially by ZEB-1.
Figure 4.
Dysregulation of FGG affects the migration and invasion of HCC cells through EMT.
Notes: (A) Western blots showed the changes in E-cadherin, N-cadherin, Slug, and ZEB-1 expression after artificial modification of FGG expression. siFGG means knockdown of FGG with small interfering RNA duplexes. (B) Quantitative analysis of EMT-associated protein expression in SK-HEP-1 cells; (C) Western blot results showed that Slug and ZEB-1 were knocked down in SK-HEP-1 cells with FGG overexpression. (D) The migration and invasion of SK-HEP-1 cells with FGG overexpression were dramatically inhibited by the knockdown of Slug and ZEB-1. In this experiment, siSlug-3 and siZEB-1–3 were used to knockdown the expression of Slug and ZEB-1, respectively. Each experiment was independently performed at least three times. Data are shown as mean ± SD. *P<0.05, **P<0.01, ***P<0.001.
Abbreviations: EMT, epithelial to mesenchymal transition; FGG, fibrinogen gamma chain; siFGG, knockdown of FGG with small interfering RNA duplexes; NC, negative control; NS, non-significant; siSlug-1, siSlug-2, and siSlug-3, knockdown of Slug with small interfering RNA duplexes-1, duplexes-2 and duplexes-3; siZEB-1-1, siZEB-1-2, and siZEB-1-3, knockdown of ZEB-1 with small interfering RNA duplexes-1, duplexes-2, and duplexes-3.
Discussion
Tumor recurrence and⁄or metastasis are the main reasons causing poor clinical outcomes of patients with advanced HCC after surgical resection.31 Hence, it is necessary to keep developing more reliable biomarkers for early prognosis evaluation of HCC and determining subsequent therapeutic strategies. Abnormal expression of FGG has been frequently observed in many cancer types, such as HCC,26 gastric cancer,10 and prostate cancer,17 which strongly indicated that FGG may play an important role in cancer progression. However, the clinical significance of FGG in HCC patients remains unknown, and the underlying molecular mechanisms in HCC progression are also required to be further clarified. In the present study, we first identified that the expression of FGG in HCC tissues was significantly higher than non-cancerous tissues. Subsequently, FGG proved to be closely associated with many clinicopathological characteristics, such as: satellite nodules (Table 1), vascular invasion (Figure 2B), and TNM stage (Figure 2A). Moreover, HCC patients with higher expression level of FGG usually suffered from shorter survival time and higher recurrence rate (Figure 2E). Additionally, Cox’s univariate and multivariate survival analyses indicated that stronger FGG staining in HCC tissues is an independent predictor of DFS in HCC patients following hepatectomy (Table 2). Finally, the phenotype study clearly demonstrated that FGG promoted the migration and invasion of SK-HEP-1 cells in vitro (Figure 3). These findings ideally explained why HCC patients with higher expression of FGG usually had poor clinical outcomes.
EMT is a well-studied process that promotes the invasiveness and metastasis of cancer cells. Although many factors could trigger EMT, whether FGG could induce EMT was undetermined. In this study, we found that the overexpression of FGG obviously increased the expression of mesenchymal marker, N-cadherin, and remarkably downregulated the expression of epithelial marker, E-cadherin, while these results were reversed as FGG is knocked down in SK-HEP-1 cells. In addition, the expressions of Slug and ZEB1 were also altered following the corresponding modification of FGG. It is well established that Slug and ZEB1 are crucial transcriptional factors to activate EMT through regulating the expressions of E-cadherin and N-cadherin.32 Furthermore, the effect of FGG on the migration and invasion was significantly attenuated by knockdown of Slug or even ZEB-1. Thus, our results adequately supported our hypothesis that FGG might modulate the migration and invasion of HCC cells by activating EMT.
Accumulating evidences showed that FGG plays a role in promoting cancer progression, as it serves as an important functional area of fibrinogen, which is a secretory protein.33,34 Nevertheless, whether FGG exerts its biological functions inside cells remains unclear. In our study, FGG was explicitly located in the cytoplasm of HCC cells, based on the observations from human specimens (Figure 5A). And extracellular FGG could be hardly detected in the medium of long-term culturing SK-HEP-1 cells with FGG overexpression (Figure 5B). Therefore, these results indicated that FGG also played roles in HCC progression in intracellular manner for the first time.
Conclusion
Overexpression of FGG is an indicator of malignant tumor tissue and is also a potential biomarker for HCC prognosis. Moreover, FGG enhances the migration and invasion of HCC cells by activating EMT. Therefore, our current study identified a novel potential prognosis biomarker for HCC and expanded our knowledge on the intracellular mechanism of FGG in regulation of HCC invasiveness and metastasis.
