Abstract
Background
Bladder cancer (BC) is among the most frequently occurring cancer types in the urinary system. In recent years, the importance of lncRNAs in BC has been acknowledged. SLCO4A1-AS1 is an oncogene in colorectal cancer. However, the role of SLCO4A1-AS1 in BC remains unknown.
Materials and methods
The expression levels of SLCO4A1-AS1 in BC tissues were analyzed by qRT-PCR. The effects of SLCO4A1-AS1 knockdown on proliferation were determined by CCK8 assay. Transwell assay was used to evaluate the role of SLCO4A1-AS1 on migration and invasion. Furthermore, xenograft assay was utilized to test the effect of SLCO4A1-AS1 on BC growth in vivo.
Results
SLCO4A1-AS1 expression was more upregulated in BC tissues than in adjacent normal tissues. Moreover, SLCO4A1-AS1 level was positively correlated with the advanced stage and metastasis in BC. The upregulation of SLCO4A1-AS1 indicates poor prognosis in BC patients. The knockdown of SLCO4A1-AS1 downregulated the proliferation, migration, and invasion of EJ and T24 cells in vitro. In addition, the loss of SLCO4A1-AS1 prevented BC growth in vivo. Mechanistic investigation showed that SLCO4A1-AS1 was the sponge for miR-335-5p, and miR-335-5p modulated OCT4 expression.
Conclusion
High SLCO4A1-AS1 expression level was associated with the progression of BC, and SLCO4A1-AS1 promoted the malignant phenotypes of BC cells through the miR-335-5p/OCT4 axis.
Keywords: bladder cancer, lncRNA, SLCO4A1-AS1, proliferation, invasion
Introduction
Bladder cancer (BC) has become the ninth most frequent cancer type around the world.1 Every year, ~400,000 patients are diagnosed with BC and 150,000 deaths are caused by BC worldwide.2 Although surgery combined with radiotherapy or chemotherapy improves the outcomes of muscle-invasive BC patients, the 5-year survival rate of this disease remains very low due to its high rates of recurrence and metastasis.3–5 Therefore, exploration of the molecular mechanism of BC progression and development of novel therapeutic strategies are urgently needed.
lncRNAs have been recently identified as a new member of noncoding RNA family. Research reveals that lncRNAs are characterized by over 200 nucleotides in length with no coding potential.6 An increasing amount of studies have demonstrated that lncRNAs play vital functions in various biological processes and are closely correlated with human diseases, including cancer.7 By participating in the regulation of cell proliferation, migration, invasion, and differentiation, lncRNAs could affect or suppress tumorigenesis.8,9 For example, lncRNA DUXAP8 promotes the progression of renal cell cancer.10 LINC00152 enhances the proliferation and metastasis of oral squamous cell cancer.11 lncRNA MNX1- AS1 has the potential to increase the proliferation, migration, and invasion of cervical cancer cells.12 Additionally, lncRNA CALML3-AS1 is essential for BC development.13 Although many lncRNAs are involved in carcinogenesis, the functions of most lncRNAs remain largely unknown.
The function of SLCO4A1-AS1 is poorly investigated. A recent study showed that SLCO4A1-AS1 could promote cell proliferation, migration, and invasion in colorectal cancer.14 SLCO4A1-AS1 expression was upregulated in BC tissues. Thus, this study aimed to explore the potential roles of SLCO4A1-AS1 in BC. Our results revealed that SLCO4A1-AS1 serves as an oncogene in BC and promotes tumorigenesis through the miR-335-5p/OCT4 signaling pathway. Moreover, SLCO4A1-AS1 might be a potential therapeutic target for BC intervention.
Materials and methods
Human bladder tissue samples
A total of 58 BC tissues and corresponding adjacent normal tissues were collected from the Qianfoshan Hospital Affiliated to Shandong University. This study was approved by the Ethics Committee of Qianfoshan Hospital Affiliated to Shandong University. Experiments involving human tissues were conducted in accordance with the Declaration of Helsinki. All patients signed the informed consent. Patients did not undergo chemotherapy or radiotherapy before surgery. The clinical features of the samples are presented in Table 1. These tissues were stored in liquid nitrogen.
Table 1.
