Abstract
In this study, we aimed to provide molecular evidence of HPV latency in humans and discuss potential challenges of conducting studies on latency. We analyzed the entire cervix of two women who underwent hysterectomy unrelated to cervical abnormality. The cervices were sectioned into 242 and 186 sets respectively, and each set was tested separately for HPV using the SPF10-PCR-DEIA-LiPA25 system. To identify whether there was any evidence of transforming or productive infection, we used the biomarkers E4 and P16INK4a to stain slides immediately adjacent to HPV-positive sections. HPV was detected in both cervices. In patient 1, 1/242 sets was positive for HPV31. In patient 2, 13/186 sets were positive for HPV18 and 1/186 was positive for HPV53. The infection was very focal in both patients, and there was no sign of a transforming or productive infection, as evaluated by the markers E4 and P16INK4a. Had we only analyzed one set from each block, the probability of detecting the infection would have been 32.3% and 2%, respectively.Our findings support the idea that HPV may be able to establish latency in the human cervix; however, the risk associated with a latent HPV infection remains unclear.
Abbreviations: HPV, (human papilloma virus); HIV, (human immunodeficiency virus); DNA, (deoxyribonucleic acid); mRNA, (messenger ribonucleic acid); HE, (hematoxylin and eosin)
Keywords: HPV, Papillomavirus, Humans, Virus latency, Uterine cervical neoplasm, Molecular biology
1. Introduction
For several years, there has been discussion as to whether or not human papillomavirus (HPV) is able to establish latency in the epithelium of the human uterine cervix [1], [2], [3], [4], with the possibility of viral reactivation during periods of immune deficiency [5]. Despite evidence from clinical studies, such as the reporting of HPV re-appearance following an HPV-negative test result [6], [7], [8], [9] in sexually abstinent women and a higher risk of HPV-related disease in immune suppressed individuals, including organ transplant recipients [10] and HIV seropositive patients [11], [12], there is still no consensus in the scientific community on whether or not HPV is able to establish latency. If HPV is able to establish latency in humans, the viral genome would be expected to be maintained in the basal epithelial cell layer of stratified epithelium, with no shedding of viral particles and without clinical evidence of disease, similar to what has been reported in the animal model [3], [5]. Moreover, the infection may be focal, and be characterized by low HPV DNA copy number and possibly also a low number of infected cells [13]. These characteristics provide a challenge for the detection of a latent HPV infection, and may explain why a latent HPV infection is not picked up by routine HPV testing of cervical cytology samples. In cervical cytology samples, only the superficial cell layer is sampled for HPV testing. The detection of a latent HPV infection is likely to be facilitated by the use of samples that include the basal epithelial cell layer, such as tissue sections from the cervix. Furthermore, an intensive sampling procedure may be required in human studies to ensure detection, as there is no tattoo ink to guide us to the site of previous infection, unlike in the animal model [3]. In the present study, we have taken these considerations into account, and have aimed to provide molecular evidence of HPV latency in the human uterine cervix, along with a discussion of the challenges of conducting studies on HPV latency in humans.
2. Material and methods
For the present study, we selected two women considered at high risk of harboring a latent HPV infection based on their previous exposure to HPV (i.e., >10 life time sex partners) and because they had a record of a previous abnormal cervical cytology (i.e., atypical squamous cells of undetermined significance or worse), which had not been surgically treated. These women were selected from a group of women who had their cervix removed as part of total hysterectomy unrelated to epithelial abnormality of the cervix such as bleeding disorders, fibromas, and prolapse at the Department of Obstetrics and Gynecology, Aarhus University Hospital, from March 1st, 2013 through April 1st, 2015. Prior to surgery, both patients had a normal cervical cytology and were HPV-negative on routine testing using COBAS 4800® (Roche Molecular Diagnostics). After surgical removal, an experienced gynecological pathologist reviewed hematoxylin and eosin (H&E) stained cervical tissue slides by routine bright field microscopy and found no evidence of HPV infection, cervical neoplasia, or other disease.