Supplementary materials
FGG plays a similar role in MHCC97L cells.
Note: (A) Transwell assay showed that overexpression of FGG enhanced the migration and invasion in MHCC97L cells; (B) wound healing assays of MHCC97L cells with or without overexpression of FGG; (C) Western blot showed that the expressions of E-cadherin, N-cadherin, Slug, and ZEB-1 were regulated by overexpression of FGG in MHCC97L cells.
Abbreviations: CTRL, control; FGG, fibrinogen gamma chain; ZEB-1, zinc finger E-box binding homeobox 1.
Table S1.
siRNA sequences for knockdown of FGG, Slug and ZEB-1
| Name | Forward sequence | Reverse sequence |
|---|---|---|
| FGG | GGUAGUUAUUGUCCAA-CUAdTdT | UAGUUGGACAAUAACUACCdTdT |
| siSlug-1 | CGGACCCACACAUUACCUUdTdT | AAGGUAAUGUGUGGGUCCGdTdT |
| siSlug-2 | GCAGGAGAGACAUUCUGGdTdT | CCAGAAUGUCUCUCCUGCdTdT |
| siSlug-3 | ACUACAGUCCAAGCUUUCAdTdT | UGAAAGCUUGGACUGUAGUdTdT |
| siZEB-1-1 | GAAGCAGGAUGUACAGUAAdTdT | UUACUGUACAUCCUGCUUCdTdT |
| siZEB-1-2 | UUGAAUUUACGAUUACACCdTdT | GGUGUAAUCGUAAAUUCAAdTdT |
| siZEB-1-3 | GGCAAGUGUUGGAGAAUAAdTdT | UUAUUCUCCAACACUUGCCdTdT |
| siNC (CTRL) | UUCUCCGAACGUGUCACGUdTdT | ACGUGACACGUUCGGAGAAdTdT |
Abbreviations: CTRL, control; FGG, fibrinogen gamma chain; NC, negative control; siSlug-1, siSlug-2, and siSlug-3, knockdown of Slug with small interfering RNA duplexes-1, duplexes-2 and duplexes-3; siZEB-1-1, siZEB-1-2, and siZEB-1-3, knockdown of ZEB-1 with small interfering RNA duplexes-1, duplexes-2, and duplexes-3; ZEB-1, zinc finger E-box binding homeobox 1.
Acknowledgments
We thank Bin Wang and Lingyun Gao for processing and analyses of tissue samples. This project was supported by National Natural Science Foundation of China (grant no. 81602102; grant no. 81672376); the Natural Science Foundation of Fujian Province (grant no. 2015J05174; grant no. 2016J01417; grant no. 2017J01266); Scientific research project of Fujian provincial health and Family Planning Commission (grant no. 2015-1-94); the Young and Middle-aged Talent Training Project of Fujian provincial health and Family Planning Commission (grant no. 2018-ZQN-76); the Joint Funds for the Innovation of Science and Technology of Fujian province (grant no. 2017Y9116).
Footnotes
Author contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Endo T, Yoshikawa M, Ebara M, et al. Immunohistochemical metallo-thionein expression in hepatocellular carcinoma: relation to tumor progression and chemoresistance to platinum agents. J Gastroenterol. 2004;39(12):1196–1201. doi: 10.1007/s00535-004-1471-1. [DOI] [PubMed] [Google Scholar]
- 3.Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. 2016;166(1):21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 4.Dongre A, Rashidian M, Reinhardt F, et al. Epithelial-to-mesenchymal transition contributes to immunosuppression in breast carcinomas. Cancer Res. 2017;77(15):3982–3989. doi: 10.1158/0008-5472.CAN-16-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karaosmanoğlu O, Banerjee S, Sivas H. Identification of biomarkers associated with partial epithelial to mesenchymal transition in the secretome of slug over-expressing hepatocellular carcinoma cells. Cell Oncol. 2018;41(4):439–453. doi: 10.1007/s13402-018-0384-6. [DOI] [PubMed] [Google Scholar]
- 6.Chua HL, Bhat-Nakshatri P, Clare SE, Morimiya A, Badve S, Nakshatri H. NF-kappaB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: potential involvement of ZEB-1 and ZEB-2. Oncogene. 2007;26(5):711–724. doi: 10.1038/sj.onc.1209808. [DOI] [PubMed] [Google Scholar]
- 7.Caja L, Kahata K, Moustakas A. Context-dependent action of transforming growth factor β family members on normal and cancer stem cells. Curr Pharm Des. 2012;18(27):4072–4086. doi: 10.2174/138161212802430459. [DOI] [PubMed] [Google Scholar]
- 8.Yang MH, Chen CL, Chau GY, et al. Comprehensive analysis of the independent effect of twist and snail in promoting metastasis of hepatocellular carcinoma. Hepatology. 2009;50(5):1464–1474. doi: 10.1002/hep.23221. [DOI] [PubMed] [Google Scholar]
