Highlights
-
•
A total of 226 isolates were screened against three strains (cfNAV, cfCHA and cf8436) of C. falcatum by dual culture technique. Selected Twenty-Six bacteria characterized of morphology, biochemical activity, PGP activity, antifungal potential and 16S rRNA gene sequence. These isolates belonged to proteobacteria (13), firmicutes (10) and bacterioidetes (03) respectively.
-
•
Ochrobactrum intermedium (TRD14), Acinetobacter sp (PK9), Bacillus sp (RSC29 and KR91) and Escherichia sp (VRE34) selected for green house study. The most promising results in suppression of the disease as well as plant growth were observed in treatment withVRE34. The plant height and stem diameter were increased from 13.27 ± 0.67 inches to 24.03 ± 1.40 inches and from 6.07 ± 0.45 mm to 9.87 ± 0.93 mm.
-
•
Isolates identified in this study could be used as an alternative to chemical fungicides to control red rot pathogen of sugarcane plants. However, detailed investigations on their inoculations in the field to confirm its growth promotion potency and biocontrol efficacy under natural environmental and soil conditions shall make these strains as important bioinoculants for integrated disease management of red rot disease in sugarcane.
Keywords: Antagonism, Biocontrol, Carbendazim, Colletotrichum falcatum, Red rot, Sugarcane
Abstract
A total of 226 sugarcane rhizosphere-associated bacterial strains from the six different cultivars were screened against three pathogenic strains of C. falcatum (cfNAV, cfCHA, and cf8436) for the suppression of red rot disease. On the basis of mycelial growth inhibition in dual culture assay, 26 bacteria were selected for further characterization of morphology, biochemical activity, plant-growth-promoting (PGP) activity, antifungal potential and molecular identity by 16S rRNA gene sequence. On the basis of the 16S rRNA gene sequencing, it was found that the isolates belonged to proteobacteria (13), Firmicutes (10), and Bacteroides (3). The antagonistic bacteria tested for PGP traits revealed that 10 strains were able to solubilize tricalcium phosphate, 11 strains were able to produce siderophore, and 14 strains were able to grow in the N-free medium. The quantitative estimation of indole-3-acetic acid production was ranged from 21.58 to 66.31 μg/mL. On the basis of PGP and biocontrol traits, five strains Ochrobactrum intermedium (TRD14), Acinetobacter sp. (PK9), Bacillus sp. (RSC29 and KR91) and Escherichia sp. (VRE34) were further chosen for pot trial under greenhouse conditions on highly susceptible variety CoC671. The results showed that the pathogen-inoculated sugarcane plants were able to germinate but died within one month. However, the CoC671 inoculated with selected biocontrol strains found protected from disease and an increase in plant growth parameters on par with carbendazim fungicides. This study proves that the isolates identified in this study could be used as an alternative to chemical fungicides to control red rot pathogen of sugarcane plants.
1. Introduction
Sugarcane (Saccharum officinarum L.) is one of the economically valuable agricultural crops grown worldwide in tropical and subtropical areas mainly for their sugar source. Among the 110 sugarcane cultivated countries, India and Brazil contribute half of global production [1]. A hemibiotrophic fungal pathogen Colletotrichum falcatum causes a major devastating disease in sugarcane [2]. The pathogen initially enters to the plants through the soil and subsequently extends to the stalk by various ways, including borer, which makes the hole in the stem, as well as by other vectors in the field [3]. Infection on leaves may not affect overall yield to a great extent but stalk infection with fungus is very severe as the sugar content is reduced after the infection. During the monsoon period, because of high humidity and lower temperature, the pathogen multiplies rapidly in sugarcane stems. By using stem sucrose for its growth, the pathogen converts sucrose into alcohol and after monsoon period the infected stem starts drying as the alcohol evaporates rapidly. This ultimately reduces the weight of the cane and affects both economies of farmers and the sugar industry. The red rot is a major problem for sugarcane production and is responsible for the abolition of numerous best varieties from the cultivation due to the constant evolution of the newer species [4]. The popular sugarcane variety CoC671 succumbed to C. falcatum during the 1980s in Tamil Nadu, Kerala, Gujarat, Andhra Pradesh, and Pondicherry states of India [5].
The pathogen C. falcatum normally resides in the soil as dormant spores and on decayed host plant parts as active saprophytes. The management of the red rot disease in the field is difficult as the genetic make of this fungus changes continuously. There are three main possible ways to control the red rot disease: (1) use of a resistant variety, (2) treatment with fungicides such as carbendazim, and (3) biological control through antagonistic microorganisms. Although the use of resistant varieties is an important approach to control against red rot, the newly released resistant varieties give up to the C. falcatum due to the recurrent emergence of its newer variants [6,7]. The use of chemicals to control the phytopathogens results in accumulation of harmful residues in soil, exerting a negative effect on beneficial organisms. In addition, pathogens could acclimatize to surmount and become resistant against these fungicides. Carbendazim is a systemic fungicide generally used to control a range of fungal diseases of agricultural crops [8]. It is moderately stable in water and soil with a half-life of up to 1 year [9]. The persistency of carbendazim in soil and its systemic prevalence in plants can lead to environmental contamination [10]. This causes serious concerns because of its cytotoxicity to mammalian endocrine cells, liver cells, and reproductive tissues [11,12].
There is an increasingly growing demand for biological fertilizers [13]. Control of phytopathogen through biological means involves various mechanisms to inhibit or slow the growth of pathogen [14]. Currently, only a limited number of biocontrol products are available on the market, which makes it desirable to search and study more biocontrol bacteria [15]. The microorganisms that are associated with sugarcane roots may show biocontrol potential against C. falcatum pathogens and can play important roles in protecting sugarcane crop. Further, it can be assumed that the crop faces many biotic and abiotic challenges during the long developmental period. The plant-growth-promoting rhizobacteria (PGPR) associated with sugarcane root may be helpful in supporting plant growth by producing various plant-growth-supporting metabolites [16]. PGPR from sugarcane rhizosphere has been reported previously not only to improve plant growth by colonizing rhizosphere but also to suppress C. falcatum [3,17]. The development of an effective biological control against C. falcatum requires the screening and evaluation of native potential antagonistic bacteria capable of reducing red rot under in vitro and in vivo conditions. Recently, various bacterial genera including Pseudomonas, Enterobacter, Burkholderia, Ochrobactrum, Gluconacetobacter and Bacillus have been found to be associated with sugarcane rhizosphere with the ability to suppress the C. falcatum [3,18,17]. Application of single inoculum, which controls the red rot and simultaneously supports the sugarcane growth, is desirable to reduce the cost of fertilizer and fungicides for farmers. In this regard, efforts have been channelized more toward the growth promotion of plant and simultaneously growth inhibition of pathogens. The aim of this work was to characterize sugarcane root-associated microorganisms for biological control of different strains of C. falcatum.