Correlation between SLCO4A1-AS1 expression level and clinical features in bladder cancer
| Characteristics | Low (n=29) | High (n=29) | P-value |
|---|---|---|---|
| Age, years | 0.431 | ||
| ≤65 | 17 | 13 | |
| >65 | 12 | 16 | |
| Gender | 0.331 | ||
| Male | 25 | 21 | |
| Female | 4 | 8 | |
| Grade | 0.144 | ||
| Low | 7 | 2 | |
| High | 22 | 27 | |
| T stage | 0.024* | ||
| Tis/Ta/T1 | 14 | 5 | |
| T2/T3/T4 | 15 | 24 | |
| Lymph node metastasis | 0.033* | ||
| Yes | 8 | 17 | |
| No | 21 | 12 |
Notes: Chi-squared test.
P<0.05.
Cell culture, transfections, and selection of stable cell lines
Human BC cell lines (EJ, T24, and RT4) and human epithelial SV40 immortalized uroepithelium cell line SV-HUC-1 were obtained from the Stem Cell Bank, Chinese Academy of Sciences in Shanghai, China. Cells were cultured in DMEM medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% FBS (Thermo Fisher Scientific) in 5% CO2 at 37°C.
For transfection, siRNA, miR-335-5p mimics, miR- 335-5p inhibitors, and negative controls were synthesized by GenePharma Co., Ltd (Shanghai, China) and transfected using the Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. For the construction of stable cell line, BC cells were infected with lentiviral-CIRBP-shRNA and lentiviral-control-shRNA (LV-NC) for 24 hours and selected using 5 µg/mL puromycin (Sigma-Aldrich Co., St Louis, MO, USA) for 7 days.
Cell proliferation
Cells were seeded into 96-well plates and cultured for specific time periods. Then, 100 µL of cell counting kit-8 (CCK-8) reagent (Dojindo Molecular Technologies, Tokyo, Japan) was added and incubated for 4 hours. The absorbance was determined at 450 nm using a microplate reader.
Transwell assay
Cell migration and invasion were measured using a 24-well Transwell system (8 µm pore; Costar, Badhoevedorp, the Netherlands) by using Transwell assay based on a previous study.9
RNA isolation and quantitative reverse transcriptase PCR (qRT-PCR)
Total RNA was obtained from BC tissues and cells by using the TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. The RNA was reverse transcribed into cDNA by using a PrimeScript RT reagent Kit with gDNA Eraser (Takara), followed by qRT-PCR by using the SYBR Premix EX Taq II kit (Takara) on Roche LightCycler 480 (Roche, Basel, Switzerland). Relative expression was normalized to U6 and was calculated based on the 2–ΔΔCt method.
Xenograft model
Samples of 4-week-old male BALB/c nude mice obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd (Beijing, China) were randomly divided into three groups (n=4 per group). Then, T24 cells were subcutaneously injected into the flank of nude mice. At specific time points, tumor volumes were measured. Tumor weight was determined 4 weeks after injection. Experiments were conducted according to the instruction of the Ethics Committee at Qianfoshan Hospital Affiliated to Shandong University. All animal experiments were approved by the Ethics Committee of Qianfoshan Hospital Affiliated to Shandong University.
Luciferase reporter assays
The sequence of SLCO4A1-AS1 or OCT4 3′-UTR containing the wild-type (Wt) or mutant (Mut) binding site for miR-335-5p was constructed into the pGL3 luciferase vector (Ambion, Inc., Austin, TX, USA). For the luciferase reporter assay, T24 cells were transfected with miR-335-5p mimics or control and the respective luciferase reporter by using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. After 48 hours, the relative luciferase was measured using the dual-Luciferase Assay Kit (Promega, Madison, WI, USA) and normalized to Renilla activity.
Statistical analyses
All experiments were analyzed using the GraphPad software (version 6; GraphPad Software, Inc., La Jolla, CA, USA). Results were expressed as mean ± SD. P-value was analyzed using Student’s t-test and one-way ANOVA. Statistical significance was considered at P<0.05.
