Abstract
Aberrant expression of zinc-finger proteins has been extensively reported to contribute to malignant progression in a variety of cancers. However, clinical significance and biological roles of ZNF280A in the field of cancer are poorly known. In this study, the expression of ZNF280A was detected in clinical colorectal cancer (CRC) tissues. Functional experiments in vitro and animal experiment in vivo were performed to measure the effect of ZNF280A on the proliferation and tumorigenesis in CRC cells. Western blot and luciferase assays were used to identify the underlying pathway mediating the biological roles of ZNF280A in CRC. Here we report that ZNF280A was upregulated in CRC tissues and cells and a high expression of ZNF280A correlated with tumor, lymph node, and metastasis (TNM) classifications, clinical stage, and predicted poor prognosis and disease progression in CRC patients. Moreover, silencing ZNF280A repressed proliferation and induced G0 and/or G1 arrest in vitro, and it inhibited tumorigenesis of CRC cells in vivo. Our results further demonstrate that silencing ZNF280A inhibited the proliferation of CRC cells by activating Hippo signaling. Therefore, our results uncover a novel mechanistic understanding of ZNF280A-mediated tumor progression in CRC, and meanwhile they provide a novel prognostic factor in CRC patients and a potential therapeutic target for the treatment of CRC.
Keywords: ZNF280A, proliferation, cell cycle, tumorigenicity, Hippo signaling, colorectal cancer
Introduction
Colorectal cancer (CRC) is the most common gastrointestinal tumor, accounting for an estimated 1.3 million cancer-related deaths annually worldwide.1 Despite substantial advances in systemic or individual treatment, the prognosis remains bleak due to late detection and diagnosis at an advanced stage of CRC.2 Therefore, the identification of novel and suitable biomarkers for early detection in CRC may significantly facilitate to improve the number of CRC-associated deaths.
ZNF280A, first identified through an integrative high-resolution whole-genome profiling in mantle cell lymphoma, is a member of the zinc-finger protein family, carrying a unique zinc-finger motif composed of two contiguous Cys2His2-type fingers.3 This Cys2His2-like fold group (C2H2) is by far the best-characterized class of zinc fingers, and it is extremely common in mammalian transcription factors.4 Although functioning in diverse biological roles by binding DNA or RNA and mediating protein-protein interactions, this class of zinc fingers is best known for its role in sequence-specific DNA-binding proteins via its typical tandem repeats, with two or more fingers comprising the DNA-binding domain of the protein that occupies the major groove of DNA.4 Numerous studies have demonstrated that different members of the zinc-finger proteins, such as ZEB2, ZNF746, PISA, and Snail1, have been reported to be involved in the development, progression, and metastasis of CRC,5, 6, 7, 8 indicating that zinc-finger proteins may play an important role in CRC. Compared with these well-documented zinc-finger proteins members, reports on the biological function and role of ZNF280A in cancers are scanty.
The Hippo-signaling pathway has been found to be frequently inactivated in multiple human cancer types.9, 10, 11 The Hippo pathway mainly consists of four core kinase cassette components, including the adaptor proteins SAV1 and MOB1 and kinases MST1/2 and LATS1/2.11 The Hippo signaling is active via tightly controlling the activity of Yes Associated Protein (YAP) and Tafazzin (TAZ) through phosphorylation-ubiquitination mechanisms.12, 13 While Hippo signaling is absent, unphosphorylated YAP1 and/or TAZ translocates to the nucleus of cells, promoting the transcriptional activity of TEA domain (TEAD) family members (TEAD1–TEAD4) as the transcriptional co-activators via transcriptionally upregulating multiple downstream effectors to exert a pleiotropic role in the progression and metastasis of cancers.14, 15, 16 Several lines of evidence have shown that the inactivation of Hippo signaling was implicated in various processes of CRC. For example, REGγ contributed to the growth of CRC cells via directly interacting with LATS1, leading to the degradation of LATS1 and inactivation of Hippo signaling;17 in addition, SCC-S2 overexpression was found to promote the invasion and metastasis of CRC via depressing Hippo signaling.18 Our previous study demonstrated that the inactivation of Hippo signaling by the overexpression of TFAP2C promoted the progression of CRC via maintaining cancer stem cell-like phenotypes.19 Thus, these studies support the notion that the inactivation of Hippo pathway plays a crucial role in the progression and metastasis of CRC.
In this study, our findings revealed for the first time that ZNF280A was dramatically upregulated in CRC tissues, which significantly correlated with advanced clinicopathological features and poor overall and progression-free survival in CRC patients. Loss-of-function experiments demonstrated that silencing ZNF280A attenuated the proliferation and retarded cell cycle progression in CRC cells in vitro and repressed the tumorigenesis of CRC cells in vivo. Mechanistic investigation further showed that silencing ZNF280A activated Hippo signaling via upregulating phophorylated MST1/2 and LATS1 expression and downregulating TAZ and YAP expression in CRC cells. Therefore, our findings indicate that ZNF280A holds promise to serve as a novel marker for early detection and diagnosis in CRC patients.
Results
ZNF280A Is Increased in CRC
To determine the expression of ZNF280A in CRC, we first analyzed the CRC RNA sequencing dataset from The Cancer Genome Atlas (TCGA). As shown in Figures 1A and 1B, ZNF280A was found to be robustly upregulated in 625 individual and 50 paired CRC tissues compared with the adjacent normal colorectal tissues, respectively. We further examined ZNF280A expression levels in our CRC tissues. Consistent with the analysis result from TCGA, our results revealed that mRNA and protein expression levels of ZNF280A in 6/8 CRC tissues were upregulated compared with the matched adjacent normal tissues (Figures 1C and 1D). Expression levels of ZNF280A in CRC cells were further measured, and the results showed that ZNF280A expression levels were differentially elevated in CRC cells compared with normal colon epithelial cell (CMEC) (Figures 1E and 1F). These results demonstrated that ZNF280A is upregulated in CRC tissues and cell lines.
Figure 1.
