Abstract
Objective
To evaluate the design characteristics of studies that evaluated the performance of artificial intelligence (AI) algorithms for the diagnostic analysis of medical images.
Materials and Methods
PubMed MEDLINE and Embase databases were searched to identify original research articles published between January 1, 2018 and August 17, 2018 that investigated the performance of AI algorithms that analyze medical images to provide diagnostic decisions. Eligible articles were evaluated to determine 1) whether the study used external validation rather than internal validation, and in case of external validation, whether the data for validation were collected, 2) with diagnostic cohort design instead of diagnostic case-control design, 3) from multiple institutions, and 4) in a prospective manner. These are fundamental methodologic features recommended for clinical validation of AI performance in real-world practice. The studies that fulfilled the above criteria were identified. We classified the publishing journals into medical vs. non-medical journal groups. Then, the results were compared between medical and non-medical journals.
Results
Of 516 eligible published studies, only 6% (31 studies) performed external validation. None of the 31 studies adopted all three design features: diagnostic cohort design, the inclusion of multiple institutions, and prospective data collection for external validation. No significant difference was found between medical and non-medical journals.
Conclusion
Nearly all of the studies published in the study period that evaluated the performance of AI algorithms for diagnostic analysis of medical images were designed as proof-of-concept technical feasibility studies and did not have the design features that are recommended for robust validation of the real-world clinical performance of AI algorithms.
Keywords: Artificial intelligence, Machine learning, Deep learning, Clinical validation, Clinical trial, Accuracy, Study design, Quality, Appropriateness, Systematic review, Meta-analysis
INTRODUCTION
The use of artificial intelligence (AI) for medicine has recently drawn much attention due to the advances in deep learning technologies (1). Notably, there is a remarkable interest in using AI for diagnostic analysis of various types of medical images, primarily through convolutional neural networks, a type of deep learning technology referred to as “computer vision” (2,3,4). As with any other medical devices or technologies, the importance of thorough clinical validation of AI algorithms before their adoption in clinical practice through adequately designed studies to ensure patient benefit and safety while avoiding any inadvertent harms cannot be overstated (5,6,7,8,9,10). Note that the term “validation” is used in this study to imply confirmation, as would be used in the medicine field, and not algorithm tuning, which is used as technical jargon in the field of machine learning (11,12). Clinical validation of AI technologies can be performed at different levels of efficacy: diagnostic performance, effects on patient outcome, and societal efficacy that considers cost-benefit and cost-effectiveness (11,13). Proper assessment of the real-world clinical performance of high-dimensional AI algorithms that analyze medical images using deep learning requires appropriately designed external validation. It is recommended for the external validation to use adequately sized datasets that are collected either from newly recruited patients or at institutions other than those that provided training data in a way to adequately represent the manifestation spectrum (i.e., all relevant variations in patient demographics and disease states) of target patients in real-world clinical settings where the AI will be applied (10,12,14,15,16,17). Furthermore, use of data from multiple external institutions is important for the validation to verify the algorithm's ability to generalize across the expected variability in a variety of hospital systems (14,16,17,18). Complex mathematical/statistical AI models such as deep learning algorithms that analyze medical images need a large quantity of data for algorithm training; producing and annotating this magnitude of medical image data is especially resource intensive and difficult (19,20). Therefore, individuals developing such AI algorithms might rely on whatever data are available (methodologically referred to as convenience case-control data), although these may be prone to selection biases and artificial disease prevalence and likely not represent real-world clinical settings well (12,19,20). Since the performance of an AI algorithm is strongly dependent upon its training data, there is a genuine risk that AI algorithms may not perform well in real-world practice and that an algorithm trained at one institution provides inaccurate outputs when applied to data at another institution (9,16,17,18,19,21,22).
