Abstract
The 26S proteasome is a highly complex 2.5 MDa molecular machine responsible for regulated protein degradation. Proteasome substrates are typically marked by ubiquitination for recognition at receptor sites contributed by Rpn1/S2/PSMD2, Rpn10/S5a, and Rpn13/Adrm1. Each receptor site can bind substrates directly by engaging conjugated ubiquitin chains or indirectly by binding to shuttle factors Rad23/HR23, Dsk2/PLIC/UBQLN, or Ddi1, which contain a ubiquitin-like domain (UBL) that adopts the ubiquitin fold. Previous structural studies have defined how each of the proteasome receptor sites bind to ubiquitin chains as well as some of the interactions that occur with the shuttle factors. Here, we define how hRpn10 binds to the UBQLN2 UBL domain, solving the structure of this complex by NMR, and determine affinities for each UIM region by a titration experiment. UBQLN2 UBL exhibits 25-fold stronger affinity for the N-terminal UIM-1 over UIM-2 of hRpn10. Moreover, we discover that UBQLN2 UBL is fine-tuned for the hRpn10 UIM-1 site over the UIM-2 site by taking advantage of the additional contacts made available through the longer UIM-1 helix. We also test hRpn10 versatility for the various ubiquitin chains to find less specificity for any particular linkage type compared to hRpn1 and hRpn13, as expected from the flexible linker region that connects the two UIMs; nonetheless, hRpn10 does exhibit some preference for K48 and K11 linkages. Altogether, these results provide new insights into the highly complex and complementary roles of the proteasome receptor sites and shuttle factors.
Keywords: Rpn10, S5a, Dsk2, ubiquilin, ubiquitin
Introduction
Regulated protein degradation in eukaryotes is performed by the ubiquitin-proteasome pathway [1, 2]. Substrates are ubiquitinated by an enzymatic cascade and recognized by ubiquitin receptors Rpn10/S5a [3], Rpn13/Adrm1 [4, 5], and Rpn1/S2/PSMD2 [6] of the proteasome 19S regulatory particle (RP). The RP abuts either or both ends of the proteasome 20S catalytic core particle (CP), a large cylindrical structure with a hollow interior where substrates are proteolyzed [7]. The three RP ubiquitin receptors have distinct modes of substrate recognition. A site in Rpn1 (toroid 1, T1) binds preferentially to K6 and K48 ubiquitin chains [6]; hRpn10 binds ubiquitin at either of two helical ubiquitin-interacting motifs (UIMs) [8, 9]; and Rpn13 binds ubiquitin through loops of a pleckstrin-like receptor for ubiquitin (Pru) domain [4, 5]. Each receptor is proximal to one of the three deubiquitinating enzymes (DUBs) of the proteasome RP. Rpn11 is located near Rpn10 [10, 11] and acts at the substrate end of ubiquitin chains to promote degradation [12, 13]. Ubp6/Usp14 binds to a second toroid site in Rpn1 (toroid 2, T2) that is proximal to the T1 site [6, 14] and prefers substrates with more than one attached ubiquitin chain [15]. Uch37/UCHL5 [16] binds to a deubiquitinase adaptor domain (DEUBAD) [17] of Rpn13 [18, 19] that in the free protein, interacts intramolecularly with the Pru domain to restrict ubiquitin binding [20]; this intramolecular interaction is displaced by the docking of Rpn13 to the proteasome [20]. The DEUBAD domain splits to envelop a unique region in Uch37 that is C-terminal to its catalytic domain [21–23]. Ubp6/Usp14 acts within ubiquitin chains [15, 24], whereas Uch37 deconjugates ubiquitin chains at a distal location relative to the substrate [16]; both can either promote or antagonize degradation [15, 16, 24–26]. As ubiquitin moieties are removed from substrates by the DUBs, a heterohexameric ring of AAA ATPase proteins (Rpt1- Rpt6) promotes substrate unfolding and transit into the CP [1, 2, 27–29]. Efforts to identify additional ubiquitin receptors in the proteasome have yielded ATPase Rpt5, which cross-links to ubiquitin chains [30], intrinsically disordered protein Sem1/Dss1/Rpn15 [31], which functions in proteasome assembly [32, 33], and the VWA domain of Rpn10 [34]. In all cases however, the impact of these proposed ubiquitin-binding sites on proteasome activity has yet to be established; in particular, sem1Δ proteasomes did not show any defect in ubiquitin-binding activity [6].
It is not yet clear whether ubiquitinated substrates first encounter the proteasome by direct binding to Rpn1, Rpn10, or Rpn13. The canonical model is that ubiquitinated substrates are shuttled to the proteasome by ubiquitin-like (UBL)-ubiquitin-associated (UBA) family members Rad23/HR23, Dsk2/PLIC/UBQLN, and Ddi1 [35, 36]. The UBA domains of such shuttle factors bind ubiquitin [37, 38], whereas their UBL domains bind to Rpn1, Rpn10, or Rpn13 [4–6, 39–47]. The shuttle factors can form heterodimers by UBA:UBL domain interactions [48, 49] and multiple shuttle factors are able to simultaneously engage a ubiquitin chain [48]. In the case of hHR23a, UBL:UBA interactions also occur intramolecularly [50]. These interactions are disrupted by UBA domain binding to ubiquitin [48] or UBL domain binding to hRpn10 [50], similar to the disruption of Rpn13 intradomain interaction upon binding to its proteasome docking site [20].
UBL-UBA proteins have different specificities for ubiquitin chains, which may be advantageous to their coordinated binding of substrates. hHR23a C-terminal UBA domain preferentially binds K48-linked chains [51] by sandwiching between neighboring ubiquitin moieties [52], whereas UBQLN1 UBA domain also binds monoubiquitin and does not exhibit notable preference for K48 versus K63 ubiquitin chains [53]. Rpn13 and Rpn1 exhibit preferential binding to UBQLN2 and Rad23 UBL domains respectively over K48 diubiquitin [6, 45]. In a ternary complex of hHR23a, hRpn10 and K48 tetraubiquitin, the hHR23a C-terminal UBA domain and hRpn10 UIM-1 bind tetraubiquitin, while hRpn10 UIM-2 binds to the hHR23a UBL domain [54]. These binding preferences likely play a role in how ubiquitinated substrates are oriented when bound to the proteasome.
There appears to be a need and multiple mechanisms to regulate the level of shuttle factor at the proteasome. Dsk2 overexpression leads to accumulation of ubiquitinated substrates in S. cerevisiae [55], where it binds Rpn10 UIM more strongly than other UBL-UBA protein [56]; in turn, extra-proteasomal hRpn10 can restrict accessibility to the proteasome [55]. Severe developmental defects and lethality results from Dsk2 overexpression in Drosophila melanogaster, which can similarly be rescued by Rpn10 [57]. Depletion of UBQLN in Drosophila caused impaired proteostasis and locomotive and learning disabilities [58]. In humans, there are five distinct genes (UBQLN1, 2, 3, 4, and L) that encode UBQLN proteins [59], with overexpression of UBQLN1 and UBQLN2 leading to decreased proteasomal degradation of p53 and IκBα [60]. UBQLN proteins target to the proteasome mislocalized mitochondrial membrane proteins, mislocalized transmembrane domain proteins, and aggregation-prone proteins [61–63]. UBQLN1 is reported to prevent the accumulation of hydrophobic mitochondrial proteins in the cytosol [64], and UBQLN2 was found to colocalize with stress granules and undergo liquid-liquid phase separation that is dissolved by binding to ubiquitin; this phenomenon is proposed to enable extraction of ubiquitinated substrates from stress granules for delivery to the proteasome [65, 66].
UBQLN family members are associated with neurodegenerative diseases [67–70], cytoprotective activity during stress [71], aggresome formation [72], targeting of aggregated proteins to autophagosomes [73, 74], and clearance of expanded polyglutamine proteins [75, 76]. Moreover, mutations in UBQLN2 and UBQLN4 have been identified in patients with familial amyotrophic lateral sclerosis (ALS) [69, 77, 78], and in vivo pooled loss-of-function genetic screens discovered that UBQLN1 sensitize tumors to immunotherapy [79].
To better understand how UBQLN proteins function mechanistically in targeted protein degradation, we used a combination of biochemical and structural biology experiments to define how UBQLN2 interacts with its proteasome binding site in hRpn10. We found each hRpn10 UIM is capable of binding to the UBQLN UBL domain, but with a marked preference for UBQLN binding to the N-terminal hRpn10 UIM. We also found that hRpn10 prefers K11 and K48 linkages in ubiquitin chains, but with less discretion for a specific chain type compared to Rpn1 and Rpn13. Collectively, these findings provide insight into complementary relationships between receptor sites and shuttle factors that favor avidity effects in the proteasome. In addition, we used NMR spectroscopy to solve the structure of each hRpn10 UIM bound to an UBQLN2 UBL domain to reveal the molecular basis of UBQLN2 binding to hRpn10 at atomic level detail.
