Abstract
The CRISPR/Cas9 system can efficiently introduce biallelic mutations in ES cells (ESCs), and its application with fluorescently-tagged ESCs enables phenotype analysis in chimeric mice. We have utilized ESCs that express EGFP in the cytosol and acrosome [EGR-G101 129S2 × (CAG/Acr-EGFP) B6] in previous studies; however, the EGFP signal in the sperm cytosol is weak and the signal in the acrosome is lost after the acrosome reaction, precluding analysis between wild type and ESC derived spermatozoa. In this study, we established an ESC line from RBGS (Red Body Green Sperm) transgenic mice [B6D2-Tg (CAG/Su9-DsRed2, Acr3-EGFP) RBGS002Osb] whose spermatozoa exhibit green fluorescence in the acrosome and red fluorescence in the mitochondria within the flagellar midpiece that is retained after the acrosome reaction. We utilized these new ESCs to analyze HYDIN, which is reported to function in sperm motility in humans. Analysis of Hydin-disrupted spermatozoa in mice is difficult as Hydin-mutant mice (hy3) die within 3 weeks, before sexual maturation, due to hydrocephaly. To circumvent the early lethality of the whole-body knockout, we disrupted Hydin in RBGS-ESCs and generated chimeric mice, which survived into sexual maturity. Hydin-disrupted spermatozoa obtained from the chimeric mice possessed short tails and were immotile. When we injected Hydin-disrupted spermatozoa into oocytes, heterozygous pups were obtained, which suggests that the genome of Hydin-disrupted spermatozoa can produce viable pups. Consequently, RBGS-ESCs can be a useful tool for screening and analysis of male-fertility related genes in chimeric mice.
Keywords: CRISPR/Cas9, embryonic stem cell, flagellum, male reproduction, mouse
Introduction
Spermatozoa comprise a head region that contains the paternal haploid genomic information and the flagellum that propels the spermatozoa to the oocyte. In addition to the haploid genome, the head contains the acrosome, a large cytoplasmic organelle containing peptidases and receptors necessary for binding to the egg. The flagellum is composed of the 9+2 axoneme along the entire length that can further be divided into three parts: the midpiece, principal piece, and endpiece [8]. The midpiece is located proximal to the head and contains tightly packed mitochondria, whereas the principle piece contains the fibrous sheath providing rigidity to the flagellum, the end piece contains only the axoneme covered by the plasma membrane [8]. Any defects in the formation of the head or flagellum structures often lead to male infertility.
As generating fully differentiated and motile spermatozoa in vitro remains difficult [7, 17, 24], gene-modified mice have been the most powerful method to analyze the function of male fertility-related genes. However, the time and cost for generating gene-modified mice using the conventional homologous recombination method [11] in embryonic stem cells (ESCs) remains high. Recently, the clustered regularly interspaced short palindromic repeats (CRISPR)/ CRISPR associated proteins 9 (Cas9) system [6, 21] has decreased the cost and shortened the time required for generating gene-modified mice. For instance, by using the CRISPR/Cas9 system in zygotes, biallelic mutations are possible to obtain knockout (KO) mice within a month [12, 21]. In addition to zygotes, the CRISPR/Cas9 system can introduce biallelic mutations efficiently into ESCs [14, 21]. By combining the CRISPR/Cas9 system with ESCs expressing fluorescent proteins, chimeric analysis in the founder generation is possible since all KO cells will be fluorescently labeled [14]. Chimeric analysis has the benefit of overcoming early lethality seen in full body KO mice due to rescue by wild-type (WT) cells derived from the host embryo, which allows phenotype analysis of differentiated cells [14].
Previously, we established an ESC line (G101) from transgenic (TG) mice [C57BL/6-Tg (CAG/Acr-EGFP) C3-N01-FJ002Osb] [2], in which EGFP accumulates in the cytosol and sperm acrosome. The spermatozoa derived from these ESCs exhibit little EGFP signal in the cytosol and lose EGFP after the acrosome reaction, an exocytotic event that releases the contents of the acrosome. Unfortunately, the disappearance of the fluorescent signal precludes distinguishing KO spermatozoa from WT spermatozoa for chimeric analysis. In this study, we overcome the limitations of the G101 cell line by establishing a novel ESC line derived from a different TG mouse strain [Red body and green sperm (RBGS); B6D2-Tg (CAG/Su9-DsRed2, Acr3-EGFP) RBGS002Osb] [4]. Spermatozoa derived from the new RBGS ESC line not only contains EGFP in the acrosome but also DsRed2 in the mitochondria, whose red signal remains after the acrosome reaction.