Despite the excitement around the use of AI for medicine, the lack of appropriate clinical validation for AI algorithms seems to be a current concern, a phenomenon referred to as “digital exceptionalism” (16,23,24). For example, computer scientists typically evaluate the performance of AI algorithms on “test” datasets; however, these are usually random subsamples of the original dataset, and thus, adequate external validation of clinical performance is not possible (10,16,20,25). To our knowledge, concrete data showing the exact extent of this perceived problem are scarce. This study aimed to evaluate the design characteristics of recently published studies reporting the performance of AI algorithms that analyze medical images and determine if the study designs were appropriate for validating the clinical performance of AI algorithms in real-world practice. The study design features addressed in this study are crucial for validating the real-world clinical performance of AI but would be excessive for proof-of-concept technical feasibility studies (14). As not every research study about the use of AI for medical diagnosis is to validate the real-world clinical performance (14), the purpose of this study was not to bluntly judge the methodologic appropriateness of the published studies.
MATERIALS AND METHODS
This study did not require Institutional Review Board approval.
Literature Search and Screening
PubMed MEDLINE and Embase databases were thoroughly searched to identify original research articles that investigated the performance of AI algorithms that analyze medical images to provide diagnostic decisions (such as either diagnosing or finding specific diseases or giving information to categorize patients with a particular disease into subgroups according to disease states, subtypes, severity levels, stages, treatment responses, prognosis, and risks). We used the following search query: (“artificial intelligence” OR “machine learning” OR “deep learning” OR “convolutional neural network”) AND (diagnosis OR diagnostic OR diagnosing) AND (accuracy OR performance OR “receiver operating” OR ROC OR AUC). We limited the search period to year 2018 to obtain timely results (literature search update until August 17, 2018). Both print publications and electronic publications ahead of print were included.
After removing overlaps between the two databases, articles were screened for eligibility by two independent reviewers. Articles with any degree of ambiguity or that generated differences in opinion between the two reviewers were re-evaluated at a consensus meeting, for which a third reviewer was invited. Case reports, review articles, editorials, letters, comments, and conference abstract/proceedings were excluded. Our search was restricted to human subjects and English-language studies. We defined medical images as radiologic images and other medical photographs (e.g., endoscopic images, pathologic photos, and skin photos) and did not consider any line art graphs that typically plot unidimensional data across time, for example, electrocardiogram and A-mode ultrasound. Studies investigating AI algorithms that combined medical images and other types of clinical data were included. AI algorithms that performed image-related tasks other than direct diagnostic decision-making, for example, image segmentation, quantitative measurements, and augmentation of image acquisition/reconstruction, were not considered.
Data Extraction
The full text of eligible articles was evaluated by two reviewers for the following information: 1) whether the study used external validation as opposed to internal validation, and in case of external validation, whether the data for validation were collected, 2) with diagnostic cohort design instead of diagnostic case-control design, 3) from multiple institutions, and 4) in a prospective manner. These are fundamental methodologic features recommended for clinical validation of AI performance in real-world practice (10,11,12,14). The more of these questions receive a “Yes” answer, the more generalizable to real-world practice the algorithm performance is. If a study validated its AI performance in multiple ways, then the study received a “Yes” answer for each of the above questions if at least one analysis used the design features. We defined “external” a bit generously and included the use of validation data from institution(s) other than the one from which training data were obtained, as well as cases where training and validation data were collected from the same institution(s) but in different time periods, even though the latter is not considered external validation in a strict sense (10,16,25). For studies in which the training and validation datasets were collected at the same institution(s), validation data were only considered external if the clinical settings and patient eligibility criteria for the validation dataset were specified separately from those of the training dataset. This was to ensure that the validation data were not just a time-split subsample of the original large dataset, as that results in a type of internal validation (25). A diagnostic cohort design was referred to as one in which the study defined the clinical setting and patient eligibility criteria first and then recruited patients consecutively or randomly to undergo a particular diagnostic procedure, such as AI algorithm application (15). In contrast, a diagnostic case-control design would involve the collection of disease-positive and disease-negative subjects separately (15). Diagnostic case-control designs are prone to spectrum bias, which can lead to an inflated estimation of the diagnostic performance, and unnatural prevalence, which creates uncertainty regarding the diagnostic performance (12,26). Additionally, we noted the subject field (e.g., radiology, pathology, and ophthalmology) of each article and classified the publishing journals into either medical or non-medical journal groups. The journals were classified primarily based on the Journal Citation Reports (JCR) 2017 edition categories. For journals not included in JCR databases, we referred to journal websites and categorized them as medical if the scope/aim of the journal included any fields of medicine or if the editor-in-chief was a medical doctor. Articles with any degree of ambiguity or that generated differences in opinion between the two independent reviewers were re-evaluated at a consensus meeting including a third reviewer.
