Abstract
Introduction
Quantitative in vivo measurement of brain amyloid burden is important for both research and clinical purposes. However, the existence of multiple imaging tracers presents challenges to the interpretation of such measurements. This study presents a direct comparison of Pittsburgh compound B–based and florbetapir-based amyloid imaging in the same participants from two independent cohorts using a crossover design.
Methods
Pittsburgh compound B and florbetapir amyloid PET imaging data from three different cohorts were analyzed using previously established pipelines to obtain global amyloid burden measurements. These measurements were converted to the Centiloid scale to allow fair comparison between the two tracers. The mean and inter-individual variability of the two tracers were compared using multivariate linear models both cross-sectionally and longitudinally.
Results
Global amyloid burden measured using the two tracers were strongly correlated in both cohorts. However, higher variability was observed when florbetapir was used as the imaging tracer. The variability may be partially caused by white matter signal as partial volume correction reduces the variability and improves the correlations between the two tracers. Amyloid burden measured using both tracers was found to be in association with clinical and psychometric measurements. Longitudinal comparison of the two tracers was also performed in similar but separate cohorts whose baseline amyloid load was considered elevated (i.e., amyloid positive). No significant difference was detected in the average annualized rate of change measurements made with these two tracers.
Discussion
Although the amyloid burden measurements were quite similar using these two tracers as expected, difference was observable even after conversion into the Centiloid scale. Further investigation is warranted to identify optimal strategies to harmonize amyloid imaging data acquired using different tracers.
Keywords: PiB, Florbetapir, Amyloid imaging, Centiloid, Positron emission tomography
1. Introduction
Amyloid pathology is a neuropathological hallmark of Alzheimer's disease (AD), and it is well established that this pathology begins to accumulate decades before clinical symptoms appear [1], [2], [3], [4], [5], [6], [7]. Positron emission tomographic (PET) imaging using amyloid tracers can measure amyloid pathology in vivo and plays an important role in research, clinical trials, diagnosis, and monitoring of AD. The first selective amyloid PET imaging tracer, [11C]-Pittsburgh compound B (PiB) [8], has been used for over a decade and generated invaluable data to improve our understanding of AD; however, owing to its short half-life (20 minutes) as a 11C-labeled radioligand, PiB imaging is limited to large research centers with an onsite cyclotron. Recently, several 18F-based radioligands, [18F]-florbetapir [9], [18F]-florbetaben [10], [18F]-flutemetamol [11], and [18F]-NAV4694 [12], were developed to enable wide application of amyloid PET imaging given the longer half-life (110 minutes) of 18F.
The availability of multiple amyloid imaging tracers, in addition to the heterogeneity in imaging analysis protocols, leads to difficulties in interpreting the amyloid burden measurements across different groups [13]. To address this issue, the Centiloid Working Group proposed to establish a common scale (the Centiloid scale), which is defined based on two anchor points: the mean amyloid burden of a young control group presumed to have no amyloid plaque in their brain (defined as 0 on the Centiloid scale) and the mean amyloid burden of an AD group (defined as 100 on the Centiloid scale) [13]. They further outlined the procedure necessary to convert tracer- and group-dependent outcome measures of amyloid burden into the Centiloid scale [13]. Following this procedure, the conversion of amyloid burden measurements using [18F]-NAV4694 and [18F]-florbetaben to the Centiloid scale have been published [14], [15]. More recently, conversion to the Centiloid scale for [18F]-florbetapir was also reported, although the underlying florbetapir data were based on 10-minute scans rather than commonly adopted 50- to 70-minute time window [16]. Further investigation is warranted to compare the different tracers to help investigators making informed decisions on which tracers to use in their study.
Here, we present a study that directly compares PiB and florbetapir data in the same participants in cohorts of autosomal dominant as well as sporadic AD, and we also compare the two tracers using longitudinal data acquired on two similar, but separate, cohorts of sporadic AD spectrum participants. All the comparisons are made using the Centiloid scale.
2. Methods
2.1. Participants
The data set used in this study came from three different cohorts. The first cohort examined 182 participants from the Dominantly Inherited Alzheimer's Network Trial Unit (DIAN-TU) [17] (https://clinicaltrials.gov/ct2/show/NCT01760005) with baseline amyloid PET imaging data using both PiB and florbetapir. A total of 194 initial participants were recruited, but 12 were either missing imaging data or the processing did not pass quality control. In brief, enrollment criteria were being at risk for an autosomal dominant AD (ADAD) mutation, a Clinical Dementia Rating (CDR) score [18] of 0, 0.5, or 1, and an estimated years to symptom onset (EYO) of −15 to +10. Among the 182 DIAN-TU participants included in this study, 50 did not carry ADAD mutations (29 of these 50 participants were younger than 45), and 132 were mutation carriers (see Table 1 for more information). To define the Centiloid conversion equation for florbetapir, we followed the procedure outlined by the Centiloid Working Group [13] using a calibration data set randomly selected from the DIAN-TU cohort including 15 noncarriers younger than 45 and 22 mutation carriers who are 45 years and older with preclinical (amyloid positive and CDR = 0) or symptomatic AD (amyloid positive and CDR > 0). Amyloid positivity was defined based on PiB imaging results using previously determined thresholds (mean cortical standardized uptake value ratio [SUVr] greater than 1.42 with regional spread function [RSF]–based partial volume correction [PVC]) [19], [20].
Table 1.
