Abstract
Ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) is considered to be the most abundant enzyme on Earth. Despite this, its full diversity and distribution across the domains of life remain to be determined. Here, we leverage a large set of bacterial, archaeal, and viral genomes recovered from the environment to expand our understanding of existing RuBisCO diversity and the evolutionary processes responsible for its distribution. Specifically, we report a new type of RuBisCO present in Candidate Phyla Radiation (CPR) bacteria that is related to the archaeal Form III enzyme and contains the amino acid residues necessary for carboxylase activity. Genome-level metabolic analyses supported the inference that these RuBisCO function in a CO2-incorporating pathway that consumes nucleotides. Importantly, some Gottesmanbacteria (CPR) also encode a phosphoribulokinase that may augment carbon metabolism through a partial Calvin–Benson–Bassham cycle. Based on the scattered distribution of RuBisCO and its discordant evolutionary history, we conclude that this enzyme has been extensively laterally transferred across the CPR bacteria and DPANN archaea. We also report RuBisCO-like proteins in phage genomes from diverse environments. These sequences cluster with proteins in the Beckwithbacteria (CPR), implicating phage as a possible mechanism of RuBisCO transfer. Finally, we synthesize our metabolic and evolutionary analyses to suggest that lateral gene transfer of RuBisCO may have facilitated major shifts in carbon metabolism in several important bacterial and archaeal lineages.
Keywords: Candidate Phyla Radiation, DPANN archaea, carbon metabolism, lateral gene transfer, RuBisCO, phosphoribulokinase, nucleotide metabolism
Introduction
Forms I and II ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) are central to carbon fixation via the Calvin–Benson–Bassham (CBB) cycle in algae, plants, and some bacteria. Forms III and II/III RuBisCO, discovered in Archaea, are believed to add CO2 to ribulose-1,5-bisphosphate (RuBP) in a two-step reaction from nucleotides like adenosine monophosphate (AMP) (Sato et al. 2007; Aono et al. 2015). These “bona fide” RuBisCO enzymes were historically considered to be domain specific. In contrast, RuBisCO-like proteins (RLPs; Form IV) in both Bacteria and Archaea perform functions distinct from carbon fixation and may be involved in methionine salvage, sulfur metabolism, and d-apiose catabolism (Tabita et al. 2008; Carter et al. 2018). Although the origin of the RuBisCO superfamily is still unclear (Ashida et al. 2005; Tabita et al. 2007; Erb and Zarzycki 2018), phylogenetic analysis and enzymatic characterization have suggested that the modern distribution of “bona fide” RuBisCO can be explained by vertical and lateral transfer from an archaeal, Form III ancestor (Tabita et al. 2007).
Recently, metagenomic studies of diverse environments have introduced additional complexity to evolutionary considerations by uncovering new RuBisCO diversity. First, new genomes from Candidate Phyla Radiation (CPR) bacteria and Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanoarchaeota, and Nanohaloarchaeota (DPANN) archaea (a group originally defined based on the lineages DPANN, but now including others) contain a hybrid II/III RuBisCO similar to that found in the archaeon Methanococcoides burtonii (Wrighton et al. 2012; Castelle et al. 2015). One version of this enzyme was shown to complement photoautotrophic growth in a bacterial RuBisCO deletion strain (Wrighton et al. 2012, 2016). An additional RuBisCO form related to the archaeal Form III, called the “III-like,” was reported from genomes of other CPR and some DPANN archaea, further expanding enzyme diversity in these groups (Castelle et al. 2015; Wrighton et al. 2016; Castelle and Banfield 2018). The III-like enzyme is also predicted to function in the CO2-incorporating AMP pathway (Wrighton et al. 2016).
The presence of RuBisCO in some CPR bacteria and DPANN archaea was interesting because these organisms have small genomes and lack many core metabolic functions. The common absence of pathways for synthesis of nucleotides, amino acids, and lipids has led to speculation that they live symbiotic or syntrophic lifestyles (Wrighton et al. 2012; Brown et al. 2015). Despite this, recent research has shown that some members have a wide range of fermentative capabilities and the potential to play roles in carbon, nitrogen, sulfur, and hydrogen cycling (Wrighton et al. 2012; Danczak et al. 2017). A similar putative ecology has been suggested for the DPANN archaea (Castelle et al. 2015). The recovery of RuBisCO, functioning in an AMP pathway, expanded possible metabolic modes for some of these organisms and suggested that CPR and DPANN encoding this enzyme could derive energy and/or resources from ribose produced by other community members (Wrighton et al. 2016; Castelle and Banfield 2018).
The discovery of the reductive hexulose pathway provided new insight into the variety of ways that RuBisCO can be configured for sugar metabolism. The reductive hexulose pathway, differing only in a few steps from the CBB cycle, also employs RuBisCO along with phosphoribulokinase (PRK) to regenerate RuBP and fix carbon dioxide in some methanogenic archaea (Kono et al. 2017). In light of these discoveries, the full diversity, distribution, and possible functionality of RuBisCO in divergent groups like the CPR/DPANN remain an open and important question. In the last few years, additional work has recovered CPR and DPANN genomes from a much wider array of environmental types, including additional groundwater locations, the deep subsurface, hydrocarbon-impacted environments, and the ocean (Hernsdorf et al. 2017; Parks et al. 2017; Tully et al. 2018). Here, we examined over 300 genomes from metagenomes from these environments to further elucidate diversity, potential functions, and the evolutionary history of RuBisCO in members of the CPR bacteria and DPANN archaea. First, we expand the distribution of forms to new phylogenetic groups and propose a new type that, in some cases, might act in concert with PRK to augment carbon metabolism. Additionally, we describe a clade of putative RLPs encoded by bacteriophage from diverse environments. Drawing on these observations and previous analyses, we suggest that lateral gene transfer may have been largely responsible for the distribution of this enzyme among the CPR/DPANN. These lateral transfers could, in the presence of genes from other pathways, introduce new RuBisCO-based metabolic capacity to genomically reduced lineages.
Results
Metagenomics Expands the Diversity of RuBisCO Forms and Reveals Putative New Enzyme Types in CPR and Phage
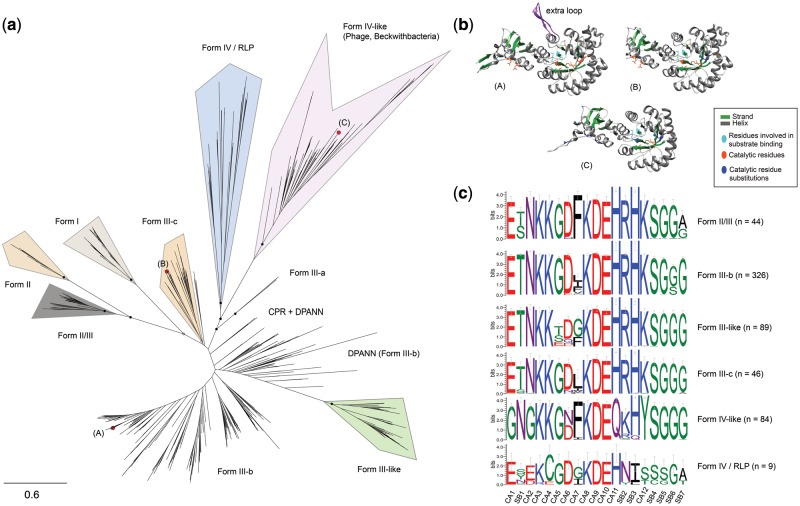
The large majority of CPR and DPANN RuBisCO sequences analyzed fell into clearly defined phylogenetic groups (fig. 1a), four of which (II/III, III, III-like, and IV) correspond to the “Forms” defined in previous literature. A small number of sequences from both the CPR and DPANN (including the generically labeled branches in fig. 1a) resolved in ambiguous phylogenetic positions, largely due to low support for internal nodes. Among the clearly defined groups, we observed the following results:
Fig. 1.