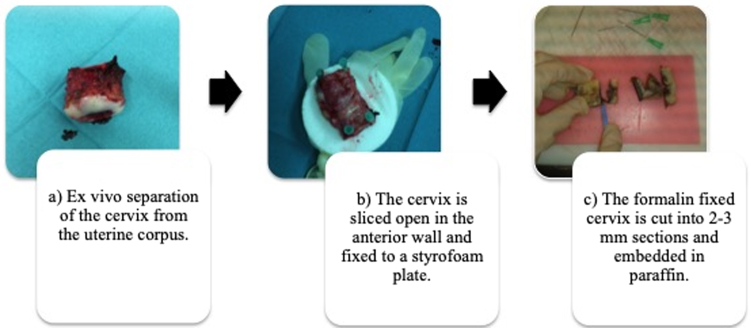
Fig. 1 illustrates the processing of cervical samples. After surgical removal, the cervix was separated from the uterine corpus (a) and sliced open in the anterior wall. The cervix was fixed to a styrofoam plate covered with a sterile glove (b) and subsequently fixed with formalin for approximately 24 h. The following day, the cervix was cut into 3-mm sections (c) and embedded in paraffin. To avoid cross-contamination, we used sterile utensils only (i.e., gloves, syringes, scalpels, etc.).
Fig. 1.
Overview of the sampling procedure.
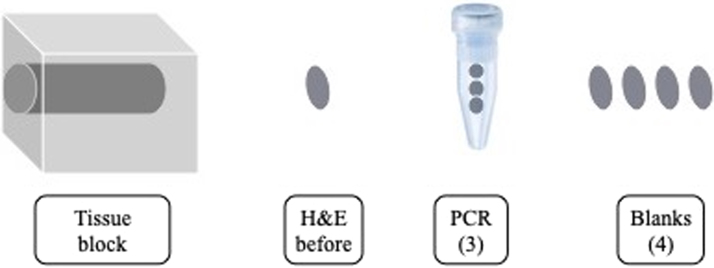
After the procedure described in Fig. 1, the entire cervix was sectioned in both patients, as illustrated in Fig. 2, resulting in up to 30 sets per formalin fixed, paraffin embedded block; a set consists of one section of 4 µm for H&E, 3 sections of 8 µm for whole tissue SPF10 PCR, and four unstained sections of 4 µm for further analyses. To check for changes characteristic of viral infection, H&E slides adjacent to HPV-positive slides underwent additional review by a pathologist experienced in routing diagnostic gynecological pathology at DDL diagnostic Laboratory.
Fig. 2.
Overview of the sectioning protocol. One set consists of an H&E slide (4 µm), a tube with 3 sections for HPV PCR (PCR 3 × 8 µm), and 4 blank slides (4 × 1 × 4 µm) for additional analysis.
DNA extraction of tissue samples was performed using a proteinase K procedure as described previously [14]. For cytological samples, DNA was isolated from 200 μL Surepath suspension (Becton Dickinson and company, Franklin, NJ, USA), containing exfoliated cervical cells, using the MagNA Pure LC instrument as described elsewhere [15]. HPV analyses were performed using the SPF10-PCR-DEIA-LiPA25 system (SPF10 HPV LiPA25 version 1; Labo Bio-Medical Products, Rijswijk, The Netherlands) as described previously [16], [17]. The DEIA is an ELISA-based assay that allows detection of at least 69 HPV types, whereas the LiPA25 allows qualitative identification of the following 25 HPV genotypes: 6, 11, 16, 18, 31, 33, 34, 35, 39, 40, 42, 43, 44, 45, 51, 52, 53, 54, 56, 58, 59, 66, 68/73, 70, and 74.
During the testing, extensive measures were taken to avoid cross-contamination. Thus, controls were incorporated in all phases of the testing and all controls were tested in the DEIA. 1) The sectioning included the cutting of an empty paraffin block at the beginning and at the end of each block, and after every 10 sets (Fig. 2). 2) Each DNA extraction run included two negative controls and one positive control. 3) The PCR included a negative and a positive control. 4) The DEIA included a negative, a borderline (to establish the cut-off for positivity), and a positive control. 5) The LiPA25 contained a positive control, which was the PCR positive control. Amplification failure or contamination was not observed. All HPV analyses were performed at DDL Diagnostic Laboratory, the Netherlands.
2.1. P16 immunohistochemistry and E4 immunofluorescence
Sections adjacent to HPV-positive slides were subject to further analysis with the purpose of localizing the infection and explore potential evidence of an HPV infection. P16INK4a staining was performed on the first adjacent slide in each HPV-positive set as this marker is commonly used as a biomarker of transforming HPV infections. The p16INK4a staining was performed using the CINtec system as described elsewhere [18]. For the present study we used p16INK4a positive tissue as internal control. Each slide contained the study tissue section as well as a section from a CIN3 (cervical intraepithelial neoplasia grade 3 or worse) lesion known to be p16INK4a positive.