Outcome Measures and Statistical Analysis
We calculated the percentage of studies that performed external validation. For studies reporting the results of external validation, the proportions of studies that involved the features of diagnostic cohort designs, inclusion of multiple institutions, and prospective data collection for external validation were identified. The results were compared between medical and non-medical journals using Fisher's exact test. A p < 0.05 was considered significant.
RESULTS
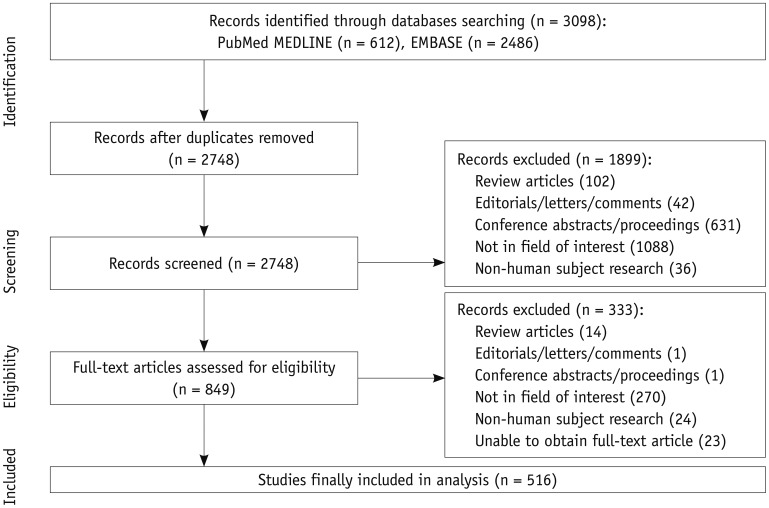
Of 2748 articles initially collected after removal of overlaps between PubMed MEDLINE and Embase, 516 articles were finally eligible (Fig. 1, Table 1). The full list of eligible articles analyzed in this study is available as an online supplement.
Fig. 1. Flow-chart of article selection based on preferred reporting items for systematic reviews and meta-analyses guidelines.
Table 1. Subject Fields of Articles Analyzed.
| Subject Fields* | Number of Articles (%) |
|---|---|
| Radiology (including nuclear medicine) | 366 (70.9) |
| Ophthalmology | 54 (10.5) |
| Pathology | 41 (7.9) |
| Dermatology | 19 (3.7) |
| Gastroenterology | 19 (3.7) |
| Other fields | 15 (2.9) |
| Combined fields | |
| Radiology and cardiology | 1 (0.2) |
| Pathology and nuclear medicine | 1 (0.2) |
| Total | 516 (100) |
*Listed in descending order of article number.
Table 2 presents the proportions of the articles that had each design feature, including breakdowns for medical vs. non-medical journals. Only 6% (31 of 516) of the studies performed external validation. None of the external validation studies adopted all three design features, namely, diagnostic cohort design, inclusion of multiple institutions, and prospective data collection. No significant difference was found between medical and non-medical journals (Table 2).