Known RuBisCO diversity is expanded by metagenomics. (a) Maximum-likelihood tree for dereplicated RuBisCO large-chain sequences, delineated by “Form.” (A), (B), and (C) show the phylogenetic position of protein sequences modeled in (b). Scale bar represents the number of substitutions per site. Closed black circles indicate bootstrap support values >70%, whereas open circles represent those >50%. For simplicity, bootstrap support below form level is not shown. (c) Aggregate sequence logos for CPR, DPANN, and phage protein sequences in each phylogenetic “Form,” describing key catalytic activity (CA) and substrate binding (SB) residues in RuBisCO sequences. Logo colors represent residues with similar chemical properties.
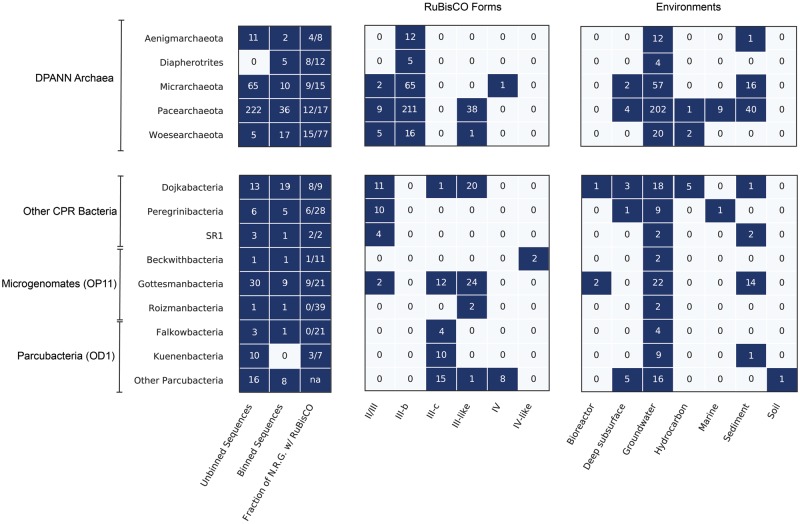
II/III. Our analysis recovered new sequences of the Form II/III RuBisCO, originally thought to be exclusively found in Archaea (Alonso et al. 2009). Here, we broaden the phylum-level distribution of the group with additional representatives from the DPANN groups Woesearchaeaota and Micrarchaeota (fig. 2). Form II/III RuBisCO of CPR and DPANN partition into two subgroups based on the presence or absence of a 29 amino acid (or longer) insertion, the biochemical implications of which are currently unknown (Wrighton et al. 2016). All but one of the full-length Micrarchaeota and Woesearchaeota sequences recovered in this study contained insertions in the expected region.
Fig. 2.
RuBisCO diversity among the CPR bacteria and DPANN archaea. Boxes represent counts of dereplicated protein sequences used in the phylogenetic analysis (Unbinned and Binned Sequences) as well as the number of those sequences that fell into described RuBisCO forms and environments. Fraction of nonredundant genomes (N.R.G.) with RuBisCO describes the proportion of genomes per phylum encoding RuBisCO after dereplication at 99% Average Nucleotide Identity (see Materials and Methods).
III. We identified archaeal Form III RuBisCO sequences in a variety of DPANN, and, potentially, CPR genomes. We refer to this as Form III-b to distinguish it from Form III-a, a divergent group described by Kono et al. (2017), and the Form III-like proteins (Wrighton et al. 2016). Most of the newly reported Form III-b sequences not only contained the critical substrate binding and catalytic residues for RuBisCO function but also largely shared residue identity with canonical archaeal versions (fig. 1c). Notably, we found this enzyme form in three DPANN groups—the Diapherotrites, the Micrarchaeota, and the Woesearchaeota—previously not known to harbor Form III-b RuBisCO, extending the presence of these enzymes beyond Pacearchaeota and Aenigmarchaeota (Castelle et al. 2015), two major groups in the DPANN (fig. 2). Approximately, 70 DPANN sequences fell outside the clade containing characterized Form III-b but were assigned to this type given low support for separating branches and overall closer relatedness to III-b than III-like sequences (DPANN [Form III-b], fig. 1a).
We identified several previously published sequences assigned to genomes from the Levybacteria and Amesbacteria phyla (CPR superphylum Microgenomates) deeply nested within the archaeal Form III-b sequences. In addition, we recovered a set of unbinned sequences with highest similarity to these same binned Levybacteria and Amesbacteria genomes. The Levybacteria sequences group loosely with sequences from Aenigmarchaeota and a reference sequence from the archaeon Methanoperedens nitroreducens (∼80% identity). In contrast, the Amesbacteria sequences grouped most closely to those from a clade from Pacearchaeota. To verify the binning of these genome fragments, we examined the top Basic Local Alignment Search Tool (BLAST) hits to well-annotated genes on each genome fragment bearing a RuBisCO gene. For the fragment putatively assigned to Amesbacteria, BLAST affiliation of nonhypothetical genes was inconclusive—some genes had only low identity with archaeal genes, others had identity to both bacterial and archaeal genes, whereas one gene encoding a serine acetyltransferase had ∼70% bacterial identity. In addition to RuBisCO, the ∼34-kb putative Levybacteria genome fragment encoded a large CRISPR-Cas locus, a novel transposase, and a phage/plasmid primase, all of which may indicate mobile genetic material. BLAST affiliation of well-annotated genes was also inconclusive for this fragment—if not of bacterial origin, the genome fragment could be phage or plasmid. We additionally recovered one small genomic fragment, possibly related to the Moisslbacteria, that also included the III-b enzyme. Ultimately, further research is needed to confirm that Form III-b RuBisCO is encoded in genomes of some CPR bacteria.
III-like. Many RuBisCO sequences fell into a deep-branching, monophyletic clade that is divergent from the archaeal Form III and referred to as “III-like” (Wrighton et al. 2016) (fig. 1a). Here, we added five Dojkabacteria (WS6, a group within the CPR) sequences from hydrocarbon-impacted environments and nearly 40 DPANN sequences from Woesearchaeota and Pacearchaeota from multiple environments (fig. 2). As reported previously (Wrighton et al. 2016), the “III-like” sequences recovered in this analysis appear to have insertions mostly 11 amino acids in length.
III-c. We report here an additional deep-branching clade of RuBisCO sequences with overall sequence similarity closest to Form III-b RuBisCO (∼50%) (fig. 1a). Although support was low for internal branches separating this clade from III-b sequences, best maximum-likelihood trees suggested a distinct phylogenetic status. This group, which we term “Form III-c,” contained nearly 50 sequences from the Dojkabacteria and several groups in the Microgenomates and Parcubacteria superphyla. Form III-c was particularly abundant in the Gottesmanbacteria and Kuenenbacteria, the latter of which appears to harbor this form exclusively (fig. 2). We identified organisms bearing this enzyme type in almost every environment included in our study, indicating that it may compose an important and previously unrecognized aspect of RuBisCO diversity. To predict the biochemical potential of this type, we examined 12 residues known to be important for carboxylase activity and 7 important for substrate binding from each sequence (Tabita et al. 2008; Saito et al. 2009). This analysis indicated that Form III-c RuBisCO sequences contain critical residues largely identical to those reported for Form III-b enzymes; however, a minority of sequences encoded modifications to the conserved aspartic acid in catalytic site #6 (CA6) that could alter its chemical properties (fig. 1c). Additionally, modeling through I-TASSER revealed that secondary structure of an III-c sequence was consistent with that of existing Form III-b templates (fig. 1b).
IV and IV-like. Form IV (RLP) proteins are a clade of highly divergent RuBisCO with low sequence similarity (∼30%) to “bona fide” RuBisCO enzymes and divergent functions (Hanson and Tabita 2001; Tabita et al. 2008). Our phylogenetic analyses identified eight Form IV (RLP) RuBisCOs in CPR genomes, all in the genomes of bacteria from the Parcubacteria superphylum (fig. 2). The Parcubacteria Form IV sequences made up of a monophyletic clade nesting within the previously described IV-Photo type RLP. However, the key catalytic/substrate binding residues were only partially conserved (fig. 1c). We recovered only one RLP from a Micrarchaeota (DPANN) genome (fig. 2), despite the fact that archaeal sequences appear to be at the root the RuBisCO/RLP superfamily (Tabita et al. 2007).
