Abstract
Development of the human gut throughout the entire life.
General features
The human gut microbiota represents an extraordinary complex microbial consortium whose activities are believed to be crucial for human health and well‐being. The formation of the human gut microbiota commences at the point of delivery of an essentially sterile baby and then further develops in terms of diversity and complexity during weaning, adolescence and into adult life. The microbial groups identified in the gut microbiota encompass representatives of all three domains of life, i.e. Archaea, Bacteria and Eukarya, whose dynamics are manoeuvred by various forces, such as viruses, which are believed to act as powerful drivers of gut microbiota homoeostasis (for a review see Ref.: Milani et al., 2017). The human gut microbiota is composed of autochthonous members that are believed to establish stable interactions with their host, as well as a transient microbial consortium whose members are mainly acquired through oral intake and who are not presumed to colonize the gut in the presence of a homoeostatic microbiota. The diverse components of the gut microbiota are known to elicit key effects in the maintenance and promotion of human health by (i) supporting the transformation of food components so as to release nutrients that would otherwise not be accessible to the host, (ii) stimulating host cell differentiation, (iii) defending the body against pathogen invasion and (iv) exerting immune modulatory activities. Several epidemiological studies have clearly demonstrated a correlation between factors disrupting the gut microbiota during childhood and the occurrence of certain immune and metabolic disorders at later stages of life (Eggesbo et al., 2003; Huh et al., 2012; Sevelsted et al., 2015), backed up by other scientific data that link long‐term health benefits to the infant gut microbiota (Relman, 2012). This scientific realization has in turn opened up the search for strategies that aim to influence the origin, development, maturation and activities of the infant microbiota by a plethora of nutraceutical/functional food products including probiotics and prebiotics.
An interesting feature of the human gut microbiota is that the development of such a microbial assemblage reaches homoeostasis, which is characterized by a compositional equilibrium of its microbial members (Tamboli et al., 2004). Various events may cause disturbances in this so‐called microbiota climax, thereby causing a state that has been referred to as dysbiosis. The concept of dysbiosis is controversial, simply because there is still a lack of knowledge and understanding of what precisely represents a ‘normal’ or healthy microbiota. Dysbiosis is usually associated with harmful effects and may have long‐term consequences leading to disorders or diseases, including obesity, diabetes and inflammatory bowel disease (IBD). However, one should realize that significant variations occur in gut microbiota composition during host development from infancy to early childhood, from young to ageing adults and during pregnancy. Even if such modifications appear to be similar to those described in certain disease conditions, in their evolving context they do not necessarily lead to illness but, rather, may enhance fitness and survival and thus are considered to be beneficial (Nuriel‐Ohayon et al., 2016).
The origin of the human gut microbiota
The establishment of the infant gut microbiota is the result of a complex process that is influenced by many environmental and host factors (Rodriguez et al., 2015) (Fig. 1). Notably, although the dogma of a sterile in utero environment, which claims that microbial colonization only commences upon birth, is still generally accepted, there is growing evidence indicating that very early colonization of humans may occur at the fetal stage (Jimenez et al., 2005; DiGiulio et al., 2008; Aagaard et al., 2014). The route by which bacteria achieve this prebirth colonization is still obscure. However, the recent discovery of a circulating microbiota that is present in human blood (Kowarsky et al., 2017) is not only reinforcing the notion that there is not any human organ that is completely sterile, but also lending further support to the hypothesis of a prenatal establishment of the gut microbiota. This perception may facilitate new research avenues and possible manipulation of the human gut microbiota through probiotic intervention during pregnancy.
Figure 1.
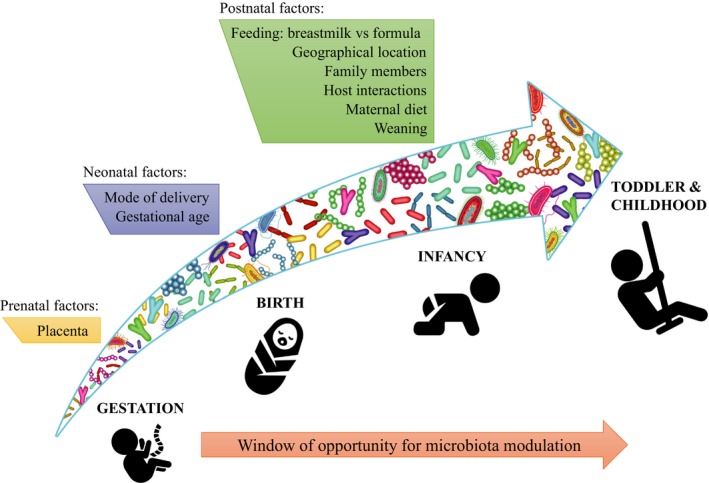
Development of the human gut throughout the entire life. The diagram describes a list of prenatal, neonatal and postnatal influences that may contribute to the establishment/maturation/succession of bacterial consortia in the human gut.
