Abstract
Motor proteins are responsible for transport of vesicles and organelles within the cell cytoplasm. They interact with the actin cytoskeleton and with microtubules to ensure communication and supply throughout the cell. Much work has been done in vitro and in silico to unravel the key players, including the dynein motor complex, the kinesin and myosin superfamilies, and their interacting regulatory complexes, but there is a clear need for in vivo data as recent evidence suggests previous models might not recapitulate physiological conditions. The zebrafish embryo provides an excellent system to study these processes in intact animals due to the ease of genetic manipulation and the optical transparency allowing live imaging. We present here the advantages of the zebrafish embryo as a system to study live in vivo processive transport in neurons and provide technical recommendations for successful analysis.
Keywords: neuronal transport, zebrafish, myosin, kinesin, dynein, in vivo
Introduction
Processive intracellular transport is essential for the distribution of organelles and cellular cargoes within the cell. In the case of neurons, such transport provides communication between different cell compartments and ensures supply to the growing synapse, clearance of detritus and serves as the support for intracellular signaling (Hirokawa et al., 2010). This process relies on the function of motor proteins and their interaction with the cell cytoskeleton, the three components of which play important roles in regulating transport. Indeed, neurofilaments provide structure and regulate axonal caliber, which influences transport metrics. Microtubules are responsible for axonal polarity, a consequence of the stereotyped orientation of their dynamic fast-growing ends, and act as the rails guiding motor proteins within the axon and dendrites. Finally, actin filaments form a structural network supporting the growth cone, pre- and postsynaptic regions, and play an important role in dendrites where they form the spines, essential for synaptic transmission.
Dynein and the kinesin superfamily are the unidirectional molecular motors responsible for transport on microtubules, both in dendrites and axon. In polarized axons, it is split according to the direction relative to the microtubule fast-growing end (+), with kinesins being responsible for ‘anterograde’ movement (toward the synapse) and dynein for ‘retrograde’ movement (toward the cell body). Their movement can in turn be categorized as ‘slow’ or ‘fast’ depending on their transport rate. Slow axonal transport is mainly used for delivery of cytoskeletal components and associated proteins, with kinetics in the range of 0.2–8 mm/day (Lasek et al., 1984). Fast axonal transport is used for organelles and vesicles, but also for mRNA granules (Maday et al., 2014), with kinetics in the range of 50–400 mm/day (Lasek et al., 1984). This type of transport, in terms of motor complex involved for different types of cargoes, motor adaptor complexes and transport metrics, has been widely studied and is well reviewed elsewhere (Maday et al., 2014).
Unconventional myosins, molecular motors of the actin cytoskeleton, are commonly associated with dynamic shaping of membranes, as well as organelle formation and transport, but their functions in neuronal transport is not well understood. Among this super-family, Myosin5a, 5b, 6 and 10 have been identified as processive transporters in neurons, and participate in local transport of intracellular cargoes over short-range distances. Processive myosins are likely part of a cooperative mechanism which is based on the coordination of actin and microtubule transporters (Wu et al., 2000). This is nicely illustrated with Myo5a binding directly to kinesins (Huang et al., 1999), suggesting that organelles transported in axons along microtubules may be transported by Myo5a in presynaptic terminals, which lack microtubules (Wu et al., 1998; Bridgman, 1999; Lalli et al., 2003; Nalavadi et al., 2012). Processive myosins are implicated in vesicle endocytosis, recycling and exocytosis, and hence participate in receptor transport and localization, regulating neuronal signaling and axonal pathfinding (Wu et al., 2002; Osterweil et al., 2005; Zhu et al., 2007; Correia et al., 2008; Nash et al., 2010; Lazo et al., 2013; Sui et al., 2015). Moreover, processive myosins take part in transport of mRNAs and RNPs in neurons, as demonstrated for Myo5a (Ohashi et al., 2002; Yoshimura et al., 2006; Balasanyan and Arnold, 2014; Calliari et al., 2014; Lindsay and Mccaffrey, 2014).
From recent evidence, it is apparent that in vivo axonal transport data do not recapitulate what has been observed in vitro (Gibbs et al., 2016; Klinman and Holzbaur, 2016; Knabbe et al., 2018), emphasizing the need for a more physiological context. With this in mind, excellent work has been published reporting in vivo axonal transport (reviewed in Sleigh et al., 2017) in models such as the Drosophila wing (Vagnoni and Bullock, 2017) and larvae (Vukoja et al., 2018), as well as the mouse brain (Knabbe et al., 2018) and sciatic nerve (Gibbs et al., 2016). All of these models have advantages and drawbacks: the mouse model is widely used and as a mammal, has a high genetic conservation of genes of interest but is not translucent and only allows access to axonal transport in a restricted area of the targeted cell population by way of surgery. The Drosophila is a model with a fantastic genetic manipulation toolbox, however, it is an invertebrate with reduced conservation to human compared to vertebrate models.
Over the last decades, zebrafish has emerged as a powerful vertebrate model to study the development of the nervous system in vivo. Adult zebrafish are small in size and produce a large number of offsprings, with a rapid external development. The embryonic zebrafish are translucent, and recent advances in genetic manipulation have made this model a great option to monitor neurodevelopment by high-resolution live imaging and at single-cell level. In addition, the zebrafish embryo is used extensively for modeling neurodegeneration (Bandmann and Burton, 2010; Kabashi et al., 2010; Santoriello and Zon, 2012; Babin et al., 2014; Patten et al., 2014; Fontana et al., 2018). Some processive motors have been associated with neurological disorders (Chen et al., 2013) and many studies have reported axonal transport defects in the context of neurodegenerative diseases (Chevalier-Larsen and Holzbaur, 2006; Goldstein, 2012; Liu et al., 2012; Millecamps and Julien, 2013), further outlining the interest of this model. In this article, we thus discuss the advantages of the zebrafish model in the study of live in vivo intracellular transport, with a particular focus on fast axonal transport.
Advantages of the Zebrafish Model
Relevance to Mammalian Models
The genome of Danio rerio is fully sequenced and presents at least one ortholog for 70% of human genes (Howe et al., 2013). In particular, kinesin, dynein and myosin molecular motors implicated in neuronal transports are extremely well conserved in eukaryotes and even more in vertebrates (Kim and Endow, 2000; Sittaramane and Chandrasekhar, 2008). These proteins have a higher conservation with the human ortholog in zebrafish compared to D. melanogaster for example. Zebrafish and drosophila dynein Dync1h1 show 91% and 72% identity (NCBI Blastp) with the human protein, respectively. Similarly, the processive Myo6 is 85% and 53% identical to the human one in zebrafish and drosophila, respectively. This high degree of conservation provides support for using zebrafish as a model system to investigate the functions of these molecular motors.