Results
Relative expression levels of SLCO4A1- AS1, miR-335-5p, and OCT4 in BC
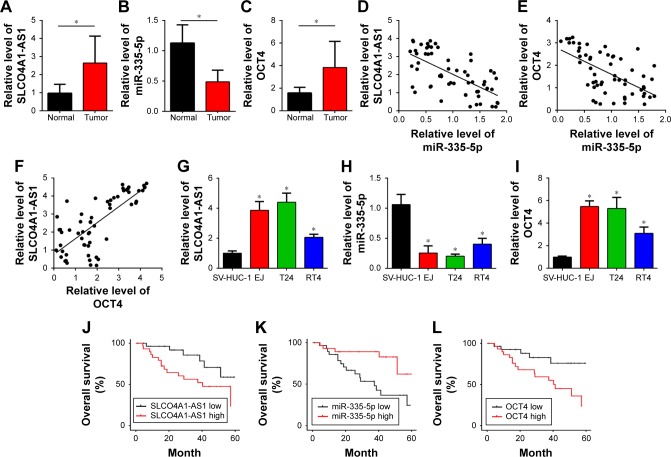
To analyze the function of SLCO4A1-AS1, we first measured its expression in 58 BC tissues and the corresponding adjacent normal tissues through qRT-PCR. We found that the level of SLCO4A1-AS1 was significantly increased in BC tissues compared with that in the adjacent normal controls (Figure 1A). lncRNAs sponge miRNAs and inhibit their expression levels in tumor.15 Bioinformatics analysis (miRDB; http://mirdb.org/miRDB/index.html) predicted that SLCO4A1-AS1 may interact with miR- 335-5p. Thus, we analyzed miR-335-5p expression, which was downregulated in BC tissues (Figure 1B). We further predicted the potential targets of miR-335-5p through bioinformatics analysis (TargetScan7; http://www.targetscan. org/vert_71/). OCT4 was identified as the most potential candidate. OCT4 expression was significantly increased in BC tissues (Figure 1C). Thus, an SLCO4A1-AS1-miR- 335-5p-OCT4 regulatory signaling pathway may exist. Interestingly, we also observed a negative correlation between SLCO4A1-AS1 and miR-335-5p or between miR-335-5p and OCT4 in BC tissues (Figure 1D and E). Furthermore, SLCO4A1-AS1 is positively correlated with OCT4 (Figure 1F). Then, we analyzed their expression in BC cell lines. Moreover, the expression levels of SLCO4A1- AS1 and OCT4 were upregulated in BC tissues, whereas that of miR-335-5p was downregulated (Figure 1G–I). Finally, Kaplan–Meier survival curves were plotted according to the median expression levels of SLCO4A1-AS1 in 45 muscle-invasive BC tissues, miR-335-5p, and OCT4, which indicated that the SLCO4A1-AS1/miR-335-5p/OCT4 axis would be a prognostic biomarker (Figure 1J–L).
Figure 1.
Relative expression levels of SLCO4A1-AS1, miR-335-5p, and OCT4 in BC.
Notes: (A–C) Relative expression levels of SLCO4A1-AS1, miR-335-5p, and OCT4 were determined by using 58 BC tissues and their corresponding adjacent normal tissues through qRT-PCR. (D–F) qRT-PCR analysis showed a negative correlation between SLCO4A1-AS1 and miR-335-5p expression or between miR-335-5p and OCT4 expression in BC tissues. It also showed a positive correlation between SLCO4A1-AS1 and OTC4 expression. (G–I) The expression levels of SLCO4A1-AS1, miR-335-5p, and OCT4 were evaluated in BC cell lines through qRT-PCR. (J–L) Kaplan–Meier survival curve was plotted according to the median expression levels of SLCO4A1-AS1, miR-335-5p, and OCT4. *P<0.05.
Abbreviations: BC, bladder cancer; qRT-PCR, quantitative reverse transcriptase PCR.