2. Materials and methods
2.1. Collection of C. falcatum
C. falcatum strains cfNAV, cfCHA, and cf8436 used in this study were maintained on potato dextrose agar (PDA) slants at 4 °C. Strains cfNAV and cfCHA were chosen according to their higher virulence shown in our previous study [19]. Cf8436 was used as a reference strain obtained from Sugarcane Breeding Institute (Coimbatore, Tamil Nadu, India).
2.2. Isolation of sugarcane rhizosphere bacteria
For the isolation of sugarcane rhizosphere bacteria (SRB), rhizosphere soils were collected from 3- to 4-month-old field-grown sugarcane cultivar (Co86032, Co86249, CoC671, Co814, and Co99004) free from any fungal infection. The composite sample of uprooted rhizosphere soil was transported to the laboratory in an ice box, and isolation of rhizobacteria was carried within 48 h of sample collection. The serial dilution was carried out (10−1–10−6) with sterile phosphate buffer (pH 6.8) and aliquots of samples were spread on nutrient agar (NA) and Hichrome Bacillus agar, and King’s B agar. All the plates were incubated at 30 ± 2 °C in an incubator for 24 h. Representative bacteria of different morphological types, for example, size, pigmentation, form, and elevation, present on all the plates were selected and purified on NA medium.
2.3. Antagonistic activity against C. falcatum by SRB
The antagonistic activity of each SRB isolate was studied by dual-culture techniques against the three isolates of C. falcatum [20]. Briefly, a small circular plug (5 mm) of each test fungi taken from an actively growing 7-day-old culture on PDA was aseptically placed at 15 mm away from the one end of a sterile 90 mm Petri plate containing PDA. Simultaneously, a loopful of individual overnight grown cultures were separately placed approximately 15 mm away from the opposite end on the same plate. The fungal culture grown on the PDA plate without any bacterial isolate served as control. The experiment was carried out in triplicate. The plates were kept in a plastic bag and incubated for the prescribed period at 30 ± 2 °C in an incubator. At the end of the incubation period, growth was measured. Growth reduction was calculated in relation to the growth of the control, which is as follows:
| FGI (%) = ((FGC―FGT)/FGC) × 100 |
where FGI is the fungal growth inhibition, FGC is the fungal growth in control, and FGT is the fungal growth in treatment.
2.4. Effect of carbendazim on C. falcatum
The effect of carbendazim on C. falcatum was assessed using poisoned food technique [21]. Carbendazim was individually amended to PDA to get fungicide concentrations of 0, 0.01, 0.05, 0.1, and 0.3 with three replications against the three strains of C. falcatum. The respective concentrations were achieved by the addition of carbendazim to the pre-autoclaved PDA medium when its temperature was about 40–45 °C. Inoculations were made with an active 5-mm mycelial disk from test isolates in 9-cm Petri plates and incubated at 30 ± 2 °C for seven days. The colony diameter was measured and the growth inhibition was recorded.
2.5. Plant-growth-promoting (PGP) traits
To find out the phosphate solubilization potential, Pikovskaya’s agar medium was inoculated at the center of the plate and incubated at 30 ± 2 °C for 96 h [22]. Positive results were observed on the basis of clear halos formed around the bacterial colony. Production of indole-3-acetic acid (IAA) was carried out according to the method given by Bric et al. [23]. SRB cultures grown for 24 h were inoculated into 10 ml sterile 0.1% tryptophan-supplemented LB broth. After incubation at 30 ± 2 °C for 96 h, IAA production was calculated in the culture supernatant using Salkowski reagent. Siderophore production was analyzed using the methodology described by Schwyn and Neilands [24]. The SRB isolates were spot-inoculated on chrome azurol S (CAS) media and incubated for 48 h at 30 ± 2 °C. Positive results were confirmed by the presence of surrounding orange halos due to iron consumption from CAS media. Nitrogen fixation was checked using Jensen media [25].
2.6. Gram nature and biochemical characterization
On the basis of biocontrol and PGP traits, selected SRB isolated were subjected to Gram nature and biochemical tests. The biochemical tests such as methyl red, Voges–Proskauer, citrate use, phenyl alanine, nitrate reduction, ammonia production, hydrolysis of casein, lipid, starch and catalase test were performed following the standard protocol [26].
2.7. Molecular characterization by 16S rRNA gene and phylogeny
Genomic DNA isolation of selected 26 SRB was carried out by modified cetyl trimethyl ammonium bromide method [27]. After DNA extraction, the integrity and quality of the DNA obtained were checked by 0.8% (w/v) agarose gel electrophoresis and by GelDoc analysis (Bio-Rad). The DNA samples were stored in 100 μl of TE buffer at ―20 °C. 16S rRNA gene amplification was carried out in a thermal cycler using universal bacterial primer set 8F: 5′-AGAGTTTGATCMTGGCTCAG-3′ and 1492R: 5′CGGTTACCTTGTTACGACTT-3′. Thereafter, 50 μl total PCR reaction mixture comprising 200 mM dNTPs, 50 mM each primer, 1 × PCR buffer, 2U Taq polymerase, and 10 ng genomic DNA were prepared. The PCR conditions involved an initial denaturation at 94 °C for 4 min, followed by 35 cycles of 94 °C for 1 min, 52 °C for 1 min, 72 °C for 2 min, and a final extension at 72 °C for 10 min. The isolates were identified for their species level using partial 16S rRNA sequence homology and data were deposited in GenBank (http://www.ncbi.nlm.nih.gov/GenBank/index.html) to obtain accession number. Molecular phylogenetic and evolutionary relationship of all SRB was studied using Mega 7 software. Further, a phylogenetic tree of the Pseudomonas and Bacilli groups of isolates was separately drawn to compare with the reference strain sequences deposited in GenBank.