Furthermore, we used a recently developed pan HPVE4 antibody (SILgrade-E4-1 kit containing XR-E4-1 monoclonal antibody, Labo Bio-medical Products, Rijswijk, The Netherlands) able to detect the E4 proteins of at least 15 types of high-risk HPV (including HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 67, and 70) [18] on the second adjacent slide. The HPV E4 protein is expressed in infected squamous cells that have initiated the productive stage of the viral life cycle [19], [20], [21], which occurs primarily in low-grade disease [22]. The E4 marker can be detected using conventional immunohistochemistry or immunofluorescence approaches [18], but in this study we used an immunofluorescence as outlined in previous studies [22], [23]. Briefly, 4 µm sections from HPV-positive sets were dried over night at 37 °C, deparaffinized in xylene, and rehydrated in a descending alcohol series. For epitope retrieval, slides were autoclaved in solution D, Ph 6.0 (Dako, Glostrup, Denmark) at 121 °C for 2 min. The antibodies against E4 (panHPVE4) were applied 1:100 and incubated over night at 4 °C. Visualization was performed with 150-fold diluted Alexa-488 (green) conjugated anti-mouse secondary antibody against E4 (Invitrogen, Paisley, UK). Finally, a nuclear counterstaining was performed using DAPI (blue) (Sigma, St. Louis, MO, USA). A CIN1 known to be caused by HPV16 was used as a positive control and gave the characteristic pattern of E4 staining as described previously [20], [22]. P16INK4a staining was performed at DDL, Diagnostic Laboratory, The Netherlands, whereas the panHPVE4 staining was performed at the Department of Pathology, University of Cambridge, UK.
We defined possible viral latency as follows: HPV detected in the tissue with no evidence of E4 and p16INK4a positivity, no clinical or histo-pathological evidence of disease, no cytological abnormality, and HPV-negative on liquid based cytology.
2.2. Statistical analysis
We used our results of the density of latent HPV detection to estimate the probability that a given cervix would yield an HPV-positive result if only a sample of tissue was tested from each specimen block. Assuming the results follow a binomial distribution, we calculated the probability of detecting HPV in a given cervix as follows:
where P (x ≥ 1) = probability that at least one sampled set tests positive, n = number of sets tested, and p = probability that a set contained a latent HPV infection based on the results of our study.
The Danish Ethics Committee (1-10-72-432-12) and the Danish Data Protection Agency (1-16-02-211-12) approved the study. Both patients signed an informed consent form prior to enrollment.
3. Results
Both patients selected for the present study had a total laparoscopic hysterectomy; patient 1 due to a bleeding disorder and patient 2 because of predisposition to ovarian cancer. Patient 1 was 52 years old and premenopausal at the time of surgery, whereas patient 2 was 67 years old and postmenopausal. Patient 1 had 12 lifetime sex partners and was in 1988 diagnosed with HSIL, for which she did not receive surgical treatment. Patient 2 was diagnosed with ASC-H in 2010, which could not be verified histologically, and she had 23 sex partners during her life. Both patients reported no current or previous use of hormone replacement therapy (Table 1). Additionally, both patients had no record of histologically verified cervical abnormality within 5 years of hysterectomy.
Table 1.
Basic characteristics of the two patients selected for analysis.
