Abstract
Due to the exponential growth of computational algorithms, artificial intelligence (AI) methods are poised to improve the precision of diagnostic and therapeutic methods in medicine. The field of radiomics in neuro-oncology has been and will likely continue to be at the forefront of this revolution. A variety of AI methods applied to conventional and advanced neuro-oncology MRI data can already delineate infiltrating margins of diffuse gliomas, differentiate pseudoprogression from true progression, and predict recurrence and survival better than methods used in daily clinical practice. Radiogenomics will also advance our understanding of cancer biology, allowing noninvasive sampling of the molecular environment with high spatial resolution and providing a systems-level understanding of underlying heterogeneous cellular and molecular processes. By providing in vivo markers of spatial and molecular heterogeneity, these AI-based radiomic and radiogenomic tools have the potential to stratify patients into more precise initial diagnostic and therapeutic pathways and enable better dynamic treatment monitoring in this era of personalized medicine. Although substantial challenges remain, radiologic practice is set to change considerably as AI technology is further developed and validated for clinical use.
Summary
Advances in artificial intelligence applied to radiomics and radiogenomics in neuro-oncologic imaging will improve our diagnostic, prognostic, and therapeutic methods, helping propel the field into an era of precision medicine.
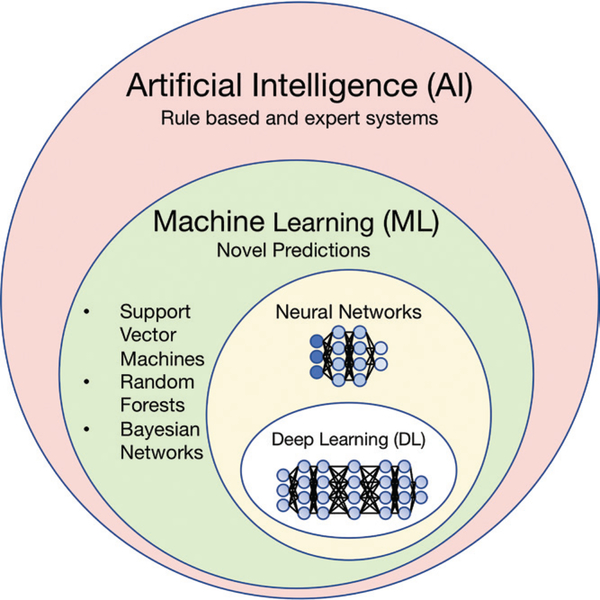
Artificial intelligence (AI) is a broad term that describes any task performed by a computer that normally requires human intelligence, including explicit rule-based systems, as well as computer algorithms that do not require hard-coded rules (Fig 1). Machine learning, which falls under the umbrella of AI, is a branch of data science that enables computers to learn from existing “training” data without explicit programming to make predictions about new data points. Deep learning is a subclass of machine learning based on neural networks, containing a large number of layers, made possible due to recent computational advances. Machine learning and deep learning methods are being increasingly adopted for radiomics research, which relies on medical imaging data as quantitative imaging biomarkers. The overarching goal of AI-based research in neuro-oncologic imaging is to better understand the complex manifestations of heterogeneous central nervous system (CNS) neoplasms in hopes of improving patient outcomes.
Figure 1:
Diagram shows overview of terms encompassed by artificial intelligence and their nested relationships with each other.
Machine Learning Methods
Most applications of machine learning in medical imaging have relied on supervised forms of machine learning, which consist of algorithms that are trained on “ground truth” labels. Labels can include different classes of diagnoses (eg, high- vs lower-grade glioma), different prognoses (eg, long vs short survival), or different classes that exist within a single set of image volumes (eg, enhancing tumor vs necrotic tissue vs edema vs normal brain tissue). When provided with sufficient examples of the different classes, algorithms “learn” how to classify novel data. Supervised machine learning methods include logistic regression, support vector machines, and random forests, as well as tools useful for clinical decision support (eg, decision trees and Bayesian networks) (1,2). In general, these traditional supervised approaches are applied to explicitly engineered intermediate features, often after a step of feature reduction, which is necessary to reduce model complexity and avoid overfitting (ie, memorizing the training sample cases rather than learning the relevant pattern)—a prevalent problem that creeps into many machine learning studies without proper “held out” validation samples. These approaches, while powerful, oftentimes require extensive, domain-specific, expert knowledge about the underlying biologic basis of the process being studied. Another frequently used category of machine learning is unsupervised algorithms, such as k-means clustering, which can generate novel groupings or categories from complex data sets and have important roles in discovery science and big data.
Deep Learning Methods
Deep learning grew out of a desire to model the tiered organization of the mammalian brain’s visual cortex, where hierarchically organized layers process increasingly complex intermediate visual features such as lines, edges, shapes, and entire visual objects (3–6). The recent growth of computing power through parallel graphical processing units and improved mathematical optimization methods enabled upscaling the architectures of these neural network models to contain many intermediate layers, differentiating deep learning from traditional neural networks, which were first conceived in the 1940s (3–6). Through an iterative process of up-dating model weights (“backpropagation”), these algorithms learn to appropriately identify lower and intermediate level image information in order to maximize classification performance. Typically, convolutional neural networks (CNNs), a class of feed-forward neural networks, have been used for image-based problems. In recent years, deep learning approaches have achieved super-human performance on visual processing tasks, as benchmarked by the ImageNet challenge, a yearly competition to test algorithms on visual recognition tasks (7,8). Interestingly, the weights derived from these networks trained on ImageNet can be adapted for new tasks, including medical images, in a process termed “transfer learning.” Although some deep learning architectures for medical image processing use transfer learning, others use custom image-naive architectures, which may perform better depending on the specific task. Although deep learning models can run quickly and do not require as much manual intervention as traditional machine learning approaches, they tend to require large amounts of labeled training data in order to be robust to data variability.
Radiomics in Neuro-Oncology
Although clinical radiology generally relies on visual assessment of images in subjective and qualitative terms, the field of radiomics extracts information from clinical images for use as quantitative imaging biomarkers (9,10). The first essential step of radiomics generally involves lesion segmentation (Fig 2, A), which is generally preceded by image preprocessing steps including skull stripping, intensity normalization, and alignment of image volumes from different modalities. A variety of methods have been used for segmentation, ranging from manual labeling and/or annotation and semiautomated methods (11) to more recent deep learning methods (12–14).
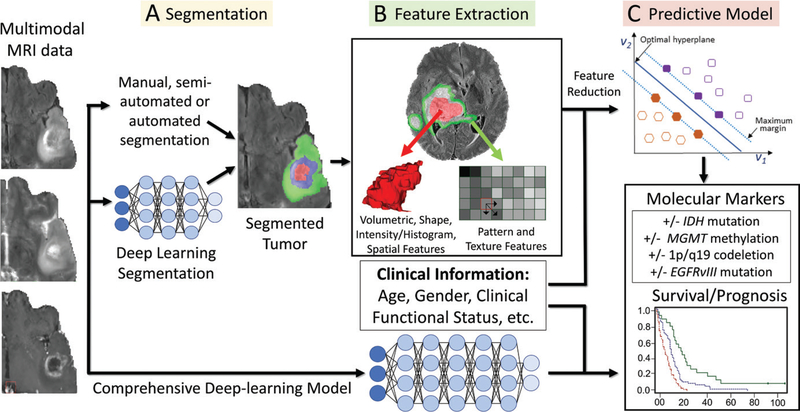
Figure 2:
Workflow of radiomics in neuro-oncology. A, After preprocessing steps, multimodal MR images are segmented by using automated or manual methods. B, This is followed by feature extraction with use of a variety of different techniques. C, Machine learning methods are then trained on the features to generate models of underlying molecular markers and predict survival. Deep learning models can be used for performing each of the described steps individually or in a more comprehensive fashion (bottom pathway of figure). EGFRvIII = epidermal growth factor receptor variable III, IDH = isocitrate dehydrogenase, MGMT = O6-methylguanine-DNA-methyltransferase.
The next step of radiomics with traditional machine learning involves the extraction of quantitative features, including basic shape, size, and intensity metrics, as well as more complex features derived from a variety of statistical approaches applied to the images, for example, histogram-based features, texture-based features, fitted biophysical models, spatial patterns, and deep learning features (Fig 2, B). A variety of different machine learning models can then be applied to the intermediate quantitative features in order to “mine” them for significant associations, allowing them to predict crucial information about a tumor, such as infiltrating tumor margins, molecular markers, and prognosis (Fig 2, C), which are relevant for therapeutic decision making. Alternatively, deep learning approaches to radiomics in neurooncology generally necessitate less domain-specific knowledge compared with the explicitly engineered features for traditional machine learning, allowing them to make predictions without explicit feature selection or reduction steps.