2.8. Greenhouse pot trial
Pot trial was carried out to select effective biocontrol agents in greenhouse facilities. For this, inoculums of selected five strains, viz., Ochrobactrum intermedium (TRD14), Acinetobacter sp. (PK9), Bacillus sp. (RSC29), Bacillus sp. (KR91), and Escherichia sp. (VRE34), were prepared by growing bacteria in nutrient broth overnight at 30 ± 2 °C in an incubator shaker. Cultures for seed inoculation were prepared in 0.85% NaCl (saline) after removal of media by centrifugation. The cells were diluted in saline to a final concentration of 1 × 106 cfu/ml. The pathogen was prepared in the form of spore suspension from 10-day-old culture on PDA. Spores were collected in distilled water (DW). Stems having single eye buds were collected from a 7-month-old CoC671 crop, which is a disease-free highly susceptible sugarcane variety. They were collected from the field and washed properly with DW to remove soil particles on them. The buds were treated with 500 μl bacterial culture and 250 μl pathogen spore suspension at the same time on both open ends of sugarcane stem and then planted in a pot containing autoclaved soil. The effects of the chemical fungicide, carbendazim, were also studied by dipping sugarcane in 0.3 ppm solution for 30 min. before planting. Further, to confirm plant-growth-promoting potential, all five strains were separately inoculated without any pathogen. The inoculated plants were maintained under greenhouse condition at 12:12 h light/dark cycle with regular irrigation. Plants inoculated with DW served as control. The trials were arranged in a randomized complete block design with three replicates. The effects of bacteria were evaluated in terms of plant growth parameter such as height, stem diameter, number of leaves, and condition of the top. Data were recorded on 30 and 60 days after plantation (DAP). The data were examined using SPSS software (SPSS Inc., Chicago, IL) to analyze the variance for a randomized complete block design.
3. Results
Although the total numbers of bacterial strains collected using spread plate technique were 226, twenty-six isolates were selected based on biocontrol activity in dual culture assay.
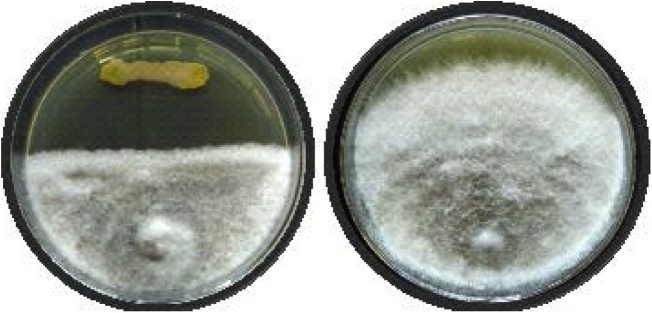
The results showed that the antagonistic activity was varied for different strains of C. falcatum. The percentage inhibition was ranged from 28.97% to 61.18% for cfNAV, 34.01% to 69.64% for cfCHA, and 28.96% to 53.48% for cf8436 (Table 1 & Fig. 1). In case of C. falcatum strain cfNAV, Ochrobactrum intermedium TRD14 and Escherichia sp. VRE34 has shown maximum inhibition i.e. 61.18% and 61.11% respectively. Escherichia sp. VRE34 has also shown the highest antagonism against cfCHA (69.64%). Reference strain cf8436 growth was found inhibited maximum when co-inoculated with Escherichia sp. VRE34. Out of 26 strains studied 23 strains have sown more than 50% inhibition against one or other C. falcatum strain. It can be speculated that higher inhibition recorded in the present study may because of metabolite released from the bacterial strains spread into the PDA continuously. It has been observed that the spread of bacterial growth on media was limited to the inoculation area (Fig. 1). While fungus growth is slow as compare to bacteria so bacterial strains may reach to stationary phase and release as much as metabolites up to seven days. In the case of chemical fungicide carbendazim, the increase in concentration resulted in 100% control (0.3 ppm) of C. falcatum mycelia growth. The same concentration was used in pot assay as a positive control.
Table 1.
Percentage inhibition by biocontrol agents and carbendazim against C falcatum isolates. Values in each column with same letter do not differ significantly at P ≤ 0.05 by Duncan’s Multiple Range Test.
| Isolate Treatment | %Inhibition against C falcatum |
||
|---|---|---|---|
| cfNAV | cfCHA | Cf8436 | |
| Stenotrophomonas acidaminiphila TRD10 | 39.52 j | 38.26kl | 34.26mn |
| Ochrobactrum anthropi TRD11 | 31.43 k | 34.1m | 28.96° |
| Ochrobactrum intermedium TRD14 | 61.18 a | 58.8b | 53.48b |
| Bacillus safensis PK1 | 53.5 b | 40.47ijk | 43.31ghi |
| Bacillus megaterium PK2 | 52.29bc | 42.67ghi | 46.05fg |
| Sphingobacterium thalpophilum PK6 | 41.37 ij | 37.65klm | 36.71lm |
| Acinetobacter sp. PK9 | 60.44 a | 51.63 de | 49.75de |
| Acinetobacter sp. PK10 | 50.46 cd | 53.32cd | 49.61 de |
| Stenotrophomonas acidaminiphila RSC6 | 51.04 cd | 41hij | 47.68ef |
| Sphingobacterium thalpophilum RSC24 | 48.64 de | 42.81gh | 43.82gh |
| Escherichia sp. RSC25 | 28.97 k | 40.06ijk | 37.54kl |
| Bacillus sp. RSC29 | 51.07 bcd | 57.05b | 50.09cd |
| Enterobacter sp. RSC32 | 44.37 gh | 44.27gh | 40.43ijk |
| Bacillus sp. KR91 | 58.65 a | 56.89bc | 53.12bc |
| Cronobacter muytjensii VRE6 | 42.58 ghi | 45.05g | 36.22lm |
| Enterobacter cloacae VRE7 | 52.38 bc | 39.16jkl | 38.67jkl |
| Pseudomonas sp. VRE8 | 41.27 ij | 45.03g | 44.42fg |
| Bacillus thuringiensis VRE11 | 45.34 fg | 46.02 fg | 36.25lm |
| Pseudomonas aeruginosa VRE12 | 54.17 b | 49.95 de | 45.38fg |
| Sphingobacterium sp. VRE29 | 48.55 de | 36.11 lm | 34.78mn |
| Escherichia sp. VRE34 | 61.11a | 69.64 a | 58.96a |
| Pseudomonas sp. S1 | 41.68 hij | 37.7klm | 32.51n |
| Pseudomonas plecoglossicida S2 | 47.27 ef | 51.93 de | 39.13jk |
| Pseudomonas sp. S4 | 45.24 fg | 38.63kl | 39.1jkl |
| Bacillus safensis B1 | 44.37gh | 45.12g | 41.06hij |
| Bacillus subtilis C1 | 50.46 cd | 48.85 ef | 51.2bc |
| *Carbendazim (0.01 ppm) | 20.22 | 22.37 | 27.98 |
| *Carbendazim (0.05 ppm) | 59.39 | 50.84 | 61.51 |
| *Carbendazim (0.1 ppm) | 80.26 | 79.15 | 83.06 |
| *Carbendazim (0.3 ppm) | 100 | 100 | 100 |
Fungicide.
Fig. 1.
Antifungal activity of Ochrobactrum intermedium TRD14 against C. falcatum strain cf8436. (Left; Treatment, Right; Control).