RESULTS
hRpn10 prefers K48 and K11 linked ubiquitins
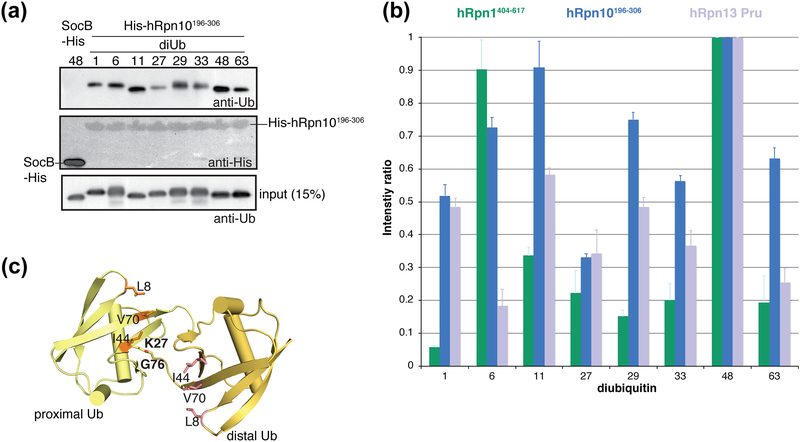
In a pull-down assay with the eight possible diubiquitin linkages, Rpn1 prefers K6 and K48 linkage types, with the latter also preferred by Rpn13 [6, 45]. We used the same assay to test for linkage preferences in hRpn10. Diubiquitin with each of the eight ubiquitin linkage types (M1, K6, K11, K27, K29, K33, K48 and K63) was incubated with Ni-NTA resin containing pre-bound His-tagged hRpn10196–306 protein and interaction detected by immunoblotting with anti-ubiquitin antibody following the removal of unbound protein (Figure 1a). Intrinsically disordered protein SocB with a His-tag [80] was used as a negative control with K48 diubiquitin. Indeed, hRpn10196–306 interacted with all ubiquitin chain types, with strongest affinity for K48 linked diubiquitin (Figure 1a), the preferred chain type also for Rpn1 and Rpn13 [6, 45]. This experiment was repeated, quantified, and the results normalized to K48 diubiquitin binding for all three proteasome substrate receptors (Figure 1b). With the exception of the K27 linkage, hRpn10 bound to each diubiquitin type within a 2-fold difference of affinity, thus demonstrating greater versatility for the various linkage types compared to Rpn1 and Rpn13. Similar to hRpn13, K11 diubiquitin is the second preference for hRpn10 (Figure 1b). hRpn10 preference for the K48 and K11 linkage was also found in a pull-down assay with tetraubiquitin chains [81]. Rpn1, Rpn10, and Rpn13 all demonstrated weak affinity for K27 diubiquitin (Figure 1b). The structure of K27 diubiquitin (Figure 1c) [82, 83] provides an explanation for this result, as compared to the other linkage types, K27 yields the lowest mobility and solvent accessibility for the canonical L8-I44-V70 binding surface [82], which is used by all three of the proteasome ubiquitin receptors [4–6, 8]. Thus, the weaker binding to this chain type is likely intrinsic to K27 diubiquitin itself.
Figure 1. hRpn10196–306 preferentially binds to K48 and K11 ubiquitin linkages.
(a) Pulldown assay with His-hRpn10196–306 and M1-, K6-, K11-, K27-, K29-, K33-, K48-, and K63-diubiquitin, as indicated. Immunoblotting was done with antibody against ubiquitin (anti-ubiquitin) (top) or polyhistidine (anti-His) (middle). Intrinsically disordered protein SocB-His [80] was used as a negative control with K48 diubiquitin, as indicated. Direct loading for 15% of the diubiquitin input for each chain type with immunoblotting by ubiquitin-specific antibody is included (bottom). (b) The pull-down assay was repeated three times and the diubiquitin signal intensities separately normalized for each receptor to the strongest signal by using ImageJ [129] and the values plotted (dark blue). The plots from pull-down assays with GST-hRpn1404–617 (green) [6] or GST-hRpn13Pru (purple) [45] for the eight diubiquitin types are also included for comparison. (c) Ribbon diagram of K27 diubiquitin (PDB 5J8P) [83] with hydrophobic amino acids L8-I44-V70 and linkage amino acids K27-G76 labeled and colored orange and pink respectively.
Interaction of UBQLN proteins with hRpn10 in HCT116 cells
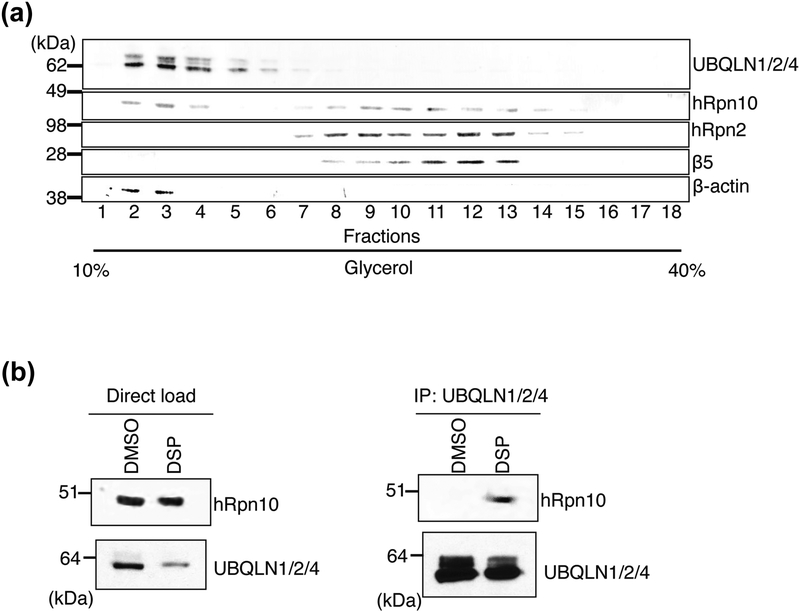
In HEK293 cells, UBQLN was found to be widely distributed in a glycerol gradient [84] designed to fractionate proteasome complexes [25]. To study further the abundance of UBQLN proteins at the proteasome, we subjected cell lysates from HCT116 cells to fractionation over a 10–40% linear glycerol gradient. The location of the 26S proteasome was identified by immunoblotting for RP component hRpn2 and CP component β5 (Figure 2a). hRpn10 was present in both proteasome-containing and proteasome-free fractions, as expected from previous studies [39, 55, 84–86]. The presence of the UBQLN proteins was revealed by immunoblotting with an antibody that recognizes UBQLN1/2/4 [87]. This experiment revealed the bulk of UBQLN1/2/4 to co-fractionate with extra-proteasomal hRpn10 with little observable presence at the proteasome (Figure 2a). The UBQLN proteins also appeared in higher molecular weight complexes that did not contain hRpn10 (Figure 2a, fractions 5 and 6).
Figure 2. UBQLN proteins interact with hRpn10 in HCT116 cells.
(a) A 10–40 % linear glycerol gradient was loaded with HCT116 cell lysate and subjected to ultracentrifugation. Gradient fractions were subjected to SDS-PAGE and immunoprobed for proteasome markers hRpn2 and β5 as well as β-actin. An antibody against UBQLN1/2/4 was used to locate these UBQLN proteins while anti-hRpn10 antibody revealed the presence of hRpn10. (b) HCT116 cell was treated with crosslinker DSP or DMSO (as a control). Cell lysates were immunoprecipitated with anti-UBQLN1/2/4 antibody, subjected to SDS-PAGE, and immunoprobed with anti-hRpn10 or anti-UBQLN1/2/4 antibodies, as indicated (right panel). Immunoblots of the cell lysates prior to immunoprecipitation for the DSP- and DMSO-treated cells is also included (left panel).
To test directly whether UBQLN1/2/4 binds hRpn10, we performed a crosslinking immunoprecipitation experiment with denaturing conditions. HCT116 cells were incubated with dithiobis(succinimidyl) propionate (DSP) for 30 minutes followed by lysis in radioimmunoprecipitation assay (RIPA) buffer. UBQLN1/2/4 was then immunoprecipitated from the whole cell lysate by anti-UBQLN1/2/4 antibodies and interaction with hRpn10 probed by anti-hRpn10 antibodies. Indeed, hRpn10 co-immunoprecipitated with UBQLN1/2/4 in HCT116 cells (Figure 2b).
UIM-1 exhibits 25-fold greater affinity for UBQLN2UBL compared to UIM-2
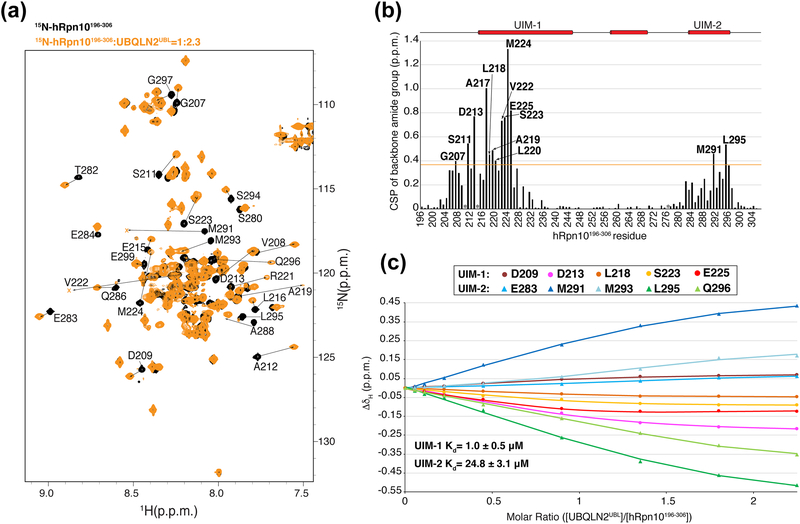
To identify the amino acids of hRpn10196–306 used to bind to UBQLN2UBL, we performed a titration experiment in which unlabeled UBQLN2UBL was added incrementally to 15N-labeled hRpn10196–306 and monitored the effects at an amino acid scale by using 2D 1H, 15N HSQC experiments. We used previously published assignments for free hRpn10196–306 [88] and assigned its UBQLN2UBL-bound state by using 15N/13C NOESY experiments acquired on 15N/13C-labeled hRpn10196–306, as described in Materials and Methods. The HSQC experiments revealed shifting for amino acids in both UIM1 and UIM2 (Figure 3a). The effects at 2.3-fold molar excess UBQLN2UBL were quantified to yield a chemical shift perturbation (CSP) plot that demonstrated a greater magnitude of shifting and number of amino acids affected for UIM-1 as compared to UIM-2 (Figure 3b).
Figure 3. UBQLN2UBL exhibits 25-fold higher affinity for hRpn10 UIM-1.