| Characteristics | Patient 1 | Patient 2 |
|---|---|---|
| Age (years) | 52 | 67 |
| Previous abnormal cervical cytology | HSIL in 1988, not surgically treated | ASC-H in 2010, normal histology |
| Number of life-time sex partners | 12 | 23 |
| Number of new sex partners by age (years) | Age 13–19: 3 | Age 13–19: 4 |
| Age 20–29: 8 | Age 20–29: 11 | |
| Age 30–39: 1 | Age 30–39: 3 | |
| Age 40–49: 0 | Age 40–49: 3 | |
| Age 50–59: 0 | Age 50–59: 1 | |
| Age 60–69: 2 | ||
| Number of blocks tested | 9 | 11 |
| Amount of tissue tested by SPF10-PCR (μm) | 4464 | 5808 |
| Number of SPF10-PCR tests performed | 186 | 242 |
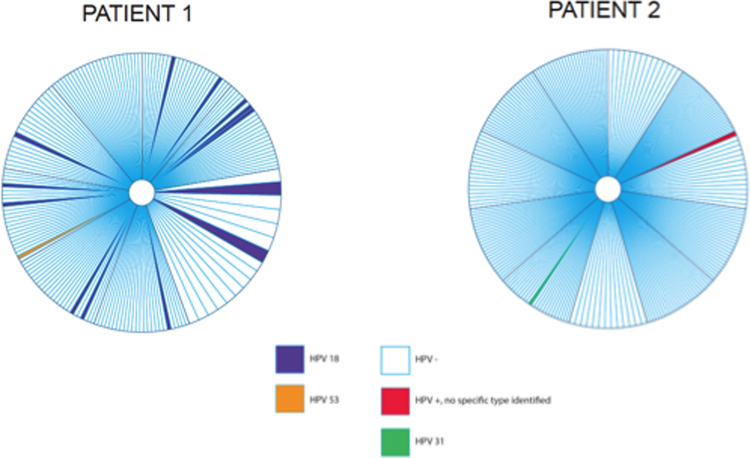
HPV was detected in both patients when the entire cervix was evaluated with the use of whole tissue PCR. In patient 1, a total of 9 blocks were sectioned into 186 sets. Seven of the nine blocks contained HPV-positive sets. HPV-positive sets were mainly adjacent to HPV-positive sets (Fig. 3). Of 186 sets tested, 13 sets (7.0%) were HPV18 positive and one set (0.5%) was positive for HPV53. In patient 2, a total of 11 blocks were sectioned into 242 sets of which one set (0.4%) was positive for HPV31, and one set was borderline positive on the DEIA. However, because the borderline positive sample was negative by reverse hybridization using the LiPA25 strip, the sample was deemed HPV negative, according to standard protocol.
Fig. 3.
Distribution of HPV in the human uterine cervix. Each pie chart illustrates the cervix seen from the vagina and the location of the HPV-positive test results. The central white circle illustrates the cervical orifice (i.e. opening to the uterine cavity). Each large pie piece represents a 3-mm block and each smaller slice represents a whole tissue section set tested by SPF10 PCR.
Fig. 3 illustrates the distribution of HPV infection across the cervix. The HPV infection was very focal in both cervices, but as a result of more HPV-positive sets in patient 1, the infection appeared more widespread in patient 1 compared to patient 2.
We carefully reviewed H&E-stained slides adjacent to HPV-positive sections and found no evidence of a productive HPV infection by bright field microscopy; i.e., no micro lesions, koilocytosis, dyskariotic cells, etc. As cervical smears obtained prior to surgery were initially tested using COBAS 4800, which has lower sensitivity, we re-tested the two cervical cytology samples obtained prior to surgery using the SPF10-PCR-DEIA-LiPA25 system, and both samples were HPV-negative.
To explore if HPV DNA positivity may be associated with a productive viral infection, we tested adjacent slides to HPV-positive sets for the presence of E4. While the positive controls stained strongly, all adjacent slides to HPV-positive section were negative for E4.
Overexpression of the cellular biomarker p16 INK4a in cervical tissue is usually associated with the presence of the viral oncogenes, E6 and E7. All adjacent slides to HPV-positive slides were negative for p16 INK4a, while all positive and negative controls did not suggest a failure in the analysis. The absence of E4 and p16 INK4a may therefore suggest that the HPV infection detected represents a non-transforming and non-productive infection, such as a latent HPV infection.
Acknowledging that the methodology applied in the present study was very costly and time consuming and not feasible for use in future studies, we used the results above to calculate the probability of detecting HPV in a cervix if only a proportion of the total tissue is sampled for PCR testing. Table 2 summarizes the probability of a cervix testing HPV-positive by a range of proportion of HPV-positive sets (rows) and proportion of the cervix analyzed (columns). In the present study, we found 7.5% of sets were HPV-positive in patient 1. Thus, the probability of an HPV-positive test result would be 32.3% had we only analyzed one set from each block. In patient 2, 0.4% of sets were HPV-positive, which equals to a 2% probability of an HPV-positive result if we only analyzed one set from each block.
Table 2.
Probability of detecting HPV in a cervix, by the proportion of cervical tissue sampled and the percentage of HPV-positive sets in a given cervix.