ZNF280A Is Upregulated in CRC Tissues and Cells
(A) ZNF280A expression level was elevated in colorectal cancer tissues compared with the adjacent normal tissues, as assessed by analyzing TCGA colorectal cancer RNA sequencing dataset (adjacent normal tissue, n = 50; colorectal cancer, n = 625). (B) ZNF280A expression level was elevated in 50 paired colorectal cancer tissues compared with the adjacent normal tissues, as assessed by analyzing TCGA colorectal cancer RNA sequencing dataset. (C and D) Real-time PCR (C) and western blotting (D) analyses of ZNF280A expression in 8 paired CRC tissues. α-Tubulin was detected as a loading control in the western blot and GAPDH was used as the endogenous control in RT-PCR. Each bar represents the mean values ± SD of three independent experiments. *p < 0.05. (E and F) Real-time PCR (E) and western blotting (F) analyses of ZNF280A expression in one normal colon epithelial cell CMEC and 7 CRC cell lines. GAPDH was used as the endogenous control in RT-PCR and α-tubulin was detected as a loading control in the western blot. Each bar represents the mean values ± SD of three independent experiments. *p < 0.05.
A High Expression of ZNF280A Predicts Poor Prognosis and Progression
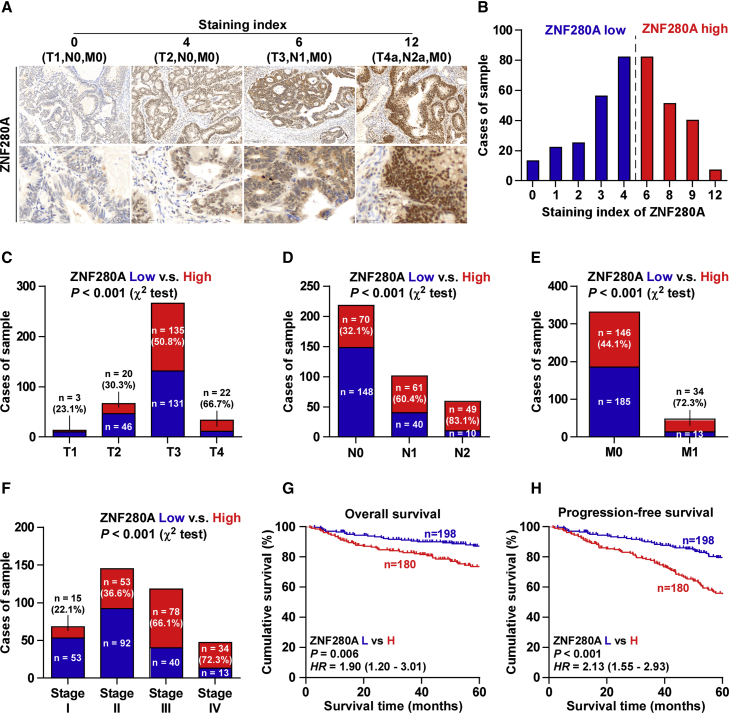
We further investigated ZNF280A expression by immunohistochemistry (IHC) analysis of 378 human CRC tissues, and four independent CRC tissues with the staining intensity (SI) scores of 0, 4, 6, and 12 were shown, respectively (Figure 2A). An SI score 4 was the median SI of all sample tissues. High and low ZNF280A expressions were stratified by the follow criteria: SI ≤ 4 was used to define tumors with low ZNF280A expression, and SI ≥ 6 was used to define tumors with high ZNF280A expression. High expression of ZNF280A was observed in 180/378 CRC tissues (47.6%) (Figure 2B). Furthermore, ZNF280A expression strongly and positively correlated with tumor (T) classification, lymph node (N) classification, metastasis (M) classification, and clinical stage in CRC patients (Figures 2C–2F; Table 1). Kaplan-Meier survival analysis demonstrated that a high expression of ZNF280A predicted poor overall and progression-free survivals (Figures 2G and 2H). Taken together, the aforementioned findings indicated that the high expression of ZNF280A is closely associated with poor prognosis and disease progression in CRC patients.
Figure 2.
Overexpression of ZNF280A Correlates with Advanced Clinicopathological Features and Poor Prognosis in CRC Patients
(A) Representative images of ZNF280A expression in CRC tissues of different scores. (B) The number of CRC tissues stratified by staining intensity of IHC. (C) Percentages and number of samples showed high or low ZNF280A expression in CRC tissues with different tumor volume. (D) Percentages and number of samples showed high or low ZNF280A expression in CRC tissues with different lymph node metastasis status. (E) Percentages and number of samples showed high or low ZNF280A expression in CRC tissues with different distant metastasis status. (F) Percentages and number of samples showed high or low ZNF280A expression in CRC tissues with different stage. (G and H) Kaplan-Meier overall survival (G) and progression-free survival (H) curves for CRC patients stratified by high and low expressions of ZNF280A.
Table 1.