(a) 1H, 15N HSQC spectra of 15N-labeled hRpn10196–306 (black) and with 2.3-fold molar excess unlabeled UBQLN2UBL (orange). hRpn10196–306 signals that shift following addition of UBQLN2UBL are labeled. The bound states of V222 and M291 are only observed at lower threshold and labeled with ‘x’. (b) CSP plot for the data depicted in (a) showing effects of 2.3-fold molar excess UBQLN2UBL addition to 15N-labeled hRpn10196–306. The orange line indicates one standard deviation above the average value and prolines are indicated with asterisks. CSP, chemical shift perturbation; p.p.m., part per million. (c) Global fit of chemical shift changes for amide protons (ΔδH) of residues in UIM-1 (D209, D213, L218, S223, and E225) and UIM-2 (E283, M291, M293, L295, and Q296) as a function of molar ratio of UBQLN2UBL to hRpn10196–306. The data were fit to a two-site binding mode to yield dissociation constants (KD) for a primary binding site (UIM-1, 1.0 ± 0.5 μM) and a secondary binding site (UIM-2, 24.8 ± 3.1 μM).
To obtain a binding affinity for each UIM, chemical shift changes of amide protons (ΔδH) for five residues in UIM-1 (D209, D213, L218, S223, and E225) and UIM-2 (E283, M291, M293, L295 and Q296) were plotted at varying molar ratio of UBQLN2UBL to hRpn10196–306 (Figure 3c). A global fit of the data to a two-site binding mode was performed as described in [89–91] to yield dissociation constants (KD). This analysis revealed a primary binding site (UIM-1) of 1.0 ± 0.5 μM and a secondary binding site (UIM-2) of 24.8 ± 3.1 μM (Figure 3c). The KD value of UIM-1 derived by using the NMR titration data is in agreement with an overall KD value of 1.1 μM measured by isothermal titration calorimetry (ITC) [84]. In addition, these results indicate a 25-fold binding preference for UBQLN2UBL binding to UIM-1, which is consistent with a previous report of hRpn10 binding to UBQLN1 through UIM-1 [92]. By contrast, ubiquitin and hHR23a/b UBL bind preferentially to UIM-2 [8, 39, 42, 43, 93].
Structure of hRpn10 UIM1 and UIM2 complexed with UBQLN2UBL
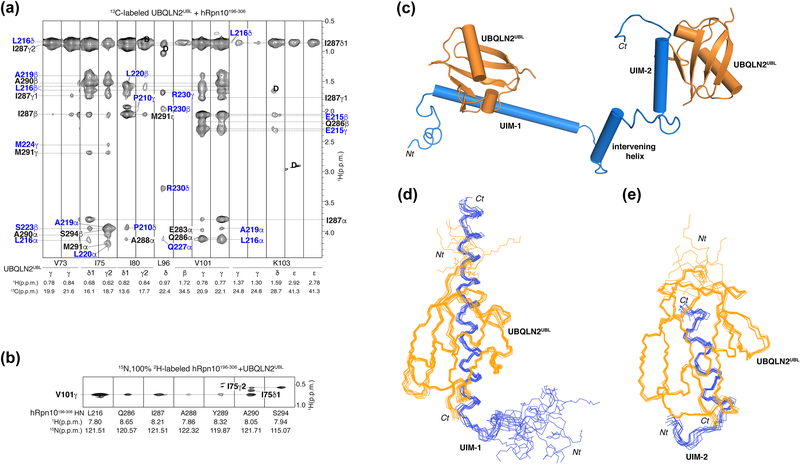
In an effort to solve the structure of hRpn10196–306 complexed with UBQLN2UBL, 13C half-filtered NOESY experiments were used to record intermolecular nuclear Overhauser effect (NOE) interactions, as described for previous complexes [94, 95]. 13C-labeled UBQLN2UBL was mixed with unlabeled hRpn10196–306 and NOE interactions detected between UBQLN2UBL residues in β3, β4, β5 and the C-terminal region (V73, I75, I80, L96, V101 and K103) and hRpn10 residues from both UIMs (Figure 4a, interactions with UIM-1 and UIM-2 indicated in blue and black respectively). NOE assignments were confirmed by analyzing a complementary 13C half-filtered NOESY spectrum recorded on a sample of 13C-labeled hRpn10196–306 mixed with unlabeled UBQLN2UBL (Figure S1). NOE interactions were only observed between UBQLN2UBL and the hRpn10 UIMs; no intermolecular NOE was observed to hRpn10 residues intervening the two UIMs. A standard 15N-dispersed NOESY experiment was also recorded on a sample of 15N, 2H-labeled hRpn10196–306 mixed with unlabeled UBQLN2UBL to assign intermolecular NOE interactions between hRpn10196–306 amide groups and UBQLN2UBL side chain atoms [96]. This approach yielded nine intermolecular NOEs between the hRpn10 UIM-1 and UIM-2 amide groups and UBQLN2UBL I75 and V101 (Figure 4b).
Figure 4. Structure of hRpn10196–306 complexed with UBQLN2UBL.
(a) Selected regions from a 1H, 13C half-filtered NOESY experiment acquired with 13C-labeled UBQLN2UBL and equimolar unlabeled hRpn10196–306 highlighting intermolecular NOE interactions. Breakthrough diagonal peaks are labeled as ‘D’. NOE interactions between residues from hRpn10 UIM-1 or UIM-2 and UBQLN2UBL are distinguished with blue and black labels respectively. (b) Selected 15N planes from a 3D 15N-dispersed NOESY spectrum recorded on a sample of 0.5 mM 2H, 15N-labeled hRpn10196–306 and 2.3-fold molar excess unlabeled UBQLN2UBL. Labels inside (bold) and outside of the strips correspond to UBQLN2UBL and hRpn10196–306, respectively. (c) A representative structure of hRpn10196–306:UBQLN2UBL is provided with hRpn10196–306 and UBQLN2UBL colored in blue or orange. (d, e) Backbone trace diagrams for the ten lowest energy structures of hRpn10 UIM-1:UBQLN2UBL (d) or UIM-2:UBQLN2UBL (e) with the secondary structural elements superimposed.
In total, 137 intermolecular NOE interactions were assigned from the three NOESY spectra (Figure 4a-b and S1) and used to derive distance restraints. These were combined with intramolecular distance restraints derived from 15N or 13C NOESY experiments acquired on either 15N or 13C-labeled hRpn10196–306 mixed with unlabeled UBQLN2UBL or 15N or 13C-labeled UBQLN2UBL mixed with unlabeled hRpn10196–306. Hydrogen bond and dihedral angle restraints were also derived, as summarized in Table 1 and described in Materials and Methods, and the hRpn10196–306: UBQLN2UBL structure calculated by using Xplor-NIH 2.47. As in free hRpn10 [8] and the hRpn10 complex with K48-linked diubiquitin [9], three helices span hRpn10 M196 – A306, including the two UIM helices and an intervening helix that extends from D257 to E269 (Figure 4c), that are not constrained relative to each other (Figure S2). All NOEs for these helical residues were assigned and no long-range NOE interaction was detected between the three helices (Figure S3).
Table 1.
Structural statistics for hRpn10196–306:UBQLN2UBL
| NOE-derived distance constraints (total) | 5647 | |
| Intermolecular | UIM-1: UBQLN2UBL | UIM-2: UBQLN2UBL |
| 63 | 87 | |
| Intramolecular | 5497 | |
| hRpn10196–306 | 1441 | |
| Intra-residue | 547 | |
| Inter-residue | 894 | |
| Sequential (|i − j| = 1) | 419 | |
| Medium-range (|i − j| ≤ 4) | 439 | |
| Long-range (|i − j| ≥ 5) | 36 | |
| UBQLN2UBL (per molecule) | 2028 | |
| Intra-residue | 614 | |
| Inter-residue | 1414 | |
| Sequential (|i − j| = 1) | 402 | |
| Medium-range (|i − j| ≤ 4) | 344 | |
| Long-range (|i − j| ≥ 5) | 668 | |
| Hydrogen bond constraints (total) | 121 | |
| hRpn10196–306 | 47 | |
| UBQLN2UBL (per molecule) | 37 | |
| Dihedral angle constraints (total) | 368 | |
| hRpn10196–306 | 104 | |
| UBQLN2UBL (per molecule) | 132 | |
| Structure statisticsa | ||
| Ramachandran plot (%) | ||
| Most-favorable region | 97.8 | |
| Additionally allowed region | 2.2 | |
| Generously allowed region | 0 | |
| Disallowed region | 0 | |
| Deviations from idealized geometry | ||
| Bond lengths (Å) | 0.003 ± 0.000 | |
| Bond angles (°) | 0.448 ± 0.014 | |
| Impropers (°) | 0.340 ± 0.011 | |
| r.m.s.d from average structure (Å) | UIM-1: UBQLN2UBL | UIM-2: UBQLN2UBL |
| Backbone atoms | 0.54 ± 0.12b | 0.49 ± 0.16c |
| Heavy atoms | 1.20 ± 0.19b | 1.03 ± 0.17c |
Statistics for II° structural elements of hRpn10196–306 and UBQLN2UBL from the 10 lowest energy structures.
Superimposing hRpn10196–306 P214-E245 and UBQLN2UBL I32–I102.
Superimposing hRpn10196–306 E283-Q296 and UBQLN2UBL I32–I102.
Backbone traces of the ten lowest energy structures superimposed for UIM-1: UBQLN2UBL and UIM-2: UBQLN2UBL are displayed in Figure 4d and 4e respectively; for regions with secondary structure, the backbone root mean square deviation (r.m.s.d.) from the average structure in each case is below 0.6 Å (Table 1).