Using the RBGS ESC line, we analyzed the function of HYDIN in chimeric mice. HYDIN is a central pair protein of the axoneme and is evolutionarily conserved from Chlamydomonas to humans. In Chlamydomonas, HYDIN is required for flagellar motility [10]. Whole-exome sequencing from patients with primary ciliary dyskinesia who display neonatal respiratory distress syndrome, recurrent productive bronchitis, early childhood pneumonia, and chronic rhinosinusitis have identified HYDIN as playing a role in these conditions [15]. The link between HYDIN and primary ciliary dyskinesia is consistent with studies of HYDIN functioning in the flagella of Chlamydomonas. Based on these findings of HYDIN in both species, it has been proposed HYDIN also functions in the flagellum of spermatozoa [15]. However, HYDIN’s role in the spermatozoa has not been analyzed using mouse models because the frameshift mutation (hy3) leads to early perinatal hydrocephaly due to impaired ciliary motility in ependymal cells and early lethality prior to sexual maturation [9]. By taking advantage of the RBGS ESC line, we utilized chimeric analysis to bypass the early lethality of hy3 and investigated the function of HYDIN in mouse spermatozoa.
Materials and Methods
Animals and embryos
129/Sv (CLEA Japan, Tokyo, Japan), ICR (Japan SLC, Shizuoka, Japan), B6D2F1 (SLC), and B6D2-Tg (CAG/Su9-DsRed2,Acr3-EGFP) RBGS002Osb mice were used. The hemizygous transgenic mice carry both transgenes (Su9-DsRed2 and Acr3-EGFP) at the same locus, without overt abnormality. To prepare embryos for ESC establishment and chimera generation, we flushed 2-cell embryos from the oviducts of 1-day pregnant super-ovulated females that had been mated with males. The 2-cell embryos were cultured in KSOM medium [5] until the blastocyst stage at 37°C under 5% CO2 in air. All animal experiments were approved by the Animal Care and Use Committee of the Research Institute for Microbial Diseases, Osaka University.
Establishment of ESCs
For derivation of ESCs, we used an equal volume mixture of KnockOut DMEM/F12 (12660-012, Thermo Fisher Scientific, Waltham, MA, USA) and Neurobasal medium (21103-049, Thermo Fisher Scientific), supplemented with 1% Penicillin-Streptomycin-Glutamine (Thermo Fisher Scientific, 21103-049), 0.1 mM 2-mercaptoethnol (21985-023, Thermo Fisher Scientific), 2% B-27 supplement (17504-044, Thermo Fisher Scientific), 1% N-2 supplement (17502-048, Thermo Fisher Scientific), 3 µM CHIR99201 (1386, Axon Medchem, Groningen, Netherland), 1 µM PD0325901 (1408, Axon Medchem), and 100 U/ml mouse Leukemia Inhibitory Factor (LIF). Different medium was used when ESCs were cultured for gene manipulation, which is written in the section “Genome editing and generation of chimeric mice”. Mouse embryonic fibroblasts (MEF) were treated with 10 µg/ml mitomycin C (134-07991, WAKO, Osaka, Japan) and used as feeder cells. ESCs were established by plating blastocysts on a feeder layer in gelatin-coated dishes. Eight days after plating, the expanded colonies derived from the inner cell mass (ICM) were obtained, dissociated with 0.25% trypsin (15090-046, Thermo Fisher Scientific), and cultured in gelatin-coated 6-well plates with feeder cells.
Chromosome number analysis and sex determination
After seeding the ESCs on 35 mm collagen-coated dishes without feeder cells, they were treated with 1% KaryoMAX Colcemid Solution in PBS (15212-012, Thermo Fisher Scientific) for 2 h to induce mitotic arrest at metaphase. Harvested ESCs were incubated with 1% sodium citrate solution for 5 min and fixed with a solution containing 75% methanol and 25% acetic acid. The cells were then spread on glass slides and stained with 10 µg/ml Hoechst 33342 (H3570, Thermo Fisher Scientific).
To determine the sex of established ESCs, genomic DNA was extracted with lysis buffer (20 mM Tris-HCl, 5 mM EDTA, 400 mM NaCl, 0.3% SDS, pH=8.0) and analyzed by PCR using the following Uba1 (ubiquitin-like modifier activating enzyme 1) (X chromosome) and Uba1y (ubiquitin-like modifier activating enzyme 1) (Y chromosome) primers, 5’-TGGTCTGGACCCAAACGCTGTCCACA-3’ and 5’-GGCAGCAGCCATCACATAATCCAGATG-3’. The size of the PCR product for Uba1 or Uba1y is 211 bp or 183 bp, respectively [1].