Genetic Manipulations
Compared especially to the mouse, the ease of stable or transient genetic manipulations has positioned the zebrafish as an ideal vertebrate model for live in vivo imaging.
Transgenesis in zebrafish is routinely and efficiently performed to express fusion proteins, mutated proteins or the gal4 transcription factor under a tissue-specific promoter thanks to the use of transposon elements. Ease of genetic manipulations in zebrafish has tremendously increased with the development of the CRISPR/Cas9 technology. The generation of knock-out mutants has become extremely powerful (Hwang et al., 2013) and using a Gal4/UAS-based restriction of Cas9 expression makes it possible to induce tissue-specific mutations and restrict the phenotype to a subset of cells (Di Donato et al., 2016). Recent advances based on the fusion of a mutated Cas9 (nickase) with an acetyl deaminase leading to the precise editing of a single nucleic acid (Komor et al., 2016) was also shown to work in zebrafish (Zhang et al., 2017). This technology makes it possible to target a specific protein domain in order to interfere with protein–protein interaction and opens the possibility of reproducing mutations associated with human diseases to elucidate the underlying pathological mechanism.
To recapitulate endogenous expression of a protein of interest, both in terms of pattern and level, bacterial artificial chromosome (BAC) transgenesis, where very large DNA sequence (up to 300 kb) can be inserted into the genome, is used in zebrafish (Lee et al., 2001; Suster et al., 2011). The CRISPR/Cas9 era has now opened the possibility of direct knock-in at a targeted locus. This strategy has been successful in zebrafish, based on the error-prone non-homologous end-joining DNA damage repair mechanism (Auer et al., 2014) and by short or long homology arm recombination (Hruscha et al., 2013; Hwang et al., 2013; Irion et al., 2014; Zhang et al., 2016). However, the efficiency of the latter technique is still low and locus-dependant. Its optimization will be an important technical advance in the field (Albadri et al., 2017), for example, to allow endogenous expression of a fusion protein of choice for visualization in vivo, without overexpression.
Pharmacological Manipulations
Zebrafish embryos are amenable to pharmacological treatment by bath application, allowing for treatment of intact, live embryos with compounds known for the modulation of cytoskeletal dynamics, for instance, targeting microtubules: Colchicine (Roche et al., 1994), vinblastine (Keiter et al., 2016; Yao et al., 2017), vincristine (Mizgirev and Revskoy, 2010; Khan et al., 2012; Holloway et al., 2016), nocodazole (Plucińska et al., 2012; Jayachandran et al., 2016) and paclitaxel (Jayachandran et al., 2016). For the actin cytoskeleton: Cytochalasin D (Nukada et al., 2015; Artelt et al., 2018) and latrunculin A (Artelt et al., 2018), jasplakinolide (Artelt et al., 2018), phalloidin oleate (Dutta and Kumar Sinha, 2015), and the inhibitor of actin–myosin interaction BDM (Norden et al., 2009) have been used with success.
Finally, zebrafish embryos are well suited to high-throughput approaches that have made them an excellent tool in drug discovery by small molecule screening (Zon and Peterson, 2005; Mathias et al., 2012; Miscevic et al., 2012; Tamplin et al., 2012; MacRae and Peterson, 2015).
Examples and Recommendations for the Analysis of In Vivo Transport in Zebrafish
To date, a few studies have taken advantage of the zebrafish model to perform in vivo axonal transport assays, generating tools to study the movement of mitochondria (Plucińska et al., 2012; Campbell et al., 2014; Paquet et al., 2014; Auer et al., 2015; Drerup et al., 2017), endosomes (Clark et al., 2011; Ponomareva et al., 2014, 2016), autophagosomes (He et al., 2009), lysosomes (Drerup and Nechiporuk, 2013), synaptophysin-containing vesicles (Auer et al., 2015) as well as motor proteins and components of their regulatory complexes (Drerup and Nechiporuk, 2013, 2016). The in vivo analysis of myosin-based transport is only starting in zebrafish neuronal development (Liu et al., 2013).
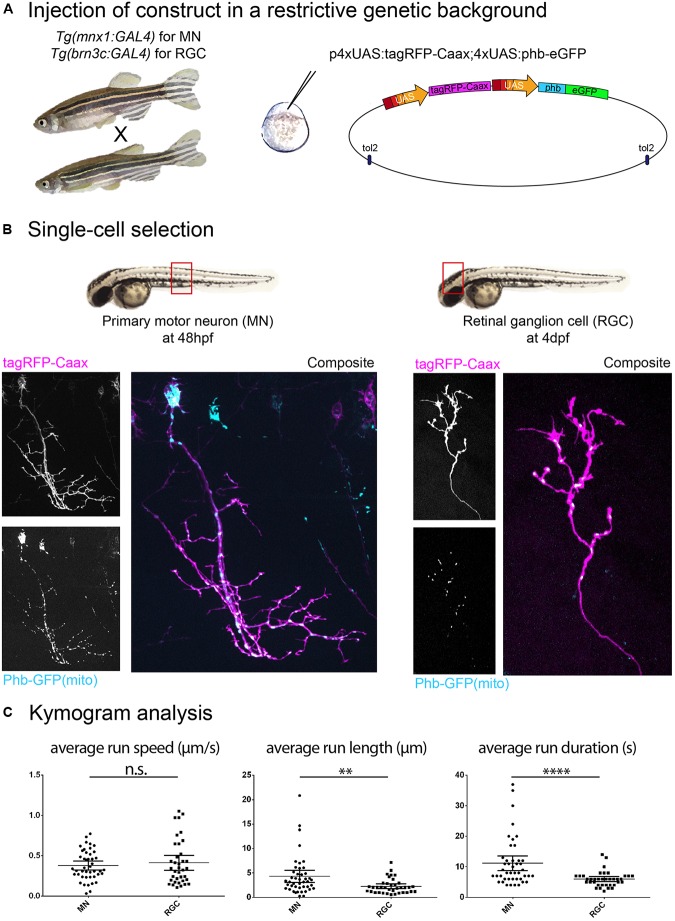
Based on published evidence, it is plain to see that the metrics reported for the same cargo visualized in vivo in zebrafish display variation between cell types and developmental stages. Indeed, we have observed metrics for mitochondrial anterograde transport in primary motor neurons (MN; axon) and in retinal ganglion cells (RGC; arbor) and while we did not find differences in average run speed, average run length and duration were significantly different in these two cell types. Furthermore, the average run speed detected was approximately 0.4 μm/s (Figure 1C), which is consistent with reported data from Campbell et al. (2014) (peripheral sensory neuron arbors, approx. 0.4 μm/s) but inconsistent with data from Plucińska et al. (2012) (peripheral Rohon-Beard sensory neuron axons, approx. 1.2 μm/s ‘moving speed’ and 0.6 μm/s ‘average speed’) and from Drerup et al. (2017) (peripheral lateral line axon, approx. 1.0 μm/s). We also found discrepancies between cell types in the transport of recycling endosomes (labeled with Rab11a-GFP), where we observed an average speed of approx. 0.5 μm/s (Figure 2B), whereas Ponomareva et al. (2014) report an average speed of approx. 0.18 μm/s/0.03 μm/s (central/peripheral Rohon-Beard sensory neuron axon).