The Relationship between ZNF280A IHC Expression Level and Clinical Pathological Characteristics in 378 Patients with Colorectal Cancer
| Parameter | Number of Cases | ZNF280A IHC Expression |
p Value |
|
|---|---|---|---|---|
| Low | High | |||
| Location | 198 | 180 | ||
| Rectum | 118 | 59 | 59 | 0.532 |
| Colon | 260 | 139 | 121 | |
| Gender | ||||
| Female | 169 | 84 | 85 | 0.349 |
| Male | 209 | 114 | 95 | |
| Age (years) | ||||
| ≤60 | 133 | 65 | 68 | 0.314 |
| >60 | 245 | 133 | 112 | |
| Grade | ||||
| G1–G2 | 341 | 184 | 157 | 0.062 |
| G3 | 37 | 14 | 23 | |
| T classification | ||||
| T1–2 | 79 | 56 | 23 | <0.001 |
| T3–4 | 299 | 142 | 157 | |
| N classification | ||||
| N0 | 218 | 148 | 70 | <0.001 |
| N1–2 | 160 | 50 | 110 | |
| M classification | ||||
| M0 | 331 | 185 | 146 | <0.001 |
| M1 | 47 | 13 | 34 | |
| Stage | ||||
| I–II | 213 | 145 | 68 | <0.001 |
| III–IV | 165 | 53 | 112 | |
| Chemotherapeutic response | ||||
| Sensitivity | 39 | 22 | 17 | 0.001 |
| Resistance | 36 | 7 | 29 | |
Silencing ZNF280A Inhibits Proliferation In Vitro
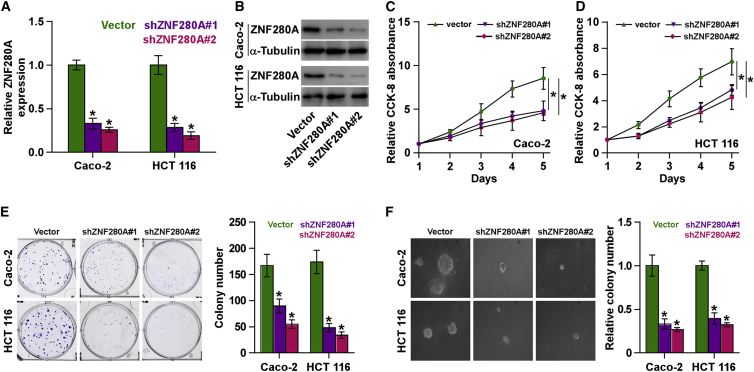
As shown above, overexpression of ZNF280A was correlated with clinical stage in CRC patients. Therefore, we further examined whether ZNF280A was implicated in the proliferation of CRC cells. We first constructed Caco-2 and HCT116 cells stably expressing ZNF280A by endogenously knocking down ZNF280A via retrovirus infection (Figures 3A and 3B), because Caco-2 and HCT116 cells expressed the highest levels of ZNF280A in all CRC cells (Figures 1E and 1F). Cell-Counting Kit-8 (CCK-8) assay was first carried out to investigate the effects of ZNF280A on CRC cells. The result indicated that silencing ZNF280A decreased the cell proliferation rate of Caco-2 and HCT116 cells (Figures 3C and 3D). Colony formation assays revealed that silencing ZNF280A repressed the colony-generating capability of Caco-2 and HCT116 cells (Figure 3E). Moreover, anchorage-independent growth assay was further performed, and we found that downregulating ZNF280A dramatically reduced anchorage-independent growth ability in Caco-2 and HCT116 cells (Figure 3F). Collectively, these results indicated that silencing ZNF280A inhibits the proliferation ability of CRC cells in vitro.
Figure 3.
Silencing ZNF280A Inhibits Proliferation in CRC Cells In Vitro
(A and B) Real-time PCR (A) and western blotting (B) analyses of ZNF280A expression in the indicated CRC cells. GAPDH was used as the endogenous control in RT-PCR, and α-tubulin was detected as a loading control in the western blot. Each bar represents the mean values ± SD of three independent experiments. *p < 0.05. (C and D) CCK-8 assay revealed that silencing ZNF280A decreased the proliferation rate in Caco-2 (C) and HCT-116 (D) cells. Each bar represents the mean values ± SD of three independent experiments. *p < 0.05. (E) Colony formation assay revealed that silencing ZNF280A reduced the colony-forming ability in Caco-2 and HCT-116 cells. (F) Anchorage-independent growth assays revealed that silencing ZNF280A reduced the anchorage-independent growth ability in Caco-2 and HCT-116 cells. Each bar represents the mean values ± SD of three independent experiments. *p < 0.05.
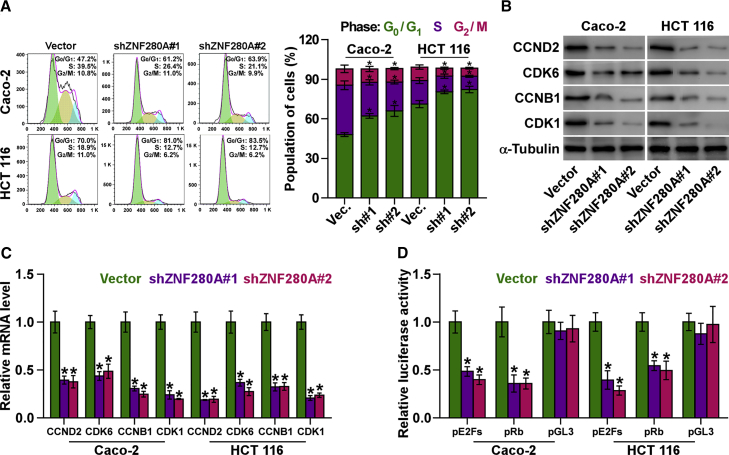
Silencing ZNF280A Retards G1 and/or S Phase Transition
To further clarify the underlying mechanism contributing to the ZNF280A downregulation-mediated proliferation inhibition in CRC cells, the cell cycle progression was analyzed. As shown in Figure 4A, flow cytometry showed that silencing ZNF280A significantly reduced the percentage of cells in the S phase, but it dramatically increased that of cells in the G0 and/or G1 phase, indicating that the inhibition of ZNF280A induced the G1 and/or S arrest in CRC cells. Furthermore, the effect of ZNF280A on multiple critical cell cycle regulators in G1 and/or S checkpoint was further investigated by western blot. As shown in Figure 4B, silencing ZNF280A decreased the protein levels of cyclin D2 (CCND2), cyclin B1 (CCNB1), cyclin-dependent kinase 1 (CDK1), and CDK6. Consistently, real-time PCR analysis demonstrated that silencing ZNF280A remarkably decreased the mRNA levels of CCND2, CCNB1, CDK1, and CKD6 (Figure 4C).
Figure 4.