Immunostaining and alkaline phosphatase detection
Alkaline phosphatase (ALP) staining was carried out with a Histofine Fuchsin Substrate kit for ALP (415161, Nichirei, Tokyo, Japan), according to the manufacturer’s instruction. Immunostaining was carried out as follows. First, ESCs were cultured for 2 days on gelatin-coated cover glasses with feeder cells. After removing the culture medium, the cover glasses were washed with PBS (DPBS; 14190-144, Thermo Fisher Scientific) twice and fixed with 4% paraformaldehyde (166-23251, WAKO). The fixed cells were then washed 3 times in PBS, stored overnight in PBS containing 1% goat serum and 0.1% Triton X-100 at 4°C, and incubated with primary antibody against OCT3/4 (PM048, MBL, Nagoya, Japan; 1:500) or NANOG (sc-33760, Santa Cruz Biotechnology, Santa Cruz, CA, USA ; 1:20) at room temprature for 1 h. After 3 washed with PBS, the cells were incubated with Alexa Fluor 488 conjugated goat anti-rabbit IgG (R37116, Thermo Fisher Scientific; 1:200) or Alexa Fluor 546 conjugated goat anti-rabbit IgG (A-11035, Thermo Fisher Scientific; 1:200). After three washes with PBS, the cells were mounted with Vectashield mounting medium containing DAPI (H-1200, Vector laboratories, Burlingame, CA, USA) and observed using Olympus (Tokyo, Japan) microscope (BX53).
Genome editing and generation of chimeric mice
For plasmid transfection and cloning of ESCs, we used KnockOut DMEM (108297-018, Thermo Fisher Scientific) supplemented with 1% Penicillin-Streptomycin-Glutamine, 55 µM 2-mercaptoethnol, 1% Non-Essential Amino Acid Solution (11140-050, Thermo Fisher Scientific), 1% Sodium Pyruvate (11360-070, Thermo Fisher Scientific), 30 µM Adenosine (A4036, Sigma-Aldrich, St. Louis, MO, USA), 30 µM Guanosine (G6264, Sigma-Aldrich), 30 µM Cytidine (C4654, Sigma-Aldrich), 30 µM Uridine (U3003, Sigma-Aldrich), 10 µM Thymidine (T1895, Sigma-Aldrich), 100 U/ml mouse LIF, and 20% FCS (51650-500, Biowest, Nuaillé, France). First, 1 × 104 ESCs were seeded on gelatin-coated 6 well plate with feeder cells. Then 6–8 h later, pX330 and pX459 plasmids [12] (total 2 µg) were transfected using Lipofectamine LTX & PLUS (15338-100, Thermo Fisher Scientific). pX459 plasmid was used for one target site because it contains puromycin resistance gene for the following drug selection. After 14–18 h transfection, the cells were selected with 0.2 µg/ml puromycin (ant-pr-1, InvivoGen, San Diego, CA, USA) for 48 h, grown for 3–4 more days without puromycin, and plated on 6-well plates (1 × 103 cells) for cloning. The plated cells were picked up after 5–6 days, transferred onto gelatin-coated 96-well plates with feeder cells, and split in duplicate for freezing and DNA harvesting 48–72 h later. After PCR amplification and direct sequencing, the positive clones were selected and expanded to analyze their karyotypes. The ESCs with normal chromosome number were injected into 8-cell ICR embryos, and the chimeric blastocysts were transplanted into the uteri of pseudopregnant females. After 17 days, pups were taken out of uteri and put with foster mothers in the cage if the pups were not delivered naturally.
Preparation of testis section
Testis were fixed with 4% paraformaldehyde and embedded with technovit 8100 (RT8100, Kulzer, Wehrheim, Germany), according to the manufacturer’s instruction. After slicing, the testis sections were put on slide glass and mounted with Vectashiled mounting medium and observed using Olympus microscope (BX53).
Sperm motility and morphology of chimeric mice
The cauda epididymal spermatozoa were collected from the chimeric mice. Spermatozoa were dispersed in TYH [20] medium to observe sperm motility and morphology using Olympus microscope (BX53).
Plasmid construction and genotyping for Hydin targeting
Plasmids expressing Cas9 and gRNA were prepared by ligating oligos into the BbsI site of pX330 or pX459. The gRNA target sequences were 5’-AGCCACACATTTGATCTTCA-3’ (near the start codon, pX330-gRNA#01) and 5’-CACTTCAGCGTGCTTAGGTG-3’ (near the stop codon, pX459-gRNA#02). Primers used for genotyping were 5’-TATTGGGACCGCAAGCAAG-3’(primer c) and 5’-TTGGGAAGCTTGTCACTCTCC-3’ (primer d) for WT allele and 5’-CACCCCACTGAATTTAGGG-3’ (primer a) and 5’-TGGGGGTGGTGACATTTAGG-3’(primer b) for KO allele (Fig. 3A).
Fig. 3.