All twenty-six isolates were characterized for their morphology and metabolic activities (Table 2). The Gram’s reaction showed that out of 26 isolates, 7 were positive and the rest was negative. The biochemical tests of antagonistic bacteria showed that none of the isolates were positive for phenyl alanine and Voges–Proskauer test. The results of other experiments varied for each isolate. However, 13 bacteria were found to be capable of reducing nitrate and 6 producing ammonia.
Table 2.
Biochemical characterization of sugarcane rhizospheric microbes. MR- Methyl Red, VP- Voges Proskauer, PA- Phenyl Alanine, NH3-Ammonia.
| SRB | Gram reaction | MR | VP | Citrate | PA | Urea | Nitrate | NH3 | Amylase | Protease | Lipase | Catalase |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TRD10 | – | – | – | – | – | – | + | + | – | – | – | – |
| TRD11 | – | – | – | – | – | + | – | – | – | + | – | – |
| TRD14 | – | – | – | – | – | + | – | + | – | – | – | + |
| PK1 | + | – | – | + | – | – | – | – | – | + | + | – |
| PK2 | + | – | – | – | – | – | + | – | + | – | – | – |
| PK6 | – | + | – | – | – | – | + | – | – | – | – | – |
| PK9 | – | – | – | – | – | – | – | + | + | + | – | – |
| PK10 | – | – | – | – | – | – | + | – | – | – | – | + |
| RSC6 | – | – | – | + | – | + | – | – | – | + | – | – |
| RSC24 | – | – | – | – | – | – | + | – | – | – | + | – |
| RSC25 | – | – | – | + | – | – | – | – | + | – | – | – |
| RSC29 | + | – | – | – | – | – | – | + | – | – | – | + |
| RSC32 | – | – | – | – | – | – | + | – | – | – | + | – |
| KR91 | + | + | – | – | – | – | + | – | – | + | – | + |
| VRE6 | – | – | – | + | – | – | – | + | – | – | – | – |
| VRE7 | – | – | – | – | – | – | + | – | – | – | – | – |
| VRE8 | – | – | – | – | – | + | – | – | – | + | + | – |
| VRE11 | + | – | – | – | – | – | – | – | + | + | – | – |
| VRE12 | – | – | – | – | – | – | + | – | – | + | – | – |
| VRE29 | – | + | – | – | – | – | + | – | – | – | – | – |
| VRE34 | – | – | – | – | – | – | – | + | – | + | – | + |
| S1 | – | – | – | – | – | + | – | – | – | + | – | – |
| S2 | – | – | – | + | – | – | – | – | + | – | – | – |
| S4 | – | + | – | – | – | – | + | – | – | – | + | – |
| B1 | + | – | – | – | – | + | – | – | + | – | – | + |
| C1 | + | – | – | + | – | – | + | – | – | + | – | – |
–= Negative += Positive.
The identification of antagonistic bacteria based on 16S rRNA gene sequencing showed the presence of 10 different genera. These isolates belonged to proteobacteria (13), Firmicutes (10), and Bacteroides (3). It is interesting to note that a diverse group of bacteria could be isolated from the rhizosphere soils of sugarcane (Table 3).
Table 3.
List of SRB isolated from sugarcane rhizosphere.
| Sr NO | Isolate code | Host Cultivar | Place of collection | % Homology | Identified as | Accession Number |
|---|---|---|---|---|---|---|
| 1 | TRD10 | Co 86032 | Mahuva | 99 | Stenotrophhomonas acidaminiphilia | MF351813 |
| 2 | TRD11 | Co 86032 | Mahuva | 99 | Ochrobactrum anthropi | KY672866 |
| 3 | TRD14 | Co 86032 | Mahuva | 99 | Ochrobactrum intermedium | MF351814 |
| 4 | PK1 | Co 86249 | Timbarva | 99 | Bacillus safensis | KU867835 |
| 5 | PK2 | Co 86249 | Timbarva | 99 | Bacillus megaterium | KU867836 |
| 6 | PK6 | Co 86249 | Timbarva | 99 | Sphingobacterium thalpophilum | KU867842 |
| 7 | PK9 | Co 86249 | Karachka | 99 | Acinetobacter sp. | KX168053 |
| 8 | PK10 | Co 86249 | Karachka | 99 | Acinetobacter sp. | KX168037 |
| 9 | RSC6 | Co 671 | Navsari | 99 | Stenotrophomonas acidaminiphila | KU867837 |
| 10 | RSC24 | Co 671 | Navsari | 99 | Sphingobacterium thalpophilum | KU867848 |
| 11 | RSC25 | Co 671 | Madhi | 99 | Escherichia sp. | KX228402 |
| 12 | RSC29 | Co 671 | Madhi | 99 | Bacillus sp. | KX181401 |
| 13 | RSC32 | Co 671 | Madhi | 99 | Enterobacter sp. | KX168052 |
| 14 | KR91 | Co 8145 | Rayam | 99 | Bacillus sp. | KX168055 |
| 15 | VRE6 | Co 94004 | Vyara | 98 | Cronobacter muytjensii | KU867847 |
| 16 | VRE7 | Co 94004 | Vyara | 99 | Enterobacter cloacae | KU867838 |
| 17 | VRE8 | Co 94004 | Varad | 99 | Pseudomonas sp. | KX168038 |
| 18 | VRE11 | Co 94004 | Vyara | 99 | Bacillus thuringiensis | KU867844 |
| 19 | VRE12 | Co 94004 | Vyara | 99 | Pseudomonas aeruginosa | KU867839 |
| 20 | VRE29 | Co 94004 | Vyara | 99 | Sphingobacterium sp. | KU867840 |
| 21 | VRE34 | Co 94004 | Varad | 99 | Escherichia sp. | KX228403 |
| 22 | S1 | Co 86002 | Rajpara | 99 | Pseudomonas sp. | KX168054 |
| 23 | S2 | Co 86002 | Timbarva | 99 | Pseudomonas plecoglossicida | KU867841 |
| 24 | S4 | Co 86002 | Timbarva | 99 | Pseudomonas sp. | KU867843 |
| 25 | B1 | Co 86002 | Timbarva | 99 | Bacillus safensis | KU867845 |
| 26 | C1 | Co 86002 | Timbarva | 99 | Bacillus subtilis | KU867846 |
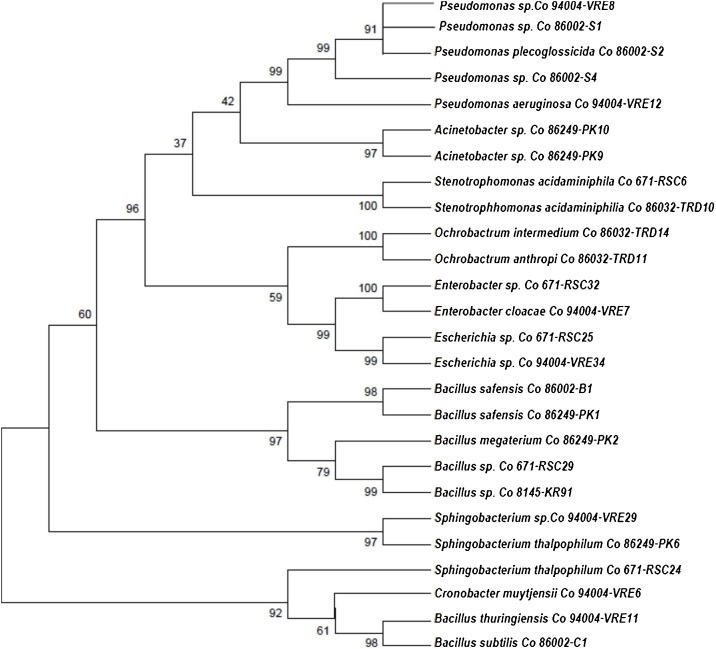