| Percentage of sets tested |
||||||
|---|---|---|---|---|---|---|
| 5 | 10 | 15 | 20 | 25 | ||
| Assumed percentage of HPV-positive sets | 0.1 | 0.5 | 1.0 | 1.5 | 2.0 | 2.5 |
| 0.5 | 2.5 | 4.9 | 7.2 | 9.5 | 11.8 | |
| 1.0 | 4.9 | 9.6 | 14.0 | 18.2 | 22.2 | |
| 2.0 | 9.6 | 18.3 | 26.1 | 33.2 | 39.7 | |
| 5.0 | 22.6 | 40.1 | 53.7 | 64.2 | 72.3 | |
| 7.5 | 32.3 | 54.1 | 68.9 | 79.0 | 85.8 | |
| 10.0 | 41.0 | 65.1 | 79.4 | 87.8 | 92.8 | |
4. Discussion
Thus far, several clinical and epidemiological studies have suggested that HPV is likely being controlled by the immune system rather than cleared [1], [4], [24], [25], [26]. Such studies include the periodical shedding of viral particles [27], the re-appearance of HPV following an HPV-negative test [8], and the increase in HPV prevalence after the initiation of immune suppression [25]. Although animal studies have shown that papillomavirus DNA can be detected in a latent stage up to one year after lesion regression, studies reporting molecular evidence of HPV latency in the human uterine cervix are still lacking.
In the present study, HPV was detected in the human uterine cervix in both patients, with no evidence of a productive or transforming infection by immunohistochemistry and immunofluorescence, and without clinical, cytological, or histopathological evidence of HPV-related disease. These findings may represent HPV infections below detectable levels or possibly molecular evidence of HPV latency. Our results are consistent with previous animal studies in which papillomavirus DNA and mRNA expression was detected in the mucosa following an incident infection without there being evidence of clinical disease [3], [28]. It is unlikely that infections in this latent state pose any immediate risk of severe disease as evidence by the lack of abnormal pathology; however, it remains unclear whether future reactivation from the latent state would result in disease risk similar to that observed in young women with newly acquired infection.
In the animal models, tattoo ink is often used at the site of inoculation, which allows for the distinctive analyses of tissue known to have been exposed to the virus [3], [5], [28]. In the human uterine cervix no such marker exists. Additionally, results from the animal model suggest that, during HPV latency, only a few basal epithelial cells may harbor HPV DNA and at a very low copy number. These factors provide a challenge for the detection of a latent HPV infection and highlight the importance of using a highly sensitive HPV assay and an extensive sampling procedure. In the animal model, viral latency was only detected using a PCR-based technique as the viral copy number during latency was too low to allow for visual detection by in situ hybridization [3] using techniques available at the time of the study.
Due to the focal nature of a latent HPV infection as reported in the animal model, we chose to perform an intensive sampling and testing procedure on two patients who were considered at high risk of harboring a latent HPV infection (i.e., ≥ 10 life time sex partners and a previous record of HPV related disease). This decision was based on previous studies reporting that HPV detection at older age is more likely attributed to previous and not recent sexual activity [9], [29], [30]. The focal nature of the HPV infections observed in the present study suggests that the infection may have been missed had we only analyzed one section from each block. As reported in Table 2, an intense sampling and testing procedure will be required to accurately determine the presence or absence of a latent HPV infection.
To the best of our knowledge, only a few studies have reported evidence of possible HPV latency in humans. Two studies analyzed normal, HPV-positive cervical tissue with no signs of viral replication, adjacent to a cervical lesion (i.e. cervical intraepithelial neoplasia or cancer) [31], [32], whereas another study analyzed morphologically normal cervices. In the study by Leonard et al., the authors reported the presence of HPV in 42% of morphologically normal cervices using in situ hybridization and PCR-based methods [33]. Because persistence of HPV in the absence of clinical disease does not preclude viral activities acquired for genome maintenance, such as replication of the viral genome below detectable levels or production of viral transcripts, p16 and E4 is often used as markers of a productive infection. However, although the authors reported the absence of p16 and E4 like in the present study, it is unclear if the findings reported by Leonard et al. reflect “true” latency as cervical cytology samples were not analyzed for the presence of HPV. Thus, it is unclear if the high HPV prevalence reported in their study may be a result of an active HPV infection (that would have been picked up by routine HPV testing of a cervical cytology specimen), rather than a latent HPV infection, or a combination. In our study, both patients were HPV-negative and had normal cytology on a liquid based cytology sample obtained prior to surgery. Additionally, we found no evidence of E4 and p16 on slides adjacent to HPV-positive sections, which suggest that the infection is non-productive and might represent a latent HPV infection. Unfortunately, we are unable to determine if the HPV detected represents a new infection in it's early stage, a regressing infection, or a “true” latent infection, as there is currently no molecular marker available that is able to distinguish between these stages. We cannot exclude that the HPV infection in patient 2 may represent an early infection because of recent new sex partners; however, we find a new early infection unlikely in patient 1, as there was no reporting of new sex partners within the preceding 10–15 years.