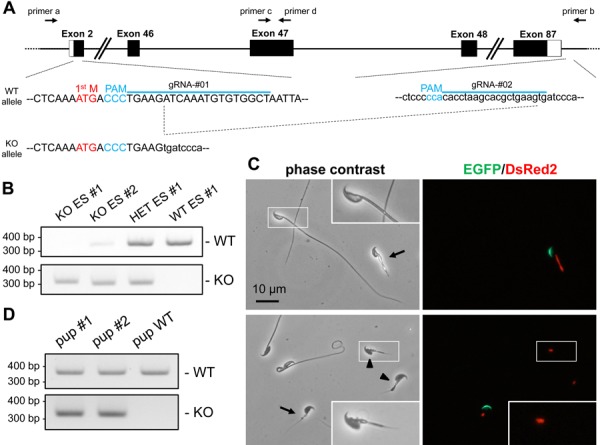
Analysis of Hydin KO spermatozoa. A) The design of gRNAs (blue line) and genotyping primers (primers a and b for KO allele and primers c and d for WT allele). B) An example of genotyping. There is a weak band for the WT allele in the KO ES line #2 probably due to the contamination from WT feeder cells. C) Spermatozoa of chimeric mice. Green or red fluorescence indicates EGFP (acrosome) or DsRed2 (mitochondria), respectively. Arrows indicate KO spermatozoa based on both green and red fluorescence. Spermatozoa indicated by arrowheads lose EGFP signal, but can be judged as KO spermatozoa because the red fluorescence remains. White rectangle region is magnified to show the misshapen head in the KO spermatozoa. D) Genotyping of pups obtained via ICSI.
Intracytoplasmic sperm injection (ICSI) of Hydin-disrupted spermatozoa
ICSI was performed as previously described with some modifications [22]. Briefly, mature oocytes were collected from superovulated B6D2F1 mice. After treatment with hyaluronidase to remove cumulus cells, oocytes were placed in KSOM medium at 37°C under 5% CO2 until ICSI. Whole spermatozoa collected from cauda epididymis were injected into the MII oocyte using a piezo manipulator (Prime Tech, Ibaraki, Japan). Two-cell embryos were transferred to pseudopregnant females the following day. Pups were genotyped at birth. Obtained mutant mice were assigned labels as follows: STOCK Hydin em1Osb.
Results
Establishment of RBGS-ESCs
To establish ESCs with transgenes CAG/Su9-DsRed2 and Acr3-EGFP, RBGS transgenic male mice (B6D2) were mated with superovulated females (129/Sv). Forty-three two-cell stage embryos were collected and cultured for 3 days until the blastocyst stage. DsRed2 positive blastocysts (12/43; Supplementary Fig. S1A) were seeded on feeder cells (murine embryonic fibroblasts; MEF) and cultured under 2i/LIF conditions [23] for 8 days. Subsequently, 6 dome-shaped colonies (#1–#6) were picked up and cultured on feeder cell layer. After 4 passages in 2i/LIF medium, chromosome numbers were counted (Supplementary Fig. S1B), with clone #1 showing a high rate of aneuploidy while the other 5 lines displayed a normal karyotype (Supplementary Table S1).
We next determined the sex of the 5 established lines (Fig. 1A), as we intended to analyze the function of male-fertility related genes. Sex chromosome content was determined by PCR using primers for Uba1 (X chromosome; PCR product=211 bp) and Uba1y (Y chromosome; PCR product=183 bp). The PCR results showed 3 out of 5 lines (#3, #5 and #6) contained an XY karyotype (Fig. 1B). We selected RBGS-ESC line #3 (hereafter, RBGS-ESC) for subsequent experiments as this line appeared to have more robust growth and maintained a normal chromosome composition even after 15 passages (Supplementary Table S1).
Fig. 1.
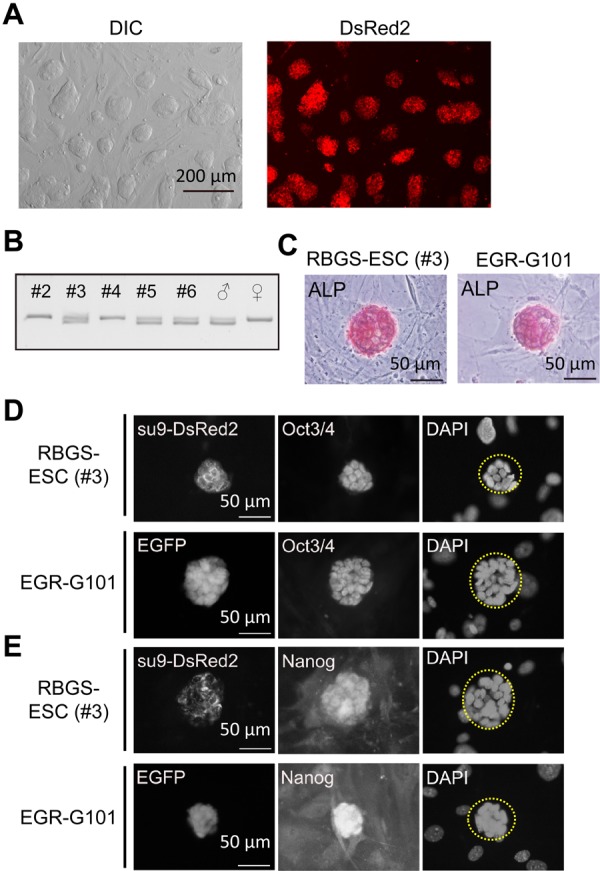
Establishment of ESCs from RBGS transgenic mice. A) RBGS-ESCs at the fifth passage cultured in serum/LIF medium. The cells exhibited DsRed2 signals. B) Sex determination of established ESCs. The upper band was amplified from the X chromosome (211 bp), and the lower band from the Y chromosome (183 bp). C) Detection of alkaline phosphatase (ALP) activity. Red staining indicates ALP reacting with the substrate. EGR-G101 ESCs were used as a positive control. D) Expression of pluripotent marker OCT3/4 in ESCs. Su9-DsRed2 localizes to the mitochondria, and OCT3/4 in the nucleus. DAPI indicates nucleus. EGR-G101 ESCs were used as a positive control. No OCT3/4 signals were observed in feeder cells that can be seen with DAPI surrounding the ESC colony (ESC colonies are marked by a yellow dashed line). E) Expression of another pluripotent marker NANOG in ESCs. NANOG localized to the nucleus. EGR-G101 ESCs were used as a positive control. No NANOG signals were observed in feeder cells that can be seen with DAPI surrounding the ESC colony.