Public Neuro-Oncology Radiomic Data and Competitions
Advances in the field of segmentation and radiomics within neuro-oncology have been supported by data made available through the Cancer Imaging Archive (TCIA) (15), which is part of a larger effort from the Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/). Since 2012, TCIA data have been further curated through the annual Multimodal Brain Tumor Image Segmentation Benchmark (BraTS) challenge (16,17), which seeks to improve the accuracy of automated glioma segmentation and survival prediction with preoperative MR images. This public data set contains multimodal images of high- and lowergrade gliomas with expert manual segmentations with five labels (healthy brain tissue, necrosis, edema, and nonenhancing and enhancing tumor). Deep learning approaches have surpassed more traditional segmentation methods to win the BraTS challenges in 2016 (12) and 2017 (14).
Current Methods in Neuro-Oncologic Imaging
Most research in neuro-oncology has focused on diffuse gliomas, World Health Organization (WHO) grade II–IV tumors, which are typically divided into lower-grade gliomas (WHO grade II and III) and glioblastoma (WHO grade IV) (18). Much work has focused on glioblastoma, given that it represents more than half of malignant primary brain tumors and has an aggressive course and grim prognosis (19). Lowergrade gliomas can sometimes progress into glioblastoma, and these are known as secondary glioblastomas. A variety of other brain tumors, including WHO grade I tumors, pediatric CNS tumors, primary CNS lymphoma, and brain metastases, encompass important areas of neuro-oncology but represent lessactive areas of research given smaller sample sizes, more disease heterogeneity, and relatively lower morbidity.
Given its excellent soft-tissue contrast, MRI is the central tool for tumor detection and characterization. Conventional MRI sequences, which include pre- and postcontrast T1-weighted imaging, T2-weighted imaging, and T2-weighted fluid-attenuated inversion recovery (FLAIR) sequences, are good at helping delineate tumor volume and morphologic characteristics. Unfortunately, contrast enhancement is nonspecific and the detection of foci of tumor infiltration within the T2-weighted FLAIR signal intensity abnormality is nearly impossible with conventional sequences (20). Most institutions rely on other advanced MRI methods that are more sensitive to this crucial aspect of tumor physiology. Diffusion-weighted imaging is a useful method for evaluating areas of high cellularity and can be extended to diffusion tensor imaging, which can help identify tissue microstructure and depict neoplastic infiltration in areas of brain that appear normal on conventional MR images (20–22). Perfusion MRI with dynamic susceptibility-weighted contrast material enhancement, dynamic contrast enhancement, or arterial spin labeling exploits the neoangiogenic properties of gliomas. MR spectroscopy, which depicts the distribution of chemical metabolites such as choline, creatine, and N-acetylaspartate, is used clinically for grading gliomas and identifying regions of tumor infiltration (23,24). Despite the potential viability of these advanced modalities, they are usually interpreted in a qualitative manner. Their widespread adoption is further complicated by variability across sites, imaging units, and postprocessing methods.
Genomics and Radiogenomics in Neuro-Oncology
Vast mutational, molecular, and microenvironment heterogeneity in CNS neoplasms substantially complicates diagnostic and treatment approaches. Diffuse gliomas typically harbor more than 60 genetic alterations, encompassing several major cellular pathways (25). Developing a better understanding of these cellular pathways is crucial for improving diagnostic methods and delivering targeted therapies. In fact, given the prognostic significance of different mutations, molecular markers have recently taken a larger role in establishing an “integrated diagnosis,” as seen in the most recent 2016 WHO classification of CNS tumors (26) (Fig 3). For example, glioblastoma is now fundamentally grouped according to the presence of mutations in isocitrate dehydrogenase (IDH), with worse survival seen in IDH wild-type gliomas (27). Seminal work analyzing gene expression data of subjects from TCGA (https://cancergenome.nih.gov/) identified prognostically influential gene expression–based subgroups, namely proneural, neural, classic, and mesenchymal, representing distinct cellular pathways with prognostic and therapeutic implications (28).
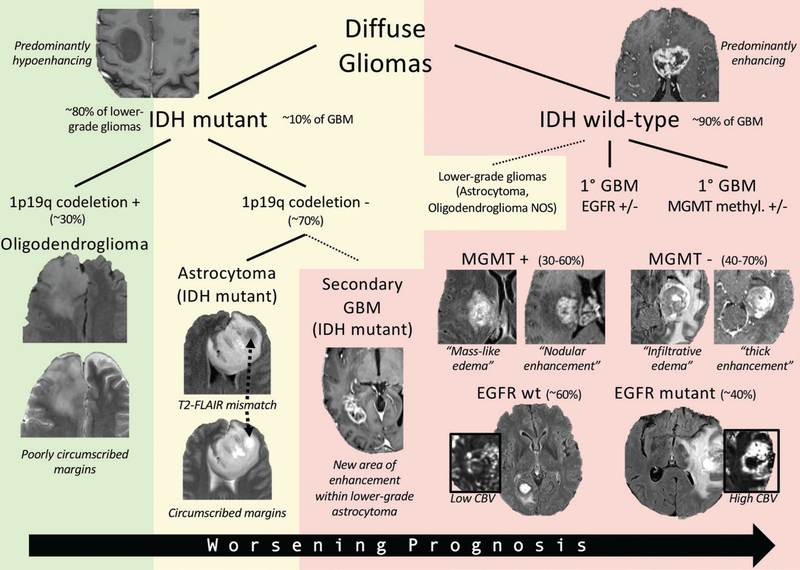
Figure 3:
Genomic and radiogenomic landscape of diffuse gliomas. Diffuse gliomas are fundamentally differentiated according to presence of IDH mutation. IDH mutant gliomas are typically lower grade (World Health Organization [WHO] grades II–III) but can sometimes be glioblastoma (GBM) (WHO grade IV), in which case they usually arise from a lower-grade astrocytoma. IDH mutant gliomas are subdivided according to presence of 1p19q codeletion, which defines an oligodendroglioma, and are associated with “poorly circumscribed” margins. The 1p19q non-codeleted tumors are characterized by “circumscribed” margins and exhibit the “T2–fluid-attenuated inversion recovery (FLAIR) mismatch” pattern. IDH wild-type gliomas are typically glioblastomas but may sometimes be a lower-grade astrocytoma or oligodendroglioma not otherwise specified (NOS). Epidermal growth factor receptor (EGFR) mutations and O6-methylguanine-DNA-methyltransferase (MGMT ) methylation status are important molecular and prognostic markers. EGFR mutant gliomas are associated with increased cerebral blood volume (CBV ). Methylated MGMT gliomas are associated with “masslike” T2-FLAIR signal intensity abnormality and heterogeneous and/or nodular enhancement pattern, whereas unmethylated MGMT gliomas are associated with “infiltrative” T2-FLAIR signal intensity abnormality and thick enhancement pattern with central necrosis. Background shading represents overall prognosis (green = best, yellow = intermediate, red = worst). wt = wild-type.
Further advancing our understanding of neuro-oncology is the nascent field of radiogenomics (imaging genomics), which correlates imaging characteristics with genetic, mutational, and expression patterns (29,30). Radiogenomics also has the power to dynamically monitor the microenvironment over the course of treatment, potentially allowing for a reduced number of repeat biopsies or resections. Glioma radiogenomics (Fig 3) has begun to characterize radiomic phenotypes of several candidate genetic alterations, including IDH mutation, O6-methylguanine-DNA-methyltransferase (MGMT) methylation, epidermal growth factor receptor (EGFR) splice variant, and the 1p/19q codeletion. Additional work in radiogenomics has attempted to develop a systems-level understanding between gene expression patterns and radiomics.