The evolutionary history and molecular diversity were inferred using the UPGMA method. The optimal tree with the sum of branch length = 4.61531710 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown next to the branches. The evolutionary distances were computed using the Kimura 2-parameter method and are in the units of the number of transitional substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1). All ambiguous positions were removed for each sequence pair. There were a total of 1544 positions in the final dataset. The results of the phylogenetic analysis showed one major group with five Pseudomonas strains (Pseudomonas sp. (VRE8), Pseudomonas sp. (S1), P. plecoglossicida (S2), Pseudomonas sp. (S4), and P. aeruginosa (VRE12)) including three from Co 86002 grouped as cluster 1. Cluster 2 presented two strains (PK9 andPK10) from Co86249. Two strain of Stenotrophomonas acidaminiphila (RSC6 and TRD10) were grouped under cluster 3. TRD14 and TRD11 were grouped in cluster 4. Enterobacter sp. and Escherichia sp. were grouped under cluster 5. Clusters 6 and 8 presented isolates belonging to genus Bacillus. Cluster 7 consisted of four isolates belonging to the order Sphingobacter (Fig. 2).
Fig. 2.
A phylogram derived from sequences of 16S rDNA region of sugarcane rhizospheric isolates collected in the present study. Numbers on nodes represent bootstrap values (%) from 500 replicates. A phylogenetic tree was constructed using MEGA 7.0.
The antagonistic bacteria tested for PGP traits showed that 10 isolates were able to solubilize tricalcium phosphate in the medium. Eleven strains were able to produce siderophore and 14 strains were able to grow in the N-free medium. The quantitative estimation showed that all strains were able to produce IAA in certain quantities, which ranged from 21.58 to 66.31 μg/ml (Table 4).
Table 4.
Plant growth promoting characters of sugarcane rhizospheric microbes.
| SRB | IAA Production (μg/ml) | P Solubilization | N2 Fixation | Siderophore |
|---|---|---|---|---|
| Stenotrophomonas acidaminiphila TRD10 | 21.58 | – | – | + |
| Ochrobactrum anthropi TRD11 | 34.28 | – | – | – |
| Ochrobactrum intermedium TRD14 | 66.31 | + | + | – |
| Bacillus safensis PK1 | 39.68 | – | – | – |
| Bacillus megaterium PK2 | 44.28 | – | + | + |
| Sphingobacterium thalpophilum PK6 | 43.73 | – | + | + |
| Acinetobacter sp. PK9 | 63.79 | + | – | + |
| Acinetobacter sp. PK10 | 59.09 | + | + | – |
| Stenotrophomonas acidaminiphila RSC6 | 56.98 | + | + | – |
| Sphingobacterium thalpophilum RSC24 | 23.74 | + | + | – |
| Escherichia sp. RSC25 | 54.28 | + | + | – |
| Bacillus sp. RSC29 | 65.66 | + | + | – |
| Enterobacter sp. RSC32 | 56.98 | + | + | – |
| Bacillus sp. KR91 | 60.36 | – | + | + |
| Cronobacter muytjensii VRE6 | 36.31 | – | + | – |
| Enterobacter cloacae VRE7 | 34.54 | – | – | – |
| Pseudomonas sp. VRE8 | 43.86 | + | – | – |
| Bacillus thuringiensis VRE11 | 27.11 | – | – | – |
| Pseudomonas aeruginosa VRE12 | 54.95 | – | – | – |
| Sphingobacterium sp. VRE29 | 25.90 | – | – | – |
| Escherichia sp. VRE34 | 51.16 | + | + | + |
| Pseudomonas sp. S1 | 53.94 | – | – | + |
| Pseudomonas plecoglossicida S2 | 42.38 | – | – | + |
| Pseudomonas sp. S4 | 47.11 | – | + | + |
| Bacillus safensis B1 | 33.40 | – | – | + |
| Bacillus subtilis C1 | 37.40 | – | + | + |
–= Negative + = Positive.
On the basis of antagonistic and PGP properties, five strains were tested in vivo against three strains of C. falcatum. The results revealed that TRD14was most effective in controlling the pathogenicity of cfNAV and promoted sugarcane height up to 18.33 ± 1.08 in. on 60 DAP compared to control (15.03 ± 2.61 in.) (Table 5). Furthermore, sugarcane treated with TRD14 also experienced an increase in stem diameter from 6.37 ± 0.50 mm (30 DAP) to 10.27 ± 0.45 mm (60 DAP), and the condition of the top was green even after two months. In the case of PK9 and RSC29, when challenged with C. falcatum strains, it was found that plant height and stem diameter were not significantly supported and sugarcane was started drying after 45 days, but in absence of any red rot pathogen both the strains helped increase the stem height up to 16.63 ± 1.2 and 16.63 ± 2.3 in., and stem diameter up to 7.27 ± 0.45 and 8.83 ± 0.70 mm. Similarly, KR91 without pathogen challenge supported maximum plant height and stem diameter up to 21.53 ± 1.60 in. and 9.07 ± 0.76 mm 60DAP and the condition of the top was green. Further, inoculation of KR91 showed a good biocontrol activity against cf8436 in which the plant height and stem diameter were more than doubled, that is, from 9.63 ± 0.85 in. (30 DAP) to 21.10 ± 1.7 in. (60 DAP). The most promising results in suppression of the disease as well as plant growth were observed in treatment withVRE34. The plant height and stem diameter were increased from 13.27 ± 0.67 in. to 24.03 ± 1.40 in. and from 6.07 ± 0.45 mm to 9.87 ± 0.93 mm. VRE34 supported good plant growth in treatment with all pathogens. In the case of carbendazim treatment, plants showed good growth but leaves almost became pigment-less on the 60th day against cfCHA and cf8436. It was observed that the sugarcane growth was almost stopped on 30 DAP when inoculated with red rot pathogens. In addition, stem diameter was found to decrease from 30 to 60 DAP because of plant death followed by dryness of tissues. This directly reflects the severity of red rot pathogen. There was no significant difference in the number of leaves, and in the majority of cases, it was 3–4 on 60 DAP, especially in pathogen-inoculated pots where plants died within a month and the number of leaves produced was only 2.