- 9.Mosesson MW. Fibrinogen and fibrin structure and functions. J Thromb Haemost. 2005;3(8):1894–1904. doi: 10.1111/j.1538-7836.2005.01365.x. [DOI] [PubMed] [Google Scholar]
- 10.Duan S, Gong B, Wang P, Huang H, Luo L, Liu F. Novel prognostic bio-markers of gastric cancer based on gene expression microarray: COL12A1, GSTA3, FGA and FGG. Mol Med Rep. 2018;18(4):3727–3736. doi: 10.3892/mmr.2018.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyman GH, Khorana AA, Cancer KAA. Cancer, clots and consensus: new understanding of an old problem. J Clin Oncol. 2009;27(29):4821–4826. doi: 10.1200/JCO.2009.22.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niu JY, Tian T, Zhu HY, et al. Hyperfibrinogenemia is a poor prognostic factor in diffuse large B cell lymphoma. Ann Hematol. 2018;97(10):1841–1849. doi: 10.1007/s00277-018-3382-x. [DOI] [PubMed] [Google Scholar]
- 13.Huang W, Wang S, Zhang H, Zhang B, Wang C. Prognostic significance of combined fibrinogen concentration and neutrophil-to-lymphocyte ratio in patients with resectable non-small cell lung cancer. Cancer Biol Med. 2018;15(1):88–96. doi: 10.20892/j.issn.2095-3941.2017.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shiose S, Hata Y, Noda Y, et al. Fibrinogen stimulates in vitro angiogenesis by choroidal endothelial cells via autocrine VEGF. Graefes Arch Clin Exp Ophthalmol. 2004;242(9):777–783. doi: 10.1007/s00417-004-0910-2. [DOI] [PubMed] [Google Scholar]
- 15.Adams GN, Rosenfeldt L, Frederick M, et al. Colon cancer growth and dissemination relies upon thrombin, stromal PAR-1, and fibrinogen. Cancer Res. 2015;75(19):4235–4243. doi: 10.1158/0008-5472.CAN-15-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao HJ, Chen YJ, Zuo D, et al. Quantitative proteomic analysis for high-throughput screening of differential glycoproteins in hepatocellular carcinoma serum. Cancer Biol Med. 2015;12(3):246–254. doi: 10.7497/j.issn.2095-3941.2015.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davalieva K, Kiprijanovska S, Maleva Kostovska I, et al. Comparative proteomics analysis of urine reveals down-regulation of acute phase response signaling and LXR/RXR activation pathways in prostate cancer. Proteomes. 2017;6(1):1. doi: 10.3390/proteomes6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen JY, Chen WN, Liu LL, et al. Hepatitis B spliced protein (HBSP) generated by a spliced hepatitis B virus RNA participates in abnormality of fibrin formation and functions by binding to fibrinogen γ chain. J Med Virol. 2010;82(12):2019–2026. doi: 10.1002/jmv.21918. [DOI] [PubMed] [Google Scholar]
- 19.Yokoyama K, Erickson HP, Ikeda Y, Takada Y. Identification of amino acid sequences in fibrinogen gamma -chain and tenascin C C-terminal domains critical for binding to integrin alpha vbeta 3. J Biol Chem. 2000;275(22):16891–16898. doi: 10.1074/jbc.M000610200. [DOI] [PubMed] [Google Scholar]
- 20.Yokoyama K, Zhang XP, Medved L, Takada Y. Specific binding of integrin alpha v beta 3 to the fibrinogen gamma and alpha E chain C-terminal domains. Biochemistry. 1999;38(18):5872–5877. doi: 10.1021/bi9827619. [DOI] [PubMed] [Google Scholar]
- 21.Nagata K, Arai S, Taira C, Sugano M, Honda T, Okumura N. A novel frameshift mutation in the fibrinogen γC terminal region, FGG c.1169_1170 del AT, leading to hypofibrinogenemia. Thromb Res. 2017;159:82–85. doi: 10.1016/j.thromres.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Liu H, Liang D, et al. Reveal the molecular signatures of hepatocellular carcinoma with different sizes by iTRAQ based quantitative proteomics. J Proteomics. 2017;150:230–241. doi: 10.1016/j.jprot.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Cai Z, Zeng Y, Xu B, et al. Galectin-4 serves as a prognostic biomarker for the early recurrence/metastasis of hepatocellular carcinoma. Cancer Sci. 2014;105(11):1510–1517. doi: 10.1111/cas.