Downregulation of ZNF280A Retards the Cell Cycle of CRC Cells
(A) Flow cytometry analysis of the indicated CRC cells. Each bar represents the mean values ± SD of three independent experiments. *p < 0.05. (B) Western blot analysis of CCNB1, CCND2, CDK1, and CDK6 expressions in the indicated cells. α-Tubulin was detected as a loading control. (C) Real-time PCR analysis of CCNB1, CCND2, CDK1, and CDK6 mRNA expressions in the indicated CRC cells. GAPDH was used as the loading control. Each bar represents the mean values ± SD of three independent experiments. *p < 0.05. (D) Relative Rb activity and E2F reporter activity in the indicated cells. Each bar represents the mean values ± SD of three independent experiments. *p < 0.05.
It has been well known that the progression of cell cycle was stimulated by E2F transcription factors. E2F can work with Rb, a negative regulator of the cell cycle, in regulating G1 and/or S checkpoint. Several lines of evidence have shown that E2F activation or Rb inactivation can induce entry of cells into S phase.20, 21 Thus, we further examined whether ZNF280A has an influence on the activity E2F-Rb complexes. As shown in Figure 4D, our results demonstrated that silencing ZNF280A suppressed the transcriptional activity of E2F and Rb via luciferase reporter analysis (Figure 4D). Collectively, these results indicated that silencing ZNF280A inhibits the cell cycle progression in CRC cells.
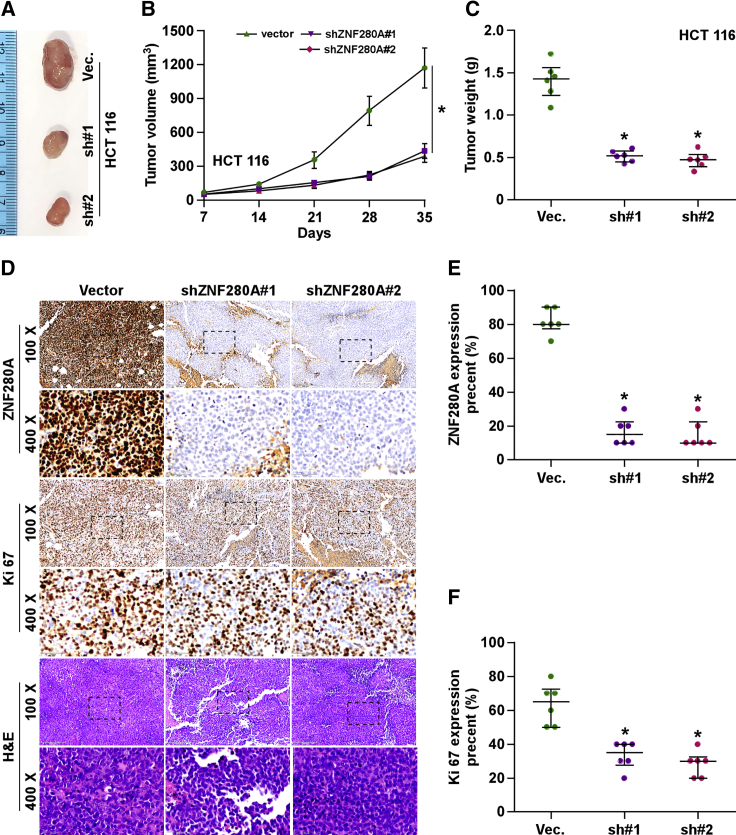
Silencing ZNF280A Inhibits the Tumorigenesis of CRC Cells In Vivo
The effect of ZNF280A on the tumorigenesis of CRC cells was further validated in vivo. Mice were randomly divided into three groups (n = 6/group), and 1 × 106 HCT116-vector cells and two ZNF280A-downregulating HCT116 cells were inoculated subcutaneously into the inguinal folds of the mice. As shown in Figures 5A–5C, the tumor volumes and weight in the mice injected with ZNF280A-downregulating HCT116 cells were dramatically reduced compared with those in the vector group. The IHC and H&E staining analyses revealed that the ZNF280A-silencing tumor tissues displayed reduced ZNF280A expression levels, as well as decreased numbers of Ki67-positive cells (Figures 5D–5F). These results demonstrated that silencing ZNF280A represses the tumorigenesis of CRC cells in vivo.
Figure 5.
Silencing ZNF280A Represses the Tumorigenesis of CRC Cells In Vivo
(A) Images of excised tumors from the BALB/c mice on day 35 after injection with the indicated cells. (B) Tumor volumes were measured every 7 days. Each bar represents the median values ± quartile values. (C) Average weight of excised tumors from the indicated mice. Each bar represents the median values ± quartile values. *p < 0.05. (D) Representative images of sections sliced from the indicated tumors and stained with anti-ZNF280A (upper panel), anti-Ki67 (middle panel), and H&E staining (lower panel), respectively. (E) The percentage of ZNF280A expression in the sections sliced from the indicated tumors. Each bar represents the median values ± quartile values. *p < 0.05. (F) The percentage of Ki67 expression in the sections sliced from the indicated tumors. Each bar represents the median values ± quartile values. *p < 0.05.
Silencing ZNF280A Activates the Hippo-Signaling Pathway
To explore the underlying signaling pathway mediating the effects of ZNF280A on proliferation in CRC cells, gene set enrichment analysis (GSEA) was performed, and the results showed that ZNF280A expression level was significantly associated with Hippo signaling, suggesting that the activity of Hippo signaling may be associated with the anti-proliferative role of ZNF280A downregulation in CRC (Figure 6A). Luciferase analysis showed that silencing ZNF280A reduced the Hippo-YAP signaling optimal promoter (HOP)-Flash, but not Hippo-YAP signaling incompetent promoter (HIP)-Flash, luciferase reporter activity, indicating that silencing ZNF280A repressed the TEAD-dependent luciferase activity in CRC cells (Figure 6B). Western blotting analysis demonstrated that silencing ZNF280A increased phosphorylated MST1/2, LATS1, and YAP expressions and reduced YAP and TAZ expressions, but had no effect on total levels of MST1 and LATS1 in CRC cells (Figure 6C). Real-time PCR analysis showed that silencing ZNF280A decreased expression levels of multiple downstream genes of Hippo pathway, including CTGF, SOX9, and PPIA in CRC cells (Figure 6D). Importantly, YAP and TAZ expressions in the xenograft tumors from mice injected with ZNF280A-downregulating HCT116 cells were dramatically reduced compared with those in the tumors from the vector mouse group (Figure 6E). Analysis of CRC datasets from TCGA revealed that expression levels of ZNF280A significantly correlated with the protein expression levels of transcriptional co-activators YAP of Hippo signaling (Figure 6F). Furthermore, our results demonstrated that YAP1-S127A that constitutively activate YAP122 significantly enhanced the proliferation ability repressed by silencing ZNF280A (Figures 6G and 6H). Thus, these results indicated that silencing ZNF280A activates the Hippo-signaling pathway in CRC cells.