Study cohorts
| Variable | CAL∗ |
DIAN-TU |
ADRC crossover |
ADRC longitudinal |
|||
|---|---|---|---|---|---|---|---|
| Noncarrier | Carrier | Noncarrier | Carrier | PiB | Florbetapir (AV45) | ||
| N | 15 | 22 | 50 (29 YC, 21 OC) | 132 | 103 | 54 | 26 |
| Age (SD) years | 39.3 (4.6) | 54.5 (6.3) | 43.3 (8.9) | 44.6 (10.1) | 67.4 (8.9) | 71.4 (7.0) | 72.1 (6.8) |
| Male (%) | 7 (46.7) | 14 (63.6) | 26 (52.0) | 62 (47.0) | 44 (42.7) | 23 (42.6) | 13 (50.0) |
| APOE ε4+ (%) | 4 (26.7) | 9 (40.9) | 16 (32.0) | 38 (28.8) | 36 (35.0) | 35 (64.8) | 15 (57.7) |
| CDR > 0 (%) | 0 (0.0) | 16 (72.7) | 3 (6.0) | 20 (15.2) | 5 (4.9) | 9 (16.7) | 4 (18.2) |
| Interval between scans (years) | - | - | - | 2.2† | 3.3† | ||
Abbreviations: ADRC, Alzheimer's Disease Research Center; APOE, apolipoprotein E; CDR, Clinical Dementia Rating; DIAN-TU, Dominantly Inherited Alzheimer's Network Trial Unit; OC, old controls; PiB, Pittsburgh compound B; SD, standard deviation; YC, young controls (<45 yrs old).
CAL (calibration data set) is a subset of DIAN-TU and is used for establishing the florbetapir Centiloid conversions.
Significantly different (P < .0001 based on the Welch two-sample t-test).
To compare the two tracers in late-onset spectrum populations, a cohort of 103 participants drawn from the Knight Alzheimer's Disease Research Center (ADRC) were enrolled in a crossover study to have both PiB and florbetapir scans within a short time window (<1 month). In addition, longitudinal PiB (N = 54) and florbetapir (N = 26) data for participants from Knight ADRC with positive baseline amyloid scans were included for further comparison of these two tracers in measuring rate of amyloid accumulation in a late-onset AD spectrum. Including amyloid-positive participants only allows more accurate assessment of the rate of amyloid accumulation, as PET measurement in people with minimal amyloid burden is mainly influenced by nonspecific binding and other factors unrelated to amyloid. A summary of cohort characteristics is presented in Table 1. For the longitudinal cohort, primary analysis was based on two time points rather than the full longitudinal data set because very few participants had more than two florbetapir scans.
For all participants across the cohorts, dementia status was assessed using the CDR and CDR sum-of-boxes (CDRSB) score. The Mini–Mental State Examination (MMSE) [21] was also performed to obtain an MMSE score. Each participant's apolipoprotein E (APOE) genotype was determined using previously described methods [22]. For the DIAN-TU cohort, EYO was calculated as the difference between the participant's age at evaluation and the age at which parental cognitive decline began [2]. The ADAD mutation status was determined using established methods [2]. Clinical evaluators in the DIAN-TU study were blind to participant mutation status.
2.1.1. Ethics statement
All assessment and imaging procedures were approved by Washington University's (WU's) Human Research Protection Office. Written informed consent was obtained from all individuals or their authorized representatives. Local institutional review boards also approved the image collection and analysis at each non-WU study site.
2.2. Imaging
For the DIAN-TU participants, all florbetapir PET scans include data between 50 and 70 minutes after injection and all PiB PET scans include data between 40 and 70 minutes after injection. The interval between florbetapir and PiB scans was less than a month (range 0–28 days). For the sporadic AD PiB-florbetapir crossover study, PiB PET includes data between 30 and 60 minutes after injection and florbetapir PET includes data between 50 and 70 minutes after injection. The PiB-florbetapir scan interval was also less than a month (range 1–18 days). For the longitudinal cohorts, same protocols were used for PiB and florbetapir as the sporadic AD crossover study. In addition to PET, 3D sagittal T1-weighted images of the head were also acquired for each participant.
2.3. Image analysis
PiB and florbetapir images were analyzed using our standard processing pipeline (PUP; https://github.com/ysu001/PUP) [23], [24]. Briefly, FreeSurfer (v5.3; Martinos Center for Biomedical Imaging, Charlestown, Massachusetts, USA; https://surfer.nmr.mgh.harvard.edu/fswiki) was used to process T1-weighted magnetic resonance imaging data and facilitate regional analysis of PET data. PUP processing includes scanner resolution harmonization filter [25], interframe motion correction, PET-to-magnetic resonance registration, regional intensity extraction, RSF-based PVC [23], [26], and SUVr analysis using the cerebellar cortex as the reference region. Although the brainstem/pons is commonly used as the reference region for the investigation of ADAD [2], [5], we used the cerebellar cortex to be consistent among cohorts, and also it has been shown recently that using the cerebellar cortex generated similar results as brainstem referencing in DIAN [27]. As the global index of amyloid burden, a mean cortical SUVr was calculated based on a selected set of cortical regions defined by FreeSurfer, that is, frontal, parietal, temporal, and precuneus [24].
To facilitate between tracer comparison, PiB and florbetapir SUVrs were both converted to the Centiloid scale. The Centiloid conversion for PiB data was reported previously [28]. For DIAN-TU data, the conversion equations for 40- to 70-minute time window were used; and for the crossover and longitudinal data set, the equations for 30- to 60-minute time window were used (see Supplementary Material for details). To establish the Centiloid conversion equations for florbetapir PET processed using PUP, we followed the recommended level-2 calibration procedure [13]. The PiB PET data from the calibration data set were analyzed using the Centiloid pipeline to obtain PiB Centiloid SUVr according to Klunk et al. [13]. Linear regression was then performed between florbetapir mean cortical SUVrs and PiB Centiloid SUVrs to obtain the florbetapir Centiloid conversion equations using the level-2 calibration procedure [13]. These equations were reported in Supplementary Material including the ones using cerebellar cortex referencing used in our primary analysis and equations derived for brainstem and white matter referencing. All PUP-based mean cortical SUVrs were then converted to the Centiloid scale using corresponding equations. In subsequent analysis, we always used the Centiloid values as the target variable, except where we determined amyloid positivity based on SUVr thresholds as discussed in our Supplementary Material.