Using manually curated Hidden Markov Models (HMMs) constructed from recovered CPR and DPANN RuBisCO sequences, we also identified approximately 80 nonredundant sequences related to Form IV RuBisCO on putative phage as well one unusually large, curated phage genome (∼200 kb) (supplementary fig. S1a, Supplementary Material online). The phage sequences appeared highly divergent from other RLPs, but most shared some key residues found in canonical Form IV RuBisCOs (fig. 1c) and scored highly on HMMs constructed from verified Form IV RuBisCO (score > 200, e-val ≪ 0.05). Additionally, modeling of one sequence revealed a secondary structure consistent with large-chain templates but missing several conserved residues, as expected for Form IV enzymes (fig. 1b). Analysis of coencoded phage terminase proteins indicated that at least some of the viral sequences were from members of the Myoviridae. Two other RLP-related sequences attributed to Beckwithbacteria grouped with those from putative/confirmed phage and together appeared as an outgroup to all other RLPs (fig. 1a). Analysis of the genes encoded on the manually curated Beckwithbacteria fragment with IV-like RuBisCO supported its bacterial origin (supplementary fig. S1b, Supplementary Material online).
Form III-c RuBisCO Is Encoded in Close Proximity to Genes of the CO2-Incorporating AMP Pathway
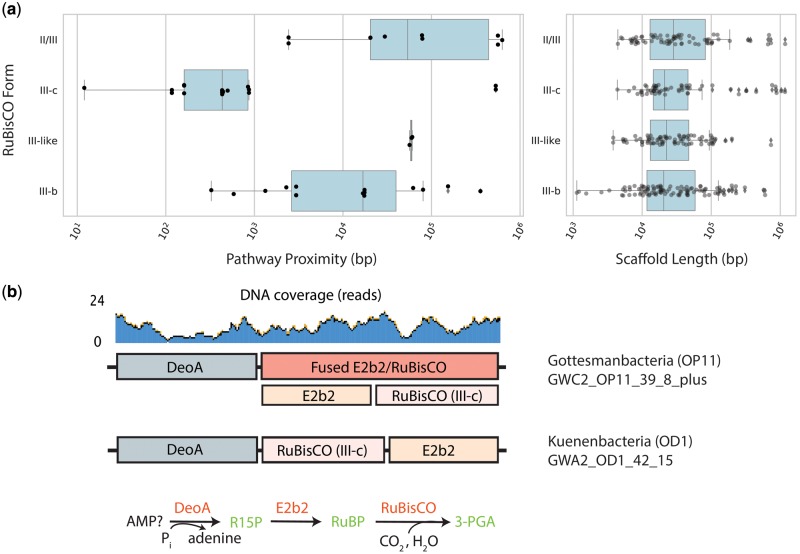
To test the hypothesis that the Form III-c RuBisCO participates in the previously described AMP pathway, we narrowed our focus on the binned genomes containing this form of the enzyme. The AMP pathway employs AMP phosphorylase (deoA) and ribose-1,5-bisphosphate (R15P) isomerase (e2b2) to provide RuBisCO with RuBP substrate, incorporating CO2 to produce two 3-phosphoglyceric acid molecules (3-PGA) which are fed into glycolysis (Sato et al. 2007; Aono et al. 2015). Thirty-nine genomes encoding Form III-c RuBisCO (67%) also contained homologs for e2b2 and deoA. Most Gottesmanbacteria (OP11) contained homologs to deoA at a threshold lower than originally used in the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, as these sequences generally had a conserved deletion of ∼80 amino acids at the start of the protein compared with other CPR proteins. Despite this, we predict that these proteins are divergent deoA homologs based on phylogenetic placement and realignment with reference sequences. Among Gottesmanbacteria genomes, the percentage containing all three genes associated with AMP metabolism was higher (∼91%). Thus, the new Form III-c RuBisCO, especially among Gottesmanbacteria, is consistently associated with deoA and e2b2 homologs, as reported previously for CPR genomes with other forms of the enzyme (Wrighton et al. 2016) (supplementary fig. S2a, Supplementary Material online).
To analyze possible function of the newly recovered RuBisCOs, we examined the genomic proximity of deoA and e2b2 homologs with RuBisCO in all genomes containing Form III-b, III-c, III-like, and II/III enzymes. Forty-four genomes contained fragments encoding all three genes on the same stretch of assembled DNA (supplementary fig. S2b, Supplementary Material online). Most fragments encoded RuBisCO between the other genes, although a minority of genomes appeared to contain rearrangements (supplementary fig. S2c, Supplementary Material online), as previously reported for a Form II/III-bearing PER-1 genome (Wrighton et al. 2016). Next, for cases where all three genes occurred on the same contig, we defined a metric called “pathway proximity” (sum of the genomic distance between the three genes, see Materials and Methods). This analysis suggested that genes involved in AMP metabolism are more frequently and more proximally colocated in the genomes of organisms bearing the Form III-c RuBisCO than in genomes with other forms, even though these RuBisCO were present on assembled fragments of similar length (fig. 3a). Two outlier Form III-c genomes from a previous study (Parks et al. 2017) encoded extremely distant deoA genes on long fragments, possibly the result of genetic rearrangement or errors in genome assembly. Interestingly, some genomes bearing Form III-c RuBisCO encoded the three genes consecutively, where in other cases the RuBisCO gene was fused to the isomerase (fig. 3b and supplementary fig. S3, Supplementary Material online). The close proximity and fusion support the conclusion that this RuBisCO form participates in the CO2-incorporating AMP pathway.
Fig. 3.
Genomic context of Form III-c RuBisCO in CPR and DPANN. (a) Proximity of genes on genome fragments encoding all three components of the CO2-incorporating AMP pathway, and the length of all binned fragments containing the specified RuBisCO form. (b) Genomic diagram of AMP components in CPR genomes with fused RuBisCO and consecutive ordering of genes on the chromosome.
RuBisCO Phylogeny Is Incongruent with That of e2b2 and deoA
The discovery of Form III-like, and now III-c, RuBisCO in the CPR bacteria suggested that lateral gene transfer may have played a role in shaping the distribution of Form III-related enzymes across the tree of life (Wrighton et al. 2016; Erb and Zarzycki 2018). To evaluate the extent of lateral gene transfer involving the genes of the AMP pathway, we compared the phylogeny of RuBisCO with that of the other pathway components (Wrighton et al. 2016). With the possible exception of the Peregrinibacteria, we found that the trees for CPR and DPANN AMP phosphorylase (deoA) and R15P isomerase (e2b2) recapitulate organism phylogeny and so were largely incongruent with that of RuBisCO (supplementary fig. S4, Supplementary Material online). Among the monophyletic Gottesmanbacteria, genomes with the same RuBisCO Form (III-c and III-like) appeared to cluster together, as would be expected if lateral gene transfer of various RuBisCO followed vertical inheritance of deoA and e2b2 by different subphylum-level lineages. Although still undersampled, there is some indication that RuBisCO-correlated phylogenetic clustering will emerge for Parcubacteria (OD1) and Dojkabacteria (WS6), which at the phylum level contain high RuBisCO diversity.