4.1. Strengths and limitations
Our data demonstrates that studies attempting to use hysterectomy samples to estimate a woman-level prevalence of latent HPV must use a very sensitive PCR-based HPV assay with intensive sampling across the tissue due to the focal nature of the infections. Sampling of even 25% of the cervix is likely to result in a false negative result if the latent infection density is <2%. In the present study we used the SPF10 PCR followed by DEIA/LIPA25, which can detect 1–44 HPV DNA copies per PCR reaction by amplifying a fragment of only 65 base pairs. However, we cannot rule out that some HPV-positive sections may have been missed, particularly because the viral copy number is likely very low during latency [3], and because we used formalin-fixed, paraffin-embedded tissue. The process of formalin fixation and paraffin embedding is known to cause DNA cross linkages, change of DNA structure, and breaks, which may result in subsequent difficulties with PCR [34], [35]. The short PCR fragment of 65 base pairs would have reduced the risk of missing positive samples because of DNA damage due to formalin fixation. Furthermore, we acknowledge the risk of contamination, especially when dealing with a low copy number HPV infection. Thus, we found it critical to rule out contamination as an explanation for our positive test results. In the present study, we therefore included negative controls in all steps from the cutting of the block to PCR, DEIA, and LiPA25. All negative controls were negative, and we therefore find contamination unlikely.
5. Conclusion
In conclusion, we found molecular evidence of a non-productive and non-transforming HPV infection in two patients using an extensive sampling and testing procedure. These results support the findings of previous clinical and epidemiological studies that HPV is able to establish latency in the human uterine cervix. Future studies are required to determine if this controlled HPV infection is able to reactivate to productive and/or transforming infection, as our data suggest that women exiting screening with a negative HPV test may indeed continue to harbor high-risk HPV infections.
Acknowledgements
The authors want to thank Dr. David Jenkins for reviewing HE-stained slides, Stephanie van Zoelen for her assistance in the laboratory, and Dr. Angelo Elmi for assistance with the statistical section.
Acknowledgments
Funding
This study was funded by the Danish Cancer Society, Aase and Einar Danielsen's's Foundation, Einar Willumsen's's Foundation, and Dagmar Marshall's foundation. The funders had no role in the study design or interpretation of study results.
Conflict of interest
The authors report no conflicts of interest.
References
- 1.Gravitt P.E. Evidence and impact of human papillomavirus latency. Open Virol. J. 2012;6:198–203. doi: 10.2174/1874357901206010198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doorbar J. Molecular biology of human papillomavirus infection and cervical cancer. Clin. Sci. 2006;110:525–541. doi: 10.1042/CS20050369. [DOI] [PubMed] [Google Scholar]
- 3.Maglennon G.A., McIntosh P., Doorbar J. Persistence of viral DNA in the epithelial basal layer suggests a model for papillomavirus latency following immune regression. Virology. 2011;414:153–163. doi: 10.1016/j.virol.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gravitt P.E., Winer R.L. Natural history of HPV infection across the lifespan: role of viral latency. Viruses. 2017;9 doi: 10.3390/v9100267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maglennon G.A., McIntosh P.B., Doorbar J. Immunosuppression facilitates the reactivation of latent papillomavirus infections. J. Virol. 2014;88:710–716. doi: 10.1128/JVI.02589-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Insinga R.P., Perez G., Wheeler C.M., Koutsky L.A., Garland S.M., Leodolter S., Joura E.A., Ferris D.G., Steben M., Brown D.R., Elbasha E.H., Paavonen J., Haupt R.M. FUTURE I investigators, incidence, duration, and reappearance of type-specific cervical human papillomavirus infections in young women. Cancer Epidemiol. Biomark. Prev. 2010;19:1585–1594. doi: 10.1158/1055-9965.EPI-09-1235. [DOI] [PubMed] [Google Scholar]
- 7.Brogaard K.A., Munk C., Iftner T., Frederiksen K., Kjaer S.K. Detection of oncogenic genital human papillomavirus (HPV) among HPV-negative older and younger women after 7 years of follow-up. J. Med. Virol. 2014;86:975–982. doi: 10.1002/jmv.23914. [DOI] [PubMed] [Google Scholar]