Candidate Genetic Alterations
Mutations in IDH1 and IDH2 are found in 70%–80% of lower-grade gliomas but only 5%–10% of glioblastomas, which are usually secondary glioblastomas, arising from lower-grade gliomas (27,31–32). IDH mutant gliomas result in the accumulation of D-2-hydroxyglutarate, an oncometabolite absent in IDH wild-type tumors. D-2-hydroxyglutarate can be detected with high specificity with MR spectroscopy (33–35). Although advanced MR spectroscopy sequences (eg, two-dimensional localized correlation spectroscopy at 7.0 T [36]) are highly reliable in the detection of 2-hydroxyglutarate, they are not readily available in most centers, which limits their utility. Interestingly, visually apparent imaging biomarkers, including indistinct margins and T2-FLAIR mismatch (regions within the tumor that are hyperintense on T2-weighted images but hypointense on FLAIR images), have been shown to be useful in the differentiation of IDH mutant from IDH wild-type gliomas (37). CNNs applied to conventional MRI modalities have been used to differentiate IDH mutant gliomas from IDH wild-type tumors with 92% accuracy, consistent with prior visual assessment and underlying pathophysiology that IDH wild-type tumors demonstrate more infiltrative, ill-defined margins (38).
Approximately 33%–57% of diffuse gliomas exhibit hypermethylation of the promoter of the MGMT gene, encoding for a DNA repair protein (39,40). MGMT promoter hypermethylation has been associated with better prognosis owing to improved sensitivity to alkylating agents (eg, temozolomide) (39,40). Radiomic studies have identified distinct imaging signatures for this molecular marker. Several groups have been able to predict MGMT methylation status with up to 88% accuracy by combining texture features with traditional supervised machine learning methods (41–43). In addition, several deep learning architectures have been shown to predict MGMT methylation status (44,45). Korfiatis et al (44) obtained up to 94.9% accuracy with only T2-weighted images and without the need for previous tumor segmentation. By performing principal component analysis on their final CNN layer, Chang et al (38) found that nodular and heterogeneous enhancement and “masslike FLAIR edema” were helpful for predicting MGMT methylation status, with up to 83% accuracy.
EGFR is a receptor tyrosine kinase that regulates normal cellular growth in epithelial cell lines (46). EGFR mutations are present in approximately 40% of glioblastomas but are rarely found in lower-grade gliomas (31). The most common extracellular EGFR mutation in glioblastoma is the splice variant III (EGFRvIII), which is found in 31% of patients (47). Radiogenomic studies performed with perfusion imaging have demonstrated a moderate relationship between EGFR amplification and tumor blood flow and volume (48,49). Support vector machine–based approaches have shown that EGFRvIII mutant gliomas exhibit deep peritumoral infiltration, which is consistent with a more aggressive and/or infiltrative phenotype (50). Further multivariate approaches incorporating a larger set of multiparametric features have shown that EGFRvIII tumors have increased neovascularization and cell density, as well as a spatial preference for frontal and parietal regions (51,52).
The codeletion of chromosome arms 1p/19q is present in approximately 30% of lower-grade gliomas (but is not present in glioblastoma) and, in combination with an IDH mutation, now defines an oligodendroglioma (26). The 1p/19q codeletion has been shown to have a protective effect on prognosis (53). Comparison of lower-grade gliomas with and without the 1p/19q codeletion showed that noncircumscribed borders, heterogeneous signal on T1- and/or T2-weighted images, and lower apparent diffusion coefficients are strongly associated with 1p/19q codeletions (37,54). More recent work with use of CNNs found that the 1p/19q codeletion is associated with increased enhancement, left frontal predominance, and ill-defined margins on FLAIR images with mass effect (33), with up to 93% accuracy (55).
Systems-Level Radiogenomic Approaches
Complementary to these candidate gene approaches, a systemslevel radiogenomic approach has been used by some groups to better understand how radiomic markers relate to gene expression patterns more globally, which is important considering that each tumor contains a combination of different mutations. An early study (56) looked at the relationship between gene expression modules with several neuroradiologist-defined MRI patterns, including contrast-to-necrosis ratio, subventricular zone involvement, contrast-to-T2 ratio, and T2 heterogeneity patterns. The investigators found that activation of specific gene expression programs can be inferred from imaging traits; for example, the hypoxia module was associated with contrast enhancement and the proliferation module was associated with mass effect. Grossmann et al (57) compared genomic pathway enrichment analyses with volumetric tumor phenotypes, finding that immune response and apoptosis cellular pathways were associated with necrosis pathways and that tumor bulk and edema were associated with homeostasis and cell cycling pathways. Zinn et al (58) evaluated the relationship between radiomic features in gliomas and nonoverlapping mutations in TP53, PTEN, and EGFR genes in 29 patients from TCGA. Although there were minimal differences in conventional MRI volumetric parameters, texture analyses yielded distinct and partially nonoverlapping radiomic feature sets and a variety of gene expression signatures uniquely associated with these three mutations (TP53 with angiogenesis, PTEN with invasion, and EGFR with immune response).
Prognostication in Neuro-Oncology
In current clinical practice, prognostication is based on histologic tumor grade and clinical models incorporating the patient’s age, sex, and functional status (eg, the Karnofsky performance status scale) (59). In addition, some of the previously discussed specific molecular markers are now central in diagnosis and prognosis. However, imaging features and radiomic metrics are not used in any widely adopted clinical prognostic model, despite their potential to capture underlying tumor biology and outcomes.
Radiomic Prediction of Prognosis
Previous radiomic studies have shown that basic imaging metrics, including maximal dimension and enhancing volume, are predictive beyond clinical models (60,61). A rule-based model combining clinical, imaging, and genetic variables resulted in the best predictive accuracy in patients in TCIA (62). Diffusion, perfusion, and MR spectroscopy measures have also been found to be prognostic (63,64), and more recent studies have used machine learning methods to predict patient survival on the basis of multiparametric MR images (65–67). Kickingereder et al (65) identified 11 crossvalidated features portending a poor prognosis, including volume, shape, texture, and wavelet features. These features were exclusively derived from FLAIR images within the contrastenhancing portion of the tumor. Macyszyn et al (66), using features explicitly extracted from traditional and advanced MRI sequences, developed a support vector machine model to predict survival group (low, medium, and high), with up to 80% accuracy in training and prospective replication cohorts. The most predictive features in this model included tumor volumes, angiogenesis (enhancing tumor volume), peritumoral infiltration (peak perfusion height), cell density (trace diffusion values), and distance to the ventricles.
Systems-Level Radiomic Approaches for Prognostication
With use of unsupervised machine learning methods, radiomic features can also be used to generate novel subgroups that may more closely align with the underlying biology of gliomas. Itakura et al (68) performed clustering of features capturing the shape, texture, and edge sharpness of 121 solitary glioblastomas with postcontrast T1-weighted MRI. They identified three clusters—premultifocal, spherical, and rim enhancing. These clusters were subsequently validated in 144 multi-institution subjects from TCIA and had significant differences in survival. The best prognosis was seen in the rim-enhancing subgroup, and the worst prognosis was seen in the premultifocal group. Rathore et al (43) also applied an unsupervised high-dimensional clustering algorithm to a comprehensive feature set derived from conventional and advanced MRI in 208 patients with glioblastoma. The resulting subtypes were remarkably similar to those found in the study by Itakura et al (68), clustering into rim-enhancing, irregular, and solid subgroups. These subgroups were associated with distinct survival estimates, with the best survival seen in the rim-enhancing subgroup. The clusters were also associated with particular anatomic locations, molecular subtypes, and genetic variations, including IDH, MGMT, and EGFRvIII. Although these subgroups are still preliminary, requiring larger sample sizes and additional validation to ensure their robustness, they show the promise of unsupervised methods in providing more precise subgrouping and prognostication than clinical models and molecular markers currently used in WHO classification.
Treatment Response Assessment in Neuro-Oncology
Current standard-of-care treatment for glioblastoma consists of maximal safe resection followed by radiation and chemotherapy with temozolomide (69), whereas lower-grade gliomas may be treated with surgery and/or chemoradiation. The addition of tumor-treating fields has more recently been shown to have an additional survival benefit in glioblastoma (19). Although not yet proven, an array of clinical trials using immunotherapy are being applied to treat patients with glioblastoma (70), including ones that target specific molecular pathways such as EGFR (71).
Clinical Response Assessment
Increasing size of T2/FLAIR signal abnormality with new or increasing areas of enhancement after combined radiation and chemotherapy, known as pseudoprogression (72), makes the evaluation of treatment response particularly challenging and is more common in MGMT methylated and IDH mutant tumors. Conversely, antiangiogenic agents (eg, bevacizumab) can result in pseudoresponse, which consists of a dramatic reduction of enhancement by altering the blood-brain barrier, but do little to alter progression of the infiltrating component—with no improvement in overall survival (73).