CPR Bacteria Bearing Form III-c RuBisCO Also Encode Phosphoribulokinase
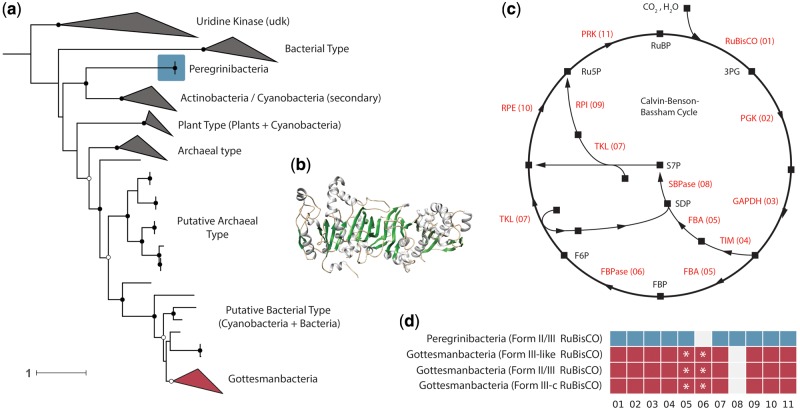
We examined whether recovered CPR RuBisCO, including the Form III-c, might participate in a form of the CBB pathway by searching all binned genomes for homologs of PRK. PRK is a key CBB marker gene that is critical for regenerating RuBP substrate. Our analysis recovered 31 genomes encoding PRK homologs among the Gottesmanbacteria and Peregrinibacteria, the first PRK homologs reported in the CPR. PRK sequences were encoded by Gottesmanbacteria harboring Form III-like and III-c RuBisCO, whereas Peregrinibacteria PRK were associated with the typical Form II/III enzyme. Phylogenetic analysis revealed that Gottesmanbacteria PRK sequences comprised a well-supported monophyletic clade nesting within a group of sequences from recently isolated cyanobacterial genomes and, more broadly, a larger clade of archaeal PRK homologs (fig. 4 and supplementary fig. S5, Supplementary Material online). These putative archaeal and cyanobacterial sequences appeared to be distinct from classical versions and have not yet been assayed for functional activity. Similarly, two recovered Peregrinibacteria PRK formed a monophyletic clade sister to additional divergent cyanobacterial sequences (fig. 4 and supplementary fig. S5, Supplementary Material online). To test whether CPR genomes containing PRK have the full genomic repertoire for the CBB cycle, we searched genomic bins for nine other genes involved in this pathway (supplementary table S2, Supplementary Material online). Several Gottesmanbacteria bins contained near-complete CBB pathways, lacking only the gene for sedoheptulose-1,7-bisphosphatase (SBPase, 08 in fig. 4). Additionally, instead of separate genes for fructose-1,6-bisphosphate aldolase (FBA) and fructose-1,6-bisphosphatase (FBPase), most Gottesmanbacteria genomes encoded an enzyme most similar to a bifunctional version found in some thermophilic, chemoautotrophic bacteria and archaea (Say and Fuchs 2010) (fig. 4 and supplementary fig S7, Supplementary Material online). One Peregrinibacteria genome contained all genes involved in the CBB cycle with the exception of FBPase (06), although additional genome sampling/reconstruction is necessary to definitively designate this enzyme as missing.
Fig. 4.
Some CPR bacteria encode a putative PRK. (a) Maximum-likelihood tree showing phylogenetic position of putative PRK in CPR phyla. Scale bar represents the number of substitutions per site. Closed black circles indicate bootstrap support values >70%, whereas open circles represent those >50%. See supplementary fig. S5, Supplementary Material online, for fully labeled tree. (b) Example protein model of putative PRK in the CPR phylum Gottesmanbacteria. (c) Schematic of the CBB cycle. Squares represent molecular intermediates, whereas arrows represent enzymatic steps. Abbreviations: PGK, phosphoglycerate kinase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; TIM, triosephosphate isomerase; FBA, fructose-bisphosphate aldolase; FPBase, fructose-1,6-bisphosphatase; TKL, transketolase; SBPase, sedoheptulose-bisphosphatase; RPI, ribose-5-phosphate isomerase; RPE, ribulose 5-phosphate 3-epimerase. (d) Genomic repertoires of CPR bacteria encoding PRK. Numbers refer to enzymatic steps in (c). Asterisks indicate genomes that harbor an enzyme with highest homology to a bifunctional FBA/FBPase instead of separate FBA and FBPase.
Discussion
New Diversity in the RuBisCO Superfamily and Its Implications for Phylogenetic Distribution and Metabolic Function among the CPR and DPANN
Through increased metagenomic sampling of diverse environments, we provide new information about the distribution of RuBisCO in major bacterial and archaeal groups and expand RuBisCO superfamily diversity. Our results also allow a quantitative assessment of RuBisCO diversity, revealing that the Pacearchaeota and Dojkabacteria in particular frequently encode various forms of the enzyme across many environmental types (fig. 2). This suggests that RuBisCO may be an important metabolic enzyme for these groups, which appear to have the most minimal metabolic and biosynthetic capacities among the DPANN and CPR radiations (Castelle and Banfield 2018).
At present, Form III-c sequences occur only in several CPR lineages, contrasting with other Form III-related enzymes with archaeal representatives. Specifically, Form III-a is only known in methanogens, Form III-b is known to occur in archaea and possibly several CPR lineages, as well as another bacterium (Ammonifex degensii) (Berg et al. 2010), and Form III-like enzymes appear to be widely (but sparsely) distributed in both DPANN archaea and CPR bacteria. However, like the other Form III-related enzymes, the association of Form III-c RuBisCO with e2b2 and deoA suggests this enzyme may also function in an AMP metabolism pathway. This pathway, originally described for the Form III-b enzyme in Thermococcus kodakarensis, relies on two proteins to provide RuBisCO with its substrate molecule, ribose-1,5-bisphosphate (Sato et al. 2007; Aono et al. 2015). First, an AMP phosphorylase encoded by the deoA gene catalyzes the release of ribose-1,5-bisphosphate (R15P) which is then subsequently converted to RuBP by a R15P isomerase encoded by e2b2 (Sato et al. 2007; Aono et al. 2015). Next, the RuBisCO incorporates H2O and CO2 with this substrate to create two molecules of 3-phosphoglycerate (3-PGA), which in turn can be diverted into central carbon metabolism (Sato et al. 2007; Aono et al. 2015). Among the CPR, this pathway is thought to provide a simple mechanism for ribose salvage that may facilitate the syntrophic ecology of these organisms (Wrighton et al. 2016; Castelle and Banfield 2018). Contig-level analyses revealed a notable spatial association and occasional fusion of genes involved in AMP metabolism in genomes bearing the Form III-c RuBisCO, supporting the association of the enzyme with this pathway. However, it is critical that future studies characterize the specific biochemistry of this new form and its possible function in CPR bacteria. Although residue analysis of the Form III-c RuBisCO suggests that these enzymes encoded the minimum set of catalytic and substrate binding sites necessary for carboxylase activity, the associated metal cation and the impact of nonactive site catalysis remain unknown.
Form I and Form II RuBisCO function in the CBB cycle, which relies on PRK to regenerate RuBP before carbon fixation by RuBisCO (fig. 4). The presence of PRK in Gottesmanbacteria raises the possibility that a CBB-like pathway may operate in carbon assimilation in these organisms. Lacking from their genomic repertoires, however, is SBPase (fig. 4). In plants, SBPase catalyzes the dephosphorylation of sedoheptulose-1,7-bisphosphate and is important for regulation of intermediate molecules in the CBB cycle (Harrison et al. 1997). Among Cyanobacteria, it has been shown that a single enzyme often functions as both an SBPase and an FBPase, catalyzing a similar reaction on fructose bisphosphate in the second branch of the cycle (fig. 4) (Gerbling et al. 1986; Feng et al. 2014). Similarly, bifunctional activity has been demonstrated for other bacterial FBPase enzymes in both the CBB (Ralstonia eutropha) and a ribulose monophosphate cycle (Bacillus methanolicus) (Yoo and Bowien 1995; Stolzenberger et al. 2013). Complicating the possibility of a bifunctional FBPase/SBPase in the Gottesmanbacteria is the observation that most of these genomes encode a single enzyme most similar to a bifunctional FBA/FBPase, instead of separate FBPase and FBA. In the archaeal and bacterial lineages in which bifunctional FBA/FBPases have been characterized, these enzymes are thought to play a role in gluconeogenesis instead of the CBB pathway (Say and Fuchs 2010). Thus, the association of this gene in the classical CBB in Gottesmanbacteria would require tripartite function as a FBPase, FBA, and SBPase. As such, the functioning of a CBB-like pathway in CPR remains uncertain.