FIGURE 1.
Construct expression and cell type selection. (A) Injection of DNA constructs coding for fusion proteins can be restricted to a single cell type by use of the Gal4/UAS system. (B) In this example, we injected a UAS construct labeling mitochondria (phb, prohibitin-GFP see schematics in A) combined with a membrane reporter (tagRFP-Caax). We obtained labeling of a single primary motor neuron (MN; in the Tg(mnx1:gal4) background) and a single retinal ganglion cell (RGC; in the Tg(brn3c:gal4) background), respectively, in the embryonic spinal cord (48 hpf) and in the larval optic tectum (4 dpf). (C) Time-lapse imaging of mitochondria (1 Hz for 10 min) was performed on these cell types, and transport dynamics were calculated from kymograms. Here, we show example of the disparity in transport metrics that can arise when comparing different cell types for a single cargo (MN n = 7 cells/44 anterograde runs; RGC n = 7 cells/37 anterograde runs). ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
FIGURE 2.
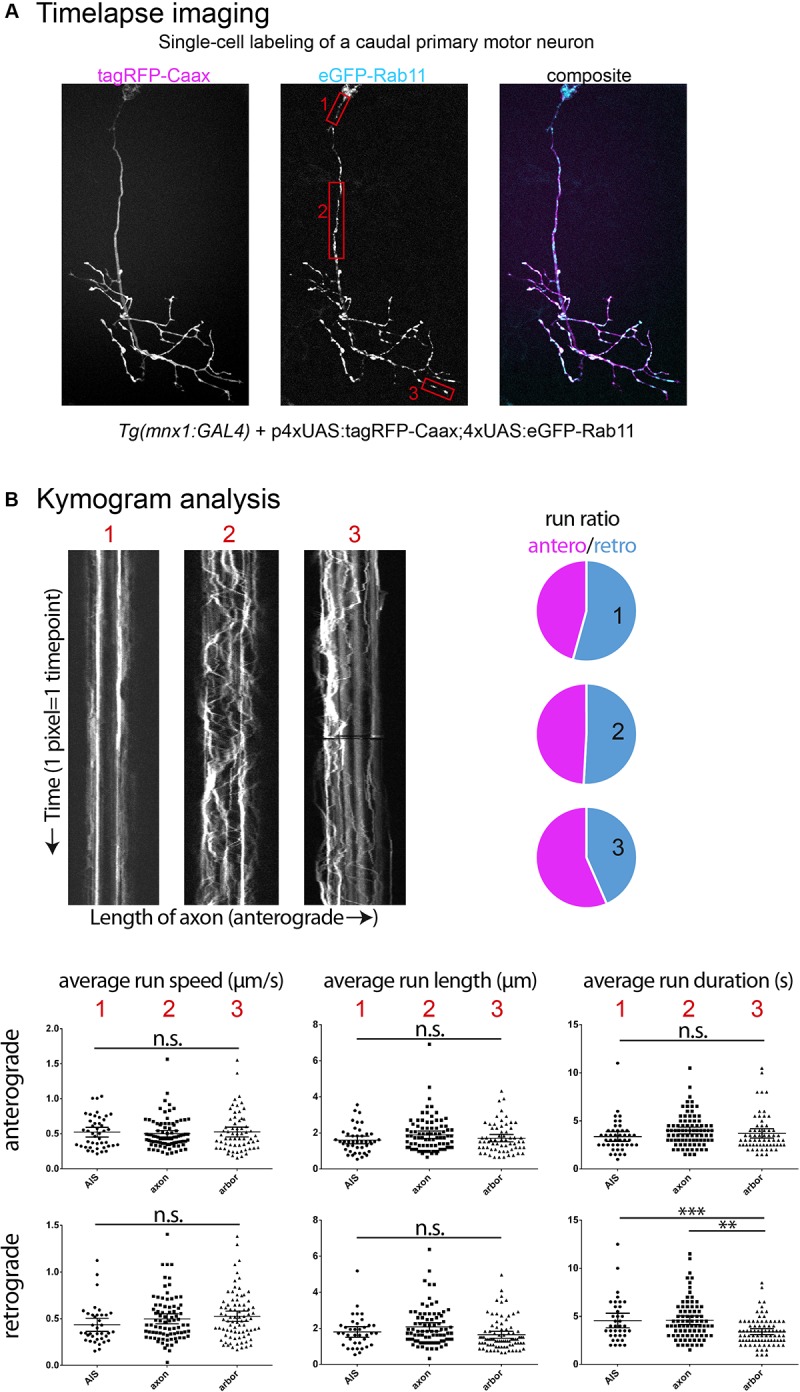
Examples of time-lapse analysis. (A) As described in Figure 1A, single cell labeling of primary motor neurons was obtained for recycling endosomes (Rab11a-eGFP), combined with a membrane reporter (tagRFP-Caax) to identify cell type. Red boxes: Three cell compartments were imaged (2 Hz for 5 min), 1-axon initial segment, 2-mid-axonal segment, 3-axonal arbor segment. (B) Kymograms were generated from the time-lapses acquired (Kymograph tool, ImageJ) and a variety of transport metrics can be calculated manually (compiled in Excel, statistics in Graphpad Prism6). In this example, significant differences between neuronal segments are detected for the transport direction ratio (anterograde/retrograde), and retrograde run duration (n = 3 cells; AIS n = 47/54 anterograde/retrograde runs; mid-axon n = 85/82 anterograde/retrograde runs; arbor n = 63/86 anterograde/retrograde runs). ∗∗p < 0.01, ∗∗∗p < 0.001.