2.4. Statistical analysis
2.4.1. Cross-sectional analysis
The mean and standard deviation of the estimated amyloid burden in the 29 young ADAD mutation noncarriers from the DIAN-TU cohort were used to assess the variability in the amyloid burden measurement in Centiloids and define amyloid positivity threshold using the specificity approach, that is, 95th percentile of the amyloid burden measurements for the young controls [29]. Multivariate linear models with unstructured covariance matrix were used to compare the means and interindividual variability of the two tracers. Similarly, multivariate linear models with unstructured covariance matrix parameterized in terms of variances and correlations were used to estimate and compare the strength of the correlations between levels of amyloid burden and clinical/cognitive outcomes by tracers. To test whether the two tracers are equivalent in inter-individual variability and correlation to other variables in the context of multivariate model, a likelihood ratio test (test statistics approximately follow distribution) was performed. A major advantage of using the multivariate-model-based test is that other covariates can be added into the model straightforwardly and be adjusted for. One of the 132 mutation carrier DIAN-TU participants was missing EYO data and excluded from this correlation analysis. These analyses were done using Proc Glimmix, SAS 9.4 (SAS Institute Inc., Cary, NC). Correlation analysis was also performed for the cross-sectional sporadic AD spectrum data to compare the two tracers.
2.4.2. Longitudinal analysis
For the longitudinal cohort of sporadic AD, baseline amyloid positivity was determined based on previously established criteria for PiB (mean cortical SUVr greater than 1.42 with RSF PVC) [19], [20] and its equivalent florbetapir cutoff of 1.19 (see Supplementary Material for the derivation of florbetapir cutoff). Multivariate linear mixed effects models with random intercepts and random slopes were used to estimate and compare the longitudinal change in amyloid burden for each tracer. Different covariance matrices for the random effects and different residual variances were assumed between tracers. These models can handle missing, unbalanced, and unevenly spaced longitudinal data and have been used in previous studies of AD [30]. These analyses were done using Proc Mixed, SAS 9.4.
3. Results
3.1. Florbetapir Centiloid
Based on the calibration data set, the Centiloid conversion equations for florbetapir SUVrs were generated and listed in Supplementary Material. Our florbetapir conversion equation (CL = 163.6 x SUVr – 181.0) (supp. Eq. 5) derived based on raw SUVrs without RSF PVC was similar to the published equations from a recent paper [16]. The difference is attributable to variation in image acquisition protocols and quantification procedures. Florbetapir mean cortical SUVrs strongly correlated with PiB Centiloid SUVrs with (r = 0.8861, P < .0001) or without RSF PVC (r = 0.8906, P < .0001, Supplementary Fig. 2).
3.2. DIAN-TU baseline analysis
The amyloid burden measured within the young mutation noncarriers and the mutation carriers within the DIAN-TU data set is reported in Table 2. Multivariate model analyses showed that for the young control group, the mean amyloid burden in the Centiloid scale was significantly different for the two tracers when RSF PVC was used (t-test, df [degree of freedom] = 29, P < .0001), but not when correction was not used (P = .10), whereas the variability was significantly different with (SD = 3.2 for PiB vs. SD = 5.4 for florbetapir, test, df = 1, P = .0006) and without (SD = 4.4 for PiB vs. SD = 14.2 for florbetapir, test, df = 1, P < .0001) RSF PVC (Table 2).
Table 2.
Interindividual variability comparisons based on the DIAN-TU baseline data
| Variable | CL_PiB_SUVr | CL_PiB_SUVr_RSF | CL_Florbetapir_SUVr | CL_Florbetapir_SUVr_RSF |
|---|---|---|---|---|
| YC mean | −1.2 | 0.3 | 2.7 | 4.1 |
| YC SD | 4.4 | 3.2 | 14.2 | 5.4 |
| MC mean | 65.6 | 66.3 | 65.2 | 63.0 |
| MC SD | 52.6 | 50.9 | 54.4 | 44.3 |
| Specificity threshold (95%) | ||||
| 6.0 | 5.5 | 26.1 | 12.9 | |
| P values | ||||
| Interindividual variability for YC | ||||
| PiB versus AV45 | <.0001 | .0006 | ||
| RSF versus non-RSF | .005 | <.0001 | ||
| Comparison of YC mean | ||||
| PiB versus AV45 | .10 | <.0001 | ||
| RSF versus non-RSF | .003 | .53 | ||
Abbreviations: ADAD, autosomal dominant Alzheimer's disease; AV45, florbetapir; MC, ADAD mutation carriers; DIAN-TU, Dominantly Inherited Alzheimer's Network Trial Unit; PiB, Pittsburgh compound B; RSF, regional spread function; SD, standard deviation; SUVr, standardized uptake value ratio; YC, young control participants (<45 years) without ADAD mutation. Statistical significant P values (P < .05) are highlighted in bold.