An alternative inference is that PRK contributes to carbon metabolism in Gottesmanbacteria by providing additional RuBP as substrate for RuBisCO functioning in the AMP pathway. The same may be true of the two Peregrinibacteria genomes encoding the PRK but missing FBPase (fig. 4). In this scenario, components of the oxidative pentose phosphate pathway could convert glucose-6P into ribulose-5-P, which could then be converted to RuBP by the PRK (supplementary fig. S6, Supplementary Material online). Going forward, it is critical that the PRK from both lineages, as well as the Gottesmanbacteria FBPase/FBA, be characterized biochemically, especially given that these sequences are divergent from well-studied enzymes (supplementary fig. S7, Supplementary Material online). In any case, the presence of PRK, RuBisCO, and a putative bifunctional FBPase/FBA in CPR genomes suggests that these organisms may have acquired fundamental components of carbon metabolism by lateral transfer. This extends the prior observations of transfer of the bifunctional FBA/FBPase to bacteria (Say and Fuchs 2010) as well as the general occurrence of transfer among CPR bacteria (Jaffe et al. 2016).
Finally, we report Form IV RLPs in the genomes of several CPR bacteria and a DPANN archaeon. Previously described RLPs fall into six clades and have distinct patterns of active site substitutions that likely affect their functionality (Tabita et al. 2007). Sequences from Parcubacteria and Micrarchaeota were mostly closely related to a known RLP clade called the IV-Photo, which is implicated in sulfur metabolism/stress response in green sulfur bacteria like Chlorobium tepidum (Hanson and Tabita 2001). Although active site residue divergence complicates the inference of function based on phylogenetic placement, it is possible that Form IV sequences found in CPR bacteria also function in oxidative stress response. Our analysis also revealed the presence of a large, highly divergent clade of RuBisCO sequences related to Form IV/RLP in the genomes of bacteriophage (fig. 1a), some of which were classified as Myoviridae. That these sequences were recovered from various environments suggests that these phage proteins may be a widespread and to date underappreciated reservoir of diversity in the RuBisCO superfamily. Sequence analysis revealed that these putative RLPs encoded key residues divergent from known type IVs, leaving possible functionality unclear. However, future work may uncover “bona fide” RLPs on phage genomes, supporting the inference that these enzymes are widely laterally transferred across lineages. Previous work has shown that marine phage can impact host carbon metabolism through auxiliary expression of other photosynthetic genes (Thompson et al. 2011; Crummett et al. 2016). Regardless of type, phage-associated proteins with homology to RuBisCO should be assessed functionally to evaluate whether they have to the potential to augment host metabolism during infection.
Sparse Distribution of the RuBisCO Superfamily Suggests That Lateral Gene Transfer Shapes the Distribution of Multiple Forms among CPR, DPANN, and Bacteriophage
Regardless of the ancestral function of RuBisCO superfamily (Ashida et al. 2005; Tabita et al. 2007; Erb and Zarzycki 2018), ancient lateral gene transfer is likely an important process underlying the current distribution of RuBisCO in bacteria and archaea. Transfers probably drove the evolution of Form IV as well as Forms I and II from the ancestral Form III (Tabita et al. 2007, 2008), ultimately resulting in diverse RuBisCO types that now occur in Archaea, Bacteria, and Eukaryotes. Results of the current study support this inference and extend it, suggesting that lateral gene transfer has also played a role in distributing recently recognized RuBisCO forms across Archaea and Bacteria. First, the discovery of Form III-c RuBisCO in CPR bacteria suggests gene transfer between CPR bacteria and archaea, as this new form is most closely related to the archaeal Form III-b. The recovery of canonical archaeal Form III-b proteins within previously published Amesbacteria and Levybacteria bins (both CPR), if verified, would support this conclusion. Additionally, previous findings of a Dojkabacteria (WS6) genome harboring both Form III-like and Form II/III RuBisCO and a divergent Form III-b RuBisCO enzyme in the Firmicute A. degensii are best explained by gene acquisition via lateral transfer (Berg et al. 2010; Hernsdorf et al. 2017).
Broader evolutionary patterning supports the idea that lateral gene transfer has played an underappreciated role in the shaping the evolution of RuBisCO among CPR and DPANN. Our results reveal a relatively wide but sparse distribution of RuBisCO across CPR/DPANN lineages, with most lineages containing very low frequencies of the enzyme (fig. 2). The noncongruency of RuBisCO phylogeny with those of deoA and e2b2, which appear to have been largely vertically transmitted in CPR lineages (supplementary fig. S4, Supplementary Material online), suggests divergent evolutionary histories of these functionally related genes. Thus, we conclude that the distribution of RuBisCO diversity in the CPR/DPANN is more likely to be explained by lateral transfer than extensive gene loss.
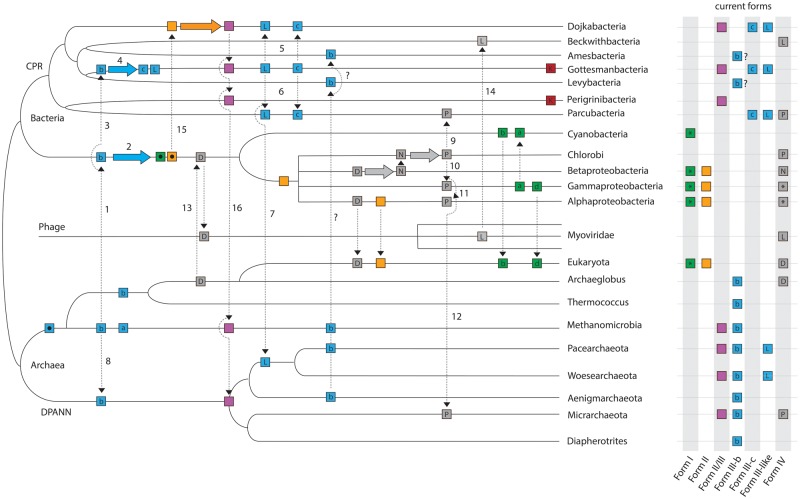
The results presented in this study give new insights on the evolution of the RuBisCO superfamily as a whole. Figure 5 is a schematic diagram that integrates ideas of Tabita et al. (2007) with inferences arising from our results, detailing one of at least several possible scenarios by which RuBisCO was distributed across the tree of life. Previous work has suggested that an archaeon, possibly an ancestor of the Methanomicrobia, laterally transferred a Form III enzyme to a bacterial ancestor (Step 1 in fig. 5) where it subsequently evolved to generate both the Form I and Form II enzymes (Step 2) (Tabita et al. 2007; Schönheit et al. 2016). The findings of the current and prior studies (Tabita et al. 2007; Wrighton et al. 2016) indicate that an ancestral Form III sequence may then have diverged from a common ancestor into at least three Form III-related types, including the Form III-b (traditional archaeal form). Specifically, an additional transfer of an ancestral Form III or III-b enzyme from Archaea to a CPR bacterium may have led to the evolution of the III-like and newly reported III-c Forms (Steps 3 and 4), as previously hypothesized (Erb and Zarzycki 2018). Subsequent transfers of both forms to the Dojkabacteria (Step 5), Parcubacteria (Step 6), and of Form III-like to the common ancestor of Pacearchaeota and Woesearchaeota (Step 7) would recapitulate the current distribution of these enzymes across the tree of life. However, with the current evidence, we cannot rule out the possibility that the III-like RuBisCO evolved within DPANN archaea and was transferred in the reverse direction to the CPR. Interestingly, Form III-like enzymes occur only in some CPR lineages and DPANN archaea, without any known representatives outside these radiations. Many DPANN lineages also encode the classical archaeal Form III-b, currently thought to have originated in the Methanomicrobia (Tabita et al. 2007). Recent phylogenomic studies of the Archaea have inferred a root in between DPANN and all other groups (Williams et al. 2017), requiring a transfer of Form III-b to DPANN from another lineage to explain this form’s extant distribution (Step 8).