To examine the stemness of RBGS-ESCs, alkaline phosphatase (ALP) activity and pluripotent marker expression were checked by staining. As shown in Figs. 1C–E, ALP activity and pluripotent marker (OCT4 and NANOG) expression were comparable to the EGR-G101 [(CAG/Acr-EGFP) B6N × (CAG/Acr-EGFP) B6N] ESC line [2].
Chimera production
To analyze their chimera-forming ability, RBGS-ESCs transfected with an empty pX459 vector (expressing spCas9 and an empty gRNA) were injected into ICR 8-cell embryos. Forty-nine treated embryos were transferred to ICR pseudopregnant females. Twenty pups were delivered with 13 showing chimeric DsRed2 fluorescence. The contribution of RBGS-ESC ranged from negligible to 95%, judged by coat color (Fig. 2A).
Fig. 2.
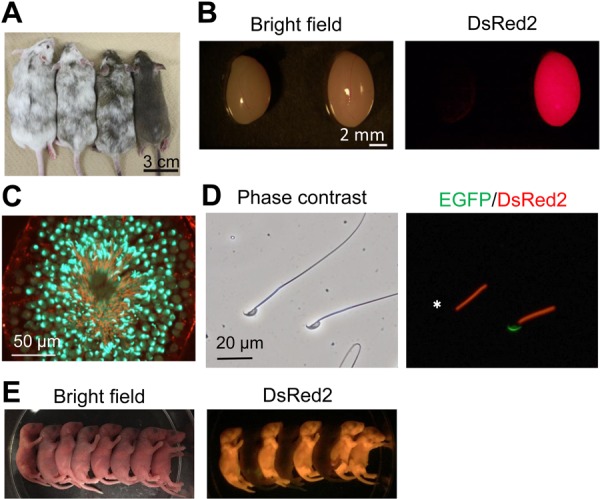
Generation of chimeric mice. A) Eight-week old chimeric mice. Contribution of ESCs can be observed via non-white coat color. B) Testis were collected from 12-week old wild-type (left) and chimeric mice (right). C) Testis section of chimeric mice from (B). Green signal indicates EGFP (acrosome) and red signal indicates DsRed2 (mitochondria). D) Spermatozoa of chimeric mice. Green or red fluorescence indicates EGFP (acrosome) or DsRed2 (mitochondria), respectively. White asterisk shows spermatozoa that have undergone the acrosome reaction. E) Two-day old pups delivered from a B6D2F1 female that was mated with an RBGS chimeric mice.
To analyze ESC contribution in more detail, we examined the red fluorescence in various mouse tissues (Fig. 2B and Supplementary Fig. S2) and determined RBGS-ESCs can contribute to all organs analyzed. Contribution to the germ-line was confirmed in testis sections showing EGFP in the acrosome and DsRed2 in the mitochondria of developing spermatozoa (Fig. 2C). The morphology of ESC-derived spermatozoa was comparable to those of host-derived spermatozoa (Fig. 2D). Using the red fluorescence in the midpiece, distinguishing between ESC-derived spermatozoa and host-derived spermatozoa was apparent even after the acrosome reaction, in contrast to previous studies using the EGR-G101 line [2].
Finally, these chimeric mice were mated with B6D2F1 female mice to confirm germline transmission. As shown in Fig. 2E, the F1 generations exhibiting DsRed2 was obtained. Collectively, established RBGS-ESCs can contribute to various tissues and be transmitted through the germline.