Based on the evidence above outlining the variability of these processes, we will highlight a few key points to take into consideration when designing experiments to characterize transport in zebrafish neurons.
Regulation of Construct Expression and Imaging
Most approaches discussed here rely on the overexpression of fusion proteins, allowing in vivo detection of the bound fluorescent protein. This can be achieved injecting DNA constructs to obtain single-cell labeling of cargoes, as shown here (Figure 1A), or creating stable transgenic lines, where restriction of expression can be achieved by a combination of Gal4- and UAS-expressing lines. While this technique produces a bright signal well suited to time-lapse imaging, overexpression of protein can lead to deleterious effects by interfering with endogenous expression and triggering stress response mechanisms (Cheng and Lee, 2010). It is therefore essential to ensure that the construct does not lead to toxicity by monitoring cell morphology and embryonic development. The acquisition parameters in time-lapse microscopy are optimized to limit bleaching of fluorescent proteins and damage of the target cell, while still observing the target movement (for instance: high frequency sampling but reduced duration). In the case of the examples presented here, time-lapse imaging of labeled cargo in neurons was performed at 2 Hz for endosomes (5 min duration; Figure 2B) and at 1 Hz for mitochondria (10 min duration; Figure 1B,C) on a spinning disk confocal microscope.
Single Cell Type
Axonal transport dynamics can be influenced by the varying expression of subunits composing the motor protein complexes or particular cargo adapters, as well as by the axon caliber, due to differences in microtubule density affecting engagement of motors (Yu et al., 2017) and due to activity- and myelination-dependent number of neurofilaments (de Waegh et al., 1992). It is therefore recommended to target one cell type (Figure 1B,C), and in the case of spinal cord neurons, to limit observation to a specific region as cell size can fluctuate along the trunk and tail owing to the rostro-caudal developmental wave.
Single Cell Compartment
Different cell compartments have different cytoskeletal composition and regulatory mechanisms that can affect the composition and expression of motor proteins and their adaptor proteins, hence influencing the regulation of axonal transport (de Waegh et al., 1992; Yu et al., 2017; Gumy and Hoogenraad, 2018). As shown here (Figure 2), time-lapse imaging in different compartments of the same neuron for the same cargo can yield significant differences in some metrics of transport dynamics, but not others. The selection of a compartment most suited to the research hypothesis and consistency in the segment imaged across individual embryos and larva appears thus crucial, as well as the analysis of a variety of metrics such as run length and duration, average speed, pause frequency, average pause duration, switching behavior, area flux and transport rates of cargoes, both in the anterograde and retrograde directions.
Perspectives for Future Research
Alternative Labeling
Nanoparticles are inorganic semiconductors representing an attractive alternative for fluorescent labeling in live imaging applications because of their high spatial resolution and photostability. In addition, it is possible to tune their emission wavelength by varying their size and chemical composition. Because of this, and their broad absorption profile, it is possible to excite multiple colors at once, which is useful to reduce sample phototoxicity (Gao et al., 2005). In contrast to genetically encoded fluorescent protein tags, however, they need to be efficiently targeted to their biologically relevant endpoint. This has so far relied on surface modifications and solubilization strategies that led to very large particles better suited to high-sensitivity detection of low number of targets, such as single-molecule detection (Pinaud et al., 2006). Of note, this approach has allowed for real-time visualization of single-molecules in living cells (Dahan et al., 2003). Conjugation to biomolecules is, however, an interesting avenue to allow precise targeting and while still requiring the expression of a genetically encoded protein adaptor (Gao et al., 2005; Howarth et al., 2005), would provide the advantageous optical properties of nanoparticles over traditional fluorescent proteins.
Microscopy Improvements
Advances in imaging technology in the last years have yielded many optimized systems applicable to the study of in vivo axonal transport in the zebrafish embryo. Indeed, a great example of this is the swept field confocal microscope, which permits higher frame-rate capture when compared with spinning disk confocal and allows the rapid acquisition of z-stack time-lapses or high speed imaging (upward of 1,000 fps) of movement in single-plane (Castellano-Muñoz et al., 2012). Other systems circumvent classic caveats to an in vivo approach, such as photodamage, single-plan and temporal restriction and low signal, for instance: 2-photon microscopy (Renninger and Orger, 2013), light-sheet microscopy (Huisken et al., 2004; Panier et al., 2013; Park et al., 2015; Tomer et al., 2015; Fu et al., 2016). In the context of in vivo imaging of axonal transport, these strategies could allow the tracking of cargo and motors with exquisite temporal resolution, while also permitting 3D tracking in a whole embryo over long periods of time; considerable advantages over in vitro and other in vivo models.
Automated Detection and Analysis
The generation of kymograms as a 2D representation of time-lapse imaging is a common tool for the analysis of axonal transport, where the tracked target often moves on a single focal plane, in a linear trajectory. When analyzing movement in more complex environment, however, single particle tracking becomes a necessity, which renders manual analysis an arduous task. In the past years, many options have become available for automated detection and tracking, both commercially (Imaris, Metamorph, Igor Pro, etc.) and via open-source programs (MATLAB, ImageJ, etc.). Still, time-lapse videos obtained in vivo from intact animals are often noisier by nature than their cell culture counterpart, and since these samples are prone to photodamage, lead to undersampled data. This in turn impedes automatic detection and requires manual check of extracted metrics, while possibly omitting crucial information. Further advances in detection algorithms, based on in vivo data estimating how cargoes should behave, will surely be of benefit to researchers facing the tedious task of manual tracking.
Conclusion
The zebrafish embryo has emerged as an excellent model to pursue the characterization of processive transport in vivo as it can meet the need for more inclusive models, where the contribution of neuronal activity, glia and the cell cytoskeleton are taken into account. We outlined here some advantages and technical hints to use the zebrafish model for this type of analysis. Considering the recent breakthroughs in genetic manipulations and imaging technologies, this vertebrate is gaining attention in the field of neurodegenerative disease modeling, where axonal transport deficits are common hallmarks. In addition, a new emerging model sharing the same subfamily as zebrafish, Danionella translucida, which remains transparent throughout its life, will further expand the possibilities of adult neuronal imaging in vivo (Schulze et al., 2018). It is thus only a matter of time before axonal transport assays in zebrafish embryos become widespread for the study of physiological and pathological conditions.