Figure 6.
ZNF280A Inactivates the Hippo-Signaling Pathway in CRC Cells
(A) GSEA showed that ZNF280A expression level was associated with Hippo signaling. (B) TEAD transcriptional activity was assessed by HOP-Flash luciferase reporter in the indicated cells. Error bars represent the mean ± SD of three independent experiments. *p < 0.05. (C) Western blot analysis of p-MST1/2, MST1, p-LATS1, LATS1, p-YAP, YAP, and TAZ expressions in the indicated cells. α-Tubulin was detected as a loading control. (D) Real-time PCR analysis of CTGF, CYR61, HOXA1, and SOX9 in the indicated cells. Error bars represent the mean ± SD of three independent experiments. *p < 0.05. (E) Western blotting of nuclear YAP and TAZ expressions in the tumor tissues from the indicated mice. The nuclear protein p84 was used as the nuclear protein marker. (F) ZNF280A expression level is positively associated with the protein expression levels of transcriptional co-activators YAP of the Hippo-signaling pathway, as assessed through the CRC dataset from TCGA. (G and H) CCK-8 assay revealed that YAP1-S127A increased the proliferation ability in ZNF280A-silenced Caco-2 (G) and HCT 116 (H). Each bar represents the mean values ± SD of three independent experiments. *p < 0.05.
Discussion
In this study, we found that ZNF280A was elevated in CRC tissues and a high expression of ZNF280A correlated with advanced clinicopathological features, poor prognosis, and disease progression in CRC patients. Furthermore, silencing ZNF280A attenuated proliferation and inhibited cell cycle in CRC cells in vitro, and it suppressed tumorigenesis in vivo. Our results also demonstrated that ZNF280A inactivated Hippo signaling, which further promoted the progression of CRC. Therefore, our findings present a novel mechanism by which ZNF280A promotes the proliferation and tumorigenesis of CRC cells.
Surprisingly, as a transcriptional factor containing the two contiguous Cys2His2 zinc-finger motif, less is understood about the biological role of ZNF280A in cancers. Through a high-density SNP array and gene expression-profiling analysis, Beà and colleagues3 have found that homozygous deletions of ZNF280A were found in mantle cell lymphoma; moreover, Gunn et al.23 used an oligonucleotide-based array comparative genome hybridiza (CGH) analysis to detect genomic imbalances in 187 chronic lymphocytic leukemia (CLL) cases, and they found that the deletion of ZNF280A was found in 28 cases (15%). Real-time qPCR showed that the mRNA expression level of ZNF280A was significantly lower in the deleted region compared to non-deleted cases.23 However, the clinical significance and functional role of ZNF280A were not investigated in these studies, even in the context of cancer. In this study, we found that ZNF280A expression was robustly increased in CRC tissues and a high expression of ZNF280A was positively associated with T, N, and M classifications; clinical stages; and poor prognosis and disease progression in CRC patients. Furthermore, our results demonstrated that silencing ZNF280A inhibited the proliferation and tumorigenesis of CRC cells in vitro and in vivo. Therefore, our findings determine the oncogenic role of ZNF280A in the development and progression of CRC.
The Hippo-signaling pathway has been found to be frequently inactivated in multiple human cancer types,9, 10, 11 including CRC.17, 18, 19 Numerous studies have reported that the downregulation of the Hippo pathway components mammalian MST1/2 and LATS1/2 or the upregulation of YAP or TAZ consistently contributed to the inactivation of Hippo signaling, which further promoted the progression of CRC.24, 25, 26 In this study, we found that silencing ZNF280A repressed the HOP-Flash, but not HIP-Flash, luciferase reporter activity, indicating that silencing ZNF280A activated Hippo signaling in CRC cells. Western blot and RT-PCR analysis further revealed that silencing ZNF280A dramatically enhanced the phosphorylated levels of MST1 and LATS1 and downregulated YAP and TAZ expressions, as well as reduced the expression levels of multiple downstream genes of the Hippo pathway in CRC cells. Thus, our findings uncover a novel mechanism that ZNF280A promotes the progression of CRC via inactivating Hippo signaling.
As mentioned above, ZNF280A was reported to be deleted in hematopoietic malignancies, including mantle cell lymphoma3 and chronic lymphocytic leukemia,23 suggesting that ZNF280A may function as a tumor suppressor in hematopoietic malignancies. Conversely, our results found that ZNF280A was significantly upregulated via our samples and TCGA analysis. Importantly, functional experiments showed that silencing ZNF280A inhibited the cell proliferation and tumorigenesis in CRC, demonstrating the oncogenic role of ZNF280A in CRC. Therefore, existing reports in combination with our findings imply that ZNF280A may play an opposite, even paradoxical role dependent on cancer type. However, the underlying mechanism responsible for ZNF280A overexpression in CRC remains unclear, which is a major drawback deserving further clarification in the future work.
In summary, our findings for the first time reveal that ZNF280A plays an oncogenic role in CRC, via regulating proliferation promotion and cell cycle transition, as well as clarify that ZNF280A inactivates Hippo signaling in CRC. Therefore, an in-depth understanding of the underlying mechanism and the function role of ZNF280A in the pathogenesis of CRC provides a novel marker for early detection and diagnosis of CRC.