Table 2. Study Design Characteristics of Articles Analyzed.
| Design Characteristic | All Articles (n = 516) | Articles Published in Medical Journals (n = 437) | Articles Published in Non-Medical Journals (n = 79) | P* |
|---|---|---|---|---|
| External validation | 1.000 | |||
| Used | 31 (6.0) | 27 (6.2) | 4 (5.1) | |
| Not used | 485 (94.0) | 410 (93.8) | 75 (94.9) | |
| In studies that used external validation | ||||
| Diagnostic cohort design | 5 (1.0) | 5 (1.1) | 0 (0) | 1.000 |
| Data from multiple institutions | 15 (2.9) | 12 (2.7) | 3 (3.8) | 0.713 |
| Prospective data collection | 4 (0.8) | 4 (0.9) | 0 (0) | 1.000 |
| Fulfillment of all of above three criteria | 0 (0) | 0 (0) | 0 (0) | 1.000 |
| Fulfillment of at least two criteria | 3 (0.6) | 3 (0.7) | 0 (0) | 1.000 |
| Fulfillment of at least one criterion | 21 (4.1) | 18 (4.1) | 3 (3.8) | 1.000 |
Data are expressed as number of articles with corresponding percentage enclosed in parentheses. *Comparison between medical and non-medical journals.
DISCUSSION
Our results reveal that most recently published studies reporting the performance of AI algorithms for diagnostic analysis of medical images did not have design features that are recommended for robust validation of the clinical performance of AI algorithms, confirming the worries that premier journals have recently raised (23,24). Our study did not consider various detailed methodologic quality measures for AI research studies (14), but simply evaluated major macroscopic study design features. Therefore, the extent of deficiency in the clinical validation of AI algorithms could likely be even more significant.
However, it should be noted that these results do not necessarily mean that the published studies were inadequately designed by all means. The four criteria used in this study–external validation and data for external validation being obtained using a diagnostic cohort study, from multiple institutions, and in a prospective manner–are fundamental requirements for studies that intend to evaluate the clinical performance of AI algorithms in real-world practice. These would be excessive for studies that merely investigate technical feasibility (14). Readers and investigators alike should distinguish between proof-of-concept technical feasibility studies and studies to validate clinical performance of AI (14) and should avoid incorrectly considering the results from studies that do not fulfill the criteria mentioned above as sound proof of clinical validation.
Some related methodologic guides have recently been published (11,12,14). We suspect that most studies that we analyzed in this study may have been conceived or executed before these methodologic guides were made available. Therefore, the design features of studies that intend to evaluate the clinical performance of AI algorithms for medicine may improve in the future.
Another issue that was not directly addressed in our study but is worth mentioning is transparency regarding a priori analysis plans and full publication of all results in studies validating the clinical performance of AI algorithms (6,11,14,27). As the performance of an AI algorithm may vary across different institutions (16,17,18), some researchers or sponsors might be inclined to selectively report favorable results, which would result in underreporting of unfavorable results. Prospective registration of studies, including a priori analysis plans, similar to the registration of clinical trials of interventions (e.g., at https://clinicaltrials.gov), would help increase the transparency of these studies (27). Prospective registration of diagnostic test accuracy studies, which include studies to validate AI performance, has already been proposed (28). The adoption of this policy by academic journals would help enhance transparency in the reporting of studies that validate the clinical performance of AI algorithms.
Our current study has some limitations. First, while the timeliness of research data is important (29), as AI is a rapidly evolving field with numerous new studies being published, the shelf life of our study results could be short. Ironically, we hope to see substantial improvements in the design of studies reporting clinical performance of AI in medicine soon. Despite such rapid changes, our research remains meaningful as the baseline against which comparisons can be made to see if any improvements are made in the future, given that most published studies that were analyzed here likely predated the recent release of related methodologic guides (11,12,14). Second, while this study only evaluated studies reporting the diagnostic performance of AI, clinical validation of AI extends to evaluating the impact of AI on patient outcomes (12,30). However, to our knowledge, studies of how AI application affects patient outcomes are scarce, and systematically reviewing published studies is not feasible.