The progress and evolution of the gut microbiota is a dynamic and non‐casual process, involving intricate interactions between key microbial components (Avershina et al., 2013, 2016). This process is affected by several perinatal conditions, such as mode of delivery, type of feeding and antibiotic usage (Milani et al., 2017). In addition, diet, mother's age and metabolic status, family genetics and lifestyle have been described to influence the infant gut microbiota composition (Milani et al., 2017).
The origin of the human gut microbiota is associated with the inheritance of an assemblage of microorganisms that are vertically transmitted from mother to her offspring. Remarkably, the mammalian newborn does not appear to passively acquire a new microbiota, but the mother seems to actively promote the transfer of specific members of her microbiota to her offspring and may thus directly influence the development of the new microbiota of her child. This selective vertical transmission of microbes is driven by the specific nourishment with prebiotic milk compounds favouring certain microorganisms, in particular bifidobacteria. Recently, metagenomic investigations of mothers and corresponding newborns have highlighted the existence of shared bifidobacterial communities, in particular bifidobacterial strains that are capable of metabolizing specific glycans contained in human milk, i.e. human milk oligosaccharides (HMOs; Milani et al., 2015; Duranti et al., 2017). This vertical transmission route involving bifidobacterial strains includes human milk as a vectoring system (Milani et al., 2015; Duranti et al., 2017). A similar scenario seems to occur in other non‐human primates as well as in non‐primate mammals, where bifidobacterial strains were found to be maternally inherited by means of mother's milk (Milani et al., 2017). These findings support the hypothesis that those bifidobacteria that are directly acquired from mothers are maintained in the adult gut, perhaps at very low levels, in order to be ultimately transferred to the next generation (Milani et al., 2015). This intriguing process may therefore reflect millennia of co‐evolution between bifidobacteria and their mammalian host.
As mentioned above, human milk represents an important selective medium for the establishment and modulation of the gut microbiota during the early stages of life. In fact, human milk, apart from representing a source of nutrients for the infant, encompasses a large amount of carbohydrates such as HMOs; i.e., 1 l of human milk contains 5–20 g of these complex glycans, which is typically more than all human milk proteins combined and which acts as a highly selective nourishment for specific members of the infant gut microbiota. Notably, HMOs encompass a diverse group of complex glycans, where five monosaccharide building blocks glucose, galactose, N‐acetylglucosamine, fucose and N‐acetyl‐neuraminic acid are arranged in a variety of combinations, which generates more than 200 structurally distinct HMOs (Bode, 2012; Milani et al., 2017). Furthermore, it should be noted that HMO content and composition depend on lactation stage, the genetics of the mother and environmental factors (McGuire et al., 2017). Therefore, human milk can be considered as a ‘personalized, mother‐specific nutrition’ and HMOs as ‘personalized prebiotics’, together promoting the arrangement of a distinct infant gut microbiome, the composition of which being determined, at least in part, by the HMOs present in mother's milk. Every mother provides a different HMO mix to her infant(s), determining distinct infant microbial assemblies with potential short‐ and long‐term effects (Milani et al., 2017).
Only a small number of bacteria, such as the Bifidobacterium longum subsp. infantis ATCC 15697 and Bifidobacterium bifidum PRL2010, have been shown to possess the enzymatic machinery to metabolize many if not all HMOs present in human milk (Sela et al., 2008; Turroni et al., 2010). In contrast, other bacteria can access and metabolize only specific elements of the HMO mixture. Nevertheless, microbial communities may be capable to act in concert, consecutively breakdown and metabolize HMO structures in a team effort through cross‐feeding activities (Egan et al., 2014a,b; Gotoh et al., 2018).