SLCO4A1-AS1 is a ceRNAfor miR-335-5p, while miR-335-5p targets OCT4 directly
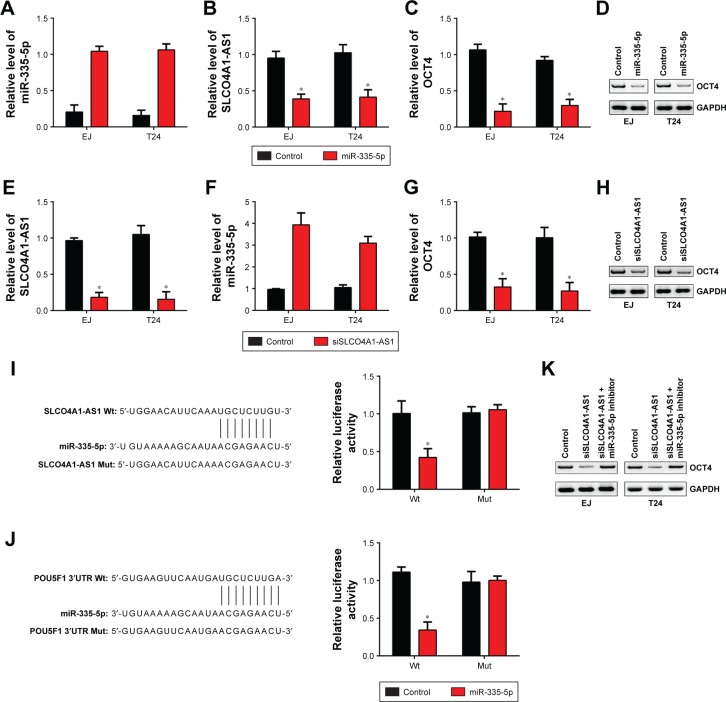
Then, we sought to demonstrate whether an SLCO4A1- AS1/miR-335-5p/OCT4 regulatory signaling pathway existed in BC. We overexpressed miR-335-5p in EJ and T24 cells (Figure 2A). qRT-PCR analysis showed that the upregulation of miR-335-5p downregulated the expression levels of SLCO4A1-AS1 and OCT4 (Figure 2B and C). Western blot analysis results also confirmed this observation (Figure 2D). Additionally, we knocked down SLCO4A1-AS1 in EJ and T24 cells (Figure 2E). SLCO4A1-AS1 silencing upregulated miR-335-5p and downregulated OCT4 in EJ and T24 cells (Figure 2F–H). To further validate whether direct interaction exists among SLCO4A1-AS1, miR-335-5p, and OCT4, we designed luciferase reporter assays. Wt and Mut reporter plasmids of SLCO4A1-AS1 or OCT4 were constructed, following which luciferase reporter assays were performed. miR-335-5p mimic transfection significantly suppressed the relative luciferase activity of both SLCO4A1-AS1 and OCT4 Wt reporters (Figure 2I and J), demonstrating that miR-335-5p directly interacted with either SLCO4A1-AS1 or OCT4. Moreover, the Western blot analysis showed that SLCO4A1-AS1 silencing downregulated OCT4 expression in EJ and T24 cells, but was reversed by the inhibition of miR-335-5p (Figure 2K). These data demonstrate that SLCO4A1- AS1 sponges miR-335-5p to upregulate OCT4 in BC.
Figure 2.
SLCO4A1-AS1 is a ceRNA for miR-335-5p, while miR-335-5p targets OCT4 directly.
Notes: (A–C) qRT-PCR analysis for the expression of miR-335-5p, SLCO4A1-AS1, and OCT4 in EJ and T24 cells. (D) Western blot analysis showed that miR-335- 5p inhibited OCT4 expression. (E–G) Relative expression levels of SLCO4A1-AS1, miR-335-5p, and OCT4 were measured in EJ and T24 cells after transfection with siSLCO4A1-AS1 or control. (H) Western blot analysis showed that SLCO4A1-AS1 silencing suppressed OCT4 expression in EJ and T24 cells. (I) Luciferase reporter assay showed that miR-335-5p mimic transfection suppressed the relative luciferase activity of SLCO4A1-AS1-Wt reporter in T24 cells. (J) Luciferase reporter assay showed that miR-335-5p mimic transfection suppressed the relative luciferase activity of the OCT4-Wt reporter in T24 cells. (K) Western blot analysis showed that the downregulation of OCT4 expression through SLCO4A1-AS1 silencing was reversed by the addition of miR-335-5p inhibitors. *P<0.05.
Abbreviations: Mut, mutant; qRT-PCR, quantitative reverse transcriptase PCR; Wt, wild type.