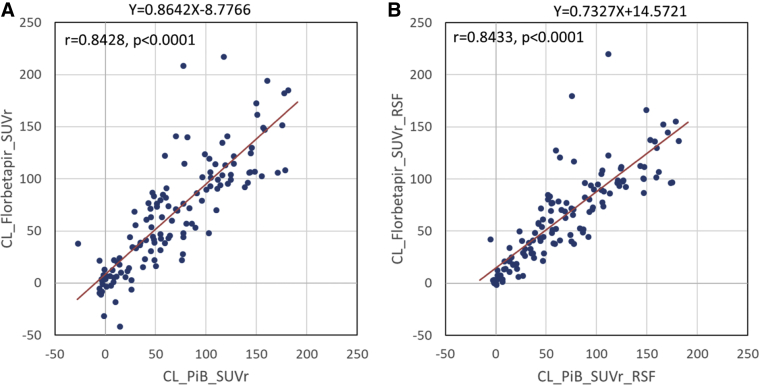
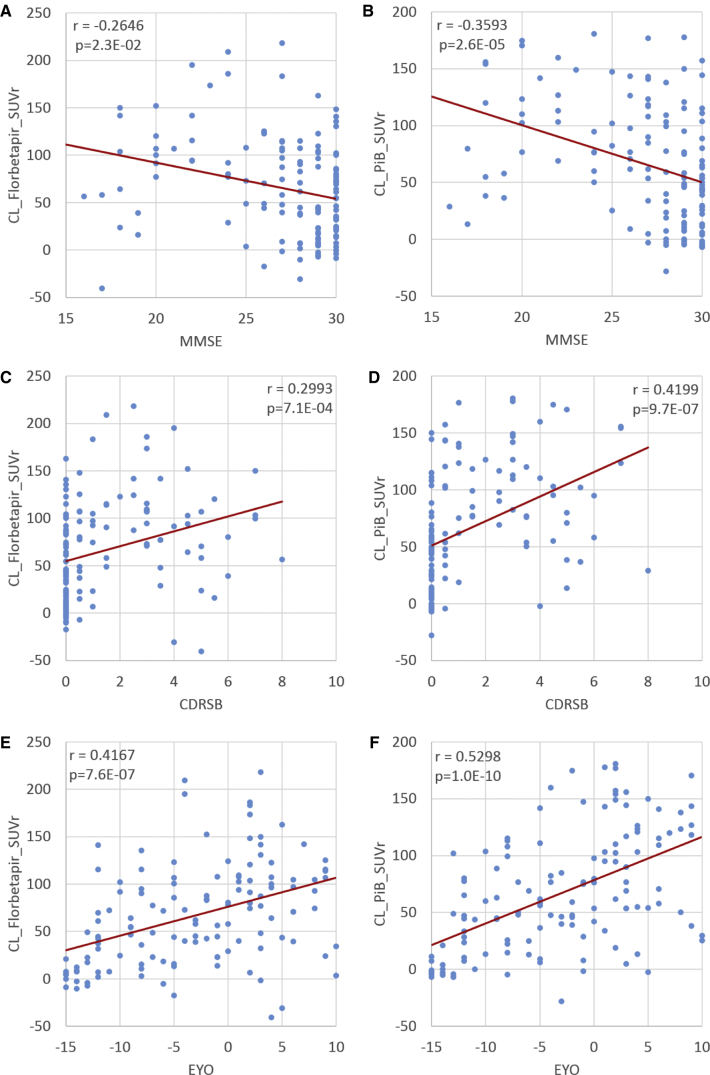
After conversion to the Centiloid scale, the amyloid burden measured with each tracer was approximately in the same range (0 to 200); however, because of the higher variability, the specificity-based amyloid positivity threshold (i.e., 95th percentile of young controls) was higher in the Centiloid scale when florbetapir was used as the imaging tracer (Table 2). For the DIAN-TU data, the amyloid burden measurements were strongly correlated between PiB- and florbetapir-based measurements with (r = 0.8433, P < .0001) and without (r = 0.8428, P < .0001) RSF PVC (Fig. 1), whereas the intraclass correlation was significant with RSF PVC (0.5616, P < .0001) but not without correction (0.3013, P = .17). For the mutation carriers, the correlations between amyloid burden and clinical variables such as MMSE, CDRSB, and EYO were significantly different for the two tracers without RSF PVC but were similar with correction (Table 3) based on the likelihood ratio tests. It was also observed that PiB-based measurements and quantification with RSF PVC tend to have smaller P values and larger r-values (Table 3). Scatter plots depicting the relationships between amyloid burden and clinical variables are shown in Fig. 2. When these relationships were assessed for APOE ε4 carriers only, similar (numerically slightly stronger) levels of associations between amyloid burden and the clinical variables were observed (Supplementary Table 1), although we no longer observe a between-tracer difference.
Fig. 1.
Comparison of amyloid burden measurements in the Centiloid scale using florbetapir without (A) and with (B) RSF PVC to PiB-based measurements in the DIAN-TU mutation carriers. Abbreviations: DIAN-TU, Dominantly Inherited Alzheimer's Network Trial Unit; PiB, Pittsburgh compound B; PVC, partial volume correction; RSF, regional spread function; SUVr, standardized uptake value ratio.
Table 3.
Pearson correlation between amyloid and clinical/cognitive outcomes by PET tracers
| Variable | Without RSF PVC |
With RSF PVC |
||||
|---|---|---|---|---|---|---|
| PiB PET, ρ (SE) | AV45 PET, ρ (SE) | P value | PiB PET, ρ (SE) | AV45 PET, ρ (SE) | P value | |
| EYO | 0.529 (0.0630) | 0.417 (0.0723) | .0075 | 0.553 (0.0607) | 0.503 (0.0654) | .218 |
| CDRSB | 0.420 (0.0721) | 0.299 (0.0797) | .007 | 0.453 (0.0695) | 0.388 (0.0744) | .134 |
| MMSE | −0.359 (0.0762) | −0.265 (0.0814) | .0392 | −0.387 (0.0744) | −0.336 (0.0776) | .259 |
Abbreviations: AV45, florbetapir; CDRSB, CDR sum of boxes; EYO, estimated years to symptom onset; MMSE, Mini–Mental State Examination; PET, positron emission tomography; PiB, Pittsburgh compound B; PVC, partial volume correction; RSF, regional spread function; SE, standard error.
Fig. 2.
Association between amyloid burden measurements and clinical variables. (A) Florbetapir SUVr versus MMSE; (B) PiB SUVr versus MMSE; (C) Florbetapir SUVr versus CDRSB; (D) PiB SUVr versus CDRSB; (E) Florbetapir SUVr versus EYO; (F) PiB SUVr versus EYO. All SUVr measurements have been converted to the Centiloid scale. Abbreviations: CDRSB, CDR sum of boxes; EYO, estimated years to symptom onset; MMSE, Mini–Mental State Examination; PET, positron emission tomography; PiB, Pittsburgh compound B; SUVr, standardized uptake value ratio.