The early microbiota as a key determinant of human health
In the course of the initial microbial colonization, the gut microbiota is still unstable and is going through continuous modifications until the infant is 2–3 years old, at which stage the microbiota reaches a composition resembling that of an adult (Yatsunenko et al., 2012). Despite this apparent instability of the infant gut microbiota, it is very well known that microbiota–host communications are particularly important during these early stages of life. In fact, during this period, microbiota–host interactions represent key events underpinning immune development of the human host, with consequent creation and conservation of the homoeostasis of the host microbiota during infancy. Remarkably, these actions have not only immediate health effects but also long‐term health consequences. In this context, the infant gut microbiota provides a strong antigenic stimulus needed for the maturation of the gut and associated immune functions (Sommer and Backhed, 2013; Gensollen et al., 2016). This effect also impacts on the development of the distal organs, by influencing the host at systemic level (Clarke et al., 2014; Neuman et al., 2015). Consequently, it is accepted that the foundation of human health is linked with the establishment and development of the microbiota (Arrieta et al., 2014; Nylund et al., 2014). In this context, it has been shown that faecal levels of IgA, which is considered to be an important marker linked to the risk of disease, may be associated with the presence of specific microbiota members (Moon et al., 2015). Thus, it may be argued that any modification from the normal neonatal microbial colonization process represents a risk factor for disease later in life. Certainly, accumulating scientific evidence indicates that gut microbiota alterations during the first stages of life are predictors of disease development (Kalliomaki et al., 2008; Fujimura et al., 2016; Simonyte Sjodin et al., 2016). In this context, it should also be noted that the early microbiota is connected to infant malnutrition and associated growth impairment (Arboleya et al., 2012; Gough et al., 2015; Reyes et al., 2015; Charbonneau et al., 2016).
There is a long list of studies involving immune pathologies such as atopic eczema and asthma, IBD, irritable bowel syndrome (IBS), type 1 diabetes (T1D), metabolic disorders, like obesity type 2 diabetes, displaying a clear correlation with alteration in the early gut microbiota.
The infant gut microbiota and its impact on the host health
An incorrect establishment of the gut microbiota during the early stages of life has been shown to be associated to disruption of immune homoeostasis, ultimately impacting on the risk of allergic diseases later in life (Johnson and Ownby, 2016). Interestingly, microbiota differences related to low loads of certain bacterial taxa such as bifidobacteria and other intestinal anaerobes such as Faecalibacterium prausnitzii and Coprococcus eutactus were identified in infants that later developed atopy or asthma (Fujimura et al., 2016). Even if these results do not connect an early microbiota profile with a decrease of atopic eczema disorder, they do indirectly highlight the protective role of a butyrate‐producing microbiota against the development of atopic eczema. In fact, it has been shown that a low relative abundance of butyrate producers is a risk predictor for the development of atopic eczema (Simonyte Sjodin et al., 2016).
Several studies have reported a clear association between early live antibiotic exposure and childhood obesity (Cox and Blaser, 2015). These findings indicate that antibiotics‐mediated disturbances of the gut microbiota, either during prenatal or postnatal periods, enhance the risk of obesity (Turta and Rautava, 2016). Recently, it has been proposed that early microbial profiles may predict overweight in children. In this context, it has been shown that the bifidobacterial loads at 6 and 12 months inversely correlate with overweight in children (Kalliomaki et al., 2008).
It has been proposed that microbiota alterations during early infancy cause a pro‐inflammatory environment that facilitates the occurrence of autoimmune disease such as T1D (Kostic et al., 2015). In particular, such microbiota perturbations include a lower diversity and significant differences in the ratio of the most abundant intestinal phyla, i.e. Firmicutes and Bacteroidetes, as well as a reduced load of the butyrate producer F. prausnitzii (Gulden et al., 2015). Notably, butyrate‐producing species have been suggested to exert a protective role in reducing the risk of developing T1D (de Goffau et al., 2014).
Several severe diseases involving intestinal inflammatory symptoms have been linked to deviations of the early life gut microbiota. The concept that the human gut immune system is educated by the early microbial colonization assumes a connection between pioneering microbial inhabitants of the human intestine and subsequent gut inflammatory disease/disorder, such as IBD and IBS. Interestingly, a number of clinical trials have indicated that early changes in the population of specific microbial groups herald the onset of intestinal inflammatory diseases at later stages of life (O'Mahony et al., 2009, 2014; Schirbel and Fiocchi, 2011; Wang et al., 2016). Nevertheless, conclusive data about the long‐lasting effects of dysbiosis in the early life and the development of IBD and IBS in later stages of life are rare and do not allow the establishment of causality.