SLCO4A1-AS1 regulates malignant behaviors of BC cells through OCT4
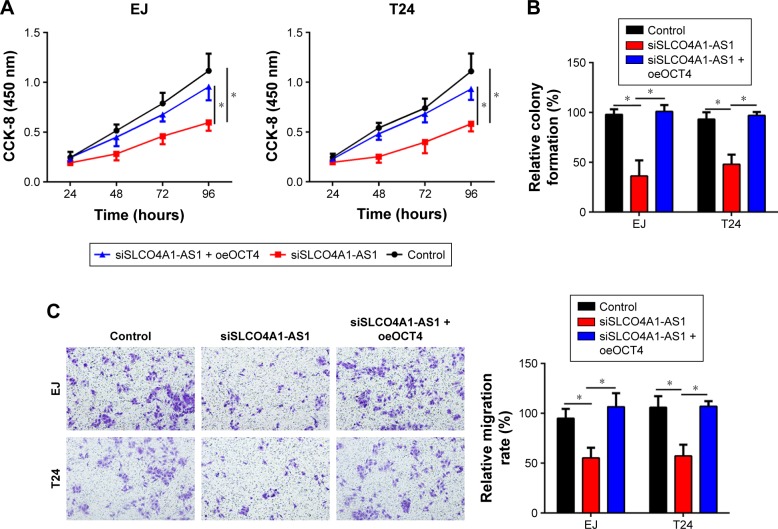
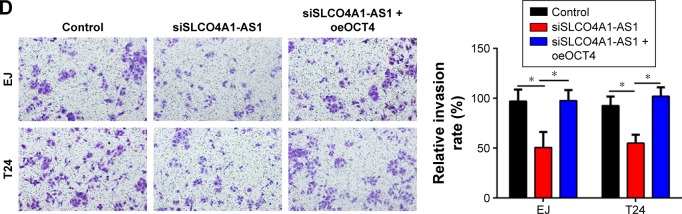
We aimed to investigate the functions of SLCO4A1-AS1 in BC. Through a series of experiments, including CCK-8, colony formation, and Transwell assays, we found that SLCO4A1-AS1 silencing significantly suppressed the proliferation, colony formation, migration, and invasion of EJ and T24 cells (Figure 3A–D). OCT4 overexpression partially rescued the abilities of proliferation, migration, and invasion in SLCO4A1-AS1-depleted EJ and T24 cells (Figure 3A–D). Overall, SLCO4A1-AS1 acted as an oncogene in BC through OCT4 expression.
Figure 3.
SLCO4A1-AS1 regulates the set of malignant behavior of BC cells through OCT4.
Notes: (A, B) CCK-8 assay and colony formation assay were carried out to analyze cell proliferation. (C, D) Transwell assay was performed to analyze cell migration and invasion (100× magnification). *P<0.05.
Abbreviations: BC, bladder cancer; CCK-8, Cell Counting Kit-8.
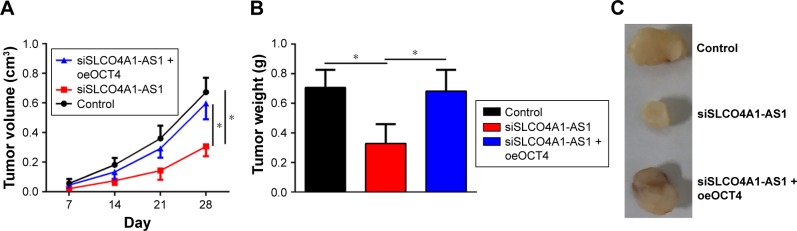
Effect of SLCO4A1-AS1 silencing on BC growth in vivo
To further demonstrate the role of SLCO4A1-AS1 in vivo, xenograft assay was performed. SLCO4A1-AS1-depleted or control T24 cells were injected into nude mice. Tumor size was measured weekly. SLCO4A1-AS1 silencing delayed tumor growth (Figure 4A). After 4 weeks, the mice were sacrificed and tumor weight was determined. SLCO4A1-AS1 silencing also decreased tumor weight (Figure 4B and C). Importantly, the rescue of OCT4 expression also restored the growth of tumors in vivo (Figure 4A–C). In summary, SLCO4A1-AS1 promotes BC growth in vivo by regulating OCT4 expression.
Figure 4.