Role of the ‘LALAL’ motif in hRpn10 binding to UBQLN2
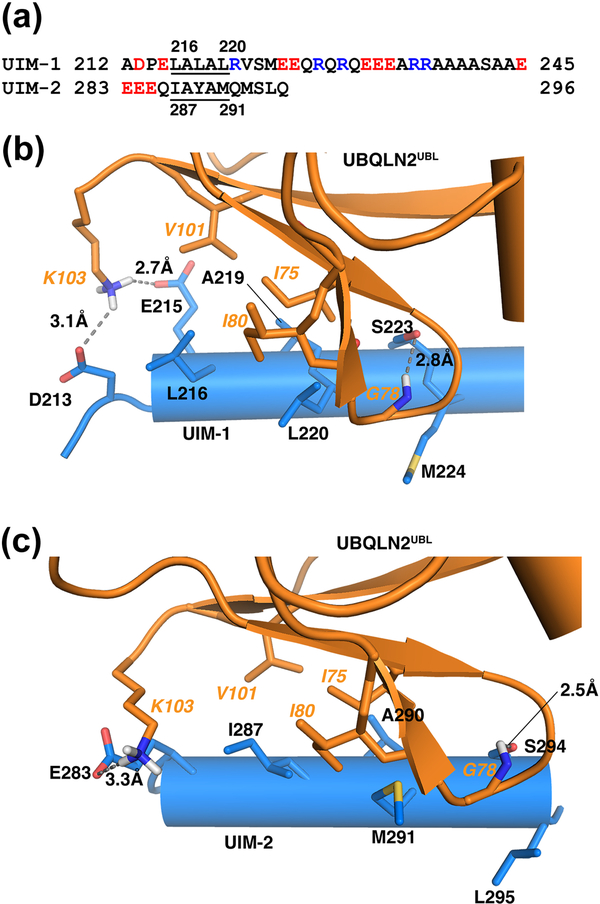
The conserved ‘LALAL’ UIM motif presents as 216LALAL220 in UIM-1 and 287IAYAM291 in UIM-2 (Figure 5a); these amino acids have previously been found to be important for interaction with ubiquitin [8, 9]. In the complexes with UBQLN2UBL, L216, A219, and L220 from UIM-1 and I287, A290, and M291 from UIM-2 contribute compact hydrophobic interactions with UBQLN2UBL I75, I80 and V101 (Figure 5b-c). These interactions are dictated by the intermolecular NOEs (Figure 4a-b and S1) and supported by CSP data for hRpn10 (Figure 3b) and UBQLN2UBL (Figure S4). V101 is strictly conserved among human UBQLN proteins and in S. cerevisiae Dsk2, as are I75 and I80 with the exception of UBQLNL, in which they are conserved as valine and leucine respectively (Figure S5).
Figure 5. UBQLN2UBL interactions involving the hRpn10 ‘LALAL’ motif and neighboring amino acids.
(a) Amino acid sequence alignment of UIM-1 and UIM-2 in hRpn10. Acidic and basic amino acids are highlighted red and blue, respectively. The hydrophobic LALAL/IAYAM residues within each UIM are underlined. (b, c) Expanded structural region to display the contact surfaces for hRpn10 UIM-1:UBQLN2UBL (b) and UIM-2:UBQLN2UBL (c). Side chains of UBQLN2UBL I75, G78, I80, V101, and K103 (orange) are displayed and labeled, as are hRpn10 UIM-1 D213, E215, L216, A219, L220, S223, and M224 (blue), and UIM-2 E283, I287, A290, M291, S294, and L295 (blue). Hydrogen bonds are displayed as grey dashed lines. Oxygen, nitrogen, and sulphur atoms are colored red, blue and yellow, respectively.
A highly conserved serine residue proximal to the ‘LALAL’ motif (S223 in UIM1 and S294 in UIM-2) forms a hydrogen bond with the backbone amide of G78 in UBQLN2UBL (Figure 5b-c). The importance of this serine has been discussed in the context of hRpn10 interaction with ubiquitin and hHR23a/b UBL [8, 42, 43]. L216/A219/L220/S223 in UIM-1 and I287/A290/M291/S294 in UIM-2 reside on the same side of the UIM α-helix and form the center of the contact surface. One helical turn from the ‘LALAL’ motif, van der Waals interactions are formed between UIM-1 M224 or UIM-2 L295 and UBQLN2UBL G78 backbone atoms (Figure 5b-c). At the other end of the ‘LALAL’ motif, acidic E215 from UIM-1 forms a hydrogen bond with UBQLN2UBL K103, which also interacts electrostatically with UIM-1 D213 and UIM-2 E283 (Figure 5b-c). This lysine in UBQLN is strictly conserved (Figure S5).
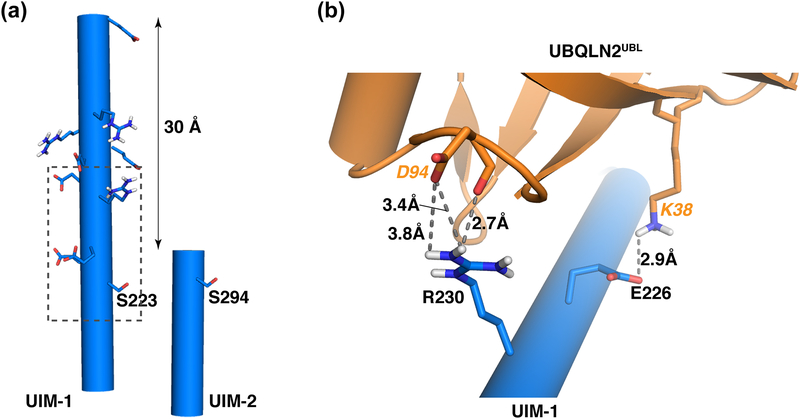
The longer UIM-1 helix provides additional hydrogen bonds to UBQLN2UBL
A key difference between the two UIMs is the additional contacts provided by the longer UIM-1 helix. UIM-1 spans residues P214 to E245 and is comprised of nine helical turns, whereas UIM2 is shorter with only 3.5 helical turns; an addition 30 Å is provided by these additional amino acids (Figure 6a). The UIM-2 α-helix ends two amino acids after S294, whereas UIM-1 contains 20 additional amino acids rich in glutamic acid, arginine and alanine amino acid type (Figure 5a) that contribute to the binding. A hydrogen bond between UIM-1 E226 and the UBQLN2UBL K38 side chain contributes to the increased UIM-1 affinity, as do interactions with R230, a helical turn away from E226 (Figure 6b); R230 forms one hydrogen bond with the UBQLN2UBL backbone oxygen atom of D94 and two additional hydrogen bonds with the D94 side chain (Figure 6b). K38 and D94 are both conserved in all human UBQLN proteins and Dsk2 (Figure S5).
Figure 6. UBQLN2UBL establishes additional interactions with the longer UIM-1 helix.
(a) hRpn10 UIM-1 and UIM-2 helices aligned based on their amino acid sequences as displayed in Figure 5a. A measurement is provided between the Cα atoms of UIM-1 E225 and E245. UIM-1 S223, UIM-2 S294, and glutamic acid and arginine residues in extended UIM-1 region are displayed. (b) Enlarged view of the UIM-1:UBQLN2UBL structure encompassed by the boxed region in (a) to illustrate the additional hydrogen bonds and electrostatic interactions provided by the longer UIM-1 helix. Hydrogen bonds and electrostatic interactions are displayed as grey dashed lines.
Discussion
Between the two hRpn10 UIM helices is a flexible region that allows a wide geometric distribution for UIM-2 relative to UIM-1 [8]. This flexibility enables the two UIMs to simultaneously bind neighboring ubiquitins in a K48-linked diubiquitin chain [9]. The results of our pulldown assay suggest that with the exception of the more restricted K27 linkage, the two UIMs can similarly engage neighboring ubiquitin moieties of the other ubiquitin chain types. Indeed, mass spectrometry studies have demonstrated that all ubiquitin chain types are capable of signaling for degradation by the proteasome [97–99]. We propose that hRpn10 in particular is well suited for this degree of versatility towards ubiquitin chains at the proteasome. Nonetheless, hRpn10 did show some preference for K11 and K48 linkages, as does hRpn13 [45]. Our results suggest that hRpn10 and hRpn13 provide the major binding sites in proteasome for these two chain types including K11/K48-branched chains. Indeed, K11/K48-branched chains were proposed to be proteasome priority signals that allow cells to rapidly clear specific proteins, including mitotic regulators and misfolded nascent polypeptides [100]. Moreover, UBQLN proteins have been identified as effectors of K11/K48-specific quality control [100], perhaps aided by their interactions with hRpn10 and hRpn13.
Our structural study of hRpn10196–306 complexed with UBQLN2UBL demonstrates both UIMs as capable of binding to UBQLN2UBL. Nonetheless, the 25-fold higher affinity for UIM-1 indicates preferred occupancy at this site, thus leaving UIM-2 free to bind ubiquitin or hHR23 proteins. Indeed, ubiquitin and hHR23 proteins prefer the UIM-2 site [8, 39, 42, 43, 93]. It is worth noting that Rpn10 is shorter in S. cerevisiae, ending before UIM-2 and retaining only the Dsk2 preferred binding site.
We find UBQLN1/2/4 to be largely outside of the proteasome in HCT116 cells, perhaps because interaction at the proteasome is transient and purposed for efficient delivery of substrates. In support of UBQLN interaction with hRpn10 being transient, we were unable to co-immunoprecipitate these proteins from cells without crosslinker. Mass spectrometry has proven sensitive to the presence of shuttle factors at the proteasome. UBQLN1 and hHR23b were both found at 26S proteasome from 293 cells under native conditions by mass spectrometry [101] and by using crosslinking assisted biomolecular tandem affinity purification combined with mass spectrometry, UBQLN2 and Ddi1 were also found at proteasomes [102].