Chimeric analysis of Hydin-disrupted spermatozoa
We applied RBGS-ESCs to analyze the phenotype of Hydin KO spermatozoa. To disrupt the gene function completely, we removed almost the entire coding region by transfecting pX330-gRNA#01 and pX459-gRNA#02 plasmids that target near the start and stop codons, respectively (Fig. 3A). Forty-eight ESC clones were selected and 7 clones with a 343 kb deletion were obtained including 2 biallelic mutant clones (Fig. 3B). The WT allele band observed in KO ES line #2 is weak compared to that of HET ES line #1 [intensity ratio=0.12 (KO ES #2/HET ES #1)], possibly due to contamination from WT feeder cells. As we intended to remove the entire coding region to ensure loss of HYDIN function, small insertion or deletion was not examined; however, we cannot exclude the possibility that such small alterations were generated. The successful excision of 343 kb is consistent with a previous study showing up to 841 kb can be efficiently excised in ESCs [14]. One biallelic mutant clone (KO ES #2) was injected into ICR 8-cell embryos because this clone exhibited a normal karyotype (Supplementary Table S2). Sixty treated embryos were transferred to ICR pseudopregnant females. After 17 days, 12 pups were delivered, with 9 showing the chimeric red fluorescence. Three pups did not show red fluorescence likely due to a low contribution of injected ESCs into blastocysts. Five chimeric mice survived up to 8 weeks old while 4 chimeric mice died within 3 weeks, reminiscent of the early lethality observed in hy3 mice [9]. The cause of death of the four pups is unknown as they were cannibalized by their mothers.
From the 5 chimeric mice that reached sexual maturity, we observed spermatozoa from the cauda epididymis. WT spermatozoa in the chimeric mouse exhibit normal sperm length whereas ESC-derived spermatozoa exhibit short-tails [average tail length=116.5 ± 1.5 µm (WT) compared to 12.4 ± 4.3 µm (KO)] (Fig. 3C and Supplementary Fig. S3A) and were immotile (30 out of 30 KO spermatozoa observed in Supplementary Fig. S3A). In addition to abnormal tail formation observed in KO spermatozoa, the mitochondrial red fluorescence signal (Su9-DsRed2) shows an aberrant midpiece. Further, misshapen heads were observed in KO spermatozoa (Fig. 3C and Supplementary Fig. S3B). As the midpiece and head morphology in WT derived spermatozoa were normal, we conclude the defect observed in KO spermatozoa is due to disruption of Hydin in germ cells and not in Sertoli cells or Leydig cells.
To determine whether the mutant RBGS spermatozoa’s genome is intact and can contribute to the next generation, the short-tail spermatozoa were injected into WT oocytes by intracytoplasmic sperm injection (ICSI). In the standard ICSI protocol in mice, only the sperm head is used; however, whole spermatozoa from Hydin KOs were injected as it was difficult to separate the head from the short flagellum. Sixty-two oocytes were treated with 26 oocytes developing to 2-cell stage embryos, which were transferred to pseudopregnant females. After 19 days, 2 heterozygous pups with the mutant allele were delivered (Fig. 3D). The heterozygous mice grew without overt abnormalities and were fertile. Our result indicate that the nuclei of KO spermatozoa have the ability to produce viable pups. Overall, our chimeric analysis reveals that HYDIN is essential for the sperm tail formation in mice.
Discussion
In the present study, we established ESC line from RBGS mice whose spermatozoa contain DsRed2 in the midpiece and EGFP in the acrosome. We confirmed expression of pluripotent markers and contribution of these ESCs to various tissues including germ cells in chimeric mice. As a proof of concept, we applied RBGS-ESCs to analyze the function of HYDIN in spermatozoa, which has previously been difficult due to the early lethality of Hydin mutant mice [9]. Several Hydin chimeric mice in our study reached sexual maturity, and by analyzing the spermatozoa collected from the cauda epididymis, we revealed that HYDIN is essential for sperm function.
Previously, we utilized G101 ESCs that express EGFP in the acrosome for chimeric analysis [14]. However, it was difficult to distinguish between host-derived and ESC-derived spermatozoa in G101 chimeric mice because the EGFP signal was lost not only when the spermatozoa underwent the acrosome reaction but also when their membrane integrity was disrupted. RBGS-ESCs that expresses DsRed2 in the mitochondria allow ESC-derived spermatozoa to be easily recognized. In addition, the morphology of mitochondrial sheath can be analyzed without staining due to the localization of DsRed2 to this organelle.
One concern associated with chimeric analysis of genes essential for male fertility is that the next generation cannot be obtained. However, as we demonstrated with ICSI using Hydin-disrupted spermatozoa, the next generation may be obtained by assisted reproductive technologies such as in vitro fertilization, ICSI, and round spermatid injection (ROSI). Once essential genes are identified by chimeric analysis, we may also be able to regenerate KO mice by genome editing in zygote. Another concern of chimeric analysis is that infertility phenotypes can be concealed by the host-derived WT cells in the case of factors that function in a cell non-autonomous manner such as secreted or membrane-bound proteins. Careful interpretation of phenotypes is necessary when analyzing non-autonomous factors in chimeras. In these cases, subsequent generations can be analyzed to confirm that host-derived WT cells do not conceal infertility phenotypes. It should be noted that conditional KO methods have a similar problem because efficiency of CRE or FLP-dependent recombination is not always 100% [3, 16], which causes mosaicism. Additionally in chimeric analysis, it is possible that phenotypes observed in the cells of interest are due to mutations in other cells. In this case, it is important to check the phenotypes of the corresponding WT cells. For example, in regards to the short-tail phenotype of Hydin KO spermatozoa in chimeric mice, we concluded that the phenotype was not due to defective Sertoli cells or Leydig cells as WT spermatozoa appear normal.