Initial guidelines for response assessment followed the Macdonald criteria (74), which incorporated only the size of enhancing components. More recently, the evaluation of treatment response has been described with the Response Assessment for Neuro-Oncology, or RANO, criteria (75), which incorporates changes in enhancing tissue and T2/FLAIR nonenhancing signal intensity abnormalities in addition to clinical status. A recent modification to the RANO criteria (76) changed the baseline assessment to be the first postradiation treatment image rather than the postresection image and provides some response assessment rubrics to identify pseudoprogression. However, RANO is still a limited tool for assessing response, especially considering that it uses two-dimensional measurements, which are subjective, and does not incorporate advanced imaging modalities such as MRI perfusion, diffusion tensor imaging, and MR spectroscopy, despite their clinical utility and use at most institutions. Newer immunotherapy agents have been shown to elicit complex inflammatory responses (77), adding additional difficulties in response assessment. The immunotherapy RANO criteria (78,79) extends the timeline for determining progression in an attempt to help account for immune inflammatory-related pseudoprogression.
Radiomic Prediction of Pseudoprogression and Progression
The differentiation of pseudoprogression from true progression remains a crucial diagnostic dilemma, for which AI methods are well suited (Fig 4). Several radiomic studies have had moderate success by evaluating diffusion-weighted imaging (80,81) and dynamic susceptibility-weighted contrast enhancement measures (82,83). Machine learning approaches that incorporate multiple measures from both diffusionweighted imaging and dynamic susceptibility-weighted contrast enhancement have also had success in predicting pseudoprogression (84,85). Notably, although most previous studies have used longitudinal clinical and radiologic follow-up to determine pseudoprogression, histologic examination of repeat resections often shows a combination of treatment-related changes and recurrent and/or residual tumor. Wang et al (86) used a multivariate logistic regression model, incorporating measures derived from dynamic susceptibility-weighted contrast-enhanced MRI and diffusion tensor imaging within the enhancing tissue to predict pseudoprogression on the basis of histologically classified cases of true progression, pseudoprogression, and mixed response, with an area under the curve of 0.90. Akbari et al (87) conducted a support vector machine–based analysis of conventional and advanced MRI features by training the model on pathologists’ scores of true progression versus pseudoprogression, demonstrating a high correlation (r = 0.86) between pathologic and radiomic scores of pseudoprogression.
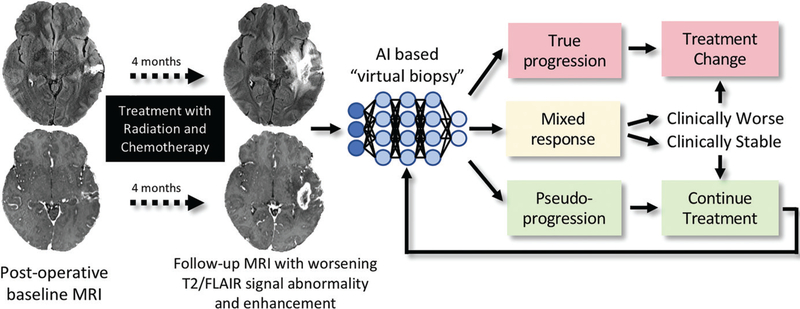
Figure 4:
Treatment response in neuro-oncologic imaging. After standard-of-care treatment with combined radiation therapy and chemotherapy, increasing T2–fluid-attenuated inversion recovery (FLAIR) signal intensity abnormality and new and/or increasing size of enhancing lesions are often seen. Artificial intelligence (AI )–based “virtual biopsy” could assist in distinguishing underlying biology and segregating treatment response into three possible categories: true progression (>75% recurrent and/or residual glioma at pathologic examination), mixed response (25%–75% recurrent and/or residual glioma at pathologic examination), and pseudoprogression (>75% treatment-related changes). Categories dictate distinct therapeutic approaches. In this example, the new enhancing lesion was found to represent 100% treatment-related changes at pathologic examination, with few atypical cells.
Radiomic Predictions of Infiltration and Recurrence
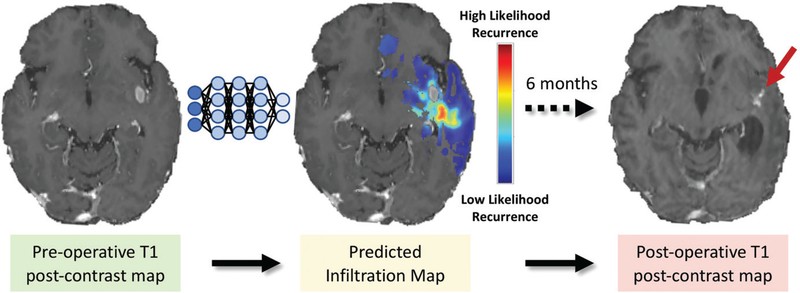
Despite the difficulty in differentiating infiltrating neoplasm from edema by using conventional qualitative approaches, there is substantial promise for machine learning methods to identify margins of infiltrative tissue on preoperative MR images. These delineations may be used to guide extended surgical resections, localized biopsies, and radiation treatment planning. With use of a voxel-wise logistic regression model, FLAIR and apparent diffusion coefficient maps were shown to be sufficient to enable prediction of areas of future tumor recurrence (88). Akbari et al (89) developed and Rathore et al (90) refined a multivariate support vector machine approach, incorporating features from conventional and advanced MRI modalities after registering areas of glioblastoma recurrences to preoperative MR images. This approach generates predictive spatial maps of infiltrated peritumoral tissue (Fig 5), with approximately 90% cross-validated accuracy.
Figure 5:
Predictive maps of tumor infiltration. Multimodal pre-operative conventional and advanced MRI data were analyzed with support vector machines to generate an estimated infiltration map overlaid on postcontrast T1-weighted image (red areas signify higher risk). Postcontrast T1-weighted MR image obtained at 6-month follow-up (right) demonstrates area of recurrence near site of highest predicted infiltration (arrow).
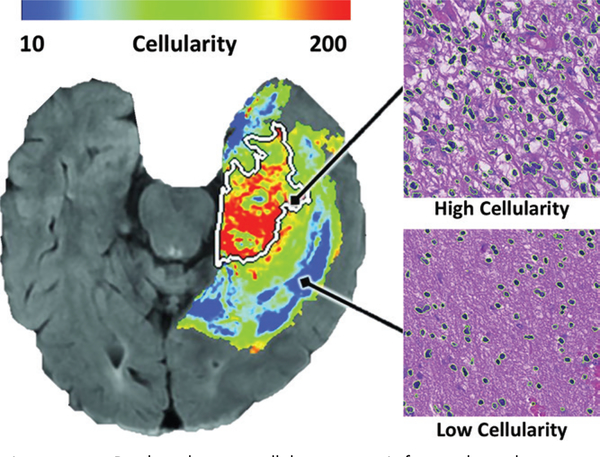
For another approach, which uses pathologic-radiologic relationships, Chang (91) developed a fully automated system to register biopsy sites from neuronavigational cross-hairs to the preoperative MR images by using a CNN in 36 patients. Multimodal imaging measures at the biopsy sites were then used to train a network on a cell density counting method applied to pathology images (Fig 6). They found an inverse relationship between cellularity and apparent diffusion coefficient and FLAIR values and a direct relationship between degree of enhancement and cellularity. This approach effectively generates noninvasive maps of cell density, which are useful for identifying the infiltrative margins of gliomas. Given that current surgical resection is guided largely by means of the enhancing tumor alone, these methods have substantial promise in developing noninvasive means to stratify patients into clinical trials and guiding more aggressive treatments. Indeed, the method by Akbari et al (89) has already spurred a clinical trial with intensified radiation to areas of infiltrative tumor for patients who have recently undergone an initial resection.
Figure 6:
Predicted tumor cellularity map. Left, voxelwise linear regression model was applied to multimodal preoperative MRI trained on automated cell counts of biopsies localized to different regions on MR image and used to generate a map of predicted cellularity (red areas signify more cells). Right, photomicrographs of biopsy specimens from regions of tumor with high and low cellularity (hematoxylin-eosin stain; original magnification, ×400). (Reprinted, with permission, from reference 91.)