In vivo effect of SLCO4A1-AS1 silencing on BC growth.
Notes: (A) Tumor volume was determined every week. (B) After 4 weeks, the tumor weights were analyzed. (C) Representative image of tumor tissues is presented. *P<0.05.
Abbreviation: BC, bladder cancer.
Discussion
In our study, lncRNA SLCO4A1-AS1 and OCT4 were conspicuously increased in BC tissues compared to that in the adjacent normal controls. Furthermore, miR-335-5p level was significantly downregulated in tumor tissues. By using the miRDB and TargetScan7 tools, we predicted that both SLCO4A1-AS1 and OCT4 have potential binding sites for miR-335-5p, and the luciferase reporter assay demonstrated their direct interaction. Additionally, SLCO4A1-AS1 inhibited miR-335-5p to augment OCT4 expression and affect the proliferation, migration, and invasion of BC cells.
The lncRNAs greatly affect the prognosis, growth, survival, epithelial-to-mesenchymal transition, and metastasis of BC cells.16,17 For instance, Yang et al showed that lncRNA UCA1 regulates the PI3K pathway to participate in the cell cycle regulation of BC cells.18 Zhu et al found that lncRNA ANRIL overexpression promotes proliferation and inhibits apoptosis in BC.19 Chen et al indicated that lncRNA-n336928 level is associated with the advanced stage, grade, and overall survival in BC, suggesting its potential as a prognostic biomarker.20 Cao et al showed that lncRNA GAS5 plays antitumor roles to suppress the proliferation and cell cycle progression of BC cells.21 In addition, Li et al showed that lncRNA UCA1 regulates the miR-195/ARL2 axis, thereby affecting the mitochondrial function of BC cells.22 These studies emphasized the importance of lncRNAs in BC and implied that lncRNAs might be a promising therapeutic target. However, the function of most lncRNAs remains unknown. lncRNA SLCO4A1-AS1 indicates poor prognosis and promotes colorectal cancer (CRC) progression.14 The mechanism of its functions in BC remains unclear. In our study, we found that SLCO4A1-AS1 expression in BC was upregulated and SLCO4A1-AS1 promoted the proliferation, migration, and invasion of BC cells.
Increasing evidence indicates that lncRNAs could act by targeting miRNAs.23 We identified that miR-335-5p might be targeted by SLCO4A1-AS1. Besides, miR-335-5p interacts with lncRNAs. For instance, Wang et al showed that lncRNA TUG1 could serve as a ceRNA for miR-335-5p in osteosarcoma.24 Wang et al indicated that lncRNA RP11- 436H11.5 functions as a ceRNA to sponge miR-335-5p in renal cell carcinoma.25 Zhang et al showed that miR-335-5p is targeted by lncRNA ZEB1-AS1 in gastric cancer.26 Additionally, lncRNA NEAT1 regulates miR-335-5p expression in gastric cancer.27 By using the luciferase reporter assay, we demonstrated that SLCO4A1-AS1 directly interacts with miR-335-5p and inhibits its reactivity. To the best of our knowledge, our study was the first to show that miR-335-5p was downregulated in BC tissues and was correlated with patient prognosis, indicating its anticancer roles in BC. Our findings were consistent with the conclusion that miR-335-5p suppresses the progression of non-small-cell lung cancer, osteosarcoma, and thyroid cancer.28–30
Several targets of miR-335-5p have been reported in cancer, such as ICAM-1, CPNE1, and BCL-W.25,28,30 However, its role and target in BC remain unknown. Through bioinformatics analysis, we found that OCT4 might be targeted by miR-335-5p and we demonstrated their interaction. We also validated the association between SLCO4A1-AS1 and OCT4 by sponging miR-335-5p. OCT4 regulates BC progression and resistance to drug.31,32 In our study, we also observed that the upregulation of OCT4 in BC indicates poor prognosis and contributes to the proliferation, migration, and invasion of BC cells.
In conclusion, our study was the first to investigate the roles of the SLCO4A1-AS1/miR-335-5p/OCT4 regulatory signaling pathway in BC progression, thereby providing a new insight on developing therapeutic targets.