Utilizing chimeric analysis, we found that Hydin KO spermatozoa exhibit abnormal tail formation in mice. Because HYDIN is localized to the central pair (CP) of the axoneme [9], it is very likely that deletion of Hydin causes abnormal tail formation; however, it should be noted that we cannot exclude possibilities of off-target mutations or deletion of non-coding RNAs localized within the Hydin locus. To reduce the likelihood off-target effects causing observed phenotypes, ESCs that do not contain integrated pX330 or pX459 or using multiple ES clones for generating chimeric mice should be explored. Rescue experiments should also be considered to determine if the gene of interest is responsible for the phenotype.
There are other CP proteins that have been found to be essential for tail formation such as SPEF2, SPAG6, CFAP54 [13, 18, 19]. Similar defects found in these KO mice suggest that correct organization of the CP apparatus may be important for flagellum formation. In addition to short tails, misshapen heads were observed in Hydin KO spermatozoa. Considering that HYDIN is located in the CP, misshapen heads may be a secondary effect caused by abnormal flagellar elongation. Alternatively, it is possible that HYDIN may play a distinct role in shaping the head as suggested in Spef2 KO mice [19]. Further experimentation is necessary to fully understand the molecular mechanism of HYDIN in spermiogenesis.
In contrast to mice, mutations of HYDIN in primary ciliary dyskinesia patients lead to impaired sperm motility but not defective flagellum formation [15]. This mild phenotype observed in humans may be because HYDIN is not completely deleted in the patients who possess a point mutation (c.3985G>T) that leads to aberrant splicing. Generating the corresponding human mutation in mice using CRISPR/Cas9 may lead to the elucidation of this discrepancy.
Our study demonstrates the benefit of using established RBGS-ESCs for investigating male fertility-related genes in chimeric mice and revealed that HYDIN is essential for the sperm tail formation. Chimeric mice can also be used to analyze the function of lethal mutations in other organs in addition to the testis. For example, using fluorescence activated cell sorting, mutated immune cells or blood cells can be isolated and analyzed. Utilizing RBGS-ESCs for chimeric analysis provides a powerful tool to analyze gene functions that are not only related to male germ cells but also somatic cells.
Supplementary Material
Acknowledgments
This research was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT)/ Japan Society for the Promotion of Science (JSPS) KAKENHI Grants (JP17H04987 to HM, JP18K14612 to TN, JP17K17852 to KS, JP17J09669 to TM, JP16K07091 to AI, JP25112007 and JP17H01394 to MI), AMED under Grant Number JP18fk0210006h0003 and JP18gm5010001h0002 (to MI), Takeda Science Foundation Grants (to HM and MI), NIH grant P01HD087157 and R01HD088412 (to MI), and The Bill & Melinda Gates Foundation (Grand Challenges Explorations grant OPP1160866) (to MI). We thank Ms. Saki Nishioka and Ms. Natsuki Furuta for technical assistance and Dr. Julio M. Castaneda for critical reading of the manuscript.
References
- 1.Chuma S., Nakatsuji N.2001. Autonomous transition into meiosis of mouse fetal germ cells in vitro and its inhibition by gp130-mediated signaling. Dev. Biol. 229: 468–479. doi: 10.1006/dbio.2000.9989 [DOI] [PubMed] [Google Scholar]
- 2.Fujihara Y., Kaseda K., Inoue N., Ikawa M., Okabe M.2013. Production of mouse pups from germline transmission-failed knockout chimeras. Transgenic Res. 22: 195–200. doi: 10.1007/s11248-012-9635-x [DOI] [PubMed] [Google Scholar]
- 3.Gallardo T., Shirley L., John G.B., Castrillon D.H.2007. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 45: 413–417. doi: 10.1002/dvg.20310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hasuwa H., Muro Y., Ikawa M., Kato N., Tsujimoto Y., Okabe M.2010. Transgenic mouse sperm that have green acrosome and red mitochondria allow visualization of sperm and their acrosome reaction in vivo. Exp. Anim. 59: 105–107. doi: 10.1538/expanim.59.105 [DOI] [PubMed] [Google Scholar]
- 5.Ho Y., Wigglesworth K., Eppig J.J., Schultz R.M.1995. Preimplantation development of mouse embryos in KSOM: augmentation by amino acids and analysis of gene expression. Mol. Reprod. Dev. 41: 232–238. doi: 10.1002/mrd.1080410214 [DOI] [PubMed] [Google Scholar]