AI Applications in Neuro-Oncologic Imaging: Non-Glioma Evaluation
Machine learning approaches have also been applied to other CNS tumors, particularly brain metastases and CNS lymphoma, with the potential to clarify diagnostically ambiguous situations and/or improve workflow efficiency and accuracy. Initially, a variety of image-processing methods, such as threedimensional template matching, were used to detect and localize brain metastases (92,93). More recent work with use of three-dimensional CNNs has shown increasing promise (94,95), with potential to aid in stereotactic radiation therapy planning. In some situations, the differentiation among brain metastases, primary CNS lymphoma, and glioblastoma is not possible with existing clinical and imaging paradigms. To address this problem, Wang et al (96) developed a decision tree and multivariate logistic regression model incorporating diffusion tensor imaging and dynamic susceptibility-weighted contrast-enhanced MRI metrics from the enhancing region to differentiate among these three entities. In another study (97), random forest analysis applied to extracted radiomic texture and wavelet features enabled the differentiation of nonnecrotic glioblastoma from CNS lymphoma, with a performance superior to that of three human readers. Sometimes patients are found to have brain metastases without a known primary site. Machine learning approaches have been applied to this clinical scenario on the assumption that underlying molecular differences and their downstream effects on local environments are likely to exhibit different radiomic features. For example, Ortiz-Ramón et al (98) were able to differentiate brain metastases due to lung cancer, melanoma, and breast cancer by using a random forest model applied to features derived from two- and three-dimensional texture analyses of T1-weighted postcontrast sequences.
Promises and Challenges of AI in Neuro-Oncologic Imaging
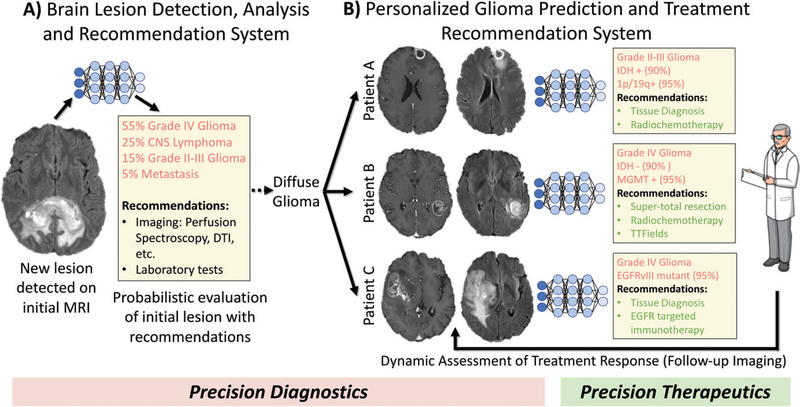
AI methods, given their ability to discern patterns and combine information in a way that humans cannot, show substantial promise for the future of radiology and precision medicine. An ideal AI-based diagnostic system for neuro-oncology would incorporate all relevant multimodal imaging data with clinical information and molecular markers to make precise predictions of biologically based and clinically relevant subtypes for a new tumor diagnosis, in line with the precision medicine movement (99). This information could help stratify patients into tailored treatments predicted to be most effective, including determining whether a patient would benefit from standard or supertotal resection and/or targeted intensification of radiation therapy to areas of infiltrative tumor, chemotherapy, or novel therapeutic agents that target specific cellular mechanisms (Fig 7).
Figure 7:
Schematic of future artificial intelligence–based neuro-oncologic imaging and clinical management workflow. A, Initial lesion detection and analysis system would generate a probabilistic differential of lesion(s) seen on patient’s initial brain MR image (precision diagnostics). It would also recommend additional useful imaging examinations, laboratory tests, or tissue sampling. B, Glioma-specific module could make personalized predictions of molecular markers, survival, and treatment responses (precision diagnostics), thereby recommending optimal treatment plan(s), which would be continuously updated on the basis of follow-up imaging (precision therapeutics). CNS = central nervous system, DTI = diffusion tensor imaging, EGFR = epidermal growth factor receptor, EGFRvIII = epidermal growth factor receptor variable III, IDH = isocitrate dehydrogenase, MGMT = O6-methylguanine-DNA-methyltransferase, TTFields = tumor-treating fields.
AI also has great potential for monitoring both standard treatments and novel treatments such as immunotherapy. Although complex inflammatory responses seen in immunotherapy (76) would require further validation of AI models that could monitor these new treatments (100), they have the potential to quickly determine treatment efficacy, thus allowing for dynamic adjustment during the course of treatment. In this regard, AI methods applied to advanced imaging could ultimately offer a personalized treatment response prediction superior to that of current methods.
Challenges
Major challenges to the promises of AI in radiology include high-quality ground truth data, generalizable and interpretable methods, and user-centric workflow integration (101). Concerns regarding the “black box” nature of these algorithms have somewhat diminished given the continuing development of methods, such as saliency maps (102) or principal component analysis (38), that can “unbox” the networks by interrogating internal algorithm feature vectors. A better mechanistic under-standing between feature patterns and underlying biology will be helpful both for clinical acceptance and for improving the biologic and treatment relevance of the patterns revealed by these methods.
One of the premier challenges in AI research is the availability of large, well-annotated data sets. Unfortunately, studies with relatively small sample sizes are prone to measurement error (103). TCIA and BraTS have made substantial progress in the creation of centralized, well-labeled data for glioma image analysis, whereas non–glioma-based research has been limited by a lack of public data sets. Nevertheless, the vast majority of available data remain siloed within individual institutions and hospital systems. In order for these algorithms to improve further, larger and more heterogeneous data sets (likely orders of magnitude larger) may be needed to improve the generalizability of an algorithm’s performance across different imaging sites, acquisition parameters, and patient populations (104). An important component of assembling such data sets is the sharing of data among institutions, similar to efforts in Alzheimer disease (105) and autism (106) research. Other ways of improving data sets include statistical techniques to harmonize the data sets and to introduce more uniform data collection by adoption of standardized neuro-oncology imaging protocols across institutions (76). Interestingly, novel deep learning methods, namely generative adversarial networks, have shown promise in improving performance by generating synthetic data (107).
An additional barrier to the development of more robust algorithms in the field of neuro-oncologic imaging, and radiomics more generally, is the lack of clear, targeted “use cases” or specific tasks against which their performance can be benchmarked. Other than the BraTS competition, the measured performance of an individual algorithm is highly taskdependent, data set–dependent, and strongly influenced by the particular scientific question, all of which limit comparison of different algorithms developed by different groups. The newly formed American College of Radiology Data Science Institute (https://acrdsi.org/) is helping define standard use cases, annotation tools, and data sets, which should greatly help with standardization and benchmarking relevant to both academic pursuits and commercial ventures.
In addition, although the U.S. Food and Drug Administration is developing pathways for approval of these emerging tools, there are many unanswered questions, including the generalizability of the methods and when retraining is appropriate.
Pathways to Clinical Implementation
Despite the growing use of AI algorithms in research settings, there are major obstacles to the efficient and consistent deployment of these sophisticated algorithms in a clinical setting. The system must be easily integrated into the radiologist’s workflow (electronic medical record system, picture archiving and communication system, and dictation software) to be adopted. Furthermore, many of the segmentation and radiomic methods require manual intervention and the use of a variety of in-house pipelines and have lengthy processing times. There has been relatively little work done to develop tools for easily translating and sharing these methods. In fact, most publications do not provide enough information to re-create their method independently. A few examples of open source tools that may facilitate sharing of different methods are Modelhub (http://modelhub.ai/), Pyradiomics (http://www.Radiomics.io/) (108), and the Cancer Imaging Phenomics Toolkit (https://med.upenn.edu/cbica/captk/), which was developed to facilitate clinical translation of these tools (109,110). Alternatively, optimal solutions for integration into routine clinical workflow may ultimately be provided through emerging commercial ventures.
The “holy grail” of AI in neuroradiology (111) might consist of a fully automated system, integrated into the radiologic workflow, that analyzes images in real time and provides a quantitative and probabilistic draft report. A more general diagnostic system for brain MR images might first assess the probability of whether a newly identified lesion represents a specific neoplasm or a neoplastic mimic (Fig 7, A). Such a system would recommend additional advanced imaging protocols and/or sequences as needed. A more specific glioma evaluation system (Fig 7, B) would then generate a personalized prediction of relevant molecular markers (precision diagnostics) and prognosis, as well as specific treatment recommendations (precision therapeutics). This automated system could also be used for monitoring treatment in “real time,” with more precise quantitative reporting tools to track changes in both conventional and advanced imaging parameters as well as patterns derived from deep learning.
Until AI methods are completely integrated into daily practice, it will be the role of the “centaur” radiologist (112), formed as a synergy of humans and computers, to integrate information from images, AI tools, and health records to improve the precision of radiology and health care.