3.3. Sporadic AD crossover analysis
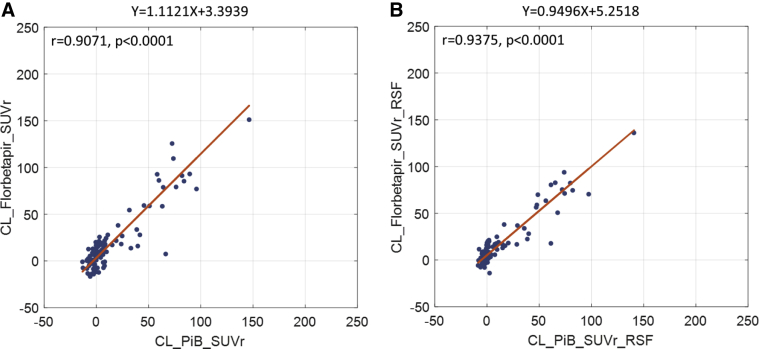
Amyloid burden measured using PiB and florbetapir strongly correlated in the sporadic AD cohort similar to the DIAN-TU cohort. The Pearson's correlation for amyloid burden in the Centiloid scale between the two tracers was 0.9071 (P < .0001) without and 0.9375 (P < .0001) with RSF PVC (Fig. 3). The Centiloid conversion was able to convert the amyloid burden measurements into a similar dynamic range (approximately between 0 and 150 in the Centiloid scale).
Fig. 3.
Comparison of amyloid burden measurements in the Centiloid scale using florbetapir without (A) and with (B) RSF PVC to PiB-based measurements in the sporadic AD crossover cohort. Abbreviations: AD, Alzheimer's disease; PiB, Pittsburgh compound B; PVC, partial volume correction; RSF, regional spread function; SUVr, standardized uptake value ratio.
3.4. Longitudinal analysis
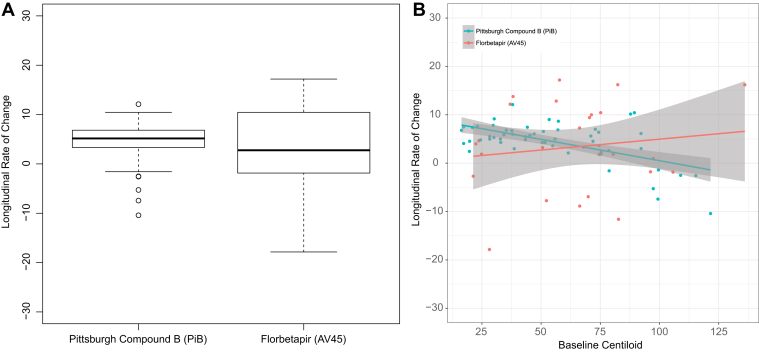
Significant accumulation of amyloid was observed in the longitudinal PiB cohort with (annual rate of change: 5.06 [0.40], P < .0001) or without PVC (annual rate of change: 4.69 [0.37], P < .0001). The longitudinal florbetapir data failed to show significant changes over time without PVC (annual rate of change: 2.62 [2.36], P = .27) but became significant when RSF PVC was performed (annual rate of change: 3.62 [1.67], P = .03); however, the annualized rate of change in amyloid burden measure did not differ by tracers with or without PVC (Fig. 4). Further sensitivity analysis using all longitudinal data yielded almost identical results (data not shown).
Fig. 4.
Annualized rate of change in measured amyloid burden using PiB and florbetapir in the longitudinal cohort. (A) Box plot of rate measurements for the two tracers. (B) Rate of change as a function of baseline amyloid burden. No difference was observed between the rate measurements from the two tracers (P = .555).
3.5. Sample sizes required to detect the same reduction in the annual rate of change by tracers
To demonstrate the effect of different tracers on the sample size required to detect the same proportion (%) of reduction in the annual rate of change, we used the rate of change and related variances of the Centiloid scale with RSF PVC to calculate the sample size. For ease of demonstration, we conceptualized a treatment versus placebo trial with 1:1 randomization ratio, 3-year duration with annual assessments, 80% power, and 5% annual dropout. The primary outcome was the annual rate of change, and the primary analysis model was the linear mixed effects model with random intercepts and random slopes. The annual rates of change for the placebo group were those estimated in Section 3.4: 5.06 for PiB and 3.62 for florbetapir; for example, a 20% reduction for the treatment group would lead to annual rates of change of 4.04 and 2.90 for PiB and florbetapir, respectively. Table 4 presented the sample size for a range of reductions in the rate of change. A larger sample size would be required in a hypothetical anti-amyloid trial if florbetapir is used as the tracer. The discrepancy was attributed to the combination of larger annual rate of change and smaller variance in the PiB amyloid burden assessments compared with AV45 (florbetapir) assessments.
Table 4.
Sample size for a 2-year trial by tracers (using RSF-PVC-based amyloid burden measurements)
| Tracer | Reduction in the annual rate of change |
|||
|---|---|---|---|---|
| 20% less | 40% less | 60% less | 80% less | |
| AV45 | 2156 | 539 | 240 | 135 |
| PiB | 305 | 77 | 34 | 20 |
Abbreviations: AV45, florbetapir; PiB, Pittsburgh compound B; PVC, partial volume correction; RSF, regional spread function.
4. Discussion
This study generated the linear conversion equations from florbetapir SUVrs to the Centiloid scale following the Centiloid Work Group guidelines [13] based on a data set of 37 participants. Florbetapir-based amyloid burden measurements strongly correlated with PiB-based measurements (R2 > 0.70). We then performed direct comparison of florbetapir against PiB-based amyloid measurements in two independent cohorts. Based on the DIAN-TU data set, which included 29 young controls who did not have ADAD mutations and therefore are presumed not to have amyloid in their brain, florbetapir imaging demonstrated considerably higher measurement variabilities than PiB imaging (Table 2). The Centiloid approach was able to convert amyloid burden measurements derived from the two tracers into a similar dynamic range. We did not observe statistically significant differences in the estimated average rate of amyloid accumulation using the two tracers in the longitudinal analysis, although the difference in measurement variability led to difference in statistical power in detecting longitudinal change and sample size needed in hypothetical clinical trials. Note that, although both PiB and florbetapir data were converted to Centiloid units for the purpose of this comparison so that the dynamic range of both tracers would be in a similar range, all results would have been quantitatively and statistically equivalent without this conversion because it is a simple linear transformation of the data.