Ethics Statement
As the EU directive 2010_63 explicitly states only “independently feeding larval forms” must be classified as animal experiments, therefore only zebrafish larvae past 120 h post fertilization should be subject to the regulations of European animal protection guidelines. For our experiments, we did not use larvae that have reached an “independent feeding” stage and therefore we did not have to submit an ethical approval to the competent local/national ethical/legal bodies.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. The Del Bene laboratory “Neural Circuits Development” is part of the Laboratoire d’Excellence (LABEX) entitled DEEP (ANR-11-LABX-0044), and of the École des Neurosciences de Paris Ile-de-France network. VB was supported by an FRSQ and CIHR Doctoral Award and was enrolled in the ENP Graduate Program. MR was supported by the Fondation pour la Recherche Médicale (FRM grant number ECO20170637481). CR was supported by a EU H2020 Marie Skłodowska-Curie Action fellowship (H2020-MSCA-IF-2014 #661527). This work has been supported by an ATIP/AVENIR program starting grant (FDB), ERC-StG #311159 (FDB), ERA-NET E-rare grant (for Research Programs on Rare Diseases, 0601165051) (FDB).
References
- Albadri S., Del Bene F., Revenu C. (2017). Genome editing using CRISPR/Cas9-based knock-in approaches in zebrafish. Methods 121–122, 77–85. 10.1016/j.ymeth.2017.03.005 [DOI] [PubMed] [Google Scholar]
- Artelt N., Ludwig T. A., Rogge H., Kavvadas P., Siegerist F., Blumenthal A., et al. (2018). The role of palladin in podocytes. J. Am. Soc. Nephrol. 29 1662–1678. 10.1681/ASN.2017091039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer T. O., Duroure K., De Cian A., Concordet J., Del Bene F. (2014). Highly efficient CRISPR / Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 24 142–153. 10.1101/gr.161638.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer T. O., Xiao T., Bercier V., Gebhardt C., Duroure K., Concordet J.-P., et al. (2015). Deletion of a kinesin I motor unmasks a mechanism of homeostatic branching control by neurotrophin-3. eLife 4:e05061. 10.7554/eLife.05061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babin P. J., Goizet C., Raldu D. (2014). Zebrafish models of human motor neuron diseases: advantages and limitations. Prog. Neurobiol. 118 36–58. 10.1016/j.pneurobio.2014.03.001 [DOI] [PubMed] [Google Scholar]
- Balasanyan V., Arnold D. B. (2014). Actin and myosin-dependent localization of mRNA to dendrites. PLoS One 9:e92349. 10.1371/journal.pone.0092349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandmann O., Burton E. A. (2010). Genetic zebrafish models of neurodegenerative diseases. Neurobiol. Dis. 40 58–65. 10.1016/j.nbd.2010.05.017 [DOI] [PubMed] [Google Scholar]
- Bridgman P. C. (1999). Myosin VA movements in normal and Dilute-Lethal axons provide support for a dual filament motor complex. J. Cell Biol. 146 1045–1060. 10.1083/jcb.146.5.1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calliari A., Farías J., Puppo A., Canclini L., Mercer J. A., Munroe D., et al. (2014). Myosin Va associates with mRNA in ribonucleoprotein particles present in myelinated peripheral axons and in the central nervous system. Dev. Neurobiol. 74 382–396. 10.1002/dneu.22155 [DOI] [PubMed] [Google Scholar]
- Campbell P. D., Shen K., Sapio M. R., Glenn T. D., Talbot W. S., Marlow F. L. (2014). Unique function of Kinesin Kif5A in localization of mitochondria in axons. J. Neurosci. 34 14717–14732. 10.1523/JNEUROSCI.2770-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano-Muñoz M., Peng A. W., Salles F. T., Ricci A. J. (2012). Swept field laser confocal microscopy for enhanced spatial and temporal resolution in live-cell imaging. Microsc. Microanal. 18 753–760. 10.1017/S1431927612000542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Tian L., Zhang F., Liu C., Lu T., Ruan Y., et al. (2013). Myosin Vb gene is associated with schizophrenia in Chinese Han population. Psychiatry Res. 207 13–18. 10.1016/j.psychres.2013.02.026 [DOI] [PubMed] [Google Scholar]
- Cheng C.-H., Lee W.-C. (2010). Protein solubility and differential proteomic profiling of recombinant Escherichia coli overexpressing double-tagged fusion proteins. Microb. Cell Fact. 9:63. 10.1186/1475-2859-9-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier-Larsen E., Holzbaur E. L. F. (2006). Axonal transport and neurodegenerative disease. Biochim. Biophys. Acta 1762 1094–1108. 10.1016/j.bbadis.2006.04.002 [DOI] [PubMed] [Google Scholar]
- Clark B., Winter M., Cohen A., Link B. (2011). Generation of Rab-based transgenic lines for in vivo studies of endosome biology in zebrafish. Dev. Dyn. 240 2452–2465. 10.1002/dvdy.22758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia S. S., Bassani S., Brown T. C., Lisé M. F., Backos D. S., El-Husseini A., et al. (2008). Motor protein-dependent transport of AMPA receptors into spines during long-term potentiation. Nat. Neurosci. 11 457–466. 10.1038/nn2063 [DOI] [PubMed] [Google Scholar]
- Dahan M., Le S., Luccardini C. (2003). Diffusion dynamics of glycine receptors revealed by single – quantum dot tracking. Science 302 442–445. 10.1126/science.1088525 [DOI] [PubMed] [Google Scholar]
- de Waegh S. M., Lee V. M. Y., Brady S. T. (1992). Local modulation of neurofilament phosphorylation, axonal caliber, and slow axonal transport by myelinating Schwann cells. Cell 68 451–463. 10.1016/0092-8674(92)90183-D [DOI] [PubMed] [Google Scholar]