Acknowledgments
We thank all patients who participated in this work.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Lu M, Ge Q, Wang G, et al. CIRBP is a novel oncogene in human bladder cancer inducing expression of HIF-1α. Cell Death Dis. 2018;9(10):1046. doi: 10.1038/s41419-018-1109-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knollman H, Godwin JL, Jain R, Wong YN, Plimack ER, Geynisman DM. Muscle-invasive urothelial bladder cancer: an update on systemic therapy. Ther Adv Urol. 2015;7(6):312–330. doi: 10.1177/1756287215607418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vartolomei MD, Ferro M, Cantiello F, et al. Validation of neutrophil- to-lymphocyte ratio in a multi-institutional cohort of patients with T1G3 non-muscle-invasive bladder cancer. Clin Genitourin Cancer. 2018;16(6):445–452. doi: 10.1016/j.clgc.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Ferro M, Vartolomei MD, Russo GI, et al. An increased body mass index is associated with a worse prognosis in patients administered BCG immunotherapy for T1 bladder cancer. World J Urol. 2018 Jul 10; doi: 10.1007/s00345-018-2397-1. Epub. [DOI] [PubMed] [Google Scholar]
- 6.Cui M, Zheng M, Sun B, Wang Y, Ye L, Zhang X. A long noncoding RNA perturbs the circadian rhythm of hepatoma cells to facilitate hepatocarcinogenesis. Neoplasia. 2015;17(1):79–88. doi: 10.1016/j.neo.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu P, Wang Y, Wu J, et al. LncBRM initiates Yap1 signalling activation to drive self-renewal of liver cancer stem cells. Nat Commun. 2016;7(1):13608. doi: 10.1038/ncomms13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang H, Bu YZ, Zhang XY, Liu J, Zhu LY, Fang Y. LINC01433 promotes hepatocellular carcinoma progression via modulating the miR-1301/STAT3 axis. J Cell Physiol. 2019;234(5):6116–6124. doi: 10.1002/jcp.27366. [DOI] [PubMed] [Google Scholar]
- 10.Huang T, Wang X, Yang X, et al. Long non-coding RNA DUXAP8 enhances renal cell carcinoma progression via downregulating miR-126. Med Sci Monit. 2018;24:7340–7347. doi: 10.12659/MSM.910054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M, Ning J, Li Z, Wang J, Zhao C, Wang L. LINC00152 promotes the growth and invasion of oral squamous cell carcinoma by regulating miR-139-5p. Onco Targets Ther. 2018;11:6295–6304. doi: 10.2147/OTT.S168807. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Liu X, Yang Q, Yan J, Zhang X, Zheng M. LncRNA MNX1-AS1 promotes the progression of cervical cancer through activating MAPK pathway. J Cell Biochem. 2019;120(3):4268–4277. doi: 10.1002/jcb.27712. [DOI] [PubMed] [Google Scholar]
- 13.Wang F, Zu Y, Huang W, Chen H, Xie H, Yang Y. LncRNA CALML3- AS1 promotes tumorigenesis of bladder cancer via regulating ZBTB2 by suppression of microRNA-4316. Biochem Biophys Res Commun. 2018;504(1):171–176. doi: 10.1016/j.bbrc.2018.08.150. [DOI] [PubMed] [Google Scholar]
- 14.Yu J, Han Z, Sun Z, Wang Y, Zheng M, Song C. LncRNA SLCO4A1-AS1 facilitates growth and metastasis of colorectal cancer through β-catenin- dependent Wnt pathway. J Exp Clin Cancer Res. 2018;37(1):222. doi: 10.1186/s13046-018-0896-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang M, Wang YR, Xu N, Zhou L, An Q. Long noncoding RNA MEG3 play an important role in osteosarcoma development through sponging microRNAs. J Cell Biochem. 2018 Oct 15; doi: 10.1002/jcb.27791. Epub. [DOI] [PubMed] [Google Scholar]