The cross-sectional comparison between PiB and florbetapir in this study was made based on a crossover design where the two scans were performed within 1 month in the same participants in both an ADAD and a sporadic AD cohort. Previously, Laudau et al. [31] compared these two tracers using florbetapir data collected approximately 1.5 years after PiB imaging and found a correlation between 0.86 and 0.95 depending on the quantification method used. More recently, Navitsky et al. [16] reported a florbetapir-to-PiB SUVr correlation of approximately 0.95. These results are in agreement with ours. The Centiloid conversion and comparison to PiB has been reported for two other 18F-tracers, that is, [18F]-NAV4694 [14] and [18F]-florbetaben [15]. An intertracer correlation of R2 = 0.99 was observed between PiB and [18F]-NAV4694, and R2 = 0.96 was observed between PiB and [18F]-florbetaben. The variance of amyloid burden measurements in young controls was SD = 3.7 for [18F]-NAV4694 and SD = 6.81 for [18F]-florbetaben [14], [15]. Recently, Navitsky et al. [16] reported an SD = 12.07 for florbetapir, and here, we observed an SD = 14.2 for the raw florbetapir SUVr-derived Centiloid measurements; however, when the RSF PVC technique was used in the quantification procedure, the variance was substantially reduced (SD = 5.4). This suggests that the variability in a pathologically free cohort may primarily be related to nonspecific uptake in the white matter, and PVC can reduce this effect. In this study, we used the cerebellar cortex as the reference region for quantification, whereas several groups have suggested that the use or inclusion of white matter as the reference region lead to lower variability and better statistical power in longitudinal studies [32], [33]. Further investigation is warranted, especially considering the reports of myelin binding of amyloid PET tracers [34], [35] and the age-related change in white matter amyloid PET signal [36].
In our previous work [28], we reported that the thresholds for amyloid positivity are dependent on the criteria used for definition and the quantification method, even after Centiloid conversion. In a multicenter European study [37], a 95% specificity-based SUVr threshold was transformed into the Centiloid scale and resulted in a cutoff of 34, which differed considerably from our previous work and the present study. Recently, postmortem neuropathology–driven thresholds were also reported [38]; however, depending on the metrics used for pathology evaluation, it also resulted in different thresholds. The current work further strengthens the observation that amyloid positivity cutoffs are sensitive to amyloid tracer, quantification method, and the underlying cohort used to derive the cutoff. A universal and physiologically/pathologically meaningful threshold remains a challenge and requires further investigation.
One limitation of this study is that the Centiloid conversion for florbetapir is established based on an ADAD cohort, which may have different patterns of amyloid pathology than the sporadic AD population. Although we have crossover data in the sporadic AD cohort, the PiB imaging protocol in that cohort only acquires data up to 60 minutes after injection and does not satisfy the 50–70 minutes requirement put forward by the Centiloid Working Group [13]. Although the optimal Centiloid conversion strategy for florbetapir-derived amyloid burden measurements remains to be determined with additional data and further research, our comparison between PiB and florbetapir is still valid as the Centiloid conversion is simply a linear transformation, which does not alter the statistical distribution of the underlying data. We do not expect the observed signal-to-noise properties to change with a different Centiloid conversion strategy. Our longitudinal comparison is limited by the fact that the PiB and florbetapir data were collected on two similar but separate cohorts; therefore, it may not have the power to detect small differences between the longitudinal performances of the two tracers. Also, the large differences in estimated sample size in hypothetical trials should also be interpreted with caution as the underlying mean and standard deviation data are not derived from the same population. Future studies are necessary to further compare different amyloid tracers in longitudinal studies.
5. Conclusion
Florbetapir-based amyloid measurements had higher variability, which may relate to its white matter nonspecific uptake and lower dynamic range before Centiloid transformation. The difference in variability also resulted in large differences in the 95% specificity-based amyloid positivity threshold and differences in ability to detect subtle amyloid burden. Further study is necessary to characterize tracer performance in longitudinal studies.
Research in Context.
-
1.
Systematic review: The literature was reviewed using PubMed to identify recent publications on the comparison of different amyloid positron emission tomographic (PET) imaging tracers and the application of the Centiloid approach for harmonization of amyloid burden measurements.
-
2.
Interpretation: This work thoroughly compares two widely used amyloid PET imaging tracers in both autosomal dominant Alzheimer's disease and late-onset Alzheimer's disease spectrum cohorts to examine their sensitivity to amyloid burden and its change over time. The comparison also adopted the Centiloid approach to bring different amyloid PET measurements into a common scale.
-
3.
Future directions: Significant differences in sensitivity to amyloid burden and its longitudinal change were observed for the two tracers even after converting to the Centiloid scale. This resulted in substantial differences in sample sizes needed for hypothetical antiamyloid trials. Further longitudinal study is needed to verify this finding and compare different amyloid PET tracers.
Acknowledgments
This research was supported by National Institute of Health grants: P50AG005681, P01AG003991, P01AG026276, U01AG042791, R01AG046179, P30NS048056 R01AG055444, and R01AG031581. Image acquisition and analysis received additional support by UL1TR000448, P30NS098577, and R01EB009352. Research was also supported by BrightFocus Foundation grants A2017272S and A2017330S, Alzheimer's Association Research grant AARG-17-532945, Arizona Alzheimer's Research Consortium, the Charles and Joanne Knight Alzheimer Disease Research Center Support Fund, the David and Betty Farrell Medical Research Fund, the Daniel J. Brennan Alzheimer Research Fund, the Fred Simmons and Olga Mohan Alzheimer Research Support Fund, the Barnes-Jewish Hospital Foundation, the Alzheimer's Association, Eli Lilly and Company, Hoffman La-Roche, Avid Radiopharmaceuticals, GHR Foundation, and an anonymous organization. The authors acknowledge the altruism of the participants and their families and contributions of the DIAN-TU research and support staff at the coordinating center and each of the participating sites for their contributions to this study.