- Di Donato V., De Santis F., Auer T. O., Testa N., Sánchez-iranzo H., Mercader N., et al. (2016). 2C-Cas9: a versatile tool for clonal analysis of gene function. Genome Res. 26 681–692. 10.1101/gr.196170.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drerup C. M., Herbert A. L., Monk K. R., Nechiporuk A. V. (2017). Regulation of mitochondria-dynactin interaction and mitochondrial retrograde transport in axons. eLife 6:e22234. 10.7554/eLife.22234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drerup C. M., Nechiporuk A. V. (2013). JNK-interacting protein 3 mediates the retrograde transport of activated c-Jun N-terminal kinase and lysosomes. PLoS Genet. 9:e1003303. 10.1371/journal.pgen.1003303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drerup C. M., Nechiporuk A. V. (2016). In vivo analysis of axonal transport in zebrafish. Methods Cell Biol. 131 311–329. 10.1016/bs.mcb.2015.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A., Kumar Sinha D. (2015). Turnover of the actomyosin complex in zebrafish embryos directs geometric remodelling and the recruitment of lipid droplets. Sci. Rep. 5:13915. 10.1038/srep13915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana B. D., Mezzomo N. J., Kalueff A. V., Rosemberg D. B. (2018). The developing utility of zebrafish models of neurological and neuropsychiatric disorders: a critical review. Exp. Neurol. 299 157–171. 10.1016/j.expneurol.2017.10.004 [DOI] [PubMed] [Google Scholar]
- Fu Q., Martin B. L., Matus D. Q., Gao L. (2016). Imaging multicellular specimens with real-time optimized tiling light-sheet selective plane illumination microscopy. Nat. Commun. 7:11088. 10.1038/ncomms11088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Yang L., Petros J. A., Marshall F. F., Simons J. W., Nie S. (2005). In vivo molecular and cellular imaging with quantum dots. Curr. Opin. Biotechnol. 16 63–72. 10.1016/j.copbio.2004.11.003 [DOI] [PubMed] [Google Scholar]
- Gibbs K. L., Kalmar B., Sleigh J. N., Greensmith L., Schiavo G. (2016). In vivo imaging of axonal transport in murine motor and sensory neurons. J. Neurosci. Methods 257 26–33. 10.1016/j.jneumeth.2015.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein L. S. B. (2012). Axonal transport and neurodegenerative disease: can we see the elephant? Prog. Neurobiol. 99 186–190. 10.1016/j.pneurobio.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumy L. F., Hoogenraad C. C. (2018). Local mechanisms regulating selective cargo entry and long-range trafficking in axons. Curr. Opin. Neurobiol. 51 23–28. 10.1016/j.conb.2018.02.007 [DOI] [PubMed] [Google Scholar]
- He C., Bartholomew C., Zhou W., Klionsky D. (2009). Assaying autophagic activity in transgenic GFP-Lc3 and GFP- Gabarap zebrafish embryos. Autophagy 5 520–526. 10.4161/auto.5.4.7768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N., Niwa S., Tanaka Y. (2010). Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68 610–638. 10.1016/j.neuron.2010.09.039 [DOI] [PubMed] [Google Scholar]
- Holloway M. P., Denardo B. D., Phornphutkul C., Nguyen K., Davis C., Jackson C., et al. (2016). An asymptomatic mutation complicating severe chemotherapy-induced peripheral neuropathy (CIPN): a case for personalised medicine and a zebrafish model of CIPN. NPJ Genom. Med. 1:16016. 10.1038/npjgenmed.2016.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth M., Takao K., Hayashi Y., Ting A. Y. (2005). Targeting quantum dots to surface proteins in living cells with biotin ligase. Proc. Natl. Acad. Sci. U.S.A. 102 7583–7588. 10.1073/pnas.0503125102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K., Clark M., Torroja C., Torrance J., Berthelot C., Muffato M., et al. (2013). The zebrafish reference genome sequence and its relationship to the human genome. Nature 496 498–503. 10.1038/nature12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruscha A., Krawitz P., Rechenberg A., Heinrich V., Hecht J., Haass C., et al. (2013). Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development 140 4982–4987. 10.1242/dev.099085 [DOI] [PubMed] [Google Scholar]
- Huang J. D., Brady S. T., Richards B. W., Stenoien D., Resau J. H., Copeland N. G., et al. (1999). Direct interaction of microtubule- and actin based transport motors. Nature 397 267–270. 10.1038/16722 [DOI] [PubMed] [Google Scholar]
- Huisken J., Swoger J., Del Bene F., Wittbrodt J., Stelzer E. H. K. (2004). Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305 1007–1009. 10.1126/science.1100035 [DOI] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Shengdar Q., Sander J. D., et al. (2013). Efficient In Vivo genome editing using RNA-guided nucleases. Nat. Biotechnol. 31 227–229. 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion U., Krauss J., Nusslein-Volhard C. (2014). Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development 141 4827–4830. 10.1242/dev.115584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayachandran P., Olmo V. N., Sanchez S. P., McFarland R. J., Vital E., Werner J. M., et al. (2016). Microtubule-associated protein 1b is required for shaping the neural tube. Neural Dev. 11 1–18. 10.1186/s13064-015-0056-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E., Champagne N., Brustein E., Drapeau P. (2010). In the swim of things: recent insights to neurogenetic disorders from zebrafish. Trends Genet. 26 373–381. 10.1016/j.tig.2010.05.004 [DOI] [PubMed] [Google Scholar]
- Keiter S., Burkhard-Medicke K., Wellner P., Kais B., Färber H., Skutlarek D., et al. (2016). Does perfluorooctane sulfonate (PFOS) act as chemosensitizer in zebrafish embryos? Sci. Total Environ. 54 317–324. 10.1016/j.scitotenv.2015.12.089 [DOI] [PubMed] [Google Scholar]
- Khan T. M., Benaich N., Malone C. F., Bernardos R. L., Russell A. R., Downes G. B., et al. (2012). Vincristine and bortezomib cause axon outgrowth and behavioral defects in larval zebrafish. J. Peripher. Nerv. Syst. 17 76–89. 10.1111/j.1529-8027.2012.00371.x [DOI] [PubMed] [Google Scholar]
- Kim A. J., Endow S. A. (2000). A kinesin family tree. J. Cell Sci. 113 3681–3682. [DOI] [PubMed] [Google Scholar]
- Klinman E., Holzbaur E. L. F. (2016). Comparative analysis of axonal transport markers in primary mammalian neurons. Methods Cell Biol. 131 409–424. 10.1016/bs.mcb.2015.06.011 [DOI] [PubMed] [Google Scholar]