Comparison of hRpn10 binding to UBQLN2UBL and ubiquitin
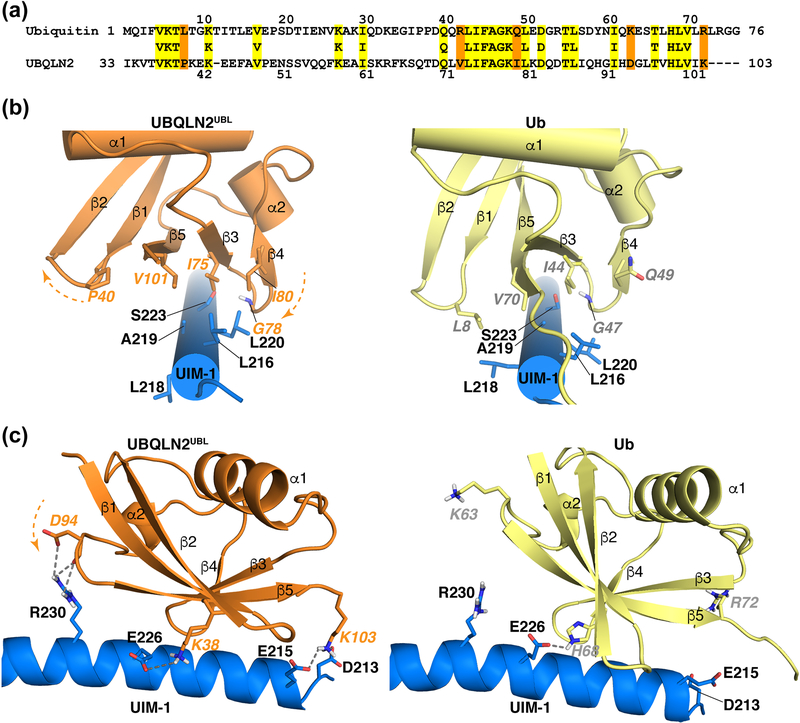
UIM-1 and UIM-2 binding to UBQLN2UBL is similar to that of ubiquitin, involving a conserved I44-A46-V70 hydrophobic interface (I75, A77, V101; Figure 7a) that centers on the ‘LALAL’ motif (Figure 7b and S6). Ubiquitin G47 is conserved in UBQLN2UBL (G78) and forms an analogous hydrogen bond to UIM-1 S223 (Figure 7b) and UIM-2 S294 (Figure S6). UIM-1, but not the shorter UIM-2, can similarly place the side chain of UBQLN2UBL K38 to mimic the hydrogen bond to UIM-1 E226 formed by ubiquitin H68 (Figure 7c). Compared to ubiquitin however, UBQLN2UBL is rotated about the ‘LALAL’ contact surface. This change in orientation places the amino acid analogous to L8 (P40) further from the UIMs (Figure 7b and S6), but provides a greater number of contacts with UBQLN2UBL I80 (ubiquitin Q49) and UBQLN2UBL V73 (ubiquitin R42). UBQLN2UBL I80 forms close contacts with UIM-1 L216 and L220 (Figure 7b) and UIM-2 I287 and M291 (Figure S6), while UBQLN2UBL V73 interacts with UIM-2 I287 (Figure S6). I80 was similarly found to be important for UBQLN2UBL binding hRpn13Pru [45] and is also present in hHR23 proteins (hHR23a I54 and hHR23b I52), where it interacts with UIM-2 M291 [42, 43]. The change in orientation also brought closer the N-terminal end of hRpn10 UIM1 and K103, which is R72 in ubiquitin. This proximity allows UBQLN2UBL K103 to form a hydrogen bond with UIM-1 E215, as well as an electrostatic interaction with UIM-1 D213 (Figure 7c). In the UIM1:ubiquitin complex, the R72 side chain is directed away from D213 and E215 (Figure 7c).
Figure 7. Comparison of hRpn10 UIM-1 binding to UBQLN2UBL and ubiquitin.
(a) Sequence alignment of UBQLN2UBL and ubiquitin by ClustalW2. Identical residues are highlighted in yellow, whereas UBQLN2UBL residues that interact uniquely with hRpn10 compared to ubiquitin are highlighted orange. (b, c) Views of the hRpn10 UIM-1 (blue):UBQLN2UBL (orange) and UIM-1(blue):ubiquitin (yellow) complexes to illustrate interactions at the contact surface. UIM1:UBQLN2UBL and UIM-1:ubiquitin are aligned by the UIM-1 helix spanning residues P214-E245. The orange arrows with dashed lines indicate rotation about the UIM-1 ‘LALAL’ contact surface relative to ubiquitin for the UBQLN2UBL β1-β2 or β3-β4 loop (b) or the α2-β5 loop (c). Key amino acids are displayed and labeled, with oxygen and nitrogen atoms colored red and blue respectively. Hydrogen bonds and electrostatic interactions are indicated with grey dashed lines.
R230 forms hydrogen bonds to the D94 side chain of UBQLN2UBL (K63 in ubiquitin) (Figure 7c), which is strictly conserved in yeast and human UBQLN proteins (Figure S5). Substitution of this aspartic acid with lysine yields a 10-fold reduction in affinity for scRpn10 [56]. Interestingly, ubiquitin is phosphorylated at S65 [103, 104], a post-translational modification linked to the activation of the E3 ligase parkin [105–108]. Superposition of S65 phosphorylated ubiquitin (PDB 4WZP) onto the UIM-1:ubiquitin complex (PDB 1YX5) demonstrates placement of the S65 phosphate group within 4 Å of the UIM-1 R230 side chain (Figure S7). Thus, phosphorylated ubiquitin may bind hRpn10 UIM-1 with higher affinity compared to unmodified ubiquitin. Indeed, ubiquitin modified by a S65 phosphomimetic caused decreased protein degradation and turnover in vivo [109], as has been observed by UBQLN over-expression [55]. Future experiments are needed to test this model.
The parkin UBL domain also interacts with hRpn10196–306 [110], and primarily through UIM-1 [111]. We generated a model structure for parkinUBL: hRpn10196–306 UIM-1, which suggested that similar interactions could be formed compared to UBQLN2UBL. For example, D62 from the parkin α2-β5 loop appears to engage UIM-1 R230 in a manner similar to D94 of UBQLN2UBL (Figure S8). An experimentally determined structure is needed however to validate this possibility.
Complementarity among the proteasome shuttle factors for receptor sites
It was previously demonstrated that loss of the Rpn1, Rpn10, and Rpn13 ubiquitin-binding sites in yeast leads to loss of Dsk2 and Rad23 interaction with the proteasome [6], suggesting that shuttle factors must compete with ubiquitin chains for access to the proteasome. Rpn1 binds preferentially to Rad23 over Dsk2 and ubiquitin [6, 45] whereas Rpn13 prefers Dsk2 [4, 45]. In mice, liver-specific deletion of both Rpn10 and Rpn13 caused severe liver injury accompanied by accumulation of ubiquitin conjugates [112]. hHR23b and UBQLN1/4 were unable to bind proteasome of these mice [112], suggesting that Rpn10 and Rpn13 are required for interaction of these shuttle factors with the proteasome.
The hRpn10 UIM-2 has long been known to bind to hHR23 [39] and here, we demonstrate complementary preference for UBQLN at hRpn10 UIM-1. Altogether, these findings indicate a binding hierarchy for UBL-UBA shuttle factors at the proteasome that would presumably dictate which receptors have accessible sites for interaction with ubiquitin. For example, our findings indicate that UBQLN proteins are more likely to bind to hRpn10 UIM-1 leaving hRpn10 UIM-2 available for interaction with ubiquitin chains attached to shuttled substrates or shuttle factor hHR23a/b.
Materials and Methods
Protein sample preparation
hRpn10196–306 and UBQLN2UBL were expressed and purified as described previously [8, 41, 45]. 15N ammonium chloride, 13C glucose, and 2H2O were used for isotope labeling. 15N, 100% 2H-labeled hRpn10196–306 was prepared by following the method described in [96]. M1, K6, K11, K48 and K63 diubiquitin were produced as described [113–116]. K27, K29, and K33 diubiquitin were purchased (UBPBio). All NMR samples were validated by LC/mass spectrometry. All NMR experiments were performed at 25°C on Bruker Avance 700, 800, or 850 MHz spectrometers equipped with cryogenically cooled probes.
His pull-down assays
His pull-down assays were performed as described in [6, 45]. Briefly, 500 pmol of purified Histagged hRpn10196–306 was added to 20 μL of pre-washed Ni-NTA agarose resin for one hour and washed once with Buffer 1 (50 mM HEPES, 50 mM NaCl, 5% (v/v) glycerol, and 15 mM 2 mercaptoethanol at pH 6.7). The resin was then incubated with 500 pmol of M1, K6, K11, K27, K29, K33, K48, and K63 diubiquitin for one hour. Intrinsically disordered protein SocB with a C-terminal His-tag [80] was used as a negative control with K48 diubiquitin. Unbound protein was removed by extensive washing in the above buffer. The resin-bound proteins were fractionated by electrophoresis and transferred to a PVDF membrane. The membrane was treated with denaturing Buffer 2 (20 mM Tris-HCl, 6 M guanidine hydrochloride, 1 mM PMSF, and 5 μM 2 -mercaptoethanol at pH 7.4) for 30 mins at 4°C, extensively washed with Trisbuffered saline +0.1% Tween, and analyzed by immunoblotting. Diubiquitin was detected with mouse anti-Ub antibody (Millipore Sigma MAB1510, 1:1000) followed by HRP-conjugated anti-mouse IgG (Millipore Sigma, 1:5000). His-tagged hRpn10196–306 or SocB-His was detected with mouse anti-His antibody (Thermo Fisher Scientific MA1–21315, 1:1000) followed by HRP-conjugated anti-mouse IgG (Millipore Sigma, 1:5000).
Glycerol gradient centrifugation and fractionation
A 10 – 40% glycerol gradient was made in a total volume of 10.5 mL, with layers of stepwise glycerol increment of 5%. Each layer was frozen in liquid nitrogen before adding the subsequent layer, as described previously [117]. The gradient was thawed overnight at 4°C prior to use. Whole cell lysate from HCT116 cells prepared in Buffer 2 (50 mM Tris-HCl, 1 mM DTT, 1 mM ATP, 5 mM MgCl2, pH 7.6) was added to the top of the gradient, which was then subjected to 131,569 g for 18 hours at 4°C. Gradient fractions were collected, resolved by SDS-PAGE (4–12% Bis-Tris gel), and immunoprobed with antibodies against UBQLN1/2/4 (Thermo Fisher Scientific 3D5E2, 1:1000), anti-hRpn10 (Cell Signaling Technology D20B2, 1:1000), anti-hRpn2 (Abcam ab2941, 1:1000), anti-β5 (Enzo Life Sciences, Inc. BML-PW 8895–0100, 1:1000) and β-actin (Cell Signaling Technology 13E5, 1:3000).