- 8.Polman N.J., Veldhuijzen N.J., Heideman D.A.M., Snijders P.J.F., Meijer C.J.L.M., Berkhof J. HPV-positive women with normal cytology remain at increased risk of CIN3 after a negative repeat HPV test. Br. J. Cancer. 2017 doi: 10.1038/bjc.2017.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rositch A.F., Burke A.E., Viscidi R.P., Silver M.I., Chang K., Gravitt P.E. Contributions of recent and past sexual partnerships on incident human papillomavirus detection: acquisition and reactivation in older women. Cancer Res. 2012;72:6183–6190. doi: 10.1158/0008-5472.CAN-12-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madeleine M.M., Finch J.L., Lynch C.F., Goodman M.T., Engels E.A. HPV-related cancers after solid organ transplantation in the United States. Am. J. Transplant. 2013;13:3202–3209. doi: 10.1111/ajt.12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heard I., Tassie J.M., Schmitz V., Mandelbrot L., Kazatchkine M.D., Orth G. Increased risk of cervical disease among human immunodeficiency virus-infected women with severe immunosuppression and high human papillomavirus load(1) Obstet. Gynecol. 2000;96:403–409. doi: 10.1016/s0029-7844(00)00948-0. [DOI] [PubMed] [Google Scholar]
- 12.Menezes L.J., Poongulali S., Tommasino M., Lin H.Y., Kumarasamy N., Fisher K.J., Saravanan S., Gheit T., Ezhilarasi C., Jeeva A., Lu B., Giuliano A.R. Prevalence and concordance of human papillomavirus infection at multiple anatomic sites among HIV-infected women from Chennai, India. Int. J. STD Aids. 2016;27:543–553. doi: 10.1177/0956462415587226. [DOI] [PubMed] [Google Scholar]
- 13.Maglennon G.A., Doorbar J. The biology of papillomavirus latency. Open Virol. J. 2012;6:190–197. doi: 10.2174/1874357901206010190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quint W.G., Scholte G., van Doorn L.J., Kleter B., Smits P.H., Lindeman J. Comparative analysis of human papillomavirus infections in cervical scrapes and biopsy specimens by general SPF(10) PCR and HPV genotyping. J. Pathol. 2001;194:51–58. doi: 10.1002/path.855. [DOI] [PubMed] [Google Scholar]
- 15.van Doorn L.J., Molijn A., Kleter B., Quint W., Colau B. Highly effective detection of human papillomavirus 16 and 18 DNA by a testing algorithm combining broad-spectrum and type-specific PCR. J. Clin. Microbiol. 2006;44:3292–3298. doi: 10.1128/JCM.00539-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleter B., van Doorn L.J., Schrauwen L., Molijn A., Sastrowijoto S., ter Schegget J., Lindeman J., ter Harmsel B., Burger M., Quint W. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J. Clin. Microbiol. 1999;37:2508–2517. doi: 10.1128/jcm.37.8.2508-2517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleter B., van Doorn L.J., ter Schegget J., Schrauwen L., van Krimpen K., Burger M., ter Harmsel B., Quint W. Novel short-fragment PCR assay for highly sensitive broad-spectrum detection of anogenital human papillomaviruses. Am. J. Pathol. 1998;153:1731–1739. doi: 10.1016/S0002-9440(10)65688-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Baars R., Griffin H., Wu Z., Soneji Y.J., van de Sandt M., Arora R., van der Marel J., Ter Harmsel B., Jach R., Okon K., Huras H., Jenkins D., Quint W., Doorbar J. Investigating diagnostic problems of CIN1 and CIN2 associated with high-risk HPV by combining the novel molecular biomarker PanHPVE4 with P16INK4a. Am. J. Surg. Pathol. 2015;39:1518–1528. doi: 10.1097/PAS.0000000000000498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Middleton K., Peh W., Southern S., Griffin H., Sotlar K., Nakahara T., El-Sherif A., Morris L., Seth R., Hibma M., Jenkins D., Lambert P., Coleman N., Doorbar J. Organization of human papillomavirus productive cycle during neoplastic progression provides a basis for selection of diagnostic markers. J. Virol. 2003;77:10186–10201. doi: 10.1128/JVI.77.19.10186-10201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doorbar J. The E4 protein; structure, function and patterns of expression. Virology. 2013;445:80–98. doi: 10.1016/j.virol.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 21.Borgogna C., Zavattaro E., De Andrea M., Griffin H.M., Dell'Oste V., Azzimonti B., Landini M.M., Peh W.L., Pfister H., Doorbar J., Landolfo S., Gariglio M. Characterization of beta papillomavirus E4 expression in tumours from epidermodysplasia verruciformis patients and in experimental models. Virology. 2012;423:195–204. doi: 10.1016/j.virol.2011.11.029. [DOI] [PubMed] [Google Scholar]