- Knabbe J., Nassal J. P., Verhage M., Kuner T. (2018). Secretory vesicle trafficking in awake and anaesthetized mice: differential speeds in axons versus synapses. J. Physiol. 596 3759–3773. 10.1113/JP276022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor A. C., Kim Y. B., Packer M. S., Zuris J. A., Liu D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533 420–424. 10.1038/nature17946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalli G., Gschmeissner S., Schiavo G. (2003). Myosin Va and microtubule-based motors are required for fast axonal retrograde transport of tetanus toxin in motor neurons. J. Cell Sci. 116 4639–4650. 10.1242/jcs.00727 [DOI] [PubMed] [Google Scholar]
- Lasek R. J., Garner J. A., Brady S. T. (1984). Axonal transport of the cytoplasmic matrix. J. Cell Biol. 99 212s–221s. 10.1083/jcb.99.1.212s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo O. M., Gonzalez A., Ascano M., Kuruvilla R., Couve A., Bronfman F. (2013). BDNF regulates Rab11-mediated recycling endosome dynamics to induce dendritic branching. J. Neurosci. 33 6112–6122. 10.1523/JNEUROSCI.4630-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. C., Yu D., Martinez De Velasco J., Tessarollo L., Swing D. A., Court D. L., et al. (2001). A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73 56–65. 10.1006/geno.2000.6451 [DOI] [PubMed] [Google Scholar]
- Lindsay A. J., Mccaffrey M. W. (2014). Myosin Va is required for the transport of fragile X mental retardation protein (FMRP) granules. Biol. Cell 106 57–71. 10.1111/boc.201200076 [DOI] [PubMed] [Google Scholar]
- Liu X., Rizzo V., Puthanveettil S. (2012). Pathologies of axonal transport in neurodegenerative diseases. Transl. Neurosci. 3 355–372. 10.2478/s13380-012-0044-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Xu X. H., Chen Q., Wang T., Deng C. Y., Song B. L., et al. (2013). Myosin Vb controls biogenesis of post-Golgi Rab10 carriers during axon development. Nat. Commun. 4:2005. 10.1038/ncomms3005 [DOI] [PubMed] [Google Scholar]
- MacRae C. A., Peterson R. T. (2015). Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 14 721–731. 10.1038/nrd4627 [DOI] [PubMed] [Google Scholar]
- Maday S., Twelvetrees A. E., Moughamian A. J., Holzbaur E. L. F. (2014). Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron 84 292–309. 10.1016/j.neuron.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias J. R., Saxena M. T., Mumm J. S. (2012). Advances in zebrafish chemical screening technologies. Future Med. Chem. 4 1811–1822. 10.4155/fmc.12.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millecamps S., Julien J.-P. (2013). Axonal transport deficits and neurodegenerative diseases. Nat. Rev. Neurosci. 14 161–176. 10.1038/nrn3380 [DOI] [PubMed] [Google Scholar]
- Miscevic F., Rotstein O., Wen X.-Y. (2012). Advances in zebrafish high content and high throughput technologies. Comb. Chem. High Throughput Screen. 15 515–521. 10.2174/138620712801619140 [DOI] [PubMed] [Google Scholar]
- Mizgirev I. V., Revskoy S. (2010). A new zebrafish model for experimental leukemia therapy. Cancer Biol. Ther. 9 895–903. 10.4161/cbt.9.11.11667 [DOI] [PubMed] [Google Scholar]
- Nalavadi V., Griffin L., Picard-Fraser P., Swanson A., Takumi T., Bassell G. (2012). Regulation of ZBP1 transport dynamics in axons by MyosinVa. J. Neurosci. 32 15133–15141. 10.1523/JNEUROSCI.2006-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash J. E., Appleby V. J., Corrêa S. A. L., Wu H., Fitzjohn S. M., Garner C. C., et al. (2010). Disruption of the interaction between myosin VI and SAP97 is associated with a reduction in the number of AMPARs at hippocampal synapses. J. Neurochem. 112 677–690. 10.1111/j.1471-4159.2009.06480.x [DOI] [PubMed] [Google Scholar]
- Norden C., Young S., Link B. A., Harris W. A. (2009). Actomyosin is the main driver of interkinetic nuclear migration in the retina. Cell 138 1195–1208. 10.1016/j.cell.2009.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nukada Y., Horie M., Fukui A., Kotani T., Yamashita M. (2015). Real-time imaging of actin filaments in the zebrafish oocyte and embryo. Cytoskeleton 72 491–501. 10.1002/cm.21253 [DOI] [PubMed] [Google Scholar]
- Ohashi S., Koike K., Omori A., Ichinose S., Ohara S., Kobayashi S., et al. (2002). Identification of mRNA/protein (mRNP) complexes containing Purα, mStaufen, Fragile X Protein, and myosin Va and their association with rough endoplasmic reticulum equipped with a kinesin motor. J. Biol. Chem. 277 37804–37810. 10.1074/jbc.M203608200 [DOI] [PubMed] [Google Scholar]
- Osterweil E., Wells D. G., Mooseker M. S. (2005). A role for myosin VI in postsynaptic structure and glutamate receptor endocytosis. J. Cell Biol. 168 329–338. 10.1083/jcb.200410091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier T., Romano S. A., Olive R., Pietri T., Sumbre G., Candelier R., et al. (2013). Fast functional imaging of multiple brain regions in intact zebrafish larvae using selective plane illumination microscopy. Front. Neural Circuits 7:65. 10.3389/fncir.2013.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet D., Plucińska G., Misgeld T. (2014). In vivo imaging of mitochondria in intact zebrafish larvae. Methods Enzymol. 547 151–164. 10.1016/B978-0-12-801415-8.00009-6 [DOI] [PubMed] [Google Scholar]
