The neonatal mammalian heart has a remarkable regenerative ability for the first few days of life, mediated by the proliferation of preexisting cardiomyocytes (CMs)1. The contribution of preexisting cardiomyocytes to neonatal heart regeneration is supported by histological analysis of cardiomyocyte mitotic indices, as well as fate mapping studies using Myh6-driven inducible Cre. However, given that this fate mapping strategy labels approximately 70–80% of cardiomyocytes, it is plausible that there is additional contribution to the unlabeled population from non-cardiomyocytes.
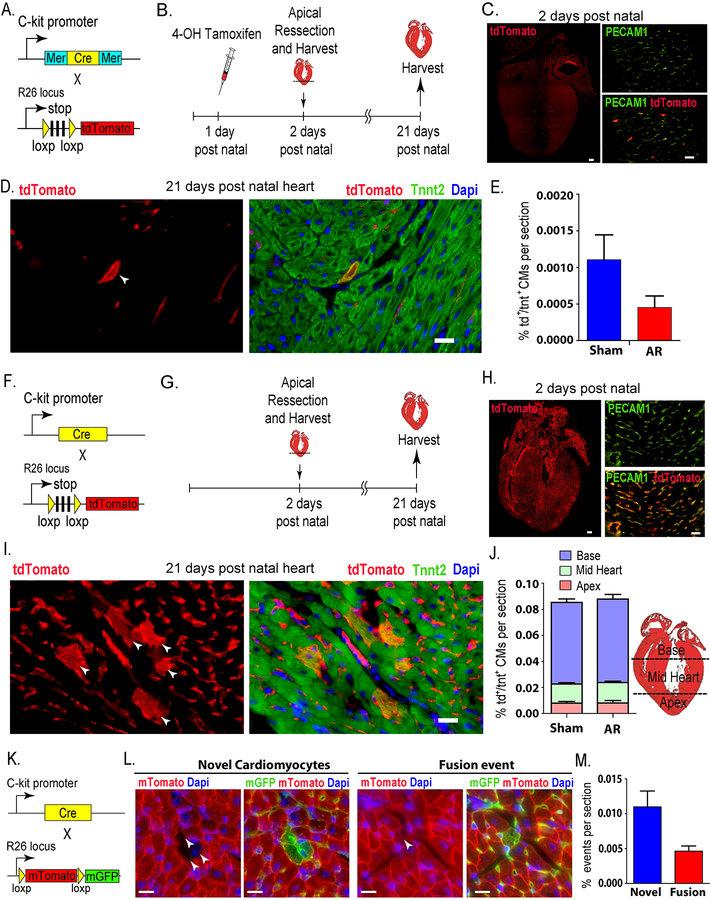
To asses the contribution of C-kit+ cells to cardiomyognesis in the neonatal heart after injury, we first crossed the inducible C-kitMerCreMer/+ mice2 with the Rosa26tdTomato/tdTomato reporter mice (Fig. 1A). One day postnatal mice (P1) were then injected with a single subcutaneous dose of 4-OH tamoxifen (100 μg/pup) to irreversibly label C-kit+ cells and their progeny. Subsequently, apical resection was performed at P2, and the hearts were harvested at P21 (Fig. 1B). All protocols were approved by the Institutional Animal Care and Use Committee of the University of Texas Southwestern Medical Center (UTSW). Aged-matched sham-operated animals were used as control. Hearts harvested at P2 showed an average of ~ 300 tdTomato+ cells per four-chamber view section (>90% of which were PECAM1+)(Fig. 1C). This pattern of colocalization with PECAM1 is consistent with previous observations which suggest that all C-kit+ cells represent a subpopulation of endothelial cells3. Similar to previous reports,1 we found that the LV apex was regenerated by P21. Hearts were then cryosectioned in a four-chamber view, and stained for cardiac troponin T (Tnnt2) and tdTomato. We found that double positive cardiomyocytes (tdTomato+/Tnnt2+) were rarely detected (Fig 1D)., whether in sham or resected hearts, with an average number of 1.08 cell/section & 0.44 cell/section in the sham and resected hearts, respectively. These values indicate that less than 0.0015% of cardiomyocytes originated from a C-kit+ cells (Fig 1E). The percentage of tdTomato+/Tnnt2+ CMs showed no statistically significant difference between sham and resected hearts, demonstrating that cardiac C-kit+ cells do not significantly contribute to cardiomyogenesis after neonatal cardiac injury. Of note, tdTomato+/Tnnt2+ CMs seemed to be preferentially located in the base of the LV rather than the apex in both the sham or the resected group.
Figure 1: Role of C-kit cells in cardiomyogenesis in the neonatal mouse heart.
(A) Schematic representing genetic mouse model of tamoxifen-dependent irreversible labeling of C-kit cells with tdTomato. (B) Time course of fate-mapping experiment. (C) Left panel: P2 whole heart showing ~ 300 tdTomato+ cell/section. Scale bar, 100μm (n=3). Right panel: more than 90% colocalization of tdTomato+ cells with PECAM1 staining. Scale bar, 20μm (D) Left panel: p21 heart (white arrowhead) showing a single tdTomato+ cardiomyocyte, right panel: showing the same cell to be tdTomato+/Tnnt2+. Scale bar, 20μm. (E) Graph showing no statistically significant difference in the percentage of be tdTomato+/Tnnt2+ CMs between sham and resected hearts (Sham: n=5 / AR: n=3). (F) Schematic representing genetic mouse model of continuous labelling of C-kit cells with tdTomato. (G) Time course of fate-mapping experiment. (H) Left panel: P2 whole heart scan showing widespread tdTomato+ signal. Scale bar, 100μm (n=10). Right panel: more than 90% colocalization of tdTomato with PECAM1 staining. Scale bar, 20μm. (I) Left panel: p21 heart showing several arrowheads pointing to several tdTomato+ cardiomyocyte in colony, right panel: showing same cells to be tdTomato+/Tnnt2+. Scale bar, 20μm. (J) Graph showing no statistically significant difference in the percentage of tdTomato+/Tnnt2+ CMs between sham and resected hearts, also differential percentage of the location of the cells in heart compartments (Sham: n=10 /AR: n=10). (K) Schematic representing the genetic cross of constitutive C-kitcre/+ with the reporter mTmG mice to enable detection of fusion events (n=10). (L) Left panels: p21 heart showing novel cardiomyocytes (tdTomato−/GFP+). Right panels: p21 heart showing fusion-derived cardiomyocytes (tdTomato−/GFP+). Scale bars, 20μm. (M) Graph showing percentage of real (novel cardiomyocytes) and fusion events.