Table 5.
Evaluation of the five SRB for their antagonistic and growth promotion potential against C. falcatum in sugarcane CoC 671 under green house conditions. Values in each column with same letter do not differ significantly at P ≤ 0.05 by Duncan’s Multiple Range Test.
| Treatment | Plant Height (inches) |
Stem Diameter (mm) |
No of Leaves |
Condition of plant (60 DAP) | |||
|---|---|---|---|---|---|---|---|
| 30 DAP | 60 DAP | 30 DAP | 60 DAP | 30 DAP | 60 DAP | ||
| Control | 10.17 d | 15.03 gh | 4.77 de | 7.17fg | 3 | 4 | G |
| TRD14 | 13.17 b | 23.10 ab | 6.37 ab | 10.27 a | 3 | 4 | G |
| TRD14 X cfNAV | 5.17 ° | 18.33 de | 4.13gh | 8.10 cd | 4 | 4 | G |
| TRD14 X cfCHA | 9.67 f | 10.43 l | 5.07 cd | 6.37gh | 2 | 3 | G |
| TRD14 X cf8436 | 3.37 p | 6.23n | 3.87 hi | 6.10ij | 3 | 4 | G |
| PK9 | 10.8 cd | 16.63 ef | 5.77 ab | 7.27fg | 3 | 5 | G |
| PK9 X cfNAV | 10.87 cd | 10.77 k | 4.93 de | 5.63 jk | 3 | 3 | G |
| PK9 X cfCHA | 8.93 i | 11.13 jk | 4.23gh | 4.53 l | 3 | 3 | G |
| PK9 X cf8436 | 8.37 j | 11.60 jk | 5.70 ab | 5.93 k | 3 | 3 | G |
| RSC29 | 13.77 b | 16.63 ef | 5.37 cd | 8.83bc | 4 | 5 | G |
| RSC29 X cfNAV | 8.27 jk | 9.40lm | 4.80de | 6.57gh | 3 | 3 | G |
| RSC29 X cfCHA | 8.83 i | 10.47l | 3.93 hi | 5.17 kl | 3 | 3 | G |
| RSC29 X cf8436 | 10.70 cd | 12.90ij | 4.20gh | 7.23fg | 3 | 3 | G |
| VRE34 | 13.27 b | 24.03 a | 6.07 ab | 9.87 a | 4 | 5 | G |
| VRE34 X cfNAV | 8.90 i | 13.30 hi | 4.27gh | 7.83 ef | 2 | 4 | G |
| VRE34 X cfCHA | 11.57 c | 14.27 hi | 3.67 i | 6.17 ij | 3 | 3 | G |
| VRE34 X cf8436 | 7.43 l | 19.33 cd | 3.23 k | 6.53gh | 3 | 4 | G |
| KR91 | 17.37 a | 21.53 bc | 6.63 a | 9.07 b | 4 | 5 | G |
| KR91 X cfNAV | 6.43n | 8.07mn | 5.77ab | 5.97 jk | 2 | 3 | G |
| KR91 X cfCHA | 7.83kl | 13.37 hi | 5.33 cd | 5.73kl | 2 | 3 | G |
| KR91 X cf8436 | 9.63g | 21.10 bc | 5.27 cd | 9.03 b | 2 | 4 | G |
| Carbendazim | 8.37j | 15.53fg | 5.47bc | 8.70bc | 3 | 4 | G |
| Carbendazim X cfNAV | 9.40 h | 14.40 gh | 4.67ef | 7.93 de | 3 | 4 | G |
| Carbendazim X cfCHA | 7.0m | 9.43 lm | 5.60bc | 6.37 i | 3 | 3 | G |
| Carbendazim X cf8436 | 11.23 cd | 14.77 gh | 5.77ab | 7.03 g | 3 | 3 | G |
| cfNAV | 7.2 lm | 7.43mn | 4.47 fg | 4.37 l | 2 | 2 | D |
| cfCHA | 3.73 p | 3.97 ° | 3.47 i | 3.40 m | 2 | 2 | D |
| Cf8436 | 4.0 op | 4.07 ° | 3.33 jk | 3.23 m | 2 | 2 | D |
*G- Green, D-Dry.
4. Discussion
The present study was successful in selecting 26 most promising bacteria from sugarcane rhizosphere that can be a useful component of integrated disease management. Microorganisms that can colonize the rhizosphere and show biocontrol potential may have an important role for crop protection against soil-borne plant pathogens [28]. Although rhizobacteria-mediated biocontrol is an easy and eco-friendly way but best results can be obtained only with its host-specific selection. In the case where the pathogen is diverse in nature, for example C. falcatum, it becomes necessary to assess a large number of biocontrol agents. The C. falcatum variants are referred as pathotypes and found to show high compatibility with the host variety. Further, there would be possibility of a variety of microorganisms associated with roots of different cultivars. In view of this, in the present study different sugarcane varieties were selected in the range of resistance to highly susceptible categories for the isolation of rhizosphere bacteria which have the ability to control highly virulent red rot strains cfCHA, cfNAV, and cf8436 [19].
According to Essghaier et al. [29] PGP bacteria can enhance plant growth through a broad range of activities such as IAA production, phosphate solubilization, siderophore production, and nitrogen fixation. Although many studies on the PGP activities of the sugarcane rhizobacteria have been reported till date, only a few of them have reported both PGP and biocontrol. In recent years, different rhizobacteria including members of the genus Acinetobacter and Klebsiella have been reported from the sugarcane to possess PGP properties [16,[30], [31], [32]].