Conclusion
The overarching goal of this line of research is to improve the outcomes of patients affected by CNS neoplasms through improvements in diagnostic and treatment methods. AI tools that combine clinical, radiomic, and genomic information into predictive models hold substantial promise for guiding and monitoring personalized therapeutics. However, many challenges exist and much work needs to be done to bring the promise of this field into fruition. Nevertheless, radiologic practice will substantially change as AI technology continues to improve to be able to enhance radiologists’ accuracy and efficiency. It is crucial for the future radiologist to understand and appropriately use these powerful tools as they become more integrated into everyday clinical practice in the coming years.
Essentials.
Artificial intelligence (AI) algorithms are driving neuro-oncology radiomics research forward by identifying complex patterns in advanced MRI with important diagnostic, prognostic, and therapeutic implications.
Radiogenomics in neuro-oncology offers insights into underlying cellular and molecular mechanisms of cancer biology.
Radiomic and radiogenomic tools provide a means of noninvasive sampling of tumor microenvironments, allowing for a dynamic and comprehensive evaluation of regionally heterogeneous central nervous system tumors.
Despite substantial challenges, targeted clinical implementation of AI methods in neuro-oncology is set to transform the field into an era of precision medicine.
Acknowledgments:
The authors thank Saima Rathore, PhD, and the Center for Biomedical Image Computing and Analytics for providing several of the images used in this review.
Abbreviations
- AI
artificial intelligence
- BraTS
Multimodal Brain Tumor Image Segmentation Benchmark
- CNN
convolutional neural networks
- CNS
central nervous system
- EGFR
epidermal growth factor receptor
- FLAIR
fluid-attenuated inversion recovery
- IDH
isocitrate dehydrogenase
- MGMT
O6-methylguanine-DNA-methyltransferase
- TCGA
the Cancer Genome Atlas
- TCIA
the Cancer Imaging Archive
- WHO
World Health Organization
Footnotes
Disclosures of Conflicts of Interest: J.D.R. disclosed no relevant relationships. A.M.R. disclosed no relevant relationships. R.N.B. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: is a founder and board member of GalileoCDS; institution receives royalties for patent licensed to GalileoCDS; has stock/stock options in GalileoCDS. Other relationships: has patents issued and held by University of Pennsylvania; has patents licensed to GalileoCDS. C.D. disclosed no relevant relationships. S.M. Activities related to the present article: institution received grants from GalileoCDS and NovoCure. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships.
Contributor Information
Jeffrey D. Rudie, Department of Radiology, Division of Neuroradiology, Perelman School of Medicine at the University of Pennsylvania, 3400 Spruce St, Philadelphia, PA 19104.
Andreas M. Rauschecker, Department of Radiology & Biomedical Imaging, University of California, San Francisco, San Francisco, Calif.
R. Nick Bryan, Department of Diagnostic Medicine, Dell Medical School, University of Texas, Austin, Tex.
Christos Davatzikos, Department of Radiology, Division of Neuroradiology, Perelman School of Medicine at the University of Pennsylvania, 3400 Spruce St, Philadelphia, PA 19104.
Suyash Mohan, Department of Radiology, Division of Neuroradiology, Perelman School of Medicine at the University of Pennsylvania, 3400 Spruce St, Philadelphia, PA 19104.
References
- 1.Chang PJ. Bayesian analysis revisited: a radiologist’s survival guide. AJR Am J Roentgenol 1989;152(4):721–727. [DOI] [PubMed] [Google Scholar]
- 2.Kahn CE Jr. Artificial intelligence in radiology: decision support systems. RadioGraphics 1994;14(4):849–861. [DOI] [PubMed] [Google Scholar]
- 3.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521(7553):436–444. [DOI] [PubMed] [Google Scholar]
- 4.Hassabis D, Kumaran D, Summerfield C, Botvinick M. Neuroscience-inspired artificial intelligence. Neuron 2017;95(2):245–258. [DOI] [PubMed] [Google Scholar]
- 5.Chartrand G, Cheng PM, Vorontsov E, et al. Deep learning: a primer for radiologists. RadioGraphics 2017;37(7):2113–2131. [DOI] [PubMed] [Google Scholar]
- 6.Goodfellow I, Bengio Y, Courville A. Deep learning. Cambridge, Mass: MIT Press, 2016. [Google Scholar]
- 7.Deng J, Dong W, Socher R, Li LJ, Li K, Li FF. ImageNet: a large-scale hierarchical image database. In: Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition New York, NY: IEEE, 2009; 248–255. [Google Scholar]
- 8.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. In: NIPS 12 Proceedings of the 25th International Conference on Neural Information Processing Systems, vol 1 Red Hook, NY: Curran Associates, 2012; 1097–1105. [Google Scholar]
- 9.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016;278(2):563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yip SS, Aerts HJ. Applications and limitations of radiomics. Phys Med Biol 2016;61(13):R150–R166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gooya A, Pohl KM, Bilello M, et al. GLISTR: glioma image segmentation and registration. IEEE Trans Med Imaging 2012;31(10):1941–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang PD. Fully convolutional deep residual neural networks for brain tumor segmentation In: Crimi A, Menze B, Maier O, Reyes M, Winzeck S, Handels H, eds. Brainlesion: glioma, multiple sclerosis, stroke and traumatic brain injuries. BrainLes 2016. Lecture notes in computer science, vol 10154. Cham, Switzerland: Springer, 2017; 108–118. [Google Scholar]
- 13.Akkus Z, Galimzianova A, Hoogi A, Rubin DL, Erickson BJ. Deep learning for brain MRI segmentation: state of the art and future directions. J Digit Imaging 2017;30(4):449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamnitsas K, Bai W, Ferrante E, et al. Ensembles of multiple models and architectures for robust brain tumour segmentation In: Crimi A, Bakas S, Kuijf H, Menze B, Reyes M, eds. Brainlesion: glioma, multiple sclerosis, stroke and traumatic brain injuries. BrainLes 2017. Lecture notes in computer science, vol 10670. Cham, Switzerland: Springer, 2018; 450–462. [Google Scholar]
- 15.Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 2013;26(6):1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menze BH, Jakab A, Bauer S, et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging 2015;34(10):1993–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakas S, Akbari H, Sotiras A, et al. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data 2017;4:170117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stupp R, Taillibert S, Kanner A, et al. Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: a randomized clinical trial. JAMA 2017;318(23):2306–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price SJ, Jena R, Burnet NG, et al. Improved delineation of glioma margins and regions of infiltration with the use of diffusion tensor imaging: an imageguided biopsy study. AJNR Am J Neuroradiol 2006;27(9):1969–1974. [PMC free article] [PubMed] [Google Scholar]
- 21.Kallenberg K, Goldmann T, Menke J, et al. Glioma infiltration of the corpus callosum: early signs detected by DTI. J Neurooncol 2013;112(2):217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stieltjes B, Schlüter M, Didinger B, et al. Diffusion tensor imaging in primary brain tumors: reproducible quantitative analysis of corpus callosum infiltration and contralateral involvement using a probabilistic mixture model. Neuroimage 2006;31(2):531–542. [DOI] [PubMed] [Google Scholar]
- 23.Deviers A, Ken S, Filleron T, et al. Evaluation of the lactate-to-N-acetyl-aspartate ratio defined with magnetic resonance spectroscopic imaging before radiation therapy as a new predictive marker of the site of relapse in patients with glioblastoma multiforme. Int J Radiat Oncol Biol Phys 2014;90(2):385–393. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, Ma L, Wang Q, Zheng X, Wu C, Xu BN. Role of magnetic resonance spectroscopy for the differentiation of recurrent glioma from radiation necrosis: a systematic review and meta-analysis. Eur J Radiol 2014;83(12):2181–2189. [DOI] [PubMed] [Google Scholar]
- 25.Belden CJ, Valdes PA, Ran C, et al. Genetics of glioblastoma: a window into its imaging and histopathologic variability. RadioGraphics 2011;31(6):1717–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol (Berl) 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 27.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verhaak RG, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuo MD, Jamshidi N. Behind the numbers: Decoding molecular phenotypes with radiogenomics--guiding principles and technical considerations. Radiology 2014;270(2):320–325. [DOI] [PubMed] [Google Scholar]
- 30.Ellingson BM. Radiogenomics and imaging phenotypes in glioblastoma: novel observations and correlation with molecular characteristics. Curr Neurol Neurosci Rep 2015;15(1):506. [DOI] [PubMed] [Google Scholar]