Fig. 5.
Conceptual diagram illustrating the role of lateral gene transfer in the evolution of RuBisCO and PRK (K). Distinct RuBisCO forms are represented by boxes of different colors, and form subtypes are indicated by a letter within the box if applicable. Asterisks indicate multiple subtypes. Form III enzymes are expanded for additional clarity and are abbreviated as follows: a, Form III-a; b, Form III-b; c, Form III-c; L, Form III-like. Form IV abbreviations are as follows: D, DeepYkr; N, NonPhoto; P, Photo (see Tabita et al., 2007); L, Form IV-like (this study). Ancestral sequences are represented by black dots inside boxes. Dotted arrows represent possible lateral gene transfers, solid colored arrows represent evolution of RuBisCO within a lineage. Step numbers are referenced in the Discussion section. Note.—This tree does not convey time-calibrated information and is arranged to optimize conceptual understanding over accurate evolutionary relationships.
Our results also suggest the importance of lateral transfer processes in shaping the distribution of Form IV RuBisCO. For example, the Parcubacteria and Micrarchaeota Form IV enzymes are similar to those in characterized green sulfur bacteria and may indicate transfer from this source (Steps 9 and 12 in fig. 5). Previous work has suggested that the IV-Photo enzyme is mobile and may have been transferred from Chlorobi to both Gammaproteobacteria and Alphaproteobacteria (Steps 10 and 11) (Tabita et al. 2007). Our results extend the breadth of Form IV-Photo RuBisCO distribution to new branches of the tree of life and add to the possible instances of its gene transfer, including one across domains. However, it is still unclear which lineage of bacteria among those that are known to bear this version of the enzyme is likely to have been the original source for the transfers to the CPR and DPANN. Similarly, we recovered RuBisCO enzymes related to the Form IV encoded in the genome of at least one Beckwithbacteria and many scaffolds of bacteriophage origin, including one unusually large, manually curated phage genome. One possibility is that a phage acquired a copy of the ancestral Form IV enzyme, termed the DeepYkr (Tabita et al. 2007), from a bacterial ancestor (Step 13), ultimately evolving into a divergent clade. One of these divergent sequences may then have been transferred to Beckwithbacteria (Step 14). The reverse scenario is also possible, in which members of the Myoviridae acquired this enzyme from Beckwithbacteria, possibly as prophages. However, to date, we know of no cases of >200-kb phage genomes integrating into small (generally <1 Mb) CPR genomes. The discovery of RuBisCO homologs in phage also provides a possible mechanism for the widespread lateral gene transfer of this enzyme observable across the tree of life (fig. 5) (Canchaya et al. 2003). However, as of yet, phage encoding “bona fide” RuBisCO forms have not been identified.
The Form II/III enzymes are presently distributed among the CPR, DPANN, and at least one methanogenic archaeal lineage (Alonso et al. 2009; Wrighton et al. 2012). Given that this form is most closely related to Form II (fig. 1a), we speculate here that the Form II/III enzyme evolved in a CPR lineage (possibly the Dojkabacteria) after transfer of a Form II sequence (Step 15). Form II/III could then be transferred to several other CPR lineages and one or more archaeal lineages (Step 16). Notably, no CPR with Form II enzymes have been reported to date.
Finally, there are two possible explanations for the apparent discordance between the phylogenetic pattern showing Forms II and II/III branching together and separate from Form I (fig. 1a) and the pathway association of these Forms (I and II in CBB vs. II/III in the AMP pathway). If Form II/III preserves its ancestral function in the AMP pathway then the CBB pathway function in Forms I and II must have arisen by convergent evolution. Alternatively, the CBB pathway function in Forms I and II shared a common ancestor and Form II/III reverted back to function in the AMP pathway, possibly due to loss of the other CBB pathway enzymes. We suggest that convergent evolution of the more complex CBB pathway (which also requires PRK and transketolase, as well as various glycolysis enzymes) is less likely than reversion due to gene loss, especially given that gene loss is likely to have been widespread in the CPR. DPANN archaea, which generally do not have PRK, and methanogens could then have acquired the Form II/III RuBisCO by lateral transfer (fig. 5).
Conclusion
In conclusion, we show that CPR bacteria, DPANN archaea, and bacteriophage harbor RuBisCO diversity that broadens our understanding of the distribution of this enzyme across the tree of life. The wide but sparse distribution of RuBisCO within the CPR and DPANN may be the consequence of extensive lateral gene transfer as well as gene loss. Further, some transfers may have catalyzed major shifts in carbon metabolism in bacterial lineages with limited metabolic repertoires. Specifically, lateral transfer of RuBisCO could have conferred a “missing puzzle piece” for organisms already bearing AMP phosphorylase and R15P isomerase, completing the genomic repertoire for the CO2-incorporating AMP metabolism present in extant lineages. Likewise, Gottesmanbacteria may have evolved a partial CBB cycle or augmentation to the AMP pathway by linking laterally acquired RuBisCO and PRK to genes in the oxidative pentose phosphate pathway (fig. 4 and supplementary fig. S6, Supplementary Material online). Clearly, metagenomic studies of diverse environments can help to shed light on phylogenetic distribution and also to extend models of evolution for even well-studied enzymes.
Materials and Methods
Genome Collection and Annotation
We gathered a set of ∼4,000 CPR/DPANN genomes from metagenomes from several previous studies of groundwater, soil, ocean, and subsurface environments. Additionally, we binned several new genomes from a sediment from Rifle, CO (Anantharaman et al. 2016) and the water column in the Baltic Sea (Asplund-Samuelsson et al. 2016). Binning methods and taxonomic assignments followed those described in Anantharaman et al. (2016). Several genome fragments were manually curated, making use of unplaced paired reads to increase their length. This was necessary to test for bacterial affiliation based on comparison of the encoded genes with genes in known CPR genomes.
Proteins were predicted for each genome using Prodigal (“meta” mode) (Hyatt et al. 2010). Preliminary functional predictions were established using a pipeline based on KEGG Orthology (Kanehisa and Goto 2000). All versus all global search of proteins in each KO from the KEGG database was performed using usearch (Edgar 2010), and protein percent identity was used as input to Markov Cluster Algorithm clustering (Van Dongen 2008) with inflation parameter of 1.1. For each resulting cluster, the proteins were aligned using Multiple Alignment using Fast Fourier Transform (MAFFT) version 7 (Katoh and Standley 2013), and HMMs were constructed using the HMMER suite (Finn et al. 2011). Predicted proteins from each CPR/DPANN genome in this study were scanned using hmmsearch (Finn et al. 2011), and annotation was assigned according to the best HMM hit, providing it was above a predefined KEGG Orthology noise cutoff.
RuBisCO Analysis
We extracted above-threshold hits for RuBisCO large chain (K01601), yielding a final set of genomes encoding the enzyme. To analyze the number of nonredundant genomes containing RuBisCO, we repeated the above analysis with a set of ∼3,000 high quality genomes from various environments. These genomes were dereplicated at 99% secondary Average Nucleotide Identity (ANI) using dRep (-comp 20) (Olm et al. 2017) and then analyzed for presence of RuBisCO.