Washington University and Dr. David Holtzman, Department Head of Neurology where the research is being conducted, may receive royalty income for an investigational drug called solanezumab, which was developed by Dr. Holtzman and licensed by Washington University to Eli Lilly & Company. Solanezumab is evaluated in this research.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.dadm.2018.12.008.
Contributor Information
Yi Su, Email: yi.su@bannerhealth.com.
Tammie L.S. Benzinger, Email: benzingert@wustl.edu.
Supplementary data
References
- 1.Holtzman D.M., Morris J.C., Goate A.M. Alzheimer's Disease: The Challenge of the Second Century. Sci Transl Med. 2011;3:77sr1. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bateman R.J., Xiong C., Benzinger T.L., Fagan A.M., Goate A., Fox N.C. Clinical and biomarker changes in dominantly inherited Alzheimer's disease. N Engl J Med. 2012;367:795–804. doi: 10.1056/NEJMoa1202753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jack C.R., Jr., Knopman D.S., Jagust W.J., Shaw L.M., Aisen P.S., Weiner M.W. Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. Lancet Neurol. 2010;9:119–128. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris J.C., Price A.L. Pathologic correlates of nondemented aging, mild cognitive impairment, and early-stage Alzheimer's disease. J Mol Neurosci. 2001;17:101–118. doi: 10.1385/jmn:17:2:101. [DOI] [PubMed] [Google Scholar]
- 5.Benzinger T.L., Blazey T., Jack C.R., Jr., Koeppe R.A., Su Y., Xiong C. Regional variability of imaging biomarkers in autosomal dominant Alzheimer's disease. Proc Natl Acad Sci U S A. 2013;110:E4502–E4509. doi: 10.1073/pnas.1317918110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jansen W.J., Ossenkoppele R., Knol D.L., Tijms B.M., Scheltens P., Verhey F.R. Prevalence of cerebral amyloid pathology in persons without dementia: a meta-analysis. JAMA. 2015;313:1924–1938. doi: 10.1001/jama.2015.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villemagne V.L., Burnham S., Bourgeat P., Brown B., Ellis K.A., Salvado O. Amyloid beta deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer's disease: a prospective cohort study. Lancet Neurol. 2013;12:357–367. doi: 10.1016/S1474-4422(13)70044-9. [DOI] [PubMed] [Google Scholar]
- 8.Klunk W.E., Engler H., Nordberg A., Wang Y., Blomqvist G., Holt D.P. Imaging brain amyloid in Alzheimer's disease with Pittsburgh Compound-B. Ann Neurol. 2004;55:306–319. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- 9.Wong D.F., Rosenberg P.B., Zhou Y., Kumar A., Raymont V., Ravert H.T. In vivo imaging of amyloid deposition in Alzheimer disease using the radioligand 18F-AV-45 (florbetapir [corrected] F 18) J Nucl Med. 2010;51:913–920. doi: 10.2967/jnumed.109.069088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rowe C.C., Ackerman U., Browne W., Mulligan R., Pike K.L., O'Keefe G. Imaging of amyloid beta in Alzheimer's disease with 18F-BAY94-9172, a novel PET tracer: proof of mechanism. Lancet Neurol. 2008;7:129–135. doi: 10.1016/S1474-4422(08)70001-2. [DOI] [PubMed] [Google Scholar]
- 11.Vandenberghe R., Van Laere K., Ivanoiu A., Salmon E., Bastin C., Triau E. 18F-flutemetamol amyloid imaging in Alzheimer disease and mild cognitive impairment: a phase 2 trial. Ann Neurol. 2010;68:319–329. doi: 10.1002/ana.22068. [DOI] [PubMed] [Google Scholar]
- 12.Cselenyi Z., Jonhagen M.E., Forsberg A., Halldin C., Julin P., Schou M. Clinical validation of 18F-AZD4694, an amyloid-beta-specific PET radioligand. J Nucl Med. 2012;53:415–424. doi: 10.2967/jnumed.111.094029. [DOI] [PubMed] [Google Scholar]
- 13.Klunk W.E., Koeppe R.A., Price J.C., Benzinger T.L., Devous M.D., Sr. The Centiloid Project: standardizing quantitative amyloid plaque estimation by PET. Alzheimers Dement. 2015;11:1–15. doi: 10.1016/j.jalz.2014.07.003. e1–e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rowe C.C., Jones G., Dore V., Pejoska S., Margison L., Mulligan R.S. Standardized Expression of 18F-NAV4694 and 11C-PiB beta-Amyloid PET Results with the Centiloid Scale. J Nucl Med. 2016;57:1233–1237. doi: 10.2967/jnumed.115.171595. [DOI] [PubMed] [Google Scholar]
- 15.Rowe C.C., Dore V., Jones G., Baxendale D., Mulligan R.S., Bullich S. (18)F-Florbetaben PET beta-amyloid binding expressed in Centiloids. Eur J Nucl Med Mol Imaging. 2017;44:2053–2059. doi: 10.1007/s00259-017-3749-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Navitsky M., Joshi A.D., Kennedy I., Klunk W.E., Rowe C.C., Wong D.F. Standardization of amyloid quantitation with florbetapir standardized uptake value ratios to the Centiloid scale. Alzheimers Dement. 2018;14:1565–1571. doi: 10.1016/j.jalz.2018.06.1353. [DOI] [PubMed] [Google Scholar]