Conclusions
We are only beginning to unveil early life interactions between microbes and their mammalian host, and, if disturbed, the resulting long‐lasting effects on human health. Consequently, it may be time to develop alternative strategies to facilitate the appropriate establishment of the infant gut microbiota in those situations in which this process is prevented, challenged or disrupted.
This perception has fuelled the development of functional food strategies involving various nutraceutical products such as probiotics and/or prebiotics capable of modulating the infant gut microbiota establishment/maturation. However, such intervention approaches require thorough assessment of the composition of the human early gut microbiota and the identification of key microbiota components that play a role in the aetiology of diseases and that can thus act as microbial biomarkers. The identification of microbial biomarkers therefore represents an important activity for future prophylactic approaches as well as for early diagnosis of diseases.
We are only starting to understand the importance of microbiota composition during pregnancy in terms of a reservoir of bacteria for the establishment of gut microbiota in newborns. However, there are many open questions about how certain bacteria are specifically transferred by the mother to their children. Furthermore, we still do not know whether bacteria may transfer from mother to child through a systemic route, e.g. through dendritic cells, blood or milk. The answers to all these questions combined with the possibility of prenatal bacterial colonization may revolutionize paediatrics in the coming years.
Conflict of interest
None declared.
Acknowledgements
This work was funded by the EU Joint Programming Initiative – A Healthy Diet for a Healthy Life (JPI HDHL, http://www.healthydietforhealthylife.eu/) and MIUR to MV. DvS is a member of The APC Microbiome Institute funded by Science Foundation Ireland (SFI), through the Irish Government's National Development Plan (Grant number SFI/12/RC/2273).
Microbial Biotechnology (2019) 12(2), 243–248
Funding Information
This work was funded by the EU Joint Programming Initiative – A Healthy Diet for a Healthy Life (JPI HDHL, http://www.healthydietforhealthylife.eu/) and MIUR to MV. DvS is a member of The APC Microbiome Institute funded by Science Foundation Ireland (SFI), through the Irish Government's National Development Plan (Grant number SFI/12/RC/2273).
References
- Aagaard, K. , Ma, J. , Antony, K.M. , Ganu, R. , Petrosino, J. , and Versalovic, J. (2014) The placenta harbors a unique microbiome. Sci Transl Med 6: 237ra265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arboleya, S. , Binetti, A. , Salazar, N. , Fernandez, N. , Solis, G. , Hernandez‐Barranco, A. , et al (2012) Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol Ecol 79: 763–772. [DOI] [PubMed] [Google Scholar]
- Arrieta, M.C. , Stiemsma, L.T. , Amenyogbe, N. , Brown, E.M. , and Finlay, B. (2014) The intestinal microbiome in early life: health and disease. Front Immunol 5: 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avershina, E. , Storro, O. , Oien, T. , Johnsen, R. , Wilson, R. , Egeland, T. , and Rudi, K. (2013) Bifidobacterial succession and correlation networks in a large unselected cohort of mothers and their children. Appl Environ Microbiol 79: 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avershina, E. , Lundgard, K. , Sekelja, M. , Dotterud, C. , Storro, O. , Oien, T. , et al (2016) Transition from infant‐ to adult‐like gut microbiota. Environ Microbiol 18: 2226–2236. [DOI] [PubMed] [Google Scholar]
- Bode, L. (2012) Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology 22: 1147–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonneau, M.R. , O'Donnell, D. , Blanton, L.V. , Totten, S.M. , Davis, J.C. , Barratt, M.J. , et al (2016) Sialylated milk oligosaccharides promote microbiota‐dependent growth in models of infant undernutrition. Cell 164: 859–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, G. , O'Mahony, S.M. , Dinan, T.G. , and Cryan, J.F. (2014) Priming for health: gut microbiota acquired in early life regulates physiology, brain and behaviour. Acta Paediatr 103: 812–819. [DOI] [PubMed] [Google Scholar]