Materials and Methods
Cell Lines and Cell Culture
The normal colon epithelial cell CMEC was purchased from Porcell and cultured in the complete medium (CM-H040, Porcell). All CRC cell lines, including CW-2, Caco-2, HCT116, HCT-8, LS 174T, LoVo, and SW480, were obtained from Shanghai Chinese Academy of Sciences cell bank (China), and they were cultured in RPMI-1640 medium (Life Technologies, Carlsbad, CA, USA), supplemented with penicillin G (100 U/mL), streptomycin (100 mg/mL), and 10% fetal bovine serum (FBS, Life Technologies) and cultured at 37°C in a humidified atmosphere with 5% CO2.
Patients and Tumor Tissues
A total of eight paired fresh CRC tissues with matched adjacent normal tissues and 378 individual paraffin-embedded, archived CRC tissues were obtained during surgery at The First Hospital of Jilin University (Changchun, China) between January 2008 and December 2011 (Tables S1 and S2). Patients were diagnosed based on clinical and pathological evidence, and the specimens were immediately snap-frozen and stored in liquid nitrogen tanks. For the use of these clinical materials for research purposes, prior patients’ consents and approval from the Institutional Research Ethics Committee were obtained.
RNA Extraction, Reverse Transcription, and Real-Time PCR
Total RNA from tissues or cells was extracted using TRIzol (Life Technologies), according to the manufacturer’s instructions. mRNA was polyadenylated using a poly-A polymerase-based First-Strand Synthesis kit (TaKaRa, DaLian, China), and reverse transcription (RT) of total mRNA was performed using a PrimeScript RT Reagent kit (TaKaRa), according to the manufacturer’s protocol. cDNA was amplified and quantified on an ABI 7500HT system (Applied Biosystems, Foster City, CA, USA) using SYBR Green I (Applied Biosystems). The primers used in the reactions are listed in Table S3. Real-time PCR was performed as described previously.27 Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the endogenous control. Relative fold expressions were calculated with the comparative threshold cycle (2−ΔΔCt) method according to the previous study.28
Plasmid and Transfection
Knockdown of endogenous ZNF280A was performed by cloning two short hairpin RNA (shRNA) oligonucleotides into the pSUPER-puro-retro vector (OligoEngine, Seattle, WA, USA). Two separate shRNA fragments of ZNF280A are listed in Table S4. The luciferase reporter system of pE2F-luc, pRb-luc, and pGL3-luc (631914, Clontech Laboratories) was used to examine the transcriptional activity of E2F and Rb. Transfection of plasmids was performed using Lipofectamine 3000 (Life Technologies), according to the manufacturer’s instructions.
Western Blotting Analysis
Nuclear and/or cytoplasmic fractionation was separated by using a Cell Fractionation Kit (Cell Signaling Technology, USA), according to the manufacturer’s instructions, and the whole-cell lysates were extracted using radioimmunoprecipitation assay (RIPA) buffer (Cell Signaling Technology). Western blot was performed according to a standard method, as described previously.29 Antibodies against CCNB1, CCND2, CDK1, CDK6, p-MST1/2, MST1, p-LATS1, LATS1, p-YAP, and YAP were purchased from Cell Signaling Technology; TAZ from Abcam; and ZNF280A from Thermo Fisher Scientific. The membranes were stripped and reprobed with an anti-α-tubulin antibody (Cell Signaling Technology) as the loading control.
Anchorage-Independent Growth Assay
A total of 500 cells was trypsinized and suspended in 2 mL complete medium containing 0.3% agar (Sigma). This experiment was performed as previously described30 and carried out three times independently for each cell line.
Cell-Counting Kit-8 Analysis
For cell-counting kit-8 analysis, cells (2 × 103) were seeded into 96-well plates, and the specific staining process and methods were performed according to the previous study.31
Colony Formation Assay
The cells were trypsinized as single cells and suspended in the media with 10% FBS. The indicated cells (300 cells/well) were seeded into a 6-well plate for ∼10–14 days. Colonies were stained with 1% crystal violet for 10 min after fixation with 10% formaldehyde for 5 min. Plating efficiency was calculated as previously described.32 Different colony morphologies were captured under a light microscope (Olympus).
Cell Cycle Analysis
Pretreatment and staining were performed using Cell Cycle Detection Kit (KeyGEN, China) as previously described.33 Briefly, cells (5 × 105) were harvested by trypsinization, washed in ice-cold PBS, and fixed in 75% ice-cold ethanol in PBS. Before staining, cells were gently resuspended in cold PBS, and ribonuclease was added into the cells’ suspension tube incubated at 37°C for 30 min, followed by incubation with propidium iodide (PI) for 20 min at room temperature. Cell samples (2 × 104) were then analyzed with a FACSCanto II flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA), and the data were analyzed using FlowJo 7.6 software (Tree Star, Ashland, OR, USA).
Tumor Xenografts
4-week-old BALB/c-nu female mice weighing 15–20 g were maintained in a standard pathogen-free environment, where the animals were housed in sterile cages under laminar flow hoods in a 20°C–26°C temperature-controlled room, with a 12-h light/12-h dark cycle, and fed autoclaved chow and water. BALB/c-nu mice at 4–6 weeks of age were randomly divided into three groups (n = 6 per group), and the indicated HCT-116 cells (1 × 106) were inoculated subcutaneously into the flank folds of the nude mice. Tumor volume was determined using an external caliper and calculated using the equation (L × W2)/2. The mice were sacrificed by inhaling CO2 on 36 days after inoculation, and the tumors were excised and subjected to pathologic examination.