To expand the breadth of our main RuBisCO set, we identified RuBisCO sequences (many of which were unbinned) from sediment and groundwater metagenomes (e.g., Anantharaman et al. 2016; Hernsdorf et al. 2017; Probst et al. 2017). We excluded sequences shorter than 200 amino acids in length to remove fragmented proteins. Phylum-level taxonomy for these sequences was assigned based on the closest affiliation of the encoded sequences. These sequences were added to those from genomes and the entire set was dereplicated (USEARCH, -id 0.99 -sort length) (Edgar 2010). Sourcing for dereplicated sequences can be found in supplementary table S1, Supplementary Material online. Next, we combined the full set with reference RuBisCO from NCBI and aligned it using MAFFT (default parameters) (Katoh and Standley 2013). Alignments were trimmed by removing columns with >95% gaps. The unmasked alignment file of dereplicated RuBisCO sequences with metadata is attached as supplementary file 1, Supplementary Material online. We next constructed a maximum-likelihood tree with RAxML-HPC BlackBox (v. 8.2.10) as implemented on cipres.org (default parameters with rapid bootstrapping) (Stamatakis et al. 2008) and subsequently assigned each RuBisCO sequence to previously identified Forms based on phylogenetic clustering with reference sequences. Binned sequences excluded from the dereplicated set were reinserted into the tree and classified for downstream analyses. Sequences in ambiguous phylogenetic positions were annotated as “unknown.” Custom HMMs were constructed using recovered sequences for each RuBisCO form with the HMMER suite (Finn et al. 2011) and were subsequently self-tested and manually refined to exclude low-scoring sequences.
Collection and Analysis of Viral Sequences
To explore the possibility that phage encode RuBisCO, we generated a database of putative phage genome fragments using sequences from IMG/VR (img.jgi.doe.gov/vr/; last accessed November 30, 2018) and several previous metagenomic studies of groundwater and subsurface environments (Anantharaman et al. 2016; Probst et al. 2017). Contigs from the latter metagenomes were assigned a putative phage origin if the majority of encoded genes had no identifiable sequence similarity to genes in bacterial (or archaea). Only sequences >10 kb in length were included to improve the confidence of phage assignments. Predicted proteins from putative phage contigs were interrogated using the above RuBisCO HMMs, and those with significant HMM hits at or above a score of 100 and e ≪ 0.05 were retained for further analysis. Genome fragments with RuBisCO-related sequences were further evaluated to confirm the presence of additional (e.g., structural) genes indicative of phage classification. Once manually verified, the putative RuBisCO proteins of phage origin were dereplicated at 99% identity and incorporated into the phylogenetic analysis. Phage terminase proteins were extracted using existing annotations (in the case of IMG/VR fragments) or BLAST-based annotations (in the case of groundwater/subsurface fragments). To establish putative identity, we then aligned recovered phage terminases with reference proteins and created a tree with RaxML. Finally, several additional phage genome fragments encoding RuBisCO-related sequences were manually curated to increase their length.
Residue Analysis and Protein Modeling
To analyze the biochemically relevant characteristics of the RuBisCO sequences included in the dereplicated set, including those in the putative phage category, we extracted 12 residues known to be important for catalytic activity and 7 important for substrate binding from each sequence (Tabita et al. 2008; Saito et al. 2009). A sequence logo of these 19 sites for each Form was constructed using WebLogo (Crooks et al. 2004). Additionally, we modeled exemplary Form III, Form III-c, Form IV-like, and PRK proteins using the I-TASSER suite (zhanglab.ccmb.med.umich.edu/I-TASSER/; last accessed June 21, 2018) (Yang et al. 2015).
Pathway and Contig-Level Analyses
Finally, we identified two sets of proteins involved in RuBisCO-mediated carbon metabolism (supplementary table S2, Supplementary Material online) within the binned genomes using the KEGG annotation results. Of particular interest were AMP phosphorylase (deoA) and R15P isomerase (e2b2), thought to be involved in AMP metabolism (Wrighton et al. 2016), and PRK, a marker gene for the CBB pathway. We examined the distribution of these genes, plus others associated with the CBB pathway, across genomes as well as their genomic context. Specifically, for the three genes likely involved in AMP metabolism, the proximity of the genes on the same contig was calculated by taking the sum the lengths of the intervals between the genes. Gene fusions were identified by noting abnormally long RuBisCO sequences and examination of the domain structure through NCBI BlastP (blast.ncbi.nlm.nih.gov/Blast.cgi; last accessed November 30, 2018). As with RuBisCO, recovered protein sequences for PRK, AMP phosphorylase (deoA), R15P isomerase (e2b2), and CBB enzymes were aligned with MAFFT and trimmed as described above. Corresponding trees were then inferred with RAxML-HPC BlackBox (v. 8.2.10) as implemented on cipres.org (default parameters with rapid bootstrapping) (Stamatakis et al. 2008) and visualized using iTOL (Letunic and Bork 2016). For PRK, NCBI reference sequences, close BLAST hits to identified CPR PRK homologs, and sequences from the close homolog uridine kinase (udk) were gathered and added to the protein set before alignment.
Data and Software Availability
The unmasked alignment file of dereplicated RuBisCO sequences with metadata is attached as supplementary file 1, Supplementary Material online. Newly reported sequences from this study, including those from the pathway analyses, are available at the European Nucleotide Archive at the following accession number: PRJEB29721. Intermediate data files, including sequence files, and custom code used for the described analyses are available in interactive Jupyter Notebook format at https://github.com/alexanderjaffe/cpr_dpann_rubisco/; last accessed November 30, 2018.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Brian Thomas, Shufei Lei, Simonetta Gribaldo, Panagiotis Adam, Lin-Xing Chen, Adi Lavy, Karthik Anantharaman, Alexander Probst, and Patrick Shih for informatics support and helpful discussions. We also thank three anonymous reviewers whose suggestions improved the manuscript. Funding was provided by the Berkeley Fellowship to A.L.J., the Innovative Genomics Institute at the UC Berkeley, and the Chan-Zuckerberg Biohub Initiative. C.L.D. was supported by the Alternative Earths NASA Astrobiology Institute.
Author Contributions
A.L.J. conducted the phylogenetic analyses, A.L.J. and C.J.C performed genomic analyses, J.F.B. carried out genome curation, and C.J.C. conducted the protein modeling. J.F.B., C.J.C., and C.L.D. developed the project. A.L.J and J.F.B. wrote the manuscript, and all authors read and made comments on the manuscript prior to submission.