- Park O. K., Kwak J., Jung Y. J., Kim Y. H., Hong H., Hwang B. J. (2015). 3D light-sheet fluorescence microscopy of cranial neurons and vasculature during zebrafish embryogenesis. Mol. Cells 38 975–981. 10.14348/molcells.2015.0160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten S. A., Armstrong G. A. B., Lissouba A., Kabashi E., Parker J. A., Drapeau P. (2014). Fishing for causes and cures of motor neuron disorders. Dis. Model. Mech. 7 799–809. 10.1242/dmm.015719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinaud F., Michalet X., Bentolila L. A., Tsay J. M., Doose S., Li J., et al. (2006). Advances in fluorescence imaging with quantum dot bioprobes. Biomaterials 27 1679–1687. 10.1016/j.biomaterials.2005.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plucińska G., Paquet D., Hruscha A., Godinho L., Haass C., Schmid B., et al. (2012). In vivo imaging of disease-related mitochondrial dynamics in a vertebrate model system. J. Neurosci. 32 16203–16212. 10.1523/JNEUROSCI.1327-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomareva O. Y., Eliceiri K. W., Halloran M. C. (2016). Charcot-Marie-Tooth 2b associated Rab7 mutations cause axon growth and guidance defects during vertebrate sensory neuron development. Neural Dev. 11:2. 10.1186/s13064-016-0058-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomareva O. Y., Holmen I. C., Sperry A. J., Eliceiri K. W., Halloran M. C. (2014). Calsyntenin-1 regulates axon branching and endosomal trafficking during sensory neuron development In Vivo. J. Neurosci. 34 9235–9248. 10.1523/JNEUROSCI.0561-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renninger S. L., Orger M. B. (2013). Two-photon imaging of neural population activity in zebrafish. Methods 62 255–267. 10.1016/j.ymeth.2013.05.016 [DOI] [PubMed] [Google Scholar]
- Roche H., Bogé G., Pérès G. (1994). Acute and chronic toxicities of colchicine in Brachydanio rerio. Bull. Environ. Contam. Toxicol. 52 69–73. 10.1007/BF00197359 [DOI] [PubMed] [Google Scholar]
- Santoriello C., Zon L. I. (2012). Science in medicine Hooked?! modeling human disease in zebrafish. J. Clin. Invest. 122 2337–2343. 10.1172/JCI60434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze L., Henninger J., Faustino A. I., Chaigne T., Kadobianskyi M., Hakiy N., et al. (2018). Transparent Danionella translucida as a genetically tractable vertebrate brain model. Nat. Methods 15 977–983. 10.1038/s41592-018-0144-6 [DOI] [PubMed] [Google Scholar]
- Sittaramane V., Chandrasekhar A. (2008). Expression of unconventional myosin genes during neuronal development in zebrafish. Gene Expr. Patterns 8 161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleigh J. N., Vagnoni A., Twelvetrees A. E., Schiavo G. (2017). Methodological advances in imaging intravital axonal transport. F1000Res. 6:200. 10.12688/f1000research.10433.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui W.-H., Huang S.-H., Wang J., Chen Q., Liu T., Chen Z.-Y. (2015). Myosin Va mediates BDNF-induced postendocytic recycling of full-length TrkB and its translocation into dendritic spines. J. Cell Sci. 128 1108–1122. 10.1242/jcs.160259 [DOI] [PubMed] [Google Scholar]
- Suster M. L., Abe G., Schouw A., Kawakami K. (2011). Transposon-mediated BAC transgenesis in zebrafish. Nat. Protoc. 6 1998–2021. 10.1038/nprot.2011.416 [DOI] [PubMed] [Google Scholar]
- Tamplin O. J., White R. M., Jing L., Kaufman C. K., Lacadie S. A., Li P., et al. (2012). Small molecule screening in zebrafish: swimming in potential drug therapies. Wiley Interdiscip. Rev. Dev. Biol. 1 459–468. 10.1002/wdev.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomer R., Lovett-barron M., Kauvar I., Broxton M., Deisseroth K. (2015). SPED light sheet microscopy: fast mapping of biological system structure and function. Cell 163 1796–1806. 10.1016/j.cell.2015.11.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnoni A., Bullock S. L. (2017). A simple method for imaging axonal transport in ageing neurons using the adult Drosophila wing. Nat. Protoc. 11 1711–1723. 10.1038/nprot.2016.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukoja A., Rey U., Petzoldt A. G., Ott C., Vollweiter D., Quentin C., et al. (2018). Presynaptic biogenesis requires axonal transport of lysosome-related vesicles. Neuron 99 1216–1232. 10.1016/j.neuron.2018.08.004 [DOI] [PubMed] [Google Scholar]
- Wu H., Nash J. E., Zamorano P., Garner C. C. (2002). Interaction of SAP97 with minus-end-directed actin motor myosin VI: implications for AMPA receptor trafficking. J. Biol. Chem. 277 30928–30934. 10.1074/jbc.M203735200 [DOI] [PubMed] [Google Scholar]
- Wu X., Bowers B., Rao K., Wei Q., Hammer J. A. (1998). Visualization of melanosome dynamics within wild-type and dilute melanocytes suggests a paradigm for myosin v function in vivo. J. Cell Biol. 143 1899–1918. 10.1083/jcb.143.7.1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Jung G., Hammer J. A., III (2000). Functions of unconventional myosins. Curr. Opin. Cell Biol. 12 42–51. 10.1016/S0955-0674(99)00055-1 [DOI] [PubMed] [Google Scholar]
- Yao Y., Sun S., Fei F., Wang J., Wang Y., Zhang R., et al. (2017). Screening in larval zebrafish reveals tissue-specific distribution of fifteen fluorescent compounds. Dis. Model. Mech. 10 1155–1164. 10.1242/dmm.028811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A., Fujii R., Watanabe Y., Okabe S., Fukui K., Takumi T. (2006). Myosin-Va facilitates the accumulation of mRNA/protein complex in dendritic Spines. Curr. Biol. 16 2345–2351. 10.1016/j.cub.2006.10.024 [DOI] [PubMed] [Google Scholar]
- Yu C. C., Reddy B. J. N., Wortman J. C., Steven P. (2017). Axonal transport: a constrained system. J. Neurol. Neuromed. 2 20–24. 10.29245/2572.942X/2017/3.1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Huang H., Zhang B., Lin S. (2016). TALEN- and CRISPR-enhanced DNA homologous recombination for gene editing in zebrafish. Methods Cell Biol. 135 107–120. 10.1016/bs.mcb.2016.03.005 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Qin W., Lu X., Xu J., Huang H., Bai H., et al. (2017). Programmable base editing of zebrafish genome using a modified CRISPR-Cas9 system. Nat. Commun. 8:118. 10.1038/s41467-017-00175-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X. J., Wang C. Z., Dai P. G., Xie Y., Song N. N., Liu Y., et al. (2007). Myosin X regulates netrin receptors and functions in axonal path-finding. Nat. Cell Biol. 9 184–192. 10.1038/ncb1535 [DOI] [PubMed] [Google Scholar]
- Zon L. I., Peterson R. T. (2005). In vivo drug discovery in the zebrafish. Nat. Rev. Drug Discov. 4 35–44. 10.1038/nrd1606 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.