- Cox, L.M. , and Blaser, M.J. (2015) Antibiotics in early life and obesity. Nat Rev Endocrinol 11: 182–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiulio, D.B. , Romero, R. , Amogan, H.P. , Kusanovic, J.P. , Bik, E.M. , Gotsch, F. , et al (2008) Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture‐based investigation. PLoS One 3: e3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duranti, S. , Lugli, G.A. , Mancabelli, L. , Armanini, F. , Turroni, F. , James, K. , et al (2017) Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome 5: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan, M. , O'Connell Motherway, M. , Ventura, M. , and van Sinderen, D. (2014a) Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl Environ Microbiol 80: 4414–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan, M. , Motherway, O.C.M. , Kilcoyne, M. , Kane, M. , Joshi, L. , Ventura, M. , and van Sinderen, D. (2014b) Cross‐feeding by Bifidobacterium breve UCC2003 during co‐cultivation with Bifidobacterium bifidum PRL2010 in a mucin‐based medium. BMC Microbiol 14: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggesbo, M. , Botten, G. , Stigum, H. , Nafstad, P. , and Magnus, P. (2003) Is delivery by cesarean section a risk factor for food allergy? J Allergy Clin Immunol 112: 420–426. [DOI] [PubMed] [Google Scholar]
- Fujimura, K.E. , Sitarik, A.R. , Havstad, S. , Lin, D.L. , Levan, S. , Fadrosh, D. , et al (2016) Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med 22: 1187–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gensollen, T. , Iyer, S.S. , Kasper, D.L. , and Blumberg, R.S. (2016) How colonization by microbiota in early life shapes the immune system. Science 352: 539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Goffau, M.C. , Fuentes, S. , van den Bogert, B. , Honkanen, H. , de Vos, W.M. , Welling, G.W. , et al (2014) Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia 57: 1569–1577. [DOI] [PubMed] [Google Scholar]
- Gotoh, A. , Katoh, T. , Sakanaka, M. , Ling, Y. , Yamada, C. , Asakuma, S. , et al (2018) Sharing of human milk oligosaccharides degradants within bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum . Sci Rep 8: 13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough, E.K. , Stephens, D.A. , Moodie, E.E. , Prendergast, A.J. , Stoltzfus, R.J. , Humphrey, J.H. , and Manges, A.R. (2015) Linear growth faltering in infants is associated with Acidaminococcus sp. and community‐level changes in the gut microbiota. Microbiome 3: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulden, E. , Wong, F.S. , and Wen, L. (2015) The gut microbiota and type 1 diabetes. Clin Immunol 159: 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, S.Y. , Rifas‐Shiman, S.L. , Zera, C.A. , Edwards, J.W.R. , Oken, E. , Weiss, S.T. , and Gillman, M.W. (2012) Delivery by caesarean section and risk of obesity in preschool age children: a prospective cohort study. Arch Dis Child 97: 610–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez, E. , Fernandez, L. , Marin, M.L. , Martin, R. , Odriozola, J.M. , Nueno‐Palop, C. , et al (2005) Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr Microbiol 51: 270–274. [DOI] [PubMed] [Google Scholar]
- Johnson, C.C. , and Ownby, D.R. (2016) Allergies and asthma: do atopic disorders result from inadequate immune homeostasis arising from infant gut dysbiosis? Expert Rev Clin Immunol 12: 379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalliomaki, M. , Collado, M.C. , Salminen, S. , and Isolauri, E. (2008) Early differences in fecal microbiota composition in children may predict overweight. Am J Clin Nutr 87: 534–538. [DOI] [PubMed] [Google Scholar]
- Kostic, A.D. , Gevers, D. , Siljander, H. , Vatanen, T. , Hyotylainen, T. , Hamalainen, A.M. , et al (2015) The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 17: 260–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowarsky, M. , Camunas‐Soler, J. , Kertesz, M. , De Vlaminck, I. , Koh, W. , Pan, W. , et al (2017) Numerous uncharacterized and highly divergent microbes which colonize humans are revealed by circulating cell‐free DNA. Proc Natl Acad Sci USA 114: 9623–9628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire, M.K. , Meehan, C.L. , McGuire, M.A. , Williams, J.E. , Foster, J. , Sellen, D.W. , et al (2017) What's normal? Oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am J Clin Nutr 105: 1086–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani, C. , Mancabelli, L. , Lugli, G.A. , Duranti, S. , Turroni, F. , Ferrario, C. , et al (2015) Exploring vertical transmission of bifidobacteria from mother to child. Appl Environ Microbiol 81: 