Next, C-kitCre/+ mice, where Cre is knocked into the C-kit locus2, were crossed with Rosa26tdTomato/tdTomato reporter (Fig. 1F) and used to continuously label C-kit positive cells and their progeny. This strategy not only allows for detection of C-kit-derived cells during mid gestation onwards, but also labels cells that potentially express C-kit in response to injury irresepective of the presence of tamoxifen. As outlined above, apical resection was performed at P2, and the hearts were harvested at P21 (Fig. 1G). Aged-matched sham-operated animals were used as a control. Hearts harvested at P2 showed widespread tdTomato+ labeling (>90% of which were PECAM1+) (Fig. 1H). This extensive tdTomato+ labeling suggests that the constitutive cre-induced recombination labeled an early endothelial progenitor cell. Moreover, the high rate of co-localization with PECAM1 confirms the endothelial identity of cardiac C-kit+ cells, similar to what was observed in earlier reports3. While there was extensive labeling of tdTomato+ cells throughout the heart sections, less than 0.1% of cardiomyocytes (89 cells and 90 cells/section in sham and resected hearts respectively) were tdTomato+/Tnnt2 CMs (Figs 1I,J). This low percentage of tdTomato+/Tnnt2+ CMs further negates a significant role of c-kit+ cells in cardiomyogenesis. Furthermore, the lack of statistical significance between sham and resected hearts indicates that the neonatal injury does not induce further contribution of C-kit cells to myocardial lineage following neonatal injury (Fig 1J). Of note, tdTomato+/Tnnt2+ CMs were found in colonies with a location preference towards the base of the LV and less frequently in the mid-heart section, and rarely in the apex (Fig 1J).
To determine whether the tdTomato+ cardiomyocytes were generated denovo from C-kit+ cells, we crossed the C-kitcre/+ mice with the membrane-targeted tandem dimer Tomato/membrane-targeted green fluorescent protein (mT/mG) mice to obtain the compound heterozygotes (C-kitcre/+; membrane-targeted tandem dimer Tomato/membrane-targeted green fluorescent protein (mT/mG)) (Fig 1K). P21 hearts were then harvested,sectioned and examined by immunofluorescence. The percentages of tdTomato−/GFP+ novel cardiomyocytes and tdTomato+/GFP+ fusion-derived cardiomyocytes were about 0.01% and 0.005% respectively (Fig 1L,M). Expectedly, the average number of both the tdTomato−/GFP+ cells (novel cardiomyocytes) together with the tdTomato+/GFP+ cells (fusion events) was modestly lower than the number of tdTomato+/Tnnt2+ CMs observed using the tdTomato reporter (Fig 1a) as previously reported3.
While a recent report suggested that C-kit cells may significantly contribute to neonatal heart regeneration4, the genetic model used in that study did not allow for lineage tracing, and the cryoinjury model used may have played a role in the discrepant results since cryoinjury is known to result in incomplete regeneration5. Taken together, our results indicate that C-kit cells do not significantly contribute to cardiomyogenesis during early postnatal cardiac growth, or during neonatal heart regeneration following apical resection in mice.
Acknowledgments
Funding Sources:
H.S. is supported by grants from the NIH (1R01HL115275 and 5R01H2131778), National Aeronautics and Space Administration (NNX-15AE06G), American Heart Association (16EIA27740034), Cancer Prevention and Research Institute of Texas (RP160520), Hamon Center for Regenerative Science and Medicine, and Fondation Leducq.
Footnotes
Data Sharing:
The data, analytic methods, and study materials will be/have been made available to other researchers for purposes of reproducing the results or replicating the procedure by contacting the corresponding author.
Disclosures:
None
References
- 1.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Berlo JH, Kanisicak O, Maillet M, Vagnozzi RJ, Karch J, Lin SC, Middleton RC, Marban E, Molkentin JD. C-kit+ cells minimally contribute cardiomyocytes to the heart. Nature. 2014;509:337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sultana N, Zhang L, Yan J, Chen J, Cai W, Razzaque S, Jeong D, Sheng W, Bu L, Xu M, Huang GY, Hajjar RJ, Zhou B, Moon A, Cai CL. Resident c-kit(+) cells in the heart are not cardiac stem cells. Nat Commun. 2015;6:8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jesty SA, Steffey MA, Lee FK, Breitbach M, Hesse M, Reining S, Lee JC, Doran RM, Nikitin AY, Fleischmann BK, Kotlikoff MI. C-kit+ precursors support postinfarction myogenesis in the neonatal, but not adult, heart. Proc Natl Acad Sci U S A. 2012;109:13380–13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lam TA, Sadek HA. Neonatal heart regeneration: Comprehensive literature review. Circulation. 2018;138:412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]