In conclusion, nearly all of the studies published in the study period that evaluated the performance of AI algorithms for diagnostic analysis of medical images were designed as proof-of-concept technical feasibility studies and did not have the design features that are recommended for robust validation of the real-world clinical performance of AI algorithms.
Footnotes
This study was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI18C1216).
Conflicts of Interest: The authors have no potential conflicts of interest to disclose.
References
- 1.Lee JG, Jun S, Cho YW, Lee H, Kim GB, Seo JB, et al. Deep learning in medical imaging: general overview. Korean J Radiol. 2017;18:570–584. doi: 10.3348/kjr.2017.18.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamashita R, Nishio M, Do RKG, Togashi K. Convolutional neural networks: an overview and application in radiology. Insights Imaging. 2018;9:611–629. doi: 10.1007/s13244-018-0639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chartrand G, Cheng PM, Vorontsov E, Drozdzal M, Turcotte S, Pal CJ, et al. Deep learning: a primer for radiologists. Radiographics. 2017;37:2113–2131. doi: 10.1148/rg.2017170077. [DOI] [PubMed] [Google Scholar]
- 4.Choy G, Khalilzadeh O, Michalski M, Do S, Samir AE, Pianykh OS, et al. Current applications and future impact of machine learning in radiology. Radiology. 2018;288:318–328. doi: 10.1148/radiol.2018171820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.SFR-IA Group; CERF; French Radiology Community. Artificial intelligence and medical imaging 2018: French Radiology Community white paper. Diagn Interv Imaging. 2018;99:727–742. doi: 10.1016/j.diii.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Greaves F, Joshi I, Campbell M, Roberts S, Patel N, Powell J. What is an appropriate level of evidence for a digital health intervention? Lancet. 2019;392:2665–2667. doi: 10.1016/S0140-6736(18)33129-5. [DOI] [PubMed] [Google Scholar]
- 7.Maddox TM, Rumsfeld JS, Payne PRO. Questions for artificial intelligence in health care. JAMA. 2019;321:31–32. doi: 10.1001/jama.2018.18932. [DOI] [PubMed] [Google Scholar]
- 8.Shortliffe EH, Sepu´lveda MJ. Clinical decision support in the era of artificial intelligence. JAMA. 2018;320:2199–2200. doi: 10.1001/jama.2018.17163. [DOI] [PubMed] [Google Scholar]
- 9.Tang A, Tam R, Cadrin-Chênevert A, Guest W, Chong J, Barfett J, et al. Canadian Association of Radiologists (CAR) Artificial Intelligence Working Group. Canadian Association of Radiologists white paper on artificial intelligence in radiology. Can Assoc Radiol J. 2018;69:120–135. doi: 10.1016/j.carj.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Park SH, Do KH, Choi JI, Sim JS, Yang DM, Eo H, et al. Principles for evaluating the clinical implementation of novel digital healthcare devices. J Korean Med Assoc. 2018;61:765–775. [Google Scholar]
- 11.Park SH, Kressel HY. Connecting technological innovation in artificial intelligence to real-world medical practice through rigorous clinical validation: what peer-reviewed medical journals could do. J Korean Med Sci. 2018;33:e152. doi: 10.3346/jkms.2018.33.e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park SH, Han K. Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology. 2018;286:800–809. doi: 10.1148/radiol.2017171920. [DOI] [PubMed] [Google Scholar]
- 13.Fryback DG, Thornbury JR. The efficacy of diagnostic imaging. Med Decis Making. 1991;11:88–94. doi: 10.1177/0272989X9101100203. [DOI] [PubMed] [Google Scholar]