In this study, we have reported the toxicity of carbendazim against C. falcatum under in vitro conditions by poison food technique. It was found that only 0.3 ppm carbendazim is sufficient to completely seize spore germination and mycelial growth. López-Herrera and Zea-Bonilla [33] found benomyl, carbendazim, and thiophanate methyl at 0.5 μg/mL totally inhibited mycelial growth of Rosellini anecatrix. Similar results were noted by Waraitch [34], who reported that carbendazim was most effective against C. falcatum. Although chemical control of C. falcatum is intensive and effective, it poses problems to the human health as well as environment. Biocontrol with PGP activity by native bacterial strain would be best inoculums for high yield of sugarcane with minimum cost. Among all the five strains studied, TRD14 was found highly effective with IAA production, phosphate solubilization, nitrogen fixation, more than 50% inhibition again all three tested pathogens, and increased growth parameters in pot assay. So, TRD14 holds immense potential for future use in the management of red rot disease. Muangthong et al. [35] isolated O. intermedium from industrial sugarcane (UT3R1) varieties and reported them to have N fixation capacity. Previously, Bajoria et al. [36] reported the antifungal activity of O. intermedium against Macrophomina phaseolina and Fusarium oxysporum. Members of the genus Bacillus are large painstaking microbial factories for the release of a monstrous array of biologically active metabolites possibly controlling the growth of phytopathogen. In this study, we have characterized seven Bacilli strains with antagonistic potential in detail, and among them, two strains viz., RSC29 and KR91, were evaluated for biological control under greenhouse condition. B. safensis (PK1) and B. megaterium (PK2) have shown more than 50% mycelial growth inhibition for cfNAV. Our results are in accordance with Hassan et al. [37] who reported two antagonistic strains B. subtilis NH-100 and Bacillus sp. NH-217 against C. falcatum. Antagonistic strains of the genus Bacillus are advantageous over other biocontrol agents in various ways, as they are omnipresent in soils, have excessive sporulation, have prolonged shelf life, and enhance plant nutrition. Their efficiency in controlling many plant diseases has repeatedly been shown by many researchers [38,39]. Further, results obtained with VRE34 also indicated the possible application as biofertilizer and biopesticide. The sugarcane treated with only pathogens showed severe disease condition and plants died within a month. Therefore, the results showed that these five bacterial isolates might have the potential to be developed as a promising commercial biological control agent in the future. Biocontrol of C. falcatum by effective antagonistic of microbes in a controlled laboratory or greenhouse conditions has been reported in many studies [3,40]. However, together with other tests need to be done, such as field experiments and assessing suitability in a fermenter for large-scale production for commercial biofertilizer, survival potential, root colonization, adequate dose, chemical compatibility, the symbiotic effect on other community and economic viability.
In conclusion, irrespective of the mechanisms underlying interactions between sugarcane plant and endophytic pathogen, native rhizobacteria studied were capable to control C. falcatum under both in vitro and in vivo conditions. However, detailed investigations on their inoculations in the field to confirm its growth promotion potency and biocontrol efficacy under natural environmental and soil conditions shall make these strains as important bioinoculants for integrated disease management of red rot disease in sugarcane.
Conflict of interest statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Acknowledgments
Authors are grateful to the Management and Director, C. G. Bhakta Institute of Biotechnology, Uka Tarsadia University, Bardoli, for providing the necessary facilities during the course of the investigation.
References
- 1.Fischer D., Pfitzner B., Schmid M., Simões-Araújo J.L., Reis V.M., Pereira W., Ormeño-Orrillo E., Hai B., Hofmann A., Schloter M. Molecular characterisation of the diazotrophic bacterial community in uninoculated and inoculated field-grown sugarcane (Saccharum sp.) Plant Soil. 2012;356:83–99. [Google Scholar]
- 2.Ashwin N., Barnabas L., Sundar A.R., Malathi P., Viswanathan R., Masi A., Agrawal G.K., Rakwal R. Comparative secretome analysis of Colletotrichum falcatum identifies a cerato-platanin protein (EPL1) as a potential pathogen-associated molecular pattern (PAMP) inducing systemic resistance in sugarcane. J. Proteom. 2017;169:2–20. doi: 10.1016/j.jprot.2017.05.020. [DOI] [PubMed] [Google Scholar]
- 3.Hassan M.N., Afghan S., Hafeez F.Y. Suppression of red rot caused by Colletotrichum falcatum on sugarcane plants using plant growth-promoting rhizobacteria. Biocontrol. 2010;55:531–542. [Google Scholar]
- 4.Malathi P., Viswanathan R., Sundar A.R., Prakasam N., Padmanaban P., Jothi R., Devi S.R., Poongothai M. Variability among Colletotrichum falcatum pathotypes used for screening red rot resistance in sugarcane. Sugar Cane Int. 2010;28:47–52. [Google Scholar]
- 5.Viswanathan R. Pathogen virulence in sugarcane red rot pathogen versus varieties in cultivation: classical case of loss in virulence in the pathotype CF06 (Cf671) Sugar Tech. 2017;19:293–299. [Google Scholar]
- 6.Kumar N., Jhang T., Sharma T.R. Molecular and pathological characterization of Colletotrichum falcatum infecting subtropical Indian sugarcane. J. Phytopathol. 2011;159:260–267. [Google Scholar]
- 7.Malathi P., Viswanathan R., Sundar A., Prakasam N. Keys to identify pathogen variability in Colletotrichum falcatum went causing red rot in sugarcane. Eur. J. Biol. Sci. 2013;6 [Google Scholar]
- 8.Chen Y., Zhou M.-G. Characterization of Fusarium graminearum isolates resistant to both carbendazim and a new fungicide JS399-19. Phytopathology. 2009;99:441–446. doi: 10.1094/PHYTO-99-4-0441. [DOI] [PubMed] [Google Scholar]
- 9.Pandey G., Dorrian S.J., Russell R.J., Brearley C., Kotsonis S., Oakeshott J.G. Cloning and biochemical characterization of a novel carbendazim (methyl-1H-benzimidazol-2-ylcarbamate)-hydrolyzing esterase from the newly isolated Nocardioides sp. strain SG-4G and its potential for use in enzymatic bioremediation. Appl. Environ. Microbiol. 2010;76:2940–2945. doi: 10.1128/AEM.02990-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang H., Wang Y., Gao C., Yan H., Dong B., Yu Y. Isolation and characterization of Pseudomonas sp. CBW capable of degrading carbendazim. Biodegradation. 2010;21:939–946. doi: 10.1007/s10532-010-9353-0. [DOI] [PubMed] [Google Scholar]