7078–7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani, C. , Mangifesta, M. , Mancabelli, L. , Lugli, G.A. , James, K. , Duranti, S. , et al (2017) Unveiling bifidobacterial biogeography across the mammalian branch of the tree of life. ISME J 11: 2834–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, C. , Baldridge, M.T. , Wallace, M.A. , Burnham, C.A. , Virgin, H.W. , and Stappenbeck, T.S. (2015) Vertically transmitted faecal IgA levels determine extra‐chromosomal phenotypic variation. Nature 521: 90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman, H. , Debelius, J.W. , Knight, R. , and Koren, O. (2015) Microbial endocrinology: the interplay between the microbiota and the endocrine system. FEMS Microbiol Rev 39: 509–521. [DOI] [PubMed] [Google Scholar]
- Nuriel‐Ohayon, M. , Neuman, H. , and Koren, O. (2016) Microbial changes during pregnancy, birth, and infancy. Front Microbiol 7: 1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylund, L. , Satokari, R. , Salminen, S. , and de Vos, W.M. (2014) Intestinal microbiota during early life – impact on health and disease. Proc Nutr Soc 73: 457–469. [DOI] [PubMed] [Google Scholar]
- O'Mahony, S.M. , Marchesi, J.R. , Scully, P. , Codling, C. , Ceolho, A.M. , Quigley, E.M. , et al (2009) Early life stress alters behavior, immunity, and microbiota in rats: implications for irritable bowel syndrome and psychiatric illnesses. Biol Psychiatry 65: 263–267. [DOI] [PubMed] [Google Scholar]
- O'Mahony, S.M. , Felice, V.D. , Nally, K. , Savignac, H.M. , Claesson, M.J. , Scully, P. , et al (2014) Disturbance of the gut microbiota in early‐life selectively affects visceral pain in adulthood without impacting cognitive or anxiety‐related behaviors in male rats. Neuroscience 277: 885–901. [DOI] [PubMed] [Google Scholar]
- Relman, D.A. (2012) The human microbiome: ecosystem resilience and health. Nutr Rev 70(Suppl 1): S2–S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes, A. , Blanton, L.V. , Cao, S. , Zhao, G. , Manary, M. , Trehan, I. , et al (2015) Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc Natl Acad Sci USA 112: 11941–11946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, J.M. , Murphy, K. , Stanton, C. , Ross, R.P. , Kober, O.I. , Juge, N. , et al (2015) The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol Health Dis 26: 26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirbel, A. , and Fiocchi, C. (2011) Targeting the innate immune system in pediatric inflammatory bowel disease. Expert Rev Gastroenterol Hepatol 5: 33–41. [DOI] [PubMed] [Google Scholar]
- Sela, D.A. , Chapman, J. , Adeuya, A. , Kim, J.H. , Chen, F. , Whitehead, T.R. , et al (2008) The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci USA 105: 18964–18969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevelsted, A. , Stokholm, J. , Bonnelykke, K. , and Bisgaard, H. (2015) Cesarean section and chronic immune disorders. Obstet Gynecol Surv 70: 303–305. [DOI] [PubMed] [Google Scholar]
- Simonyte Sjodin, K. , Vidman, L. , Ryden, P. , and West, C.E. (2016) Emerging evidence of the role of gut microbiota in the development of allergic diseases. Curr Opin Allergy Clin Immunol 16: 390–395. [DOI] [PubMed] [Google Scholar]
- Sommer, F. , and Backhed, F. (2013) The gut microbiota–masters of host development and physiology. Nat Rev Microbiol 11: 227–238. [DOI] [PubMed] [Google Scholar]
- Tamboli, C.P. , Neut, C. , Desreumaux, P. , and Colombel, J.F. (2004) Dysbiosis in inflammatory bowel disease. Gut 53: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni, F. , Bottacini, F. , Foroni, E. , Mulder, I. , Kim, J.H. , Zomer, A. , et al (2010) Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host‐derived glycan foraging. Proc Natl Acad Sci USA 107: 19514–19519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turta, O. , and Rautava, S. (2016) Antibiotics, obesity and the link to microbes – what are we doing to our children? BMC Med 14: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F. , Kaplan, J.L. , Gold, B.D. , Bhasin, M.K. , Ward, N.L. , Kellermayer, R. , et al (2016) Detecting microbial dysbiosis associated with pediatric crohn disease despite the high variability of the gut microbiota. Cell Rep 14: 945–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko, T. , Rey, F.E. , Manary, M.J. , Trehan, I. , Dominguez‐Bello, M.G. , Contreras, M. , et al (2012) Human gut microbiome viewed across age and geography. Nature 486: 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]