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song DG, Ye Q, Carpenito C, et al. In vivo persistence, tumor localization, and antitumor activity of CAR-engineered T cells is enhanced by costimulatory signaling through CD137 (4-1BB) Cancer Res. 2011;71(13):4617–4627. doi: 10.1158/0008-5472.CAN-11-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao B, Zhao W, Wang Y, et al. Connexin32 regulates hepatoma cell metastasis and proliferation via the p53 and Akt pathways. Oncotarget. 2015;6(12):10116–10133. doi: 10.18632/oncotarget.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu WL, Fan BL, Liu DL, Zhu WX. Abnormal expression of fibrinogen gamma (FGG) and plasma level of fibrinogen in patients with hepatocellular carcinoma. Anticancer Res. 2009;29(7):2531–2534. [PubMed] [Google Scholar]
- 27.Weisel JW, Litvinov RI. Fibrin formation, structure and properties. Subcell Biochem. 2017;82:405–456. doi: 10.1007/978-3-319-49674-0_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells – an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5(9):744–749. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 29.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang P, Sun Y, Ma L. ZEB1: at the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle. 2015;14(4):481–487. doi: 10.1080/15384101.2015.1006048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia L, Huang W, Tian D, et al. Overexpression of forkhead box C1 promotes tumor metastasis and indicates poor prognosis in hepatocellular carcinoma. Hepatology. 2013;57(2):610–624. doi: 10.1002/hep.26029. [DOI] [PubMed] [Google Scholar]
- 32.Peinado H, Olmeda D, Cano A, Snail CA. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7(6):415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 33.Honda KI, Asada R, Kageyama K, et al. Protein complex of fibrinogen gamma chain and complement factor H in ovarian cancer patient plasma. Anticancer Res. 2017;37(6):2861–2866. doi: 10.21873/anticanres.11638. [DOI] [PubMed] [Google Scholar]
- 34.Schneider G, Bryndza E, Poniewierska-Baran A, et al. Evidence that vitronectin is a potent migration-enhancing factor for cancer cells chaperoned by fibrinogen: a novel view of the metastasis of cancer cells to low-fibrinogen lymphatics and body cavities. Oncotarget. 2016;7(43):69829–69843. doi: 10.18632/oncotarget.12003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FGG plays a similar role in MHCC97L cells.
Note: (A) Transwell assay showed that overexpression of FGG enhanced the migration and invasion in MHCC97L cells; (B) wound healing assays of MHCC97L cells with or without overexpression of FGG; (C) Western blot showed that the expressions of E-cadherin, N-cadherin, Slug, and ZEB-1 were regulated by overexpression of FGG in MHCC97L cells.
Abbreviations: CTRL, control; FGG, fibrinogen gamma chain; ZEB-1, zinc finger E-box binding homeobox 1.
Table S1.
siRNA sequences for knockdown of FGG, Slug and ZEB-1
| Name | Forward sequence | Reverse sequence |
|---|---|---|
| FGG | GGUAGUUAUUGUCCAA-CUAdTdT | UAGUUGGACAAUAACUACCdTdT |
| siSlug-1 | CGGACCCACACAUUACCUUdTdT | AAGGUAAUGUGUGGGUCCGdTdT |
| siSlug-2 | GCAGGAGAGACAUUCUGGdTdT | CCAGAAUGUCUCUCCUGCdTdT |
| siSlug-3 | ACUACAGUCCAAGCUUUCAdTdT | UGAAAGCUUGGACUGUAGUdTdT |
| siZEB-1-1 | GAAGCAGGAUGUACAGUAAdTdT | UUACUGUACAUCCUGCUUCdTdT |
| siZEB-1-2 | UUGAAUUUACGAUUACACCdTdT | GGUGUAAUCGUAAAUUCAAdTdT |
| siZEB-1-3 | GGCAAGUGUUGGAGAAUAAdTdT | UUAUUCUCCAACACUUGCCdTdT |
| siNC (CTRL) | UUCUCCGAACGUGUCACGUdTdT | ACGUGACACGUUCGGAGAAdTdT |
Abbreviations: CTRL, control; FGG, fibrinogen gamma chain; NC, negative control; siSlug-1, siSlug-2, and siSlug-3, knockdown of Slug with small interfering RNA duplexes-1, duplexes-2 and duplexes-3; siZEB-1-1, siZEB-1-2, and siZEB-1-3, knockdown of ZEB-1 with small interfering RNA duplexes-1, duplexes-2, and duplexes-3; ZEB-1, zinc finger E-box binding homeobox 1.