- 6.Hsu P.D., Lander E.S., Zhang F.2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157: 1262–1278. doi: 10.1016/j.cell.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishikura Y., Yabuta Y., Ohta H., Hayashi K., Nakamura T., Okamoto I., Yamamoto T., Kurimoto K., Shirane K., Sasaki H., Saitou M.2016. In vitro derivation and propagation of spermatogonial stem cell activity from mouse pluripotent stem cells. Cell Reports 17: 2789–2804. doi: 10.1016/j.celrep.2016.11.026 [DOI] [PubMed] [Google Scholar]
- 8.Toshimori K., Eddy E.M.2014. The Spermatozoon. pp.99–148. In: Physiology of Reproduction 4th edition (Knobil, E., Neil, J.D. eds.), Academic Press, London. [Google Scholar]
- 9.Lechtreck K.F., Delmotte P., Robinson M.L., Sanderson M.J., Witman G.B.2008. Mutations in Hydin impair ciliary motility in mice. J. Cell Biol. 180: 633–643. doi: 10.1083/jcb.200710162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lechtreck K.F., Witman G.B.2007. Chlamydomonas reinhardtii hydin is a central pair protein required for flagellar motility. J. Cell Biol. 176: 473–482. doi: 10.1083/jcb.200611115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capecchi M.R.1989. Altering the genome by homologous recombination. Science 244: 1288–1292. doi: 10.1126/science.2660260 [DOI] [PubMed] [Google Scholar]
- 12.Mashiko D., Fujihara Y., Satouh Y., Miyata H., Isotani A., Ikawa M.2013. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci. Rep. 3: 3355. doi: 10.1038/srep03355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKenzie C.W., Craige B., Kroeger T.V., Finn R., Wyatt T.A., Sisson J.H., Pavlik J.A., Strittmatter L., Hendricks G.M., Witman G.B., Lee L.2015. CFAP54 is required for proper ciliary motility and assembly of the central pair apparatus in mice. Mol. Biol. Cell 26: 3140–3149. doi: 10.1091/mbc.e15-02-0121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oji A., Noda T., Fujihara Y., Miyata H., Kim Y.J., Muto M., Nozawa K., Matsumura T., Isotani A., Ikawa M.2016. CRISPR/Cas9 mediated genome editing in ES cells and its application for chimeric analysis in mice. Sci. Rep. 6: 31666. doi: 10.1038/srep31666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olbrich H., Schmidts M., Werner C., Onoufriadis A., Loges N.T., Raidt J., Banki N.F., Shoemark A., Burgoyne T., Al Turki S., Hurles M.E., UK10K Consortium, Köhler G., Schroeder J., Nürnberg G., Nürnberg P., Chung E.M., Reinhardt R., Marthin J.K., Nielsen K.G., Mitchison H.M., Omran H.2012. Recessive HYDIN mutations cause primary ciliary dyskinesia without randomization of left-right body asymmetry. Am. J. Hum. Genet. 91: 672–684. doi: 10.1016/j.ajhg.2012.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sadate-Ngatchou P.I., Payne C.J., Dearth A.T., Braun R.E.2008. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis 46: 738–742. doi: 10.1002/dvg.20437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saitou M., Miyauchi H.2016. Gametogenesis from Pluripotent Stem Cells. Cell Stem Cell 18: 721–735. doi: 10.1016/j.stem.2016.05.001 [DOI] [PubMed] [Google Scholar]
- 18.Sapiro R., Kostetskii I., Olds-Clarke P., Gerton G.L., Radice G.L., Strauss J.F., III2002. Male infertility, impaired sperm motility, and hydrocephalus in mice deficient in sperm-associated antigen 6. Mol. Cell. Biol. 22: 6298–6305. doi: 10.1128/MCB.22.17.6298-6305.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sironen A., Kotaja N., Mulhern H., Wyatt T.A., Sisson J.H., Pavlik J.A., Miiluniemi M., Fleming M.D., Lee L.2011. Loss of SPEF2 function in mice results in spermatogenesis defects and primary ciliary dyskinesia. Biol. Reprod. 85: 690–701. doi: 10.1095/biolreprod.111.091132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toyoda Y., Yokoyama M., Hoshi T.1971. Studies on the fertilization of mouse eggs in vitro. Jpn. J. Anim. Reprod. 16: 147–151(In Japanese). [Google Scholar]
- 21.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R.2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153: 910–918. doi: 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura Y., Yanagimachi R.1995. Mouse oocytes injected with testicular spermatozoa or round spermatids can develop into normal offspring. Development 121: 2397–2405. [DOI] [PubMed] [Google Scholar]
- 23.Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A.2008. The ground state of embryonic stem cell self-renewal. Nature 453: 519–523. doi: 10.1038/nature06968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Q., Wang M., Yuan Y., Wang X., Fu R., Wan H., Xie M., Liu M., Guo X., Zheng Y., Feng G., Shi Q., Zhao X.Y., Sha J., Zhou Q.2016. Complete Meiosis from Embryonic Stem Cell-Derived Germ Cells In Vitro. Cell Stem Cell 18: 330–340. doi: 10.1016/j.stem.2016.01.017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