- 31.Ostrom QT, Bauchet L, Davis FG, et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol 2014;16(7):896–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen AL, Holmen SL, Colman H. IDH1 and IDH2 mutations in gliomas. Curr Neurol Neurosci Rep 2013;13(5):345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi C, Ganji SK, DeBerardinis RJ, et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med 2012;18(4):624–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andronesi OC, Kim GS, Gerstner E, et al. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci Transl Med 2012;4(116):116ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Branzoli F, Di Stefano AL, Capelle L, et al. Highly specific determination of IDH status using edited in vivo magnetic resonance spectroscopy. Neuro Oncol 2018;20(7):907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verma G, Mohan S, Nasrallah MP, et al. Non-invasive detection of 2-hydroxy-glutarate in IDH-mutated gliomas using two-dimensional localized correlation spectroscopy (2D L-COSY) at 7 Tesla. J Transl Med 2016;14(1):274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel SH, Poisson LM, Brat DJ, et al. T2-FLAIR mismatch, an imaging biomarker for IDH and 1p/19q status in lower-grade gliomas: a TCGA/TCIA project. Clin Cancer Res 2017;23(20):6078–6085. [DOI] [PubMed] [Google Scholar]
- 38.Chang P, Grinband J, Weinberg BD, et al. Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. AJNR Am J Neuroradiol 2018;39(7):1201–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rivera AL, Pelloski CE, Gilbert MR, et al. MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro Oncol 2010;12(2):116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reifenberger G, Hentschel B, Felsberg J, et al. Predictive impact of MGMT promoter methylation in glioblastoma of the elderly. Int J Cancer United States, 2012;131(6):1342–1350. [DOI] [PubMed] [Google Scholar]
- 41.Drabycz S, Roldán G, de Robles P, et al. An analysis of image texture, tumor location, and MGMT promoter methylation in glioblastoma using magnetic resonance imaging. Neuroimage 2010;49(2):1398–1405. [DOI] [PubMed] [Google Scholar]
- 42.Kanas VG, Zacharaki EI, Thomas GA, Zinn PO, Megalooikonomou V, Colen RR. Learning MRI-based classification models for MGMT methylation status prediction in glioblastoma. Comput Methods Programs Biomed 2017;140:249–257. [DOI] [PubMed] [Google Scholar]
- 43.Rathore S, Akbari H, Rozycki M, et al. Radiomic MRI signature reveals three distinct subtypes of glioblastoma with different clinical and molecular characteristics, offering prognostic value beyond IDH1. Sci Rep 2018; 8(1):5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korfiatis P, Kline TL, Lachance DH, Parney IF, Buckner JC, Erickson BJ. Residual deep convolutional neural network predicts MGMT methylation status. J Digit Imaging 2017;30(5):622–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han L, Kamdar MR. MRI to MGMT: predicting methylation status in glioblastoma patients using convolutional recurrent neural networks. Pac Symp Biocomput 2018;23:331–342. [PMC free article] [PubMed] [Google Scholar]
- 46.Gan HK, Cvrljevic AN, Johns TG. The epidermal growth factor receptor variant III (EGFRvIII): where wild things are altered. FEBS J 2013;280(21):5350–5370. [DOI] [PubMed] [Google Scholar]
- 47.Heimberger AB, Hlatky R, Suki D, et al. Prognostic effect of epidermal growth factor receptor and EGFRvIII in glioblastoma multiforme patients. Clin Cancer Res 2005;11(4):1462–1466. [DOI] [PubMed] [Google Scholar]
- 48.Tykocinski ES, Grant RA, Kapoor GS, et al. Use of magnetic perfusion-weighted imaging to determine epidermal growth factor receptor variant III expression in glioblastoma. Neuro Oncol 2012;14(5):613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kickingereder P, Bonekamp D, Nowosielski M, et al. Radiogenomics of glioblastoma: machine learning–based classification of molecular characteristics by using multiparametric and multiregional MR imaging features. Radiology 2016;281(3):907–918. [DOI] [PubMed] [Google Scholar]
- 50.Bakas S, Akbari H, Pisapia J, et al. In vivo detection of EGFRvIII in glioblastoma via perfusion magnetic resonance imaging signature consistent with deep peritumoral infiltration: the φ-index. Clin Cancer Res 2017;23(16):4724–4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akbari H, Bakas S, Pisapia JM, et al. In vivo evaluation of EGFRvIII mutation in primary glioblastoma patients via complex multiparametric MRI signature. Neuro Oncol 2018;20(8):1068–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bilello M, Akbari H, Da X, et al. Population-based MRI atlases of spatial distribution are specific to patient and tumor characteristics in glioblastoma. Neuroimage Clin 2016;12:34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu N, Richards R, Jensen R. Role of chromosomal 1p/19q co-deletion on the prognosis of oligodendrogliomas: A systematic review and meta-analysis. Interdiscip Neurosurg 2016;5:58–63. [Google Scholar]
- 54.Johnson DR, Diehn FE, Giannini C, et al. Genetically defined oligodendroglioma is characterized by indistinct tumor borders at MRI. AJNR Am J Neuroradiol 2017;38(4):678–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akkus Z, Ali I, Sedlář J, et al. Predicting deletion of chromosomal arms 1p/19q in low-grade gliomas from MR images using machine intelligence. J Digit Imaging 2017;30(4):469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diehn M, Nardini C, Wang DS, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A 2008;105(13):5213–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grossmann P, Gutman DA, Dunn WD Jr, Holder CA, Aerts HJ. Imaging-genomics reveals driving pathways of MRI derived volumetric tumor phenotype features in Glioblastoma. BMC Cancer 2016;16(1):611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zinn PO, Singh SK, Kotrotsou A, et al. Distinct radiomic phenotypes define glioblastoma TP53-PTEN-EGFR mutational landscape. Neurosurgery 2017;64(CN_suppl_1):203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Curran WJ Jr, Scott CB, Horton J, et al. Recursive partitioning analysis of prognostic factors in three Radiation Therapy Oncology Group malignant glioma trials. J Natl Cancer Inst 1993;85(9):704–710. [DOI] [PubMed] [Google Scholar]
- 60.Gutman DA, Cooper LA, Hwang SN, et al. MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology 2013;267(2):560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zinn PO, Sathyan P, Mahajan B,et al. A novel volume-age-KPS (VAK) glioblastoma classification identifies a prognostic cognate microRNA-gene signature. PLoS One 2012;7(8):e41522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nicolasjilwan M, Hu Y, Yan C, et al. Addition of MR imaging features and genetic biomarkers strengthens glioblastoma survival prediction in TCGA patients. J Neuroradiol 2015;42(4):212–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murakami R, Sugahara T, Nakamura H, et al. Malignant supratentorial astrocytoma treated with postoperative radiation therapy: prognostic value of pretreatment quantitative diffusion-weighted MR imaging. Radiology 2007;243(2):493–499. [DOI] [PubMed] [Google Scholar]
- 64.Law M, Young RJ, Babb JS, et al. Gliomas: predicting time to progression or survival with cerebral blood volume measurements at dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. Radiology 2008;247(2):490–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kickingereder P, Burth S, Wick A, et al. Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology 2016;280(3):880–889. [DOI] [PubMed] [Google Scholar]
- 66.Macyszyn L, Akbari H, Pisapia JM, et al. Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro Oncol 2016;18(3):417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Q, Bai H, Chen Y, et al. A fully-automatic multiparametric radiomics model: towards reproducible and prognostic imaging signature for prediction of overall survival in glioblastoma multiforme. Sci Rep 2017;7(1):14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Itakura H, Achrol AS, Mitchell LA, et al. Magnetic resonance image features identify glioblastoma phenotypic subtypes with distinct molecular pathway activities. Sci Transl Med 2015;7(303):303ra138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stupp R, Mason WP, van den Bent MJ, et al. ; National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 70.Sampson JH, Maus MV, June CH. Immunotherapy for brain tumors. J Clin Oncol 2017;35(21):2450–2456. [DOI] [PubMed] [Google Scholar]
- 71.O’Rourke DM, Nasrallah MP, Desai A, et al. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci Transl Med 2017;9(399):eaaa0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Wit MC, de Bruin HG, Eijkenboom W, Sillevis Smitt PA, van den Bent MJ. Immediate post-radiotherapy changes in malignant glioma can mimic tumor progression. Neurology 2004;63(3):535–537. [DOI] [PubMed] [Google Scholar]