- 17.Mills S.M., Mallmann J., Santacruz A.M., Fuqua A., Carril M., Aisen P.S. Preclinical trials in autosomal dominant AD: implementation of the DIAN-TU trial. Rev Neurol. 2013;169:737–743. doi: 10.1016/j.neurol.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris J.C. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43:2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- 19.Vlassenko A.G., McCue L., Jasielec M.S., Su Y., Gordon B.A., Xiong C. Imaging and cerebrospinal fluid biomarkers in early preclinical alzheimer disease. Ann Neurol. 2016;80:379–387. doi: 10.1002/ana.24719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sutphen C.L., Jasielec M.S., Shah A.R., Macy E.M., Xiong C., Vlassenko A.G. Longitudinal Cerebrospinal Fluid Biomarker Changes in Preclinical Alzheimer Disease During Middle Age. JAMA Neurol. 2015;72:1029–1042. doi: 10.1001/jamaneurol.2015.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Folstein M.F., Folstein S.E., McHugh P.R. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 22.Pastor P., Roe C.M., Villegas A., Bedoya G., Chakraverty S., Garcia G. Apolipoprotein Eepsilon4 modifies Alzheimer's disease onset in an E280A PS1 kindred. Ann Neurol. 2003;54:163–169. doi: 10.1002/ana.10636. [DOI] [PubMed] [Google Scholar]
- 23.Su Y., Blazey T.M., Snyder A.Z., Raichle M.E., Marcus D.S., Ances B.M. Partial volume correction in quantitative amyloid imaging. Neuroimage. 2015;107:55–64. doi: 10.1016/j.neuroimage.2014.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su Y., D'Angelo G.M., Vlassenko A.G., Zhou G., Snyder A.Z., Marcus D.S. Quantitative analysis of PiB-PET with FreeSurfer ROIs. PLoS One. 2013;8:e73377. doi: 10.1371/journal.pone.0073377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joshi A., Koeppe R.A., Fessler J.A. Reducing between scanner differences in multi-center PET studies. Neuroimage. 2009;46:154–159. doi: 10.1016/j.neuroimage.2009.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rousset O.G., Collins D.L., Rahmim A., Wong D.F. Design and implementation of an automated partial volume correction in PET: application to dopamine receptor quantification in the normal human striatum. J Nucl Med. 2008;49:1097–1106. doi: 10.2967/jnumed.107.048330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordon B.A., Blazey T.M., Su Y., Hari-Raj A., Dincer A., Flores S. Spatial patterns of neuroimaging biomarker change in individuals from families with autosomal dominant Alzheimer's disease: a longitudinal study. Lancet Neurol. 2018;17:241–250. doi: 10.1016/S1474-4422(18)30028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su Y., Flores S., Hornbeck R.C., Speidel B., Vlassenko A.G., Gordon B.A. Utilizing the Centiloid scale in cross-sectional and longitudinal PiB PET studies. NeuroImage: Clin. 2018;19:406–416. doi: 10.1016/j.nicl.2018.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jack C.R., Jr., Wiste H.J., Weigand S.D., Therneau T.M., Lowe V.J., Knopman D.S. Defining imaging biomarker cut points for brain aging and Alzheimer's disease. Alzheimers Dement. 2017;13:205–216. doi: 10.1016/j.jalz.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiong C., Jasielec M.S., Weng H., Fagan A.M., Benzinger T.L., Head D. Longitudinal relationships among biomarkers for Alzheimer disease in the Adult Children Study. Neurology. 2016;86:1499–1506. doi: 10.1212/WNL.0000000000002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landau S.M., Breault C., Joshi A.D., Pontecorvo M., Mathis C.A., Jagust W.J. Amyloid-beta Imaging with Pittsburgh Compound B and Florbetapir: Comparing Radiotracers and Quantification Methods. J Nucl Med. 2013;54:70–77. doi: 10.2967/jnumed.112.109009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen K., Roontiva A., Thiyyagura P., Lee W., Liu X., Ayutyanont N. Improved power for characterizing longitudinal amyloid-beta PET changes and evaluating amyloid-modifying treatments with a cerebral white matter reference region. J Nucl Med. 2015;56:560–566. doi: 10.2967/jnumed.114.149732. [DOI] [PubMed] [Google Scholar]
- 33.Landau S.M., Fero A., Baker S.L., Koeppe R., Mintun M., Chen K. Measurement of Longitudinal beta-Amyloid Change with 18F-Florbetapir PET and Standardized Uptake Value Ratios. J Nucl Med. 2015;56:567–574. doi: 10.2967/jnumed.114.148981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bodini B., Veronese M., Turkheimer F., Stankoff B. Benzothiazole and stilbene derivatives as promising positron emission tomography myelin radiotracers for multiple sclerosis. Ann Neurol. 2016;80:166–167. doi: 10.1002/ana.24667. [DOI] [PubMed] [Google Scholar]
- 35.Veronese M., Bodini B., Garcia-Lorenzo D., Battaglini M., Bongarzone S., Comtat C. Quantification of [(11)C]PIB PET for imaging myelin in the human brain: a test-retest reproducibility study in high-resolution research tomography. J Cereb Blood Flow Metab. 2015;35:1771–1782. doi: 10.1038/jcbfm.2015.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowe V.J., Lundt E.S., Senjem M.L., Schwarz C.G., Min H.K., Przybelski S.A. White matter reference region in PET studies of (11)C-Pittsburgh compound B uptake: Effects of age and Amyloid-beta deposition. J Nucl Med. 2018;59:1583–1589. doi: 10.2967/jnumed.117.204271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leuzy A., Chiotis K., Hasselbalch S.G., Rinne J.O., de Mendonca A., Otto M. Pittsburgh compound B imaging and cerebrospinal fluid amyloid-beta in a multicentre European memory clinic study. Brain. 2016;139:2540–2553. doi: 10.1093/brain/aww160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.La Joie R., Ayakta N., Seeley W.W., Borys E., Boxer A.L., DeCarli C. Multisite study of the relationships between antemortem [(11)C]PIB-PET Centiloid values and postmortem measures of Alzheimer's disease neuropathology. Alzheimers Dement. 2019;15:205–216. doi: 10.1016/j.jalz.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.