- 11.Farag A., Ebrahim H., ElMazoudy R., Kadous E. Developmental toxicity of fungicide carbendazim in female mice. Birth Defects Res. B Dev. Reprod. Toxicol. 2011;92:122–130. doi: 10.1002/bdrb.20290. [DOI] [PubMed] [Google Scholar]
- 12.Selmanoğlu G., Barlas N., Songür S., KocSkaya E. Carbendazim-induced haematological, biochemical and histopathological changes to the liver and kidney of male rats. Hum. Exp. Toxicol. 2001;20:625–630. doi: 10.1191/096032701718890603. [DOI] [PubMed] [Google Scholar]
- 13.Raja N. Biopesticides and biofertilizers: ecofriendly sources for sustainable agriculture. J. Biofertil. Biopestic. 2013;4 [Google Scholar]
- 14.Campbell R. Cambridge University Press; 1989. Biological Control of Microbial Plant Pathogens. [Google Scholar]
- 15.Whipps J.M. Elsevier; 1997. Developments in the Biological Control of Soil-borne Plant Pathogens. Advances in Botanical Research; pp. 1–134. [Google Scholar]
- 16.Bhardwaj G., Shah R., Joshi B., Patel P. Klebsiella pneumoniae VRE36 as a PGPR isolated from Saccharum officinarum cultivar Co99004. J. Appl. Biol. Biotechnol. 2017;5:047–052. [Google Scholar]
- 17.Katiyar D., Hemantaranjan A., Singh B., Malakar A.K. Isolation and characterization of plant growth promoting rhizobacteria Enterobacter hormaechei and their suppression efficacy against Colletotrichum falcatum in combination with Chitosan. Int. J. Plant Soil Sci. 2017;14:1–12. [Google Scholar]
- 18.Hassan M.N., Afghan S., ul Hassan Z., Hafeez F.Y. Biopesticide activity of sugarcane associated rhizobacteria: Ochrobactrum intermedium strain NH-5 and Stenotrophomonas maltophilia strain NH-300 against red rot under field conditions. Phytopathol. Mediterr. 2014:229–239. [Google Scholar]
- 19.Prittesh P., Amaresan N., Rushabh S., Krishnamurthy R., Bhasker V. Isolation and pathogenic variability of Colletotrichum falcatum causing red rot in sugarcane. J. Plant Dis. Prot. 2016;123:273–277. [Google Scholar]
- 20.Dennis C., Webster J. Antagonistic properties of species-groups of Trichoderma: III. Hyphal interaction. Trans. Br. Mycol. Soc. 1971;57 363–362. [Google Scholar]
- 21.Sharvelle E.G. Burges Publication Company; Minnesota, USA: 1960. The Nature and Uses of Modern Fungicides; p. 308. [Google Scholar]
- 22.Pikovskaya R. Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologiya. 1948;17:362–370. [Google Scholar]
- 23.Bric J.M., Bostock R.M., Silverstone S.E. Rapid in situ assay for indoleacetic acid production by bacteria immobilized on a nitrocellulose membrane. Appl. Environ. Microbiol. 1991;57:535–538. doi: 10.1128/aem.57.2.535-538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwyn B., Neilands J. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 25.Jensen H. Nonsymbiotic nitrogen fixation. Soil Nitrogen. 1965:436–480. [Google Scholar]
- 26.Harrigan W.F., McCance M.E. Academic press; 2014. Laboratory Methods in Microbiology. [Google Scholar]
- 27.Rogers S.O., Bendich A.J. Springer; 1989. Extraction of DNA From Plant Tissues. Plant Molecular Biology Manual; pp. 73–83. [Google Scholar]
- 28.El-Hassan S., Gowen S. Formulation and delivery of the bacterial antagonist Bacillus subtilis for management of lentil vascular wilt caused by Fusarium oxysporum f. sp. lentis. J. Phytopathol. 2006;154:148–155. [Google Scholar]
- 29.Essghaier B., Dhieb C., Rebib H., Ayari S., Boudabous A.R.A., Sadfi-Zouaoui N. Antimicrobial behavior of intracellular proteins from two moderately halophilic bacteria: strain J31 of Terribacillus halophilus and strain M3-23 of Virgibacillus marismortui. J. Plant Pathol. Microbiol. 2014;5:1. [Google Scholar]
- 30.Kuan K.B., Othman R., Rahim K.A., Shamsuddin Z.H. Plant growth-promoting rhizobacteria inoculation to enhance vegetative growth, nitrogen fixation and nitrogen remobilisation of maize under greenhouse conditions. PLoS One. 2016;11 doi: 10.1371/journal.pone.0152478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Modi K., Patel P. Isolation and characterization of plant growth promoting rhizobacteria associated with Saccharum officinarum L. Curr. Synth. Syst. Biol. 2017;5 2332–0737. [Google Scholar]
- 32.Patel P., Shah R., Modi K. Isolation and characterization of plant growth promoting potential of Acinetobacter sp. rsc7 isolated from Saccharum officinarum cultivar Co 671. J. Exp. Biol. Agric. Sci. 2017;5:483–491. [Google Scholar]
- 33.López-Herrera C., Zea-Bonilla T. Effects of benomyl, carbendazim, fluazinam and thiophanate methyl on white root rot of avocado. Crop. Prot. 2007;26:1186–1192. [Google Scholar]
- 34.Waraitch K. Disease build up in sugarcane setts treated with different fungicides. SISSTA Sugar J. 1989;15:50–53. [Google Scholar]
- 35.Muangthong A., Youpensuk S., Rerkasem B. Isolation and characterisation of endophytic nitrogen fixing bacteria in sugarcane. Trop. Life Sci. Res. 2015;26:41. [PMC free article] [PubMed] [Google Scholar]
- 36.Bajoria S., Varshney A.K., Pareek R.P., Mohan M.K., Ghosh P. Screening and characterization of antifungal clusterbean (Cyamopsis tetragonoloba) rhizobacteria. Biocontrol Sci. Technol. 2008;18:139–156. [Google Scholar]
- 37.Hassan M.N., Shah S.Z.-U.-H., Afghan S., Hafeez F.Y. Suppression of red rot disease by Bacillus sp. based biopesticide formulated in non-sterilized sugarcane filter cake. Biocontrol. 2015;60:691–702. [Google Scholar]
- 38.Gajbhiye A., Rai A.R., Meshram S.U., Dongre A. Isolation, evaluation and characterization of Bacillus subtilis from cotton rhizospheric soil with biocontrol activity against Fusarium oxysporum. World J. Microbiol. Biotechnol. 2010;26:1187–1194. doi: 10.1007/s11274-009-0287-9. [DOI] [PubMed] [Google Scholar]
- 39.Siddiqui S., Siddiqui Z.A., Ahmad I. Evaluation of Fluorescent Pseudomonads and Bacillus isolates for the biocontrol of a wilt disease complex of pigeonpea. World J. Microbiol. Biotechnol. 2005;21:729–732. [Google Scholar]
- 40.Mehnaz S., Bauer J.S., Gross H. Complete genome sequence of the sugar cane endophyte Pseudomonas aurantiaca PB-St2, a disease-suppressive bacterium with antifungal activity toward the plant pathogen Colletotrichum falcatum. Genome Announc. 2014;2:e01108–01113. doi: 10.1128/genomeA.01108-13. [DOI] [PMC free article] [PubMed] [Google Scholar]