- 22.Griffin H., Soneji Y., Van Baars R., Arora R., Jenkins D., van de Sandt M., Wu Z., Quint W., Jach R., Okon K., Huras H., Singer A., Doorbar J. Stratification of HPV-induced cervical pathology using the virally encoded molecular marker E4 in combination with p16 or MCM. Mod. Pathol. 2015;28:977–993. doi: 10.1038/modpathol.2015.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffin H., Doorbar J. Detection of papillomavirus gene expression patterns in tissue sections. Curr. Protoc. Microbiol. 2016;41 doi: 10.1002/cpmc.6. [DOI] [PubMed] [Google Scholar]
- 24.Gravitt P.E. The known unknowns of HPV natural history. J. Clin. Investig. 2011;121:4593–4599. doi: 10.1172/JCI57149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinten F., Hilbrands L.B., Meeuwis K.A.P., IntHout J., Quint W.G.V., Hoitsma A.J., Massuger L.F.A.G., Melchers W.J.G., de Hullu J.A. Reactivation of latent HPV infections after renal transplantation. Am. J. Transplant. 2017;17:1563–1573. doi: 10.1111/ajt.14181. [DOI] [PubMed] [Google Scholar]
- 26.Strickler H.D., Burk R.D., Fazzari M., Anastos K., Minkoff H., Massad L.S., Hall C., Bacon M., Levine A.M., Watts D.H., Silverberg M.J., Xue X., Schlecht N.F., Melnick S., Palefsky J.M. Natural history and possible reactivation of human papillomavirus in human immunodeficiency virus-positive women. J. Natl. Cancer Inst. 2005;97:577–586. doi: 10.1093/jnci/dji073. [DOI] [PubMed] [Google Scholar]
- 27.Liu S.H., Cummings D.A., Zenilman J.M., Gravitt P.E., Brotman R.M. Characterizing the temporal dynamics of human papillomavirus DNA detectability using short-interval sampling. Cancer Epidemiol. Biomark. Prev. 2014;23:200–208. doi: 10.1158/1055-9965.EPI-13-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amella C.A., Lofgren L.A., Ronn A.M., Nouri M., Shikowitz M.J., Steinberg B.M. Latent infection induced with cottontail rabbit papillomavirus. A model for human papillomavirus latency. Am. J. Pathol. 1994;144:1167–1171. [PMC free article] [PubMed] [Google Scholar]
- 29.Gravitt P.E., Rositch A.F., Silver M.I., Marks M.A., Chang K., Burke A.E., Viscidi R.P. A cohort effect of the sexual revolution may be masking an increase in human papillomavirus detection at menopause in the United States. J. Infect. Dis. 2013;207:272–280. doi: 10.1093/infdis/jis660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winer R.L., Hughes J.P., Feng Q., Xi L.F., Lee S.K., O'Reilly S.F., Kiviat N.B., Koutsky L.A. Prevalence and risk factors for oncogenic human papillomavirus infections in high-risk mid-adult women. Sex. Transm. Dis. 2012;39:848–856. doi: 10.1097/OLQ.0b013e3182641f1c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalantari M., Garcia-Carranca A., Morales-Vazquez C.D., Zuna R., Montiel D.P., Calleja-Macias I.E., Johansson B., Andersson S., Bernard H.- Laser capture microdissection of cervical human papillomavirus infections: copy number of the virus in cancerous and normal tissue and heterogeneous DNA methylation. Virology. 2009;390:261–267. doi: 10.1016/j.virol.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vinokurova S., von Knebel Doeberitz M. Differential methylation of the HPV 16 upstream regulatory region during epithelial differentiation and neoplastic transformation. PLoS One. 2011;6:e24451. doi: 10.1371/journal.pone.0024451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leonard S.M., Pereira M., Roberts S., Cuschieri K., Nuovo G., Athavale R., Young L., Ganesan R., Woodman C.B. Evidence of disrupted high-risk human papillomavirus DNA in morphologically normal cervices of older women. Sci. Rep. 2016;6:20847. doi: 10.1038/srep20847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karlsen F., Kalantari M., Chitemerere M., Johansson B., Hagmar B. Modifications of human and viral deoxyribonucleic acid by formaldehyde fixation. Lab. Investig. 1994;71:604–611. [PubMed] [Google Scholar]
- 35.Greer C.E., Peterson S.L., Kiviat N.B., Manos M.M. PCR amplification from paraffin-embedded tissues. Effects of fixative and fixation time. Am. J. Clin. Pathol. 1991;95:117–124. doi: 10.1093/ajcp/95.2.117. [DOI] [PubMed] [Google Scholar]