References
- Alonso H, Blayney MJ, Beck JL, Whitney SM.. 2009. Substrate-induced assembly of Methanococcoides burtoniid-ribulose-1,5-bisphosphate carboxylase/oxygenase dimers into decamers. J Biol Chem. 28449: 33876–33882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman K, Brown CT, Hug LA, Sharon I, Castelle CJ, Probst AJ, Thomas BC, Singh A, Wilkins MJ, Karaoz U, et al. 2016. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Commun. 7:13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aono R, Sato T, Imanaka T, Atomi H.. 2015. A pentose bisphosphate pathway for nucleoside degradation in Archaea. Nat Chem Biol. 115: 355–360. [DOI] [PubMed] [Google Scholar]
- Ashida H, Danchin A, Yokota A.. 2005. Was photosynthetic RuBisCO recruited by acquisitive evolution from RuBisCO-like proteins involved in sulfur metabolism? Res Microbiol. 156(5–6): 611–618. [DOI] [PubMed] [Google Scholar]
- Asplund-Samuelsson J, Sundh J, Dupont CL, Allen AE, McCrow JP, Celepli NA, Bergman B, Ininbergs K, Ekman M.. 2016. Diversity and expression of bacterial metacaspases in an aquatic ecosystem. Front Microbiol. 7:1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg IA, Kockelkorn D, Ramos-Vera WH, Say RF, Zarzycki J, Hügler M, Alber BE, Fuchs G.. 2010. Autotrophic carbon fixation in archaea. Nat Rev Microbiol. 86: 447–460. [DOI] [PubMed] [Google Scholar]
- Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, Wilkins MJ, Wrighton KC, Williams KH, Banfield JF.. 2015. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 5237559: 208–211. [DOI] [PubMed] [Google Scholar]
- Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brüssow H.. 2003. Phage as agents of lateral gene transfer. Curr Opin Microbiol. 64: 417–424. [DOI] [PubMed] [Google Scholar]
- Carter MS, Zhang X, Huang H, Bouvier JT, Francisco BS, Vetting MW, Al-Obaidi N, Bonanno JB, Ghosh A, Zallot RG, et al. 2018. Functional assignment of multiple catabolic pathways for d-apiose. Nat Chem Biol. 147: 696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelle CJ, Banfield JF.. 2018. Major new microbial groups expand diversity and alter our understanding of the tree of life. Cell 1726: 1181–1197. [DOI] [PubMed] [Google Scholar]
- Castelle CJ, Wrighton KC, Thomas BC, Hug LA, Brown CT, Wilkins MJ, Frischkorn KR, Tringe SG, Singh A, Markillie LM, et al. 2015. Genomic expansion of domain archaea highlights roles for organisms from new phyla in anaerobic carbon cycling. Curr Biol. 256: 690–701. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J-M, Brenner SE.. 2004. WebLogo: a sequence logo generator. Genome Res. 146: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crummett LT, Puxty RJ, Weihe C, Marston MF, Martiny JBH.. 2016. The genomic content and context of auxiliary metabolic genes in marine cyanomyoviruses. Virology 499:219–229. [DOI] [PubMed] [Google Scholar]
- Danczak RE, Johnston MD, Kenah C, Slattery M, Wrighton KC, Wilkins MJ.. 2017. Members of the Candidate Phyla Radiation are functionally differentiated by carbon- and nitrogen-cycling capabilities. Microbiome 51: 112.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2619: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Erb TJ, Zarzycki J.. 2018. A short history of RubisCO: the rise and fall (?) of Nature’s predominant CO2 fixing enzyme. Curr Opin Biotechnol. 49:100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Sun Y, Deng H, Li D, Wan J, Wang X, Wang W, Liao X, Ren Y, Hu X.. 2014. Structural and biochemical characterization of fructose-1,6/sedoheptulose-1,7-bisphosphatase from the cyanobacterium Synechocystis strain 6803. FEBS J. 2813: 916–926. [DOI] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR.. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39(Suppl): W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbling KP, Steup M, Latzko E.. 1986. Fructose 1,6-bisphosphatase form B from Synechococcus leopoliensis hydrolyzes both fructose and sedoheptulose bisphosphate. Plant Physiol. 803: 716–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson TE, Tabita FR.. 2001. A ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO)-like protein from Chlorobium tepidum that is involved with sulfur metabolism and the response to oxidative stress. Proc Natl Acad Sci U S A. 988: 4397–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison EP, Willingham NM, Lloyd JC, Raines CA.. 1997. Reduced sedoheptulose-1,7-bisphosphatase levels in transgenic tobacco lead to decreased photosynthetic capacity and altered carbohydrate accumulation. Planta 2041: 27–36. [Google Scholar]
- Hernsdorf AW, Amano Y, Miyakawa K, Ise K, Suzuki Y, Anantharaman K, Probst A, Burstein D, Thomas BC, Banfield JF.. 2017. Potential for microbial H2 and metal transformations associated with novel bacteria and archaea in deep terrestrial subsurface sediments. ISME J. 118: 1915–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ.. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AL, Corel E, Pathmanathan JS, Lopez P, Bapteste E.. 2016. Bipartite graph analyses reveal interdomain LGT involving ultrasmall prokaryotes and their divergent, membrane-related proteins. Environ Microbiol. 1812: 5072–5081. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S.. 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 281: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 304: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono T, Mehrotra S, Endo C, Kizu N, Matusda M, Kimura H, Mizohata E, Inoue T, Hasunuma T, Yokota A, et al. 2017. A RuBisCO-mediated carbon metabolic pathway in methanogenic archaea. Nat Commun. 8:14007.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1): W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olm MR, Brown CT, Brooks B, Banfield JF.. 2017. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 1112: 2864–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Rinke C, Chuvochina M, Chaumeil P-A, Woodcroft BJ, Evans PN, Hugenholtz P, Tyson GW.. 2017. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 211: 1533–1542. [DOI] [PubMed] [Google Scholar]
- Probst AJ, Castelle CJ, Singh A, Brown CT, Anantharaman K, Sharon I, Hug LA, Burstein D, Emerson JB, Thomas BC, et al. 2017. Genomic resolution of a cold subsurface aquifer community provides metabolic insights for novel microbes adapted to high CO2 concentrations. Environ Microbiol. 192: 459–474. [DOI] [PubMed] [Google Scholar]
- Saito Y, Ashida H, Sakiyama T, de Marsac NT, Danchin A, Sekowska A, Yokota A.. 2009. Structural and functional similarities between a ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO)-like protein from Bacillus subtilis and photosynthetic RuBisCO. J Biol Chem. 28419: 13256–13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Atomi H, Imanaka T.. 2007. Archaeal type III RuBisCOs function in a pathway for AMP metabolism. Science 3155814: 1003–1006. [DOI] [PubMed] [Google Scholar]
- Say RF, Fuchs G.. 2010. Fructose 1,6-bisphosphate aldolase/phosphatase may be an ancestral gluconeogenic enzyme. Nature 4647291: 1077–1081. [DOI] [PubMed] [Google Scholar]
- Schönheit P, Buckel W, Martin WF.. 2016. On the origin of heterotrophy. Trends Microbiol. 241: 12–25. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J.. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 575: 758–771. [DOI] [PubMed] [Google Scholar]
- Stolzenberger J, Lindner SN, Persicke M, Brautaset T, Wendisch VF.. 2013. Characterization of fructose 1,6-bisphosphatase and sedoheptulose 1,7-bisphosphatase from the facultative ribulose monophosphate cycle methylotroph Bacillus methanolicus. J Bacteriol. 19522: 5112–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabita FR, Hanson TE, Li H, Satagopan S, Singh J, Chan S.. 2007. Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol Mol Biol Rev. 714: 576–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabita FR, Satagopan S, Hanson TE, Kreel NE, Scott SS.. 2008. Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J Exp Bot. 597: 1515–1524. [DOI] [PubMed] [Google Scholar]
- Thompson LR, Zeng Q, Kelly L, Huang KH, Singer AU, Stubbe J, Chisholm SW.. 2011. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc Natl Acad Sci U S A. 10839: E757–E764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully BJ, Graham ED, Heidelberg JF.. 2018. The reconstruction of 2,631 draft metagenome-assembled genomes from the global oceans. Sci Data. 5:170203.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dongen S. 2008. Graph clustering via a discrete uncoupling process. SIAM J Matrix Anal Appl. 301: 121–141. [Google Scholar]
- Williams TA, Szöllősi GJ, Spang A, Foster PG, Heaps SE, Boussau B, Ettema TJG, Embley TM.. 2017. Integrative modeling of gene and genome evolution roots the archaeal tree of life. Proc Natl Acad Sci U S A. 11423: E4602–E4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton KC, Castelle CJ, Varaljay VA, Satagopan S, Brown CT, Wilkins MJ, Thomas BC, Sharon I, Williams KH, Tabita FR, et al. 2016. RubisCO of a nucleoside pathway known from Archaea is found in diverse uncultivated phyla in bacteria. ISME J. 1011: 2702–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton KC, Thomas BC, Sharon I, Miller CS, Castelle CJ, VerBerkmoes NC, Wilkins MJ, Hettich RL, Lipton MS, Williams KH, et al. 2012. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 3376102: 1661–1665. [DOI] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y.. 2015. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 121: 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo JG, Bowien B.. 1995. Analysis of the cbbF genes from Alcaligenes eutrophus that encode fructose-1,6-/sedoheptulose-1,7-bisphosphatase. Curr Microbiol. 311: 55–61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The unmasked alignment file of dereplicated RuBisCO sequences with metadata is attached as supplementary file 1, Supplementary Material online. Newly reported sequences from this study, including those from the pathway analyses, are available at the European Nucleotide Archive at the following accession number: PRJEB29721. Intermediate data files, including sequence files, and custom code used for the described analyses are available in interactive Jupyter Notebook format at https://github.com/alexanderjaffe/cpr_dpann_rubisco/; last accessed November 30, 2018.