- 16.Crea F, Clermont PL, Parolia A, Wang Y, Helgason CD. The non-coding transcriptome as a dynamic regulator of cancer metastasis. Cancer Metastasis Rev. 2014;33(1):1–16. doi: 10.1007/s10555-013-9455-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest. 2016;126(8):2775–2782. doi: 10.1172/JCI84421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang C, Li X, Wang Y, Zhao L, Chen W. Long non-coding RNA UCA1 regulated cell cycle distribution via CREB through PI3-K dependent pathway in bladder carcinoma cells. Gene. 2012;496(1):8–16. doi: 10.1016/j.gene.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 19.Zhu H, Li X, Song Y, Zhang P, Xiao Y, Xing Y. Long non-coding RNA ANRIL is up-regulated in bladder cancer and regulates bladder cancer cell proliferation and apoptosis through the intrinsic pathway. Biochem Biophys Res Commun. 2015;467(2):223–228. doi: 10.1016/j.bbrc.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Chen T, Xie W, Xie L, et al. Expression of long noncoding RNA lncRNA-n336928 is correlated with tumor stage and grade and overall survival in bladder cancer. Biochem Biophys Res Commun. 2015;468(4):666–670. doi: 10.1016/j.bbrc.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 21.Cao Q, Wang N, Qi J, Gu Z, Shen H. Long non-coding RNA-GAS5 acts as a tumor suppressor in bladder transitional cell carcinoma via regulation of chemokine (C-C motif) ligand 1 expression. Mol Med Rep. 2016;13(1):27–34. doi: 10.3892/mmr.2015.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li HJ, Sun XM, Li ZK, et al. LncRNA UCA1 promotes mitochondrial function of bladder cancer via the MiR-195/ARL2 signaling pathway. Cell Physiol Biochem. 2017;43(6):2548–2561. doi: 10.1159/000484507. [DOI] [PubMed] [Google Scholar]
- 23.He JH, Han ZP, Zou MX, et al. Analyzing the lncRNA, miRNA, and mRNA regulatory network in prostate cancer with bioinformatics software. J Comput Biol. 2018;25(2):146–157. doi: 10.1089/cmb.2016.0093. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Yang T, Zhang Z, et al. Long non-coding RNA TUG1 promotes migration and invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells. Cancer Sci. 2017;108(5):859–867. doi: 10.1111/cas.13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K, Jin W, Song Y, Fei X. LncRNA RP11-436H11.5, functioning as a competitive endogenous RNA, upregulates BCL-W expression by sponging miR-335-5p and promotes proliferation and invasion in renal cell carcinoma. Mol Cancer. 2017;16(1):166. doi: 10.1186/s12943-017-0735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang LL, Zhang LF, Guo XH, Zhang DZ, Yang F, Fan YY. Downregulation of miR-335-5p by long noncoding RNA ZEB1-AS1 in gastric cancer promotes tumor proliferation and invasion. DNA Cell Biol. 2018;37(1):46–52. doi: 10.1089/dna.2017.3926. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Zhang M, Sun G. Long non-coding RNA NEAT1 regulates the proliferation, migration and invasion of gastric cancer cells via targeting miR-335-5p/ROCK1 axis. Pharmazie. 2018;73(3):150–155. doi: 10.1691/ph.2018.7877. [DOI] [PubMed] [Google Scholar]
- 28.Tang H, Zhu J, Du W, et al. CPNE1 is a target of miR-335-5p and plays an important role in the pathogenesis of non-small cell lung cancer. J Exp Clin Cancer Res. 2018;37(1):131. doi: 10.1186/s13046-018-0811-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Zeng X, Wang N, et al. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17(1):89. doi: 10.1186/s12943-018-0837-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo L, Xia L, Zha B, et al. miR-335-5p targeting ICAM-1 inhibits invasion and metastasis of thyroid cancer cells. Biomed Pharmacother. 2018;106:983–990. doi: 10.1016/j.biopha.2018.07.046. [DOI] [PubMed] [Google Scholar]
- 31.Zhou J, Dong D, Cheng R, et al. Aberrant expression of KPNA2 is associated with a poor prognosis and contributes to Oct4 nuclear transportation in bladder cancer. Oncotarget. 2016;7(45):72767–72776. doi: 10.18632/oncotarget.11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu CS, Shieh GS, Wang CT, et al. Chemotherapeutics-induced Oct4 expression contributes to drug resistance and tumor recurrence in bladder cancer. Oncotarget. 2017;8(19):30844–30858. doi: 10.18632/oncotarget.9602. [DOI] [PMC free article] [PubMed] [Google Scholar]