- 14.England JR, Cheng PM. Artificial intelligence for medical image analysis: a guide for authors and reviewers. AJR Am J Roentgenol. 2018 Dec 17; doi: 10.2214/AJR.18.20490. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 15.Park SH. Diagnostic case-control versus diagnostic cohort studies for clinical validation of artificial intelligence algorithm performance. Radiology. 2019;290:272–273. doi: 10.1148/radiol.2018182294. [DOI] [PubMed] [Google Scholar]
- 16.Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS Med. 2018;15:e1002683. doi: 10.1371/journal.pmed.1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ting DSW, Cheung CY, Lim G, Tan GSW, Quang ND, Gan A, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017;318:2211–2223. doi: 10.1001/jama.2017.18152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Zhang S, Zhang Q, Wei X, Pan Y, Zhao J, et al. Diagnosis of thyroid cancer using deep convolutional neural network models applied to sonographic images: a retrospective, multicohort, diagnostic study. Lancet Oncol. 2019;20:193–201. doi: 10.1016/S1470-2045(18)30762-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nsoesie EO. Evaluating artificial intelligence applications in clinical settings. JAMA Netw Open. 2018;1:e182658. doi: 10.1001/jamanetworkopen.2018.2658. [DOI] [PubMed] [Google Scholar]
- 20.Zou J, Schiebinger L. AI can be sexist and racist - it's time to make it fair. Nature. 2018;559:324–326. doi: 10.1038/d41586-018-05707-8. [DOI] [PubMed] [Google Scholar]
- 21.Gianfrancesco MA, Tamang S, Yazdany J, Schmajuk G. Potential biases in machine learning algorithms using electronic health record data. JAMA Intern Med. 2018;178:1544–1547. doi: 10.1001/jamainternmed.2018.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.AlBadawy EA, Saha A, Mazurowski MA. Deep learning for segmentation of brain tumors: impact of cross-institutional training and testing. Med Phys. 2018;45:1150–1158. doi: 10.1002/mp.12752. [DOI] [PubMed] [Google Scholar]
- 23.The Lancet. Is digital medicine different? Lancet. 2018;392:95. doi: 10.1016/S0140-6736(18)31562-9. [DOI] [PubMed] [Google Scholar]
- 24.AI diagnostics need attention. Nature. 2018;555:285. doi: 10.1038/d41586-018-03067-x. [DOI] [PubMed] [Google Scholar]
- 25.Luo W, Phung D, Tran T, Gupta S, Rana S, Karmakar C, et al. Guidelines for developing and reporting machine learning predictive models in biomedical research: a multidisciplinary view. J Med Internet Res. 2016;18:e323. doi: 10.2196/jmir.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rutjes AW, Reitsma JB, Vandenbroucke JP, Glas AS, Bossuyt PM. Case-control and two-gate designs in diagnostic accuracy studies. Clin Chem. 2005;51:1335–1341. doi: 10.1373/clinchem.2005.048595. [DOI] [PubMed] [Google Scholar]
- 27.Gill J, Prasad V. Improving observational studies in the era of big data. Lancet. 2018;392:716–717. doi: 10.1016/S0140-6736(18)31619-2. [DOI] [PubMed] [Google Scholar]
- 28.Korevaar DA, Hooft L, Askie LM, Barbour V, Faure H, Gatsonis CA, et al. Facilitating prospective registration of diagnostic accuracy studies: a STARD initiative. Clin Chem. 2017;63:1331–1341. doi: 10.1373/clinchem.2017.272765. [DOI] [PubMed] [Google Scholar]
- 29.Kang JH, Kim DH, Park SH, Baek JH. Age of data in contemporary research articles published in representative general radiology journals. Korean J Radiol. 2018;19:1172–1178. doi: 10.3348/kjr.2018.19.6.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.INFANT Collaborative Group. Computerised interpretation of fetal heart rate during labour (INFANT): a randomised controlled trial. Lancet. 2017;389:1719–1729. doi: 10.1016/S0140-6736(17)30568-8. [DOI] [PMC free article] [PubMed] [Google Scholar]