Crosslinking, immunoprecipitation and immunoblotting
HCT116 cell line was purchased from the American Type Culture Collection (ATCC CCL-247). Cells were grown in McCoy’s 5A modified medium (ATCC), supplemented with 10% fetal bovine serum (Atlanta Biologicals, Inc.) in a 37oC humidified atmosphere of 5% CO2. HCT116 cells were crosslinked using 1 mM DSP (Thermo Fisher Scientific) or DMSO as a control, and incubated at room temperature for 30 mins. The reaction was quenched for 15 minutes with 20 mM Tris-HCl (pH 7.5). Cells were collected, washed with PBS, and lysed in RIPA buffer (Thermo Fisher Scientific) supplemented with protease inhibitor cocktail (Roche). Total protein concentration was determined by Pierce bicinchoninic acid (BCA) Protein Assay Kit (Thermo Fisher Scientific). Lysates were precleared with protein G sepharose (Millipore Sigma) for an hour, incubated with anti-UBQLN1/2/4 antibodies (Thermo Fisher Scientific 3D5E2, 1:100) overnight at 4 ° C, and then for 3 hours with protein G sepharose. After extensive washing, proteins bound to protein G sepharose were eluted and analyzed by immunoblotting against hRpn10 (Cell Signaling Technology D20B2, 1:500), and UBQLN1/2/4 (Thermo Fisher Scientific 3D5E2, 1:500).
NMR titration experiments
1H, 15N HSQC experiments [118, 119] were recorded on 15N-labeled samples (hRpn10196–306 or UBQLN2UBL) with increasing molar ratios of unlabeled ligands (UBQLN2UBL or hRpn10196–306) as indicated. NMRPipe [120] was used for data processing and the resulting spectra visualized with XEASY [121]. The amide nitrogen and hydrogen chemical shift perturbations (CSP) were mapped for each amino acid according to Equation 1.
| (Eq. 1) |
ΔδH, change in amide proton value (in parts per million); ΔδN, change in amide nitrogen value (in parts per million).
KD values were determined by using 1H, 15N HSQC spectra recorded on 0.15 mM of 15N-labeled hRpn10196–306 with increasing molar ratio of unlabeled UBQLN2UBL (hRpn10196–306: UBQLN2UBL at 1:0, 1:0.056, 1:0.1125, 1:0.225, 1:0.45, 1:0.9, 1:1.35, 1:1.8, 1:2.25 as indicated). ΔδH for UIM-1 residues (D209, D213, L218, S223, and E225) and UIM-2 residues (E283, M291, M293, L295 and Q296) were plotted against varying molar ratio of UBQLN2UBL to hRpn10196–306. These data were inputted into the Bindfit v0.5 software [90, 91] and fit to a two-site binding mode [89].
NOESY experiments for the hRpn10196–306:UBQLN2UBL complex
All spectra were conducted in Buffer 4 (20 mM NaPO4, 50 mM NaCl, 2 mM DTT, 0.1% NaN3, and 5% 2H2O / 95% 1H2O at pH 6.5), except for 2D 1H-13C HSQC, 3D HCCH-TOCSY and 13Cedited NOESY experiments for which the sample was dissolved in 2H2O. Chemical shift assignments of hRpn10196–306 and UBQLN2UBL in free states were available from previous work [8, 41]. To solve the structure of the hRpn10196–306:UBQLN2UBL, 15N or 13C NOESY experiments [122, 123] were acquired on 15N or 13C-labeled hRpn10196–306 mixed with 2.3-fold molar excess of unlabeled UBQLN2UBL to assign hRpn10196–306 in its bound state. Similarly, hRpn10196–306-bound UBQLN2UBL was assigned by 15N or 13C NOESY experiments acquired on 15N or 13C-labeled UBQLN2UBL mixed with unlabeled hRpn10196–306 at equimolar ratio. These spectra were also used to generate intramolecular NOE distance constraints for use in the structure calculations. Intermolecular distance constraints were determined by 13C-half-filtered 3D NOESY experiments (100 ms mixing time) performed on samples with 13C-labeled hRpn10196–306 mixed with 2.3-fold molar excess of unlabeled UBQLN2UBL and 13C-labeled UBQLN2UBL mixed with equimolar unlabeled hRpn10196–306. Additional intermolecular NOE interactions between hRpn10196–306 and UBQLN2UBL were obtained through a 15N-dispersed NOESY spectrum (200 ms mixing time) [123] recorded on a sample containing 15N, 100% 2H-labeled hRpn10196–306 mixed with 2.3-fold molar excess of unlabeled UBQLN2UBL [96]. Spin diffusion is greatly reduced in deuterated proteins, allowing for the longer NOESY mixing time and in turn, distance measurements greater than 5 Å [124, 125]. In this spectrum, all amide to aliphatic NOE crosspeaks are exclusively inter-molecular [94–96].
Structure determination of hRpn10196–306:UBQLN2UBL complex
The structure of hRpn10196–306 with two UBQLN2UBL molecules bound was determined with XPLOR-NIH 2.47 [126] on a Linux operating system by using NOE and hydrogen bond constraints as well as backbone ϕ and Ψ torsion angle constraints derived from TALOS+ [127] (Table 1); the resulting TALOS+ plot is included for hRpn10196–306 (Figure S9). Hydrogen bonds were generated by using secondary structure assignments and NOE connectivities with defined distances from the acceptor oxygen to the donor hydrogen and nitrogen of 1.8–2.1 Å and 2.5– 2.9 Å, respectively. Hydrogen bonds restraints were not included in the initial calculation, but were in the final round of structure calculations. When calculating the structures of hRpn10196–306:UBQLN2UBL, intermolecular distance constraints determined from the 13C-half-filtered 3D NOESY experiments were used, in addition to intramolecular constraints for hRpn10196–306 and UBQLN2UBL that were generated from 15N or 13C NOESY spectra acquired on the complexes (Table 1). All complexed structures were calculated from 50 linear starting structures of hRpn10196–306 and two UBQLN2UBL molecules, which were subjected to 2,000 steps of initial energy minimization to ensure full spatial sampling and appropriate coordinate geometry. The structures were next confined according to the inputted data by subjecting them to 55,000 simulated annealing steps of 0.005 ps at 3,000 K, followed by 5,000 cooling steps of 0.005 ps. 5,000 steps of energy minimization were applied to produce the final structures, which were recorded as coordinate files. The resulting structures had no distance or dihedral angle violation greater than 0.3 Å or 5°, respectively. The ten lowest energy structures were chosen for visualization and statistical analyses.
Model structure of UIM-1 complexed with S65-phosphorylated ubiquitin or parkinUBL
To generate a model structure of hRpn10 UIM-1: S65-phosphorylated ubiquitin, the structure of S65-phosphorylated ubiquitin (PDB 4WZP) was superimposed onto ubiquitin of the UIM-1:ubiquitin complex (PDB 1YX5) in PyMOL. A model structure of hRpn10 UIM-1:parkinUBL was similarly generated by superimposing the structure of parkinUBL (PDB 1IYF) onto UBQLN2UBL of the hRpn10196–306 UIM-1:UBQLN2UBL complex in PyMOL. These model structures were subsequently energy minimized by Shrödinger Maestro [128].
ACCESSION NUMBERS
Atomic coordinates for hRpn10196–306:UBQLN2UBL have been deposited in the Protein Data Bank (PDB) with accession number 6MUN. Chemical shift assignments have been deposited in the Biological Magnetic Resonance Data Bank (BMRB) with accession number 30528.
Supplementary Material
Highlights.