- 73.Tipping M, Eickhoff J, Ian Robins H. Clinical outcomes in recurrent glioblastoma with bevacizumab therapy: An analysis of the literature. J Clin Neurosci 2017;44:101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Macdonald DR, Cascino TL, Schold SC Jr, Cairncross JG. Response criteria for phase II studies of supratentorial malignant glioma. J Clin Oncol 1990;8(7):1277–1280. [DOI] [PubMed] [Google Scholar]
- 75.Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol 2010;28(11):1963–1972. [DOI] [PubMed] [Google Scholar]
- 76.Ellingson BM, Wen PY, Cloughesy TF. Modified criteria for radiographic response assessment in glioblastoma clinical trials. Neurotherapeutics 2017;14(2):307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roth P, Valavanis A, Weller M. Long-term control and partial remission after initial pseudoprogression of glioblastoma by anti-PD-1 treatment with nivolumab. Neuro Oncol 2017;19(3):454–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Okada H, Weller M, Huang R, et al. Immunotherapy response assessment in neuro-oncology: a report of the RANO working group. Lancet Oncol 2015;16(15):e534–e542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wen PY, Chang SM, Van den Bent MJ, Vogelbaum MA, Macdonald DR, Lee EQ. Response assessment in neuro-oncology clinical trials. J Clin Oncol 2017;35(21):2439–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chu HH, Choi SH, Ryoo I, et al. Differentiation of true progression from pseudoprogression in glioblastoma treated with radiation therapy and concomitant temozolomide: comparison study of standard and high-b-value diffusion-weighted imaging. Radiology 2013;269(3):831–840. [DOI] [PubMed] [Google Scholar]
- 81.Qian X, Tan H, Zhang J, Zhao W, Chan MD, Zhou X. Stratification of pseudoprogression and true progression of glioblastoma multiform based on longitudinal diffusion tensor imaging without segmentation. Med Phys 2016;43(11):5889–5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Suh CH, Kim HS, Choi YJ, Kim N, Kim SJ. Prediction of pseudoprogression in patients with glioblastomas using the initial and final area under the curves ratio derived from dynamic contrast-enhanced T1-weighted perfusion MR imaging. AJNR Am J Neuroradiol 2013;34(12):2278–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yun TJ, Park CK, Kim TM, et al. Glioblastoma treated with concurrent radiation therapy and temozolomide chemotherapy: differentiation of true progression from pseudoprogression with quantitative dynamic contrastenhanced MR imaging. Radiology 2015;274(3):830–840. [DOI] [PubMed] [Google Scholar]
- 84.Hu X, Wong KK, Young GS, Guo L, Wong ST. Support vector machine multiparametric MRI identification of pseudoprogression from tumor recurrence in patients with resected glioblastoma. J Magn Reson Imaging 2011;33(2):296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cha J, Kim ST, Kim HJ, et al. Differentiation of tumor progression from pseudoprogression in patients with posttreatment glioblastoma using multiparametric histogram analysis. AJNR Am J Neuroradiol 2014;35(7):1309–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang S, Martinez-Lage M, Sakai Y, et al. Differentiating tumor progression from pseudoprogression in patients with glioblastomas using diffusion tensor imaging and dynamic susceptibility contrast MRI. AJNR Am J Neuroradiol 2016;37(1):28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Akbari H, Bakas S, Martinez-Lage M, et al. Quantitative radiomics and machine learning to distinguish true progression from pseudoprogression in patients with GBM. Presented at the 56th annual meeting of the American Society for Neuroradiology, Vancouver, BC, Canada, June 2–7, 2018. [Google Scholar]
- 88.Chang PD, Chow DS, Yang PH, Filippi CG, Lignelli A. Predicting glioblastoma recurrence by early changes in the apparent diffusion coefficient value and signal intensity on FLAIR images. AJR Am J Roentgenol 2017;208(1):57–65. [DOI] [PubMed] [Google Scholar]
- 89.Akbari H, Macyszyn L, Da X, et al. Imaging surrogates of infiltration obtained via multiparametric imaging pattern analysis predict subsequent location of recurrence of glioblastoma. Neurosurgery 2016;78(4):572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rathore S, Akbari H, Doshi J, et al. Radiomic signature of infiltration in peritumoral edema predicts subsequent recurrence in glioblastoma: implications for personalized radiotherapy planning. J Med Imaging (Bellingham) 2018;5(2):021219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chang PD, Malone HR, Bowden SG, et al. A multiparametric model for mapping cellularity in glioblastoma using radiographically localized biopsies. AJNR Am J Neuroradiol 2017;38(5):890–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ambrosini RD, Wang P, O’Dell WG. Computer-aided detection of metastatic brain tumors using automated three-dimensional template matching. J Magn Reson Imaging 2010;31(1):85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pérez-Ramírez Ú, Arana E, Moratal D. Brain metastases detection on MR by means of three-dimensional tumor-appearance template matching. J Magn Reson Imaging 2016;44(3):642–652. [DOI] [PubMed] [Google Scholar]
- 94.Charron O, Lallement A, Jarnet D, Noblet V, Clavier JB, Meyer P. Automatic detection and segmentation of brain metastases on multimodal MR images with a deep convolutional neural network. Comput Biol Med 2018;95:43–54. [DOI] [PubMed] [Google Scholar]
- 95.Liu Y, Stojadinovic S, Hrycushko B, et al. A deep convolutional neural network-based automatic delineation strategy for multiple brain metastases stereotactic radiosurgery. PLoS One 2017;12(10):e0185844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang S, Kim S, Chawla S, et al. Differentiation between glioblastomas, solitary brain metastases, and primary cerebral lymphomas using diffusion tensor and dynamic susceptibility contrast-enhanced MR imaging. AJNR Am J Neuroradiol 2011;32(3):507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Suh HB, Choi YS, Bae S, et al. Primary central nervous system lymphoma and atypical glioblastoma: Differentiation using radiomics approach. Eur Radiol 2018;28(9):3832–3839. [DOI] [PubMed] [Google Scholar]
- 98.Ortiz-Ramón R, Larroza A, Ruiz-España S, Arana E, Moratal D. Classifying brain metastases by their primary site of origin using a radiomics approach based on texture analysis: a feasibility study. Eur Radiol 2018;28(11):4514–4523. [DOI] [PubMed] [Google Scholar]
- 99.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 2015;372(9):793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Skolnik AD, Wang S, Gopal PP, Mohan S. Commentary: pitfalls in the neuroimaging of glioblastoma in the era of antiangiogenic and immuno/ targeted therapy. Front Neurol 2018;9:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Choy G, Khalilzadeh O, Michalski M, et al. Current applications and future impact of machine learning in radiology. Radiology 2018;288(2):318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zeiler MD, Fergus R. Visualizing and understanding convolutional networks In: Fleet D, Pajdla T, Schiele B, Tuytelaars T, eds. Computer vision—ECCV 2014. ECCV 2014. Lecture notes in computer science, vol 8689. Cham, Switzerland: Springer International Publishing, 2014; 818–833. [Google Scholar]
- 103.Loken E, Gelman A. Measurement error and the replication crisis. Science 2017;355(6325):584–585. [DOI] [PubMed] [Google Scholar]
- 104.AlBadawy EA, Saha A, Mazurowski MA. Deep learning for segmentation of brain tumors: Impact of cross-institutional training and testing. Med Phys 2018;45(3):1150–1158. [DOI] [PubMed] [Google Scholar]
- 105.Jack CR Jr, Bernstein MA, Fox NC, et al. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging 2008;27(4):685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Di Martino A, Yan CG, Li Q, et al. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol Psychiatry 2014;19(6):659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shin HC, Tenenholtz NA, Rogers JK, et al. Medical image synthesis for data augmentation and anonymization using generative adversarial networks In: Gooya A, Goksel O, Obuz I, Burgos N, eds. Simulation and synthesis in medical imaging. New York, NY: Springer International Publishing, 1–11. [Google Scholar]
- 108.van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res 2017;77(21):e104–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Davatzikos C, Rathore S, Bakas S, et al. Cancer imaging phenomics toolkit: quantitative imaging analytics for precision diagnostics and predictive modeling of clinical outcome. J Med Imaging (Bellingham) 2018;5(1):011018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rathore S, Bakas S, Pati S, et al. Brain cancer imaging phenomics toolkit (brain-CaPTk): an interactive platform for quantitative analysis of glioblastoma. Brainlesion (2017) 2018;10670:133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zaharchuk G, Gong E, Wintermark M, Rubin D, Langlotz CP. Deep learning in neuroradiology. AJNR Am J Neuroradiol 2018;39(10):1776–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dreyer KJ, Geis JR. When machines think: radiology’s next frontier. Radiology 2017;285(3):713–718. [DOI] [PubMed] [Google Scholar]