IHC
The IHC procedure and scoring of ZNF280A expression levels were performed as previously described.34 Scores given by two independent investigators were averaged for further comparative evaluation of ZNF280A expression. The proportion of tumor cells was scored as follows: 0, no positive tumor cells; 1, <10% positive tumor cells; 2, 10%–35% positive tumor cells; 3, 35%–70% positive tumor cells; and 4, >70% positive tumor cells. The SI was graded according to the following criteria: 0, no staining; 1, weak staining, light yellow; 2, moderate staining, yellow brown; and 3, strong staining, brown. The SI was calculated as the product of the SI score and the proportion of positive tumor cells. Using this method of assessment, we evaluated ZNF280A expression in CRC samples by determining SI, with scores of 0, 1, 2, 3, 4, 6, 8, 9, or 12.
Luciferase Assay
Cells (4 × 104) were seeded in triplicate in 24-well plates and cultured for 24 h, and the luciferase reporter assay was performed as previously described.35 Cells were transfected with 100 ng HOP-Flash (83467, Addgene) or HIP-Flash luciferase reporter plasmid (83466, Addgene), plus 5 ng pRL-TK Renilla plasmid (Promega) using Lipofectamine 3000 (Invitrogen), according to the manufacturer’s recommendation. Luciferase and Renilla signals were measured 36 h after transfection using a Dual Luciferase Reporter Assay Kit (Promega), according to the manufacturer’s protocol.
Statistical Analysis
All values are presented as means ± SD. Significant differences were determined using GraphPad 5.0 software (USA). Student’s t test was used to determine statistical differences between two groups. One-way ANOVA was used to determine statistical differences among multiple testing. The chi-square test was used to analyze the relationship between ZNF280A expression and clinicopathological characteristics. Survival curves were plotted using the Kaplan-Meier method and compared by log-rank test. p < 0.05 was considered significant. All the experiments were repeated three times.
Author Contributions
L.W. developed ideas and drafted the manuscript. X.W. conducted the experiments and contributed to the analysis of data. D.S. contributed to the analysis of data. S.C. conducted the experiments. S.H. contributed to the analysis of data and revised the manuscript. J.T. edited the manuscript. All authors contributed to revision of the manuscript and approved the final version for publication.
Conflicts of Interest
No conflicts of interest were declared.
Acknowledgments
This study was supported by the Finance Department Foundation of Jilin Province (3D5178963428).
Footnotes
Supplemental Information includes four tables and can be found with this article online at https://doi.org/10.1016/j.omto.2019.01.002.
Supplemental Information
References
- 1.Ferlay J., Shin H.R., Bray F., Forman D., Mathers C., Parkin D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J. Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Beà S., Salaverria I., Armengol L., Pinyol M., Fernández V., Hartmann E.M., Jares P., Amador V., Hernández L., Navarro A. Uniparental disomies, homozygous deletions, amplifications, and target genes in mantle cell lymphoma revealed by integrative high-resolution whole-genome profiling. Blood. 2009;113:3059–3069. doi: 10.1182/blood-2008-07-170183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pabo C.O., Peisach E., Grant R.A. Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 5.Jägle S., Busch H., Freihen V., Beyes S., Schrempp M., Boerries M., Hecht A. SNAIL1-mediated downregulation of FOXA proteins facilitates the inactivation of transcriptional enhancer elements at key epithelial genes in colorectal cancer cells. PLoS Genet. 2017;13:e1007109. doi: 10.1371/journal.pgen.1007109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang S., Peng Z., Wang S., Yang L., Chen Y., Kong X., Song S., Pei P., Tian C., Yan H. KRAB-type zinc-finger proteins PITA and PISA specifically regulate p53-dependent glycolysis and mitochondrial respiration. Cell Res. 2018;28:572–592. doi: 10.1038/s41422-018-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jung J.H., Jung D.B., Kim H., Lee H., Kang S.E., Srivastava S.K., Yun M., Kim S.H. Zinc finger protein 746 promotes colorectal cancer progression via c-Myc stability mediated by glycogen synthase kinase 3β and F-box and WD repeat domain-containing 7. Oncogene. 2018;37:3715–3728. doi: 10.1038/s41388-018-0225-0. [DOI] [PubMed] [Google Scholar]
- 8.Ren D., Wang M., Guo W., Huang S., Wang Z., Zhao X., Du H., Song L., Peng X. Double-negative feedback loop between ZEB2 and miR-145 regulates epithelial-mesenchymal transition and stem cell properties in prostate cancer cells. Cell Tissue Res. 2014;358:763–778. doi: 10.1007/s00441-014-2001-y. [DOI] [PubMed] [Google Scholar]
- 9.Pan D. The hippo signaling pathway in development and cancer. Dev. Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halder G., Johnson R.L. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harvey K.F., Zhang X., Thomas D.M. The Hippo pathway and human cancer. Nat. Rev. Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 12.Lei Q.Y., Zhang H., Zhao B., Zha Z.Y., Bai F., Pei X.H., Zhao S., Xiong Y., Guan K.L. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol. Cell. Biol. 2008;28:2426–2436. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oka T., Mazack V., Sudol M. Mst2 and Lats kinases regulate apoptotic function of Yes kinase-associated protein (YAP) J. Biol. Chem. 2008;283:27534–27546. doi: 10.1074/jbc.M804380200. [DOI] [PubMed] [Google Scholar]