Although versatile for Ub chains, hRpn10 prefers K48 and K11 linkages
UBQLN1/2/4 is largely extra-proteasomal in HCT116 cells but crosslinks to hRpn10
UBQLN UBL has a 25-fold binding preference for hRpn10 UIM-1 over UIM-2
Interactions beyond the ‘LALAL’ motif contribute to UBQLN2 UBL preference for UIM-1
UBQLN UBL is rotated about the hRpn10 ‘LALAL’ compared to ubiquitin
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program through the Center for Cancer Research, National Cancer Institute (NCI), National Institutes of Health (1 ZIA BC011490). We are grateful to Janusz Koscielniak for maintaining the NMR facility.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Finley D, Chen X, Walters KJ. Gates, Channels, and Switches: Elements of the Proteasome Machine. Trends Biochem Sci. 2016;41:77–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ehlinger A, Walters KJ. Structural insights into proteasome activation by the 19S regulatory particle. Biochemistry. 2013;52:3618–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–61. [PubMed] [Google Scholar]
- [4].Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, et al. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Shi Y, Chen X, Elsasser S, Stocks BB, Tian G, Lee BH, et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science. 2016;351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–71. [DOI] [PubMed] [Google Scholar]
- [8].Wang Q, Young P, Walters KJ. Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J Mol Biol. 2005;348:727–39. [DOI] [PubMed] [Google Scholar]
- [9].Zhang N, Wang Q, Ehlinger A, Randles L, Lary JW, Kang Y, et al. Structure of the s5a:k48linked diubiquitin complex and its interactions with rpn13. Mol Cell. 2009;35:280–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lasker K, Forster F, Bohn S, Walzthoeni T, Villa E, Unverdorben P, et al. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci U S A. 2012;109:1380–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–5. [DOI] [PubMed] [Google Scholar]
- [13].Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–7. [DOI] [PubMed] [Google Scholar]
- [14].Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, et al. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. [DOI] [PubMed] [Google Scholar]
- [15].Lee BH, Lu Y, Prado MA, Shi Y, Tian G, Sun S, et al. USP14 deubiquitinates proteasome-bound substrates that are ubiquitinated at multiple sites. Nature. 2016;532:398–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–40. [DOI] [PubMed] [Google Scholar]
- [17].Sanchez-Pulido L, Kong L, Ponting CP. A common ancestry for BAP1 and Uch37 regulators. Bioinformatics. 2012;28:1953–6. [DOI] [PubMed] [Google Scholar]
- [18].Hamazaki J, Iemura S, Natsume T, Yashiroda H, Tanaka K, Murata S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 2006;25:4524–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L, Goldberg AL. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006;25:5742–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chen X, Lee BH, Finley D, Walters KJ. Structure of proteasome ubiquitin receptor hRpn13 and its activation by the scaffolding protein hRpn2. Mol Cell. 2010;38:404–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chen X, Walters KJ. Structural plasticity allows UCH37 to be primed by RPN13 or locked down by INO80G. Molecular cell. 2015;57:767–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sahtoe DD, van Dijk WJ, El Oualid F, Ekkebus R, Ovaa H, Sixma TK. Mechanism of UCH-L5 activation and inhibition by DEUBAD domains in RPN13 and INO80G. Molecular cell. 2015;57:887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].VanderLinden RT, Hemmis CW, Schmitt B, Ndoja A, Whitby FG, Robinson H, et al. Structural basis for the activation and inhibition of the UCH37 deubiquitylase. Molecular cell. 2015;57:901–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. [DOI] [PubMed] [Google Scholar]
- [25].Koulich E, Li X, DeMartino GN. Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol Biol Cell. 2008;19:1072–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lee B-H, Lee MJ, Park S, Oh D-C, Elsasser S, Chen P-C, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Molecular cell. 2007;27:731–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gillette TG, Kumar B, Thompson D, Slaughter CA, DeMartino GN. Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome. J Biol Chem. 2008;283:31813–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Molecular cell. 2008;30:360–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature. 2002;416:763–7. [DOI] [PubMed] [Google Scholar]
- [31].Paraskevopoulos K, Kriegenburg F, Tatham MH, Rosner HI, Medina B, Larsen IB, et al. Dss1 is a 26S proteasome ubiquitin receptor. Mol Cell. 2014;56:453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tomko RJ Jr., Hochstrasser M The intrinsically disordered Sem1 protein functions as a molecular tether during proteasome lid biogenesis. Mol Cell. 2014;53:433–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bohn S, Sakata E, Beck F, Pathare GR, Schnitger J, Nagy I, et al. Localization of the regulatory particle subunit Sem1 in the 26S proteasome. Biochemical and biophysical research communications. 2013;435:250–4. [DOI] [PubMed] [Google Scholar]
- [34].Keren-Kaplan T, Zeev Peters L, Levin-Kravets O, Attali I, Kleifeld O, Shohat N, et al. Structure of ubiquitylated-Rpn10 provides insight into its autoregulation mechanism. Nat Commun. 2016;7:12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Finley D Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Walters KJ, Goh AM, Wang Q, Wagner G, Howley PM. Ubiquitin family proteins and their relationship to the proteasome: a structural perspective. Biochim Biophys Acta. 2004;1695:73–87. [DOI] [PubMed] [Google Scholar]
- [37].Bertolaet BL, Clarke DJ, Wolff M, Watson MH, Henze M, Divita G, et al. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat Struct Biol. 2001;8:417–22. [DOI] [PubMed] [Google Scholar]
- [38].Wilkinson CR, Seeger M, Hartmann-Petersen R, Stone M, Wallace M, Semple C, et al. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat Cell Biol. 2001;3:939–43. [DOI] [PubMed] [Google Scholar]
- [39].Hiyama H, Yokoi M, Masutani C, Sugasawa K, Maekawa T, Tanaka K, et al. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J Biol Chem. 1999;274:28019–25. [DOI] [PubMed] [Google Scholar]
- [40].Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Müller B, et al. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–30. [DOI] [PubMed] [Google Scholar]
- [41].Walters KJ, Kleijnen MF, Goh AM, Wagner G, Howley PM. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry. 2002;41:1767–77. [DOI] [PubMed] [Google Scholar]
- [42].Mueller TD, Feigon J. Structural determinants for the binding of ubiquitin-like domains to the proteasome. EMBO J. 2003;22:4634–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Fujiwara K, Tenno T, Sugasawa K, Jee JG, Ohki I, Kojima C, et al. Structure of the ubiquitin-interacting motif of S5a bound to the ubiquitin-like domain of HR23B. The Journal of biological chemistry. 2004;279:4760–7. [DOI] [PubMed] [Google Scholar]
- [44].Rosenzweig R, Bronner V, Zhang D, Fushman D, Glickman MH. Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. J Biol Chem. 2012;287:14659–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chen X, Randles L, Shi K, Tarasov SG, Aihara H, Walters KJ. Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome. Structure. 2016;24:1257–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gomez TA, Kolawa N, Gee M, Sweredoski MJ, Deshaies RJ. Identification of a functional docking site in the Rpn1 LRR domain for the UBA-UBL domain protein Ddi1. BMC Biol. 2011;9:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Chojnacki M, Mansour W, Hameed DS, Singh RK, El Oualid F, Rosenzweig R, et al. Polyubiquitin-Photoactivatable Crosslinking Reagents for Mapping Ubiquitin Interactome Identify Rpn1 as a Proteasome Ubiquitin-Associating Subunit. Cell Chem Biol. 2017;24:443–57 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kang Y, Vossler RA, Diaz-Martinez LA, Winter NS, Clarke DJ, Walters KJ. UBL/UBA ubiquitin receptor proteins bind a common tetraubiquitin chain. Journal of molecular biology. 2006;356:1027–35. [DOI] [PubMed] [Google Scholar]
- [49].Kang Y, Zhang N, Koepp DM, Walters KJ. Ubiquitin receptor proteins hHR23a and hPLIC2 interact. Journal of molecular biology. 2007;365:1093–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Walters KJ, Lech PJ, Goh AM, Wang Q, Howley PM. DNA-repair protein hHR23a alters its protein structure upon binding proteasomal subunit S5a. Proc Natl Acad Sci U S A. 2003;100:12694–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Raasi S, Pickart CM. Rad23 ubiquitin-associated domains (UBA) inhibit 26 S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J Biol Chem. 2003;278:8951–9. [DOI] [PubMed] [Google Scholar]
- [52].Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Molecular cell. 2005;18:687–98. [DOI] [PubMed] [Google Scholar]
- [53].Zhang D, Raasi S, Fushman D. Affinity makes the difference: nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J Mol Biol. 2008;377:162–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kang Y, Chen X, Lary JW, Cole JL, Walters KJ. Defining how ubiquitin receptors hHR23a and S5a bind polyubiquitin. Journal of molecular biology. 2007;369:168–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Matiuhin Y, Kirkpatrick DS, Ziv I, Kim W, Dakshinamurthy A, Kleifeld O, et al. Extraproteasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol Cell. 2008;32:415–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zhang D, Chen T, Ziv I, Rosenzweig R, Matiuhin Y, Bronner V, et al. Together, Rpn10 and Dsk2 can serve as a polyubiquitin chain-length sensor. Mol Cell. 2009;36:1018–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lipinszki Z, Pal M, Nagy O, Deak P, Hunyadi-Gulyas E, Udvardy A. Overexpression of Dsk2/dUbqln results in severe developmental defects and lethality in Drosophila melanogaster that can be rescued by overexpression of the p54/Rpn10/S5a proteasomal subunit. FEBS J. 2011;278:4833–44. [DOI] [PubMed] [Google Scholar]
- [58].Jantrapirom S, Lo Piccolo L, Yoshida H, Yamaguchi M. A new Drosophila model of Ubiquilin knockdown shows the effect of impaired proteostasis on locomotive and learning abilities. Exp Cell Res. 2018;362:461–71. [DOI] [PubMed] [Google Scholar]
- [59].Marin I The ubiquilin gene family: evolutionary patterns and functional insights. BMC Evol Biol. 2014;14:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kleijnen MF, Shih AH, Zhou P, Kumar S, Soccio RE, Kedersha NL, et al. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Molecular cell. 2000;6:409–19. [DOI] [PubMed] [Google Scholar]
- [61].Itakura E, Zavodszky E, Shao S, Wohlever ML, Keenan RJ, Hegde RS. Ubiquilins Chaperone and Triage Mitochondrial Membrane Proteins for Degradation. Molecular cell. 2016;63:21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Suzuki R, Kawahara H. UBQLN4 recognizes mislocalized transmembrane domain proteins and targets these to proteasomal degradation. EMBO reports. 2016;17:842–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hjerpe R, Bett JS, Keuss MJ, Solovyova A, McWilliams TG, Johnson C, et al. UBQLN2 Mediates Autophagy-Independent Protein Aggregate Clearance by the Proteasome. Cell. 2016;166:935–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Whiteley AM, Prado MA, Peng I, Abbas AR, Haley B, Paulo JA, et al. Ubiquilin1 promotes antigen-receptor mediated proliferation by eliminating mislocalized mitochondrial proteins. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Subudhi I, Shorter J. Ubiquilin 2: Shuttling Clients Out of Phase? Molecular cell. 2018;69:919–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Dao TP, Kolaitis RM, Kim HJ, O'Donovan K, Martyniak B, Colicino E, et al. Ubiquitin Modulates Liquid-Liquid Phase Separation of UBQLN2 via Disruption of Multivalent Interactions. Molecular cell. 2018;69:965–78 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Mah AL, Perry G, Smith MA, Monteiro MJ. Identification of ubiquilin, a novel presenilin interactor that increases presenilin protein accumulation. J Cell Biol. 2000;151:847–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Hiltunen M, Lu A, Thomas AV, Romano DM, Kim M, Jones PB, et al. Ubiquilin 1 modulates amyloid precursor protein trafficking and Abeta secretion. J Biol Chem. 2006;281:32240–53. [DOI] [PubMed] [Google Scholar]