- 14.Zhang H., Liu C.Y., Zha Z.Y., Zhao B., Yao J., Zhao S., Xiong Y., Lei Q.Y., Guan K.L. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J. Biol. Chem. 2009;284:13355–13362. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu L., Li Y., Kim S.M., Bossuyt W., Liu P., Qiu Q., Wang Y., Halder G., Finegold M.J., Lee J.S., Johnson R.L. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc. Natl. Acad. Sci. USA. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imajo M., Miyatake K., Iimura A., Miyamoto A., Nishida E. A molecular mechanism that links Hippo signalling to the inhibition of Wnt/β-catenin signalling. EMBO J. 2012;31:1109–1122. doi: 10.1038/emboj.2011.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q., Gao X., Yu T., Yuan L., Dai J., Wang W., Chen G., Jiao C., Zhou W., Huang Q. REGγ Controls Hippo Signaling and Reciprocal NF-κB-YAP Regulation to Promote Colon Cancer. Clin. Cancer Res. 2018;24:2015–2025. doi: 10.1158/1078-0432.CCR-17-2986. [DOI] [PubMed] [Google Scholar]
- 18.Yang C., Xu W., Meng X., Zhou S., Zhang M., Cui D. SCC-S2 Facilitates Tumor Proliferation and Invasion via Activating Wnt Signaling and Depressing Hippo Signaling in Colorectal Cancer Cells and Predicts Poor Prognosis of Patients. J. Histochem. Cytochem. 2019;67:65–75. doi: 10.1369/0022155418799957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X., Sun D., Tai J., Chen S., Yu M., Ren D., Wang L. TFAP2C promotes stemness and chemotherapeutic resistance in colorectal cancer via inactivating hippo signaling pathway. J. Exp. Clin. Cancer Res. 2018;37:27. doi: 10.1186/s13046-018-0683-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Donnell K.A., Wentzel E.A., Zeller K.I., Dang C.V., Mendell J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 21.Sylvestre Y., De Guire V., Querido E., Mukhopadhyay U.K., Bourdeau V., Major F., Ferbeyre G., Chartrand P. An E2F/miR-20a autoregulatory feedback loop. J. Biol. Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 22.Liu J., Ye L., Li Q., Wu X., Wang B., Ouyang Y., Yuan Z., Li J., Lin C. Synaptopodin-2 suppresses metastasis of triple-negative breast cancer via inhibition of YAP/TAZ activity. J. Pathol. 2018;244:71–83. doi: 10.1002/path.4995. [DOI] [PubMed] [Google Scholar]
- 23.Gunn S.R., Bolla A.R., Barron L.L., Gorre M.E., Mohammed M.S., Bahler D.W., Mellink C.H., van Oers M.H., Keating M.J., Ferrajoli A. Array CGH analysis of chronic lymphocytic leukemia reveals frequent cryptic monoallelic and biallelic deletions of chromosome 22q11 that include the PRAME gene. Leuk. Res. 2009;33:1276–1281. doi: 10.1016/j.leukres.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Fang L., Teng H., Wang Y., Liao G., Weng L., Li Y., Wang X., Jin J., Jiao C., Chen L. SET1A-Mediated Mono-Methylation at K342 Regulates YAP Activation by Blocking Its Nuclear Export and Promotes Tumorigenesis. Cancer Cell. 2018;34:103–118.e9. doi: 10.1016/j.ccell.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Zhou D., Conrad C., Xia F., Park J.S., Payer B., Yin Y., Lauwers G.Y., Thasler W., Lee J.T., Avruch J., Bardeesy N. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren A., Yan G., You B., Sun J. Down-regulation of mammalian sterile 20-like kinase 1 by heat shock protein 70 mediates cisplatin resistance in prostate cancer cells. Cancer Res. 2008;68:2266–2274. doi: 10.1158/0008-5472.CAN-07-6248. [DOI] [PubMed] [Google Scholar]
- 27.Ren D., Yang Q., Dai Y., Guo W., Du H., Song L., Peng X. Oncogenic miR-210-3p promotes prostate cancer cell EMT and bone metastasis via NF-κB signaling pathway. Mol. Cancer. 2017;16:117. doi: 10.1186/s12943-017-0688-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai Y., Ren D., Yang Q., Cui Y., Guo W., Lai Y., Du H., Lin C., Li J., Song L., Peng X. The TGF-β signalling negative regulator PICK1 represses prostate cancer metastasis to bone. Br. J. Cancer. 2017;117:685–694. doi: 10.1038/bjc.2017.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren D., Wang M., Guo W., Zhao X., Tu X., Huang S., Zou X., Peng X. Wild-type p53 suppresses the epithelial-mesenchymal transition and stemness in PC-3 prostate cancer cells by modulating miR-145. Int. J. Oncol. 2013;42:1473–1481. doi: 10.3892/ijo.2013.1825. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X., Zhang L., Lin B., Chai X., Li R., Liao Y., Deng X., Liu Q., Yang W., Cai Y. Phospholipid Phosphatase 4 promotes proliferation and tumorigenesis, and activates Ca2+-permeable Cationic Channel in lung carcinoma cells. Mol. Cancer. 2017;16:147. doi: 10.1186/s12943-017-0717-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X., Liu F., Lin B., Luo H., Liu M., Wu J., Li C., Li R., Zhang X., Zhou K., Ren D. miR-150 inhibits proliferation and tumorigenicity via retarding G1/S phase transition in nasopharyngeal carcinoma. Int. J. Oncol. 2017;50:1097–1108. doi: 10.3892/ijo.2017.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang M., Ren D., Guo W., Huang S., Wang Z., Li Q., Du H., Song L., Peng X. N-cadherin promotes epithelial-mesenchymal transition and cancer stem cell-like traits via ErbB signaling in prostate cancer cells. Int. J. Oncol. 2016;48:595–606. doi: 10.3892/ijo.2015.3270. [DOI] [PubMed] [Google Scholar]
- 33.Ren D., Lin B., Zhang X., Peng Y., Ye Z., Ma Y., Liang Y., Cao L., Li X., Li R. Maintenance of cancer stemness by miR-196b-5p contributes to chemoresistance of colorectal cancer cells via activating STAT3 signaling pathway. Oncotarget. 2017;8:49807–49823. doi: 10.18632/oncotarget.17971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo W., Ren D., Chen X., Tu X., Huang S., Wang M., Song L., Zou X., Peng X. HEF1 promotes epithelial mesenchymal transition and bone invasion in prostate cancer under the regulation of microRNA-145. J. Cell. Biochem. 2013;114:1606–1615. doi: 10.1002/jcb.24502. [DOI] [PubMed] [Google Scholar]
- 35.Ren D., Dai Y., Yang Q., Zhang X., Guo W., Ye L., Huang S., Chen X., Lai Y., Du H. Wnt5a induces and maintains prostate cancer cells dormancy in bone. J. Exp. Med. 2018 doi: 10.1084/jem.20180661. Published online December 28, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.