- [69].Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Stieren ES, El Ayadi A, Xiao Y, Siller E, Landsverk ML, Oberhauser AF, et al. Ubiquilin-1 is a molecular chaperone for the amyloid precursor protein. J Biol Chem. 2011;286:35689–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Lu A, Hiltunen M, Romano DM, Soininen H, Hyman BT, Bertram L, et al. Effects of ubiquilin 1 on the unfolded protein response. J Mol Neurosci. 2009;38:19–30. [DOI] [PubMed] [Google Scholar]
- [72].Heir R, Ablasou C, Dumontier E, Elliott M, Fagotto-Kaufmann C, Bedford FK. The UBL domain of PLIC-1 regulates aggresome formation. EMBO reports. 2006;7:1252–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].N'Diaye EN, Kajihara KK, Hsieh I, Morisaki H, Debnath J, Brown EJ. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO reports. 2009;10:173–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Rothenberg C, Srinivasan D, Mah L, Kaushik S, Peterhoff CM, Ugolino J, et al. Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum Mol Genet. 2010;19:3219–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Wang H, Lim PJ, Yin C, Rieckher M, Vogel BE, Monteiro MJ. Suppression of polyglutamine-induced toxicity in cell and animal models of Huntington's disease by ubiquilin. Hum Mol Genet. 2006;15:1025–41. [DOI] [PubMed] [Google Scholar]
- [76].Wang H, Monteiro MJ. Ubiquilin interacts and enhances the degradation of expanded-polyglutamine proteins. Biochem Biophys Res Commun. 2007;360:423–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Williams KL, Warraich ST, Yang S, Solski JA, Fernando R, Rouleau GA, et al. UBQLN2/ubiquilin 2 mutation and pathology in familial amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33:2527 e3–10. [DOI] [PubMed] [Google Scholar]
- [78].Edens BM, Yan J, Miller N, Deng HX, Siddique T, Ma YC. A novel ALS-associated variant in UBQLN4 regulates motor axon morphogenesis. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017;547:413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Nowicka U, Hoffman M, Randles L, Shi X, Khavrutskii L, Stefanisko K, et al. Mycobacterium tuberculosis copper-regulated protein SocB is an intrinsically disordered protein that folds upon interaction with a synthetic phospholipid bilayer. Proteins. 2016:193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Kristariyanto YA, Abdul Rehman SA, Weidlich S, Knebel A, Kulathu Y. A single MIU motif of MINDY-1 recognizes K48-linked polyubiquitin chains. EMBO reports. 2017;18:392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Castaneda CA, Dixon EK, Walker O, Chaturvedi A, Nakasone MA, Curtis JE, et al. Linkage via K27 Bestows Ubiquitin Chains with Unique Properties among Polyubiquitins. Structure. 2016;24:423–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Pan M, Gao S, Zheng Y, Tan X, Lan H, Tan X, et al. Quasi-Racemic X-ray Structures of K27-Linked Ubiquitin Chains Prepared by Total Chemical Synthesis. Journal of the American Chemical Society. 2016;138:7429–35. [DOI] [PubMed] [Google Scholar]
- [84].Piterman R, Braunstein I, Isakov E, Ziv T, Navon A, Cohen S, et al. VWA domain of S5a restricts the ability to bind ubiquitin and Ubl to the 26S proteasome. Mol Biol Cell. 2014;25:398–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Isasa M, Katz EJ, Kim W, Yugo V, Gonzalez S, Kirkpatrick DS, et al. Monoubiquitination of RPN10 regulates substrate recruitment to the proteasome. Molecular cell. 2010;38:733–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Zuin A, Bichmann A, Isasa M, Puig-Sarries P, Diaz LM, Crosas B. Rpn10 monoubiquitination orchestrates the association of the ubiquilin-type DSK2 receptor with the proteasome. Biochem J. 2015;472:353–65. [DOI] [PubMed] [Google Scholar]
- [87].Safren N, Chang L, Dziki KM, Monteiro MJ. Signature changes in ubiquilin expression in the R6/2 mouse model of Huntington's disease. Brain Res. 2015;1597:37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Wang Q, Walters KJ. Chemical shift assignments of the (poly)ubiquitin-binding region of the proteasome subunit S5a. Journal of biomolecular NMR. 2004;30:231–2. [DOI] [PubMed] [Google Scholar]
- [89].Arai M, Ferreon JC, Wright PE. Quantitative analysis of multisite protein-ligand interactions by NMR: binding of intrinsically disordered p53 transactivation subdomains with the TAZ2 domain of CBP. Journal of the American Chemical Society. 2012;134:3792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Thordarson P Determining association constants from titration experiments in supramolecular chemistry. Chem Soc Rev. 2011;40:1305–23. [DOI] [PubMed] [Google Scholar]
- [91].Brynn Hibbert D, Thordarson P. The death of the Job plot, transparency, open science and online tools, uncertainty estimation methods and other developments in supramolecular chemistry data analysis. Chem Commun (Camb). 2016;52:12792–805. [DOI] [PubMed] [Google Scholar]
- [92].Ko HS, Uehara T, Tsuruma K, Nomura Y. Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett. 2004;566:110–4. [DOI] [PubMed] [Google Scholar]
- [93].Young P, Deveraux Q, Beal RE, Pickart CM, Rechsteiner M. Characterization of two polyubiquitin binding sites in the 26 S protease subunit 5a. The Journal of biological chemistry. 1998;273:5461–7. [DOI] [PubMed] [Google Scholar]
- [94].Chen X, Walters KJ. Identifying and studying ubiquitin receptors by NMR. Methods Mol Biol. 2012;832:279–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Walters KJ, Ferentz AE, Hare BJ, Hidalgo P, Jasanoff A, Matsuo H, et al. Characterizing protein-protein complexes and oligomers by nuclear magnetic resonance spectroscopy. Methods in enzymology. 2001;339:238–58. [DOI] [PubMed] [Google Scholar]
- [96].Walters KJ, Matsuo H, Wagner G. A simple method to distinguish intermonomer nuclear Overhauser effects in homodimeric proteins with C2 symmetry. J Am Chem Soc. 1997;119:5958–9. [Google Scholar]
- [97].Kirkpatrick DS, Hathaway Na, Hanna J, Elsasser S, Rush J, Finley D, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–10. [DOI] [PubMed] [Google Scholar]
- [98].Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Yau RG, Doerner K, Castellanos ER, Haakonsen DL, Werner A, Wang N, et al. Assembly and Function of Heterotypic Ubiquitin Chains in Cell-Cycle and Protein Quality Control. Cell. 2017;171:918–33 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Wang X, Chen CF, Baker PR, Chen PL, Kaiser P, Huang L. Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry. 2007;46:3553–65. [DOI] [PubMed] [Google Scholar]
- [102].Yu C, Yang Y, Wang X, Guan S, Fang L, Liu F, et al. Characterization of Dynamic UbR-Proteasome Subcomplexes by In vivo Cross-linking (X) Assisted Bimolecular Tandem Affinity Purification (XBAP) and Label-free Quantitation. Mol Cell Proteomics. 2016;15:2279–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–48. [DOI] [PubMed] [Google Scholar]
- [104].Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–6. [DOI] [PubMed] [Google Scholar]
- [106].Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, Hofmann K, et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J. 2014;460:127–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Wauer T, Swatek KN, Wagstaff JL, Gladkova C, Pruneda JN, Michel MA, et al. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. The EMBO journal. 2015;34:307–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Swaney DL, Rodriguez-Mias RA, Villen J. Phosphorylation of ubiquitin at Ser65 affects its polymerization, targets, and proteome-wide turnover. EMBO reports. 2015;16:1131–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Sakata E, Yamaguchi Y, Kurimoto E, Kikuchi J, Yokoyama S, Yamada S, et al. Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain. EMBO reports. 2003;4:301–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Safadi SS, Shaw GS. Differential interaction of the E3 ligase parkin with the proteasomal subunit S5a and the endocytic protein Eps15. The Journal of biological chemistry. 2010;285:1424–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Hamazaki J, Hirayama S, Murata S. Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis. PLoS Genet. 2015;11:e1005401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Raasi S, Pickart CM. Ubiquitin chain synthesis. Methods Mol Biol. 2005;301:47–55. [DOI] [PubMed] [Google Scholar]
- [114].Reyes-Turcu FE, Shanks JR, Komander D, Wilkinson KD. Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J Biol Chem. 2008;283:19581–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Bremm A, Komander D. Synthesis and analysis of K11-linked ubiquitin chains. Methods Mol Biol. 2012;832:219–28. [DOI] [PubMed] [Google Scholar]
- [116].Hospenthal MK, Freund SM, Komander D. Assembly, analysis and architecture of atypical ubiquitin chains. Nat Struct Mol Biol. 2013;20:555–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Luthe DS. A simple technique for the preparation and storage of sucrose gradients. Anal Biochem. 1983;135:230–2. [DOI] [PubMed] [Google Scholar]
- [118].Mori S, Abeygunawardana C, Johnson MO, Vanzijl PCM. Improved Sensitivity of HSQC Spectra of Exchanging Protons at Short Interscan Delays Using a New Fast HSQC (FHSQC) Detection Scheme That Avoids Water Saturation. J Magn Reson B. 1995;108:94–8. [DOI] [PubMed] [Google Scholar]
- [119].Czisch M, Boelens R. Sensitivity enhancement in the TROSY experiment. Journal of magnetic resonance. 1998;134:158–60. [DOI] [PubMed] [Google Scholar]
- [120].Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:27793. [DOI] [PubMed] [Google Scholar]
- [121].Bartels C, Xia TH, Billeter M, Guntert P, Wuthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR. 1995;6:1–10. [DOI] [PubMed] [Google Scholar]
- [122].Davis AL, Keeler J, Laue ED, Moskau D. Experiments for recording pure-absorption heteronuclear correlation spectra using pulsed field gradients. J Magn Reson. 1992;98:207–16. [Google Scholar]
- [123].Sklenar V, Piotto M, Leppik R, Saudek V. Gradient-Tailored Water Suppression for 1H15N HSQC Experiments Optimized to Retain Full Sensitivity. J Magn Reson A. 1993;102:241–5. [Google Scholar]
- [124].Venters RA, Metzler WJ, Spicer LD, Mueller L, Farmer BT, II. Use of 1HN-1HN NOEs to Determine Protein Global Folds in Perdeuterated Proteins. J Am Chem Soc. 1995;117:9592–3. [Google Scholar]
- [125].Grzesiek S, Wingfield P, Stahl S, Kaufman JD, Bax A. Four-Dimensional 15N-Separated NOESY of Slowly Tumbling Perdeuterated 15N-Enriched Proteins. Application to HIV-1 Nef. J Am Chem Soc. 1995;117:9594–5. [Google Scholar]
- [126].Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. [DOI] [PubMed] [Google Scholar]
- [127].Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Schrödinger Release 2017–4: Maestro. New York, NY: Schrödinger, LLC; 2017. [Google